Introduction
The field of angiogenesis and its potential as a
target to slow cancer development is well established and reviewed
within the literature. Angiogenesis is an essential process
required for normal physiological events such as wound healing,
reproduction, development and immunity. However, imbalance of this
process is often seen in disease states such as cancer progression
and metastatic spread, where enhanced tumour vasculature
facilitates rapid tumour growth and provides access for
metastasising cells (1,2). Numerous pro-angiogenic factors have
been identified. One such molecule is hepatocyte growth factor
(HGF), which has long been established as a factor that can enhance
the aggressive nature of cancer cells through its ability to
promote pro-metastatic traits such as motogenesis, mitogenesis,
morphogenesis and angiogenesis (3). HGF is able to promote angiogenesis
directly through its motogenic and morphogenic impact on
endothelial cells and also indirectly, through its ability to
enhance the production of other pro-angiogenic factors such as
vascular endothelial growth factor (VEGF) (3,4).
Previous studies have demonstrated the anti-angiogenic potential of
targeting HGF in breast and prostate cancer model systems (5,6). The
present study aimed to identify gene expression in endothelial
cells that could be regulated by HGF which may subsequently
contribute to the process of HGF-promoted angiogenesis, identifying
repulsive guidance molecule b (RGMb) as one such molecule.
Following the establishment of RGM as a gene
involved in the guidance of chick retinal axons (7), three mouse orthologues (termed RGMa,
RGMb and RGMc) were isolated and characterised in separate studies,
displaying predominant expression in the developing and adult
nervous system (RGMa and RGMb) and skeletal muscles (RGMc)
(8–10). An independent study, conducted at
the same time, also identified DRAGON (RGMb) using a genome binding
strategy to screen for DRG11 regulated genes (11).
RGMb shares structural similarities with the other
members of the RGM family, including a N-terminal signal peptide, a
proteolytic cleavage site, a von Willebrand factor (vWF) type D
domain and a glycophosphatidylinositol (GPI) anchor, and is
expressed in the developing and adult nervous system (7–11).
RGMb has also been shown to be involved in promoting mouse dorsal
root ganglion (DRG) neuron adhesion (11), has been implicated in axonal
regeneration after injury (12)
and in response to spinal cord injury, where it is found to be
upregulated (13). In addition to
its roles in the nervous system, RGMb has also been shown to
contribute to bone morphogenetic protein (BMP) signalling, where it
has been identified, together with RGMa and RGMc, as a BMP
co-receptor (14–17). Additionally, RGMb may play an
important role in reproduction through the enhancement of BMP
signalling (18) and a potential
role for RGMb has also been demonstrated in the immune system where
it can inhibit IL-6 expression in macrophages through the p38 MAPK
and ERK1/2 pathway, but not the Smad 1/5/8 pathway, in a
BMP-ligand-dependent manner (19).
A number of recent studies from our laboratories
have explored the importance of the members of the RGM family in
breast and prostate cancer (20–22).
RGM levels were examined in a breast cancer cohort in conjunction
with clinical pathological patient data. This study highlighted an
aberrant expression profile of the RGMs in breast cancer,
indicating potential links between RGMa and RGMb and patient
prognosis (20). Further work
explored the impact of targeting RGMb in breast cancer cell lines
in vitro. This study demonstrated a potential role for RGMb
in breast cancer proliferation, matrix-adhesion and migration where
targeting this molecule enhanced these aggressive traits and
implicated links with Smad-dependent and Smad-independent pathways
(22). Similarly, RGMb appears to
play a role in prostate cancer progression. Knockdown of RGMb in
the PC-3 prostate cancer cell line enhanced cell growth and
migratory rates and also increased cell-matrix adhesion.
Additionally, RGMb may be linked to Smad signalling in this cell
line as knockdown of RGMb appeared to affect the levels of
activated Smad 1 and 3, and also enhanced ID1 expression (21). Taken together these studies suggest
important roles for RGMb in breast and prostate cancer progression
and also imply an involvement in BMP and Smad signalling.
In the present study, the potential involvement of
RGMb was explored in the process of tumour angiogenesis through the
targeting of RGMb in HECV endothelial cells. Suppression of RGMb in
HECV cells did not result in significant changes to a number of
traits associated with the angiogenic process. However, we report
that suppression of RGMb in HECV cells can act to decrease or
inhibit the pro-angiogenic effect brought about by HGF and, more
substantially, BMP-7 on tubule formation and migration rates. This
study highlights the potential importance of RGMb in propagating
pro-angiogenic effects of HGF and BMP-7.
Materials and methods
Reagents, cell lines and culture
conditions
Human HECV endothelial cells were purchased from
Interlab Cell Line Collection (ICLC, Naples, Italy). The MCF-7
breast cancer cell line and the PC-3 human prostate cancer cell
line were purchased from the American Type Culture Collection
(ATCC, Rockville, MD, USA). Hepatocyte growth factor (HGF) was a
kind gift from Dr T. Nakamura (Osaka University Medical School,
Osaka, Japan) and bone morphogenetic protein-7 (BMP-7) was
purchased from Sigma (Dorset, UK). All cells were maintained in
Dubecco’s modified Eagle’s medium (DMEM) (PAA Laboratories Ltd.,
Somerset, UK), supplemented with penicillin, streptomycin and 10%
fetal calf serum (PAA Laboratories Ltd.) and incubated at 37°C, 5%
CO2 and 95% humidity.
Generation of human HECV endothelial
cells displaying suppressed RGMb expression
RGMb expression was targeted in human HECV
endothelial cells using a ribozyme transgene specifically generated
to target and cleave RGMb transcript. This methodology has been
previously reported (23,24). Plasmids containing ribozyme
transgenes had previously been developed within the laboratory
(21,22). Briefly, ribozyme transgene
sequences were designed based on the predicted secondary structure
of the RGMb transcript using Zukers RNA Mfold program (25) and were synthesised by Invitrogen
(Paisley, UK). Ribozyme transgenes were subsequently cloned into a
pEF6/V5-His-TOPO plasmid vector (Invitrogen). Both control pEF6
plasmids containing no insert, and plasmids containing the RGMb
ribozyme transgene were transfected into HECV cells using
electroporation. Following transfection, these cells underwent a
selection period and subsequent verification of RGMb knockdown.
Cells containing the RGMb ribozyme transgene were termed
HECVRGMb KO and were compared throughout the study to
control HECV cells containing closed plasmids alone, termed
HECVpEF6.
RNA extraction and reverse
transcription-polymerase chain reaction (RT-PCR)
Cells were grown to confluence in a
25-cm2 flask before RNA was extracted using total RNA
isolation (TRI) reagent (Sigma) in accordance with the supplied
protocol. RNA was subsequently quantified using a spectrophotometer
(WPA UV 1101, Biotech Photometer, Cambridge, UK), standardised to a
concentration of 500 ng and used as a template to generate cDNA
using an iScript cDNA synthesis kit (Bio-Rad Laboratories Ltd.,
Hemel Hempstead, UK). Following cDNA synthesis, sample quality and
uniformity was checked, using β-actin primers, before assessing
RGMb expression using specific primers for RGMb transcript (full
primer details are shown in Table
I). PCR was performed using REDTaq® ReadyMix™ PCR
Reaction Mix (Sigma). The following reaction conditions were set up
in a T-Cy Thermocycler (Creacon Technologies Ltd., The
Netherlands); denaturing at 94°C for 40 sec, annealing at 55°C for
40 sec and extension at 72°C for 60 sec. PCR was conducted over 34
cycles following an initial 5-min denaturing step (94°C) and
concluded with a final 10-min extension step (72°C). Amplified
products were loaded onto an agarose gel, separated
electrophoretically, stained in ethidium bromide and visualised
under ultraviolet light.
 | Table IPrimers used in the study. |
Table I
Primers used in the study.
| Primer set | Sense | Anti-sense |
|---|
| β-actin probe |
ATGATATCGCCGCGCTCG |
CGCTCGGTGAGGATCTTCA |
| RGMb probe |
GGATCCAGTGCTACTGCTAC |
GTAAAGTTGGCATCACCAGT |
| GAPDH qPCR |
CTGAGTACGTCGTGGAGTC |
ACTGAACCTGACCGTACACAGAGATGAT
GACCCTTTTG |
| RGMb qPCR |
TTCAGTTCAAGTGACAAACG |
ACTGAACCTGACCGTACATCATCTGTCACAGCTTGGTA |
Microarray analysis of gene
expression
Two sets of triplicate HECV cell flasks were treated
with either 40 ng/ml hepatocyte growth factor (HGF) for 4 h or
remained untreated and RNA extracted as described above using TRI
reagent (Sigma). Extracted RNA was quantified before being sent to
the Cardiff University Central Biotechnology Services (CBS)
microarray facility for labelling and hybridisation to a GeneChip
Human Genome U133 Plus 2.0 array (Affymetrix UK Ltd., High Wycombe,
UK).
Quantitative polymerase chain reaction
(qPCR)
Quantitative PCR was used to assess RGMb transcript
levels in control and transfected cell lines following a previously
reported method (26,27). Briefly, the iCycler IQ system was
used to detect and quantify RGMb transcript expression in each
sample. Transcript copy number was calculated based on an internal
standard. Samples were normalised against GAPDH expression (see
Table I for primer details). The
Amplifluor system (Intergen Inc., New York, NY, USA) was utilised
together with qPCR Master Mix (ABgene, Surrey, UK). Conditions for
qPCR were; 15-min initial 95°C period followed by 60 cycles of 95°C
for 15 sec, 55°C for 60 sec and 72°C for 20 sec.
SDS-PAGE and western blotting
Protein was extracted from a confluent
75-cm2 tissue culture flask. Cells were detached and
lysed in HCMF buffer containing 0.5% SDS, 1% Triton X-100, 2 mM
CaCl2, 100 μg/ml phenylmethylsulfonyl fluoride, 1 mg/ml
leupeptin, 1 mg/ml aprotinin and 10 mM sodium orthovanadate on a
rotor wheel for 1 h before removal of insolubles through
centrifugation at 13,000 g. The Bio-Rad DC Protein assay kit
(Bio-Rad Laboratories, CA, USA) was used to quantify protein levels
in the samples. Samples were subsequently standardised to 2 mg/ml
and diluted in Laemmli 2X concentrate sample buffer (Sigma) before
being boiled for 5 min. Samples were loaded onto a 10% acrylamide
gel and separated electrophoretically. Following separation the
proteins were blotted onto a Hybond-C Extra nitrocellulose membrane
(Amersham Biosciences UK Ltd., Bucks, UK) and blocked in 10% milk.
RGMb expression was detected using anti-RGMb antibody (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA). In addition to this,
GAPDH expression was also detected, to assess sample uniformity,
using an anti-GAPDH antibody (Santa Cruz Biotechnology, Inc.).
Following binding of the primary antibody, the membranes were
probed with peroxidase conjugated anti-rabbit (RGMb) or anti-mouse
(GAPDH) secondary antibodies (Sigma). Expression was then
visualised through the Supersignal West Dura Extended Duration
substrate chemi-luminescent system (Perbio Science UK Ltd.,
Cramlington, UK) and detected using a UVIProChem camera system
(UVItec Ltd., Cambridge, UK).
In vitro cell growth assay
An in vitro growth assay was used to examine
the impact of RGMb on cell growth. Cells were seeded into 96-well
plates at a density of 3,000 cells/well and triplicate plates were
set up to allow overnight, 3- and 5-day incubation periods.
Following incubation, the plates were fixed in 4% formaldehyde
(v/v) and stained with 0.5% (w/v) crystal violet. Subsequently, 10%
acetic acid (v/v) was used to extract the crystal violet stain and
cell density was detected through spectrophotmeric analysis using a
Bio-Tek ELx800 multi-plate reader (Bio-Tek Instruments Inc., VT,
USA).
In vitro cell migration/wounding
assay
Cellular migration was assessed using a
migration/wounding assay modified from a previously described
method (28). Briefly, cells were
cultured to confluence in a 24-well plate before scratching the
cell monolayer with a pointed plastic pipette tip. Subsequently,
wound closure, through the migration of cells, was tracked and
photographed at 15-min interval time-points over a 90-min period
using an inverted microscope and GXcapture software. The distance
between the two wound fronts was calculated at several consistent
points over all the time intervals using ImageJ software and
average cellular migration calculated.
In vitro tubule formation assay
The potential of RGMb to contribute to the process
of HECV tubule formation was assessed in vitro using a
Matrigel endothelial cell tubule formation assay modified from a
previously reported study (29).
Briefly, 500 μg of Matrigel, diluted in serum-free medium, was
seeded into a 96-well plate and incubated for a minimum of 40 min
to allow setting of the Matrigel. Once set, 35,000 HECV endothelial
cells (HECVpEF6 or HECVRGMb KO cells) were
seeded onto the Matrigel layer and incubated for 4–5 h. Tubule
formation occurring over the incubation period was visualised under
low magnification and images captured for analysis. Total tubule
perimeter per field was subsequently quantified using ImageJ
software. Where appropriate, treatments consisting of either 40
ng/ml BMP-7 or 40 ng/ml HGF were added to the cell medium following
seeding.
In vivo tumour development assay
The impact of RGMb suppression in vivo was
examined using a previously described in vivo angiogenesis
model (29). Briefly, a 100-μl
suspension containing 1×106 cancer cells (either
MCF-7pEF6 breast cancer cells or PC-3pEF6
prostate cancer cells) were subcutaneously injected into the left
and right flanks of 4–6-week old athymic nude mice (CD-1; Charles
River Laboratories, Kent, UK) together with either 1×106
HECVpEF6 or 1×106 HECVRGMb KO
cells in a 0.5 mg/ml Matrigel solution. The tumours were allowed to
develop over the course of the experiment and were measured twice
weekly using vernier callipers under sterile conditions. The mice
were housed in filter top units and were treated humanely in
accordance with United Kingdom Home Office and the United Kingdom
Coordinating Committee on Cancer Research (UKCCCR) guidelines. All
in vivo work undertaken in this study was conducted under
the project license (PPL 30/2591) of the British Home Office.
Animals were dispatched humanely if severity limits were reached or
at the experimental end point using a schedule 1 method. Tumour
volume was calculated for each time-point using the following
formula: Tumour volume = 0.523 × width2 × length.
Statistical analysis
The Minitab 14 statistical package was used to test
for statistical differences between RGMb knockdown HECV cells and
the pEF6 vector control HECV cells using a two sample, two tailed
t-test. Experimental procedures were repeated a minimum of three
independent times. Data represents mean values ± SEM, values of
p<0.05 were regarded as statistically significant.
Results
Hepatocyte growth factor can regulate
repulsive guidance molecule b expression
A microarray study was conducted to examine how gene
expression within the HECV human endothelial cell line was affected
following 4-h treatment with 40 ng/ml HGF. Treatment with HGF
caused a range of differential gene expression within this
endothelial cell line. Repulsive guidance molecule b (RGMb)
expression was significantly increased following HGF treatment
(p=0.004 vs. untreated control cells) and the expression of RGMb
was found to be enhanced ~2.5 times by HGF treatment (Fig. 1).
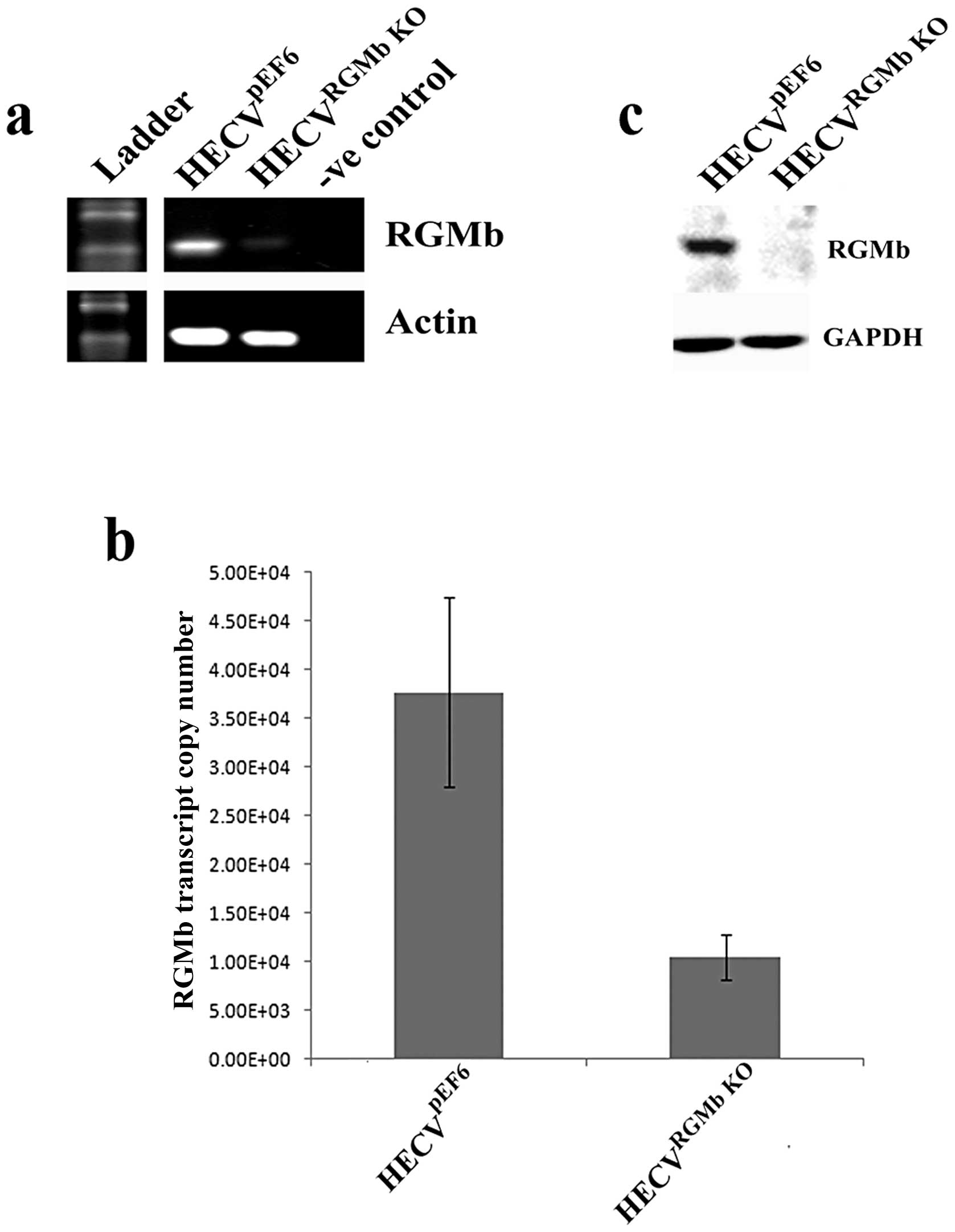
Suppression of RGMb expression using a
ribozyme transgene system
RGMb expression was successfully knocked down
following transfection of HECV cells with a pEF6 plasmid containing
a ribozyme transgene specifically targeted to RGMb transcript.
Reduced RGMb transcript expression can be seen in the HECVRGMb
KO cells in comparison to empty plasmid control
HECVpEF6 cells using RT-PCR and quantitative PCR
(Fig. 2A and B). Similarly,
reduced RGMb protein levels were observed, using western blot
analysis, in HECV cells transfected with the ribozyme transgene in
comparison to control cells (Fig.
2C).
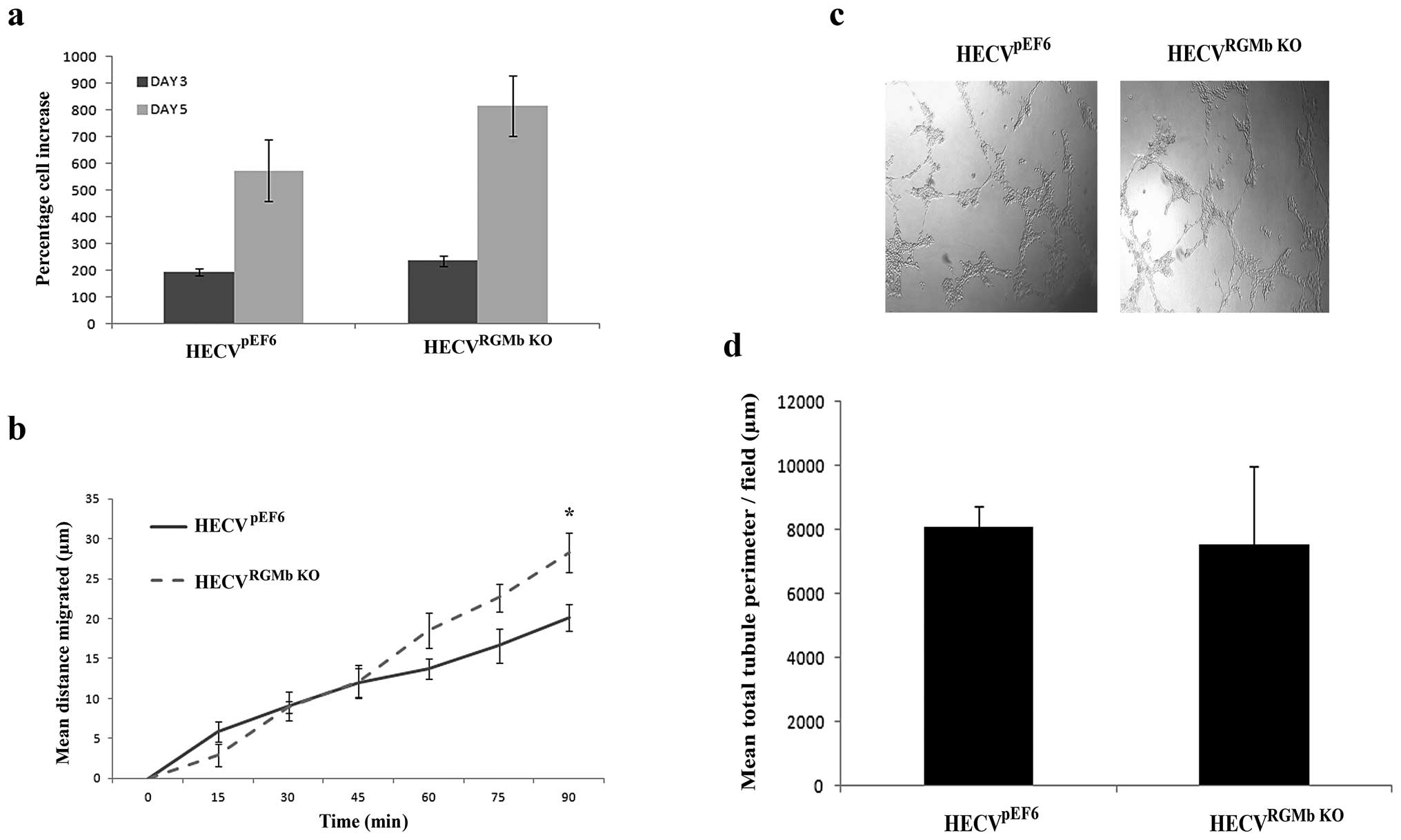
Impact of RGMb suppression on endothelial
in vitro cell traits
Following successful targeting of RGMb, the impact
of this knockdown on endothelial cell functions was examined in
vitro. Knockdown of RGMb did not significantly alter the rate
of cell growth in HECV cells (Fig.
3A), however, the suppression did tend to increase growth rates
over both a 3- and 5-day incubation period (HECVpEF6 vs.
HECVRGMb KO, p=0.08, 3-day incubation; 5-day incubation
p=0.15). Suppression of RGMb appeared to impact later stages of
HECV migration (Fig. 3B), with
enhanced levels of migration being observed in HECVRGMb
KO cells compared to HECVpEF6 control cells at 75
min (p=0.069) and 90 min (p=0.035). Our results also suggest that
knockdown of RGMb has little effect on HECV tubule formation and
thus angiogenic potential in vitro (Fig. 3C and D), where using this
angiogenic assay little difference in levels of tubule formation
was observed between control HECVpEF6 and HECVRGMb
KO cells. No significant difference between quantified total
tubule perimeter levels was seen between HECVpEF6 and
HECVRGMb KO cells (p=0.813).
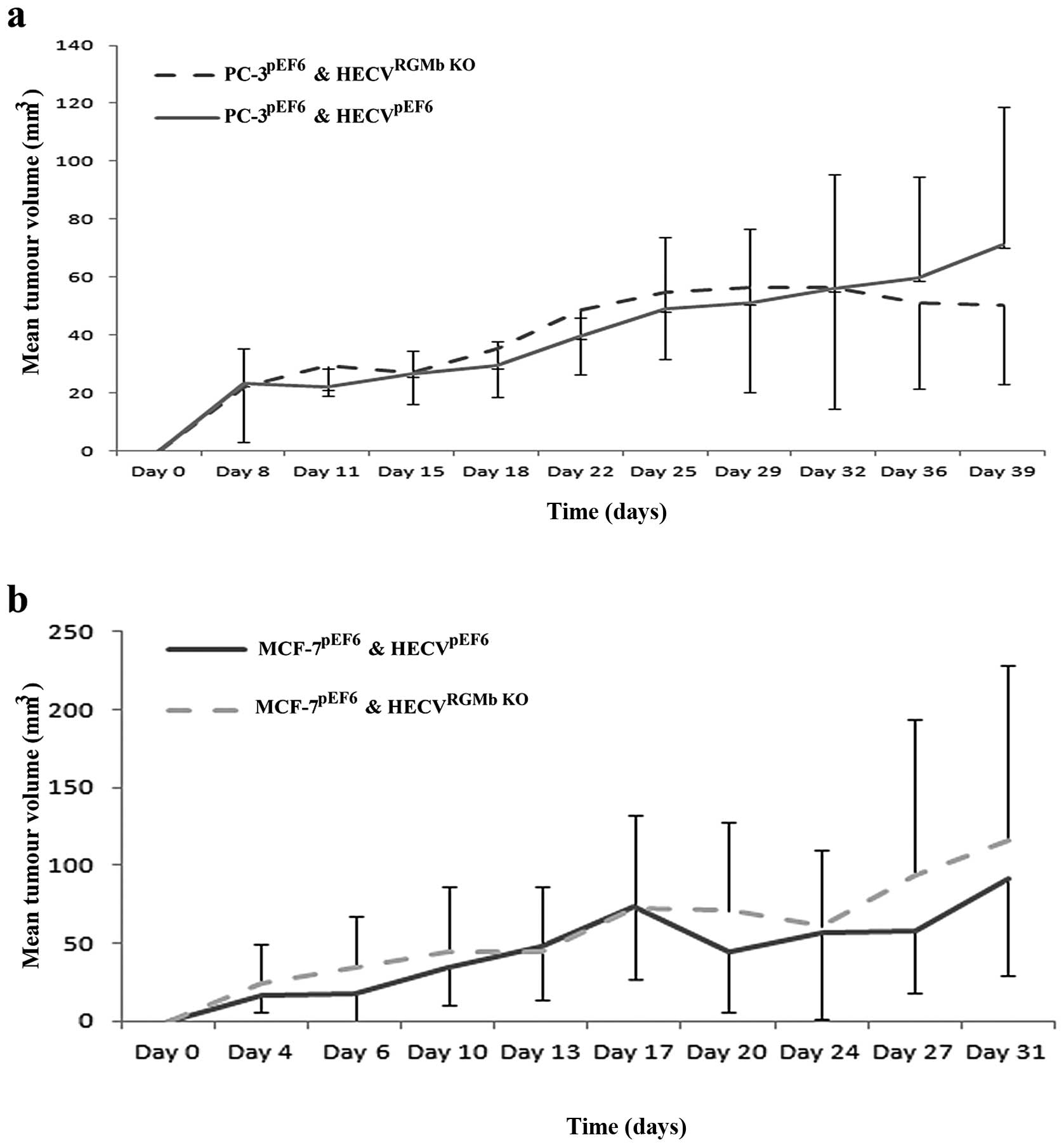
Suppression of RGMb has limited impact in
vivo
We next investigated the role of RGMb knockdown
using an in vivo angiogenesis model, whereby endothelial
cells, either control or RGMb suppressed, were inoculated alongside
cancer cells to examine their potential to impact on the
development of the cancer cell tumour. This was tested in a
prostate cancer model (Fig. 4A),
where PC-3 cells were used and a breast cancer model (Fig. 4B), where MCF-7 cells were used. In
both cases little difference was observed between tumour
development involving control HECVpEF6 or HECVRGMb
KO cells, with neither model yielding statistically different
levels of tumour development (p>0.05 in both cases).
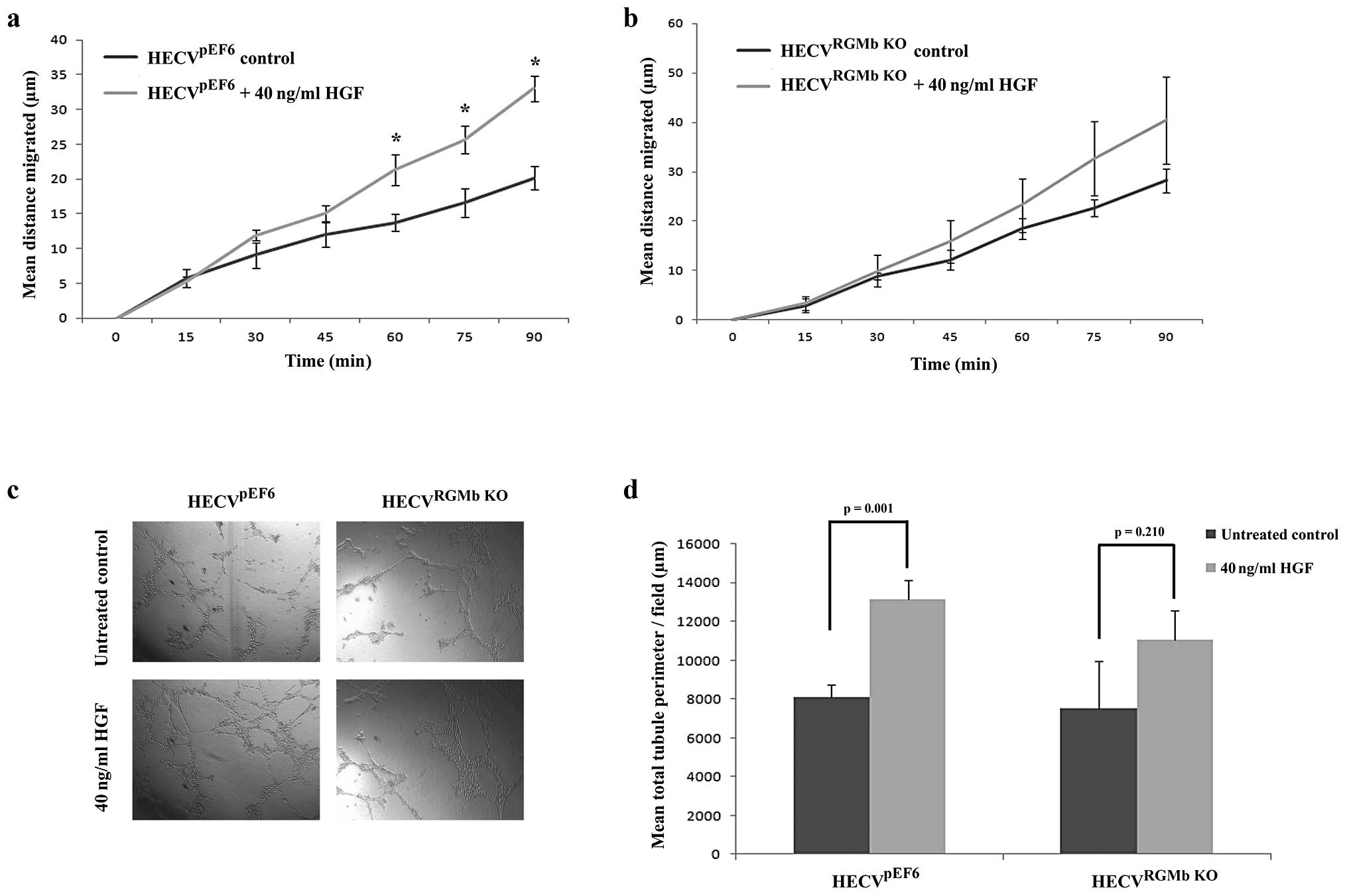
RGMb suppression can limit pro-angiogenic
effects of HGF
Following on from establishing the involvement of
RGMb in traits associated with the angiogenic cascade, we next
looked to identify its importance in how HGF could enhance these
angiogenic traits (Fig. 5).
Following treatment with 40 ng/ml HGF, a significant increase was
seen in HECVpEF6 cell migration rates (Fig. 5A). Treatment with HGF significantly
enhanced the distance migrated by HECVpEF6 cells
following 60- (p=0.02), 75- (p=0.02) and 90-min (p=0.001)
time-points. In contrast, when RGMb was targeted in this
endothelial cell line, treatment with 40 ng/ml HGF did not appear
to have as great a pro-migratory effect (Fig. 5B). General increases in migration
rates were observed in the later time-points of the experiment but
these were not found to be statistically significant (p=0.44 at 60
min, p=0.24 at 75 min and p=0.23 at 90 min). Similar trends were
observed with tubule formation capacity (Fig. 5C and D). In keeping with its
established pro-angiogenic role, HGF significantly enhanced the
level of tubule formation in this in vitro angiogenic assay
in the control HECVpEF6 cell line (untreated
HECVpEF6 vs. 40 ng/ml HGF treated HECVpEF6,
p=0.001). However, when HECVRGMb KO cells were treated
with HGF the increase in tubule formation was not as great and no
significant difference was observed between untreated and 40 ng/ml
HGF treated HECVRGMb KO cells (p=0.210).
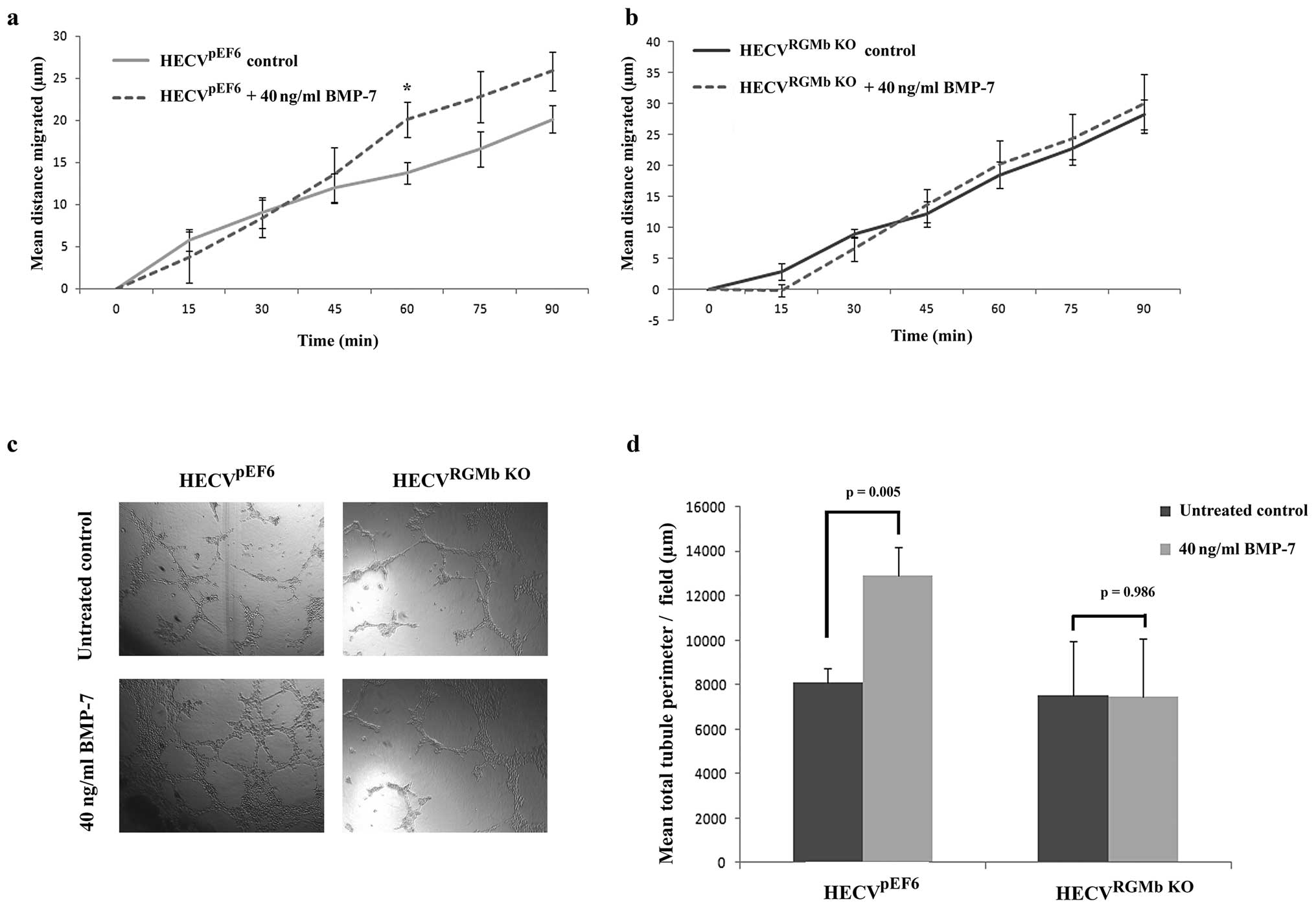
RGMb suppression inhibits BMP-7
pro-angiogenic response
As the RGM family have been identified as BMP
co-receptors, we also examined the impact of suppressing RGMb
expression on HECV BMP promoted responses, in particular BMP-7. In
control HECVpEF6 cells, treatment with BMP-7 caused a
notable increase in cell migration (Fig. 6A), which was apparent in the later
stages of the experiment (untreated HECVpEF6 vs. 40
ng/ml BMP-7 treated HECVpEF6; 60 min, p=0.037; 75 min,
p=0.144; 90 min, p=0.089). In contrast to this, suppression of RGMb
in HECV endothelial cells removed the migratory response of this
cell line to BMP-7 with very little difference being observed
between the migration pattern of HECVRGMb KO cells
treated with 40 ng/ml BMP-7 and control, untreated HECVRGMb
KO cells (Fig. 6B). A
similar trend was also apparent in the tubule formation experiments
(Fig. 6C and D). In control
HECVpEF6 cells, treatment with 40 ng/ml BMP-7 caused an
increase in the tubule formation potential of these endothelial
cells and treatment of HECVpEF6 cells with 40 ng/ml
BMP-7 brought about a significant increase in total tubule
perimeter (p=0.005). As with cell migration, suppression of RGMb in
HECV cells removed the cells responsiveness to BMP-7 treatment and
no significant difference in total tubule perimeter was observed
between untreated HECVRGMb KO and 40 ng/ml BMP-7 treated
HECVRGMb KO cells (p=0.986).
Discussion
In the present study we used a microarray approach
to identify gene expression that could be regulated by HGF and
identified RGMb as a gene whose expression was enhanced ~2.5-fold
following 4-h treatment with 40 ng/ml HGF. This data implicates
RGMb as a HGF regulated gene in HECV human endothelial cells.
Subsequently, we examined the potential of RGMb to impact on
angiogenesis related factors in vitro and in vivo.
RGMb expression was successfully targeted, using a ribozyme
transgene system, in HECV cells resulting in a strong reduction in
RGMb transcript and protein levels. Suppression of RGMb alone,
however, did not appear to have substantial effects on HECV cells
and only appeared to significantly influence cell migratory rates
in the later stages of a scratch wounding migration assay. In
vitro growth and tubule formation revealed no significant
differences in RGMb suppressed compared to control HECV cells,
though growth rates were elevated somewhat in the HECVRGMb
KO cell line. Similar trends were observed in an in
vivo model involving the co-inoculation of HECV endothelial
cells and breast or prostate tumour cells suggesting that, solely,
RGMb may contribute little to the angiogenic process. The results
observed here regarding the role of RGMb in the basic function of
endothelial cells are somewhat in line with previous work from our
laboratories focusing on breast and prostate cancer. In both of
these studies, knockout of RGMb resulted in enhanced migratory
rates of prostate (PC-3) and breast (MDA-MB-231, MCF-7) cancer cell
lines and enhanced cell growth rates in PC-3 and MDA-MB-231 cells,
though not MCF-7 cells (21,22).
Together with our results the data suggest RGMb is involved in the
regulation of cell migration and growth of certain cancer and to
some extent endothelial cells.
Despite RGMb having minimal effect independently on
these angiogenic traits, a role for RGMb was discovered in
facilitating pro-angiogenic effects brought about by HGF. HECV
cells displaying reduced RGMb expression were less able to respond
in a pro-angiogenic manner to HGF treatment, and angiogenic traits
such as cell migration and tubule formation capacity were not
significantly enhanced by HGF treatment in HECVRGMb KO
cells as they were in HGF treated HECVpEF6 cells. Taken
together with the establishment of HGFs capacity to regulate RGMb
gene expression, our data provides evidence that RGMb may act as a
potential mechanism to bring about HGFs pro-angiogenic effects.
In addition to examining the potential of RGMb to
contribute to progressing the pro-angiogenic effects of HGF, and in
light of the established roles of members of the RGM family as BMP
co-receptors (14–17) we also aimed to explore the
potential of RGMb to facilitate the pro-angiogenic effects of
BMP-7. The BMPs are members of the transforming growth factor β
(TGF-β) family, exerting their signals through type I and II
transmembrane serine/theronine kinase receptors to influence a
plethora of biological processes including angiogenesis. BMP-2, -4,
-6 and -7 have been implicated in both the activation phase, where
they can enhance endothelial cell proliferation and migration, and
also the maturation phase, where they can influence vascular smooth
muscle cells (reviewed in refs. 30 and 31). Interestingly, suppression of RGMb
in HECV cells, whilst reducing the pro-angiogenic effects of HGF,
was also found to substantially inhibit BMP-7 promoted cell
migration and tubule formation. The pro-migratory impact of BMP-7
itself was not as great as that of HGF, a well established motogen
(3), with BMP-7 enhancement of
migration rates not showing as significant increases in migration
rates of control HECV cells as HGF. However, suppression of RGMb in
HECV cells could substantially remove any pro-migratory effect of
BMP-7 with migration rates showing similar levels to that of
untreated HECVRGMb KO controls. In keeping with this
trend a similar pattern was seen on tubule formation capacity. In
control HECVpEF6 cells, treatment with 40 ng/ml BMP-7
brought about a significant increase in tubule formation levels.
Suppression of RGMb again inhibited this pro-tubule formation
effect with mean tubule formation levels comparable to untreated
controls and no significant difference being observed between
untreated and treated HECVRGMb KO cells. The data appear
to be in line with the current literature, identifying RGMs as BMP
co-receptors (14–17) and this study demonstrates the
importance of this in contributing to the angiogenic process.
It is noteworthy that HGF has previously been
demonstrated to enhance the expression of BMP-7 and the expression
of the BMPR-IB and BMPR-II BMP receptors in prostate cancer cells
(32,33). A similar trend has also been
observed briefly in the HECV endothelial cells used in this study,
where treatment with HGF was found, using qPCR, to enhance BMP-7
levels in a time course experiment (data not shown). It may be
worth considering that crosstalk between these two pathways may
present a partial means to explain the results obtained in our
study. A weaker response was apparent, particularly in the tubule
formation experiments, in RGMb knockdown cells to BMP-7 rather than
HGF. It is possible that HGF may partially act to enhance BMP
signalling, possibly through enhancement of BMP-7 and/or BMP
receptor levels in these cells, which in turn could be interrupted
through RGMb suppression. This however, requires further scientific
examination before conclusions can be drawn.
Other studies conducted in our laboratories have
implicated members of the RGM family in breast and prostate cancer.
Knockdown of RGMb was found to enhance PC-3 prostate cancer cell
growth, cell-matrix adhesion and migration. Additionally, knockdown
of RGMb enhanced levels of activated Smad 3 and ID1 expression, a
trend that was increased through treatment with BMP-7, but
generally reduced activated levels of Smad 1 (21). Similar to the prostate cancer
study, knockdown of RGMb in MDA-MB-231 breast cancer cells was also
found to enhance cell growth, matrix adhesion and migration.
Knockdown of RGMb was also found to facilitate survival from
apoptosis under serum starvation. Alterations, following RGMb
knockdown, were observed in the regulation of c-myc, caspase-3,
SNAIL, TWIST, FAK and paxillin. This study also implicated RGMb
knockdown to contribute to the Smad-dependent pathway, enhancing
levels of activated Smad 1 and 3 in untreated cells, with greater
responses seen following BMP-7 treatment and a switching to Smad 1
activation following inhibition of Smad 3, whereas phosphorylation
of JNK, ILP and TAK was inhibited following RGMb knockdown
suggesting suppression of the Smad-independent pathway (22).
In the present study, knockdown of RGMb in human
endothelial cells enhanced migration and also appeared to suppress
the pro-angiogenic response to HGF and BMP-7. Taken together, this
suggests that, while loss of RGMb may enhance aggressive traits in
prostate and breast cancer cells, suppression of RGMb may also act
to suppress HGF and BMP-7 mediated tumour angiogenesis. Also, in
contrast to the previous prostate and breast cancer studies, where
treatment of RGMb knockdown cells with BMP-7 enhances activity of
Smad 1 or 3, our present study implies that in HECV cells RGMb
knockdown may act to suppress signalling resulting from BMP-7
treatment. RGMb has been identified as a BMP co-receptor, directly
binding BMP-2, and -4 but not -7 or TGF-β ligands and associating
with the ALK2, 3 and 6, BMP type I and the ActRII and ActRIIB BMP
type II receptors (14). Since its
discovery a number of studies have shown this molecule to play a
role in enhancing BMP signalling (12, 14,
18). However, RGMb has also
demonstrated the ability to inhibit BMP signalling in C2C12
myoblasts (34). These
observations, taken with the data presented here and the previous
prostate and breast cancer studies (21,22)
suggest a complex relationship between RGMb and BMP signalling.
Further scientific study is required to establish fully the
downstream effectors of BMP-7 and HGF treatment in endothelial
cells. The data presented in this study have, for the first time,
raised the implication that RGMb may play some complex role(s) in
the process of tumour angiogenesis mediated by HGF and BMP-7 and
further links RGMb to tumour progression.
Acknowledgements
The authors wish to thank Cancer Research Wales and
the Henry Fong Family Foundation for supporting this study.
References
|
1
|
Folkman J: Angiogenesis: an organizing
principle for drug discovery? Nat Rev Drug Discov. 6:273–286. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Potente M, Gerhardt H and Carmeliet P:
Basic and therapeutic aspects of angiogenesis. Cell. 146:873–887.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jiang WG, Martin TA, Parr C, Davies G,
Matsumoto K and Nakamura T: Hepatocyte growth factor, its receptor,
and their potential value in cancer therapies. Crit Rev Oncol
Hematol. 53:35–69. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wojta J, Kaun C, Breuss JM, et al:
Hepatocyte growth factor increases expression of vascular
endothelial growth factor and plasminogen activator inhibitor-1 in
human keratinocytes and the vascular endothelial growth factor
receptor flk-1 in human endothelial cells. Lab Invest. 79:427–438.
1999.
|
|
5
|
Davies G, Mason MD, Martin TA, et al: The
HGF/SF antagonist NK4 reverses fibroblast- and HGF-induced prostate
tumor growth and angiogenesis in vivo. Int J Cancer. 106:348–354.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martin TA, Parr C, Davies G, et al: Growth
and angiogenesis of human breast cancer in a nude mouse tumour
model is reduced by NK4, a HGF/SF antagonist. Carcinogenesis.
24:1317–1323. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Monnier PP, Sierra A, Macchi P, et al: RGM
is a repulsive guidance molecule for retinal axons. Nature.
419:392–395. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oldekamp J, Kramer N, Alvarez-Bolado G and
Skutella T: Expression pattern of the repulsive guidance molecules
RGM A, B and C during mouse development. Gene Expr Patterns.
4:283–288. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schmidtmer J and Engelkamp D: Isolation
and expression pattern of three mouse homologues of chick Rgm. Gene
Expr Patterns. 4:105–110. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Niederkofler V, Salie R, Sigrist M and
Arber S: Repulsive guidance molecule (RGM) gene function is
required for neural tube closure but not retinal topography in the
mouse visual system. J Neurosci. 24:808–818. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Samad TA, Srinivasan A, Karchewski LA, et
al: DRAGON: a member of the repulsive guidance molecule-related
family of neuronal- and muscle-expressed membrane proteins is
regulated by DRG11 and has neuronal adhesive properties. J
Neurosci. 24:2027–2036. 2004. View Article : Google Scholar
|
|
12
|
Ma CH, Brenner GJ, Omura T, et al: The BMP
coreceptor RGMb promotes while the endogenous BMP antagonist noggin
reduces neurite outgrowth and peripheral nerve regeneration by
modulating BMP signaling. J Neurosci. 31:18391–18400. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu X, Hashimoto M, Horii H, Yamaguchi A,
Naito K and Yamashita T: Repulsive guidance molecule b inhibits
neurite growth and is increased after spinal cord injury. Biochem
Biophys Res Commun. 382:795–800. 2009. View Article : Google Scholar
|
|
14
|
Samad TA, Rebbapragada A, Bell E, et al:
DRAGON, a bone morphogenetic protein co-receptor. J Biol Chem.
280:14122–14129. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Babitt JL, Zhang Y, Samad TA, et al:
Repulsive guidance molecule (RGMa), a DRAGON homologue, is a bone
morphogenetic protein co-receptor. J Biol Chem. 280:29820–29827.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Babitt JL, Huang FW, Wrighting DM, et al:
Bone morphogenetic protein signaling by hemojuvelin regulates
hepcidin expression. Nat Genet. 38:531–539. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Halbrooks PJ, Ding R, Wozney JM and Bain
G: Role of RGM coreceptors in bone morphogenetic protein signaling.
J Mol Signal. 2:42007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xia Y, Sidis Y, Mukherjee A, et al:
Localization and action of Dragon (repulsive guidance molecule b),
a novel bone morphogenetic protein coreceptor, throughout the
reproductive axis. Endocrinology. 146:3614–3621. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xia Y, Cortez-Retamozo V, Niederkofler V,
et al: Dragon (repulsive guidance molecule b) inhibits IL-6
expression in macrophages. J Immunol. 186:1369–1376. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li J, Ye L, Mansel RE and Jiang WG:
Potential prognostic value of repulsive guidance molecules in
breast cancer. Anticancer Res. 31:1703–1711. 2011.PubMed/NCBI
|
|
21
|
Li J, Ye L, Kynaston HG and Jiang WG:
Repulsive guidance molecules, novel bone morphogenetic protein
co-receptors, are key regulators of the growth and aggressiveness
of prostate cancer cells. Int J Oncol. 40:544–550. 2012.PubMed/NCBI
|
|
22
|
Li J, Ye L, Sanders AJ and Jiang WG:
Repulsive guidance molecule B (RGMB) plays negative roles in breast
cancer by coordinating BMP signaling. J Cell Biochem.
113:2523–2531. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sanders AJ, Parr C, Mason MD and Jiang WG:
Suppression of hepatocyte growth factor activator inhibitor-1 leads
to a more aggressive phenotype of prostate cancer cells in
vitro. Int J Mol Med. 20:613–619. 2007.
|
|
24
|
Yuan Z, Sanders AJ, Ye L, Wang Y and Jiang
WG: Knockdown of human antigen R reduces the growth and invasion of
breast cancer cells in vitro and affects expression of
cyclin D1 and MMP-9. Oncol Rep. 26:237–245. 2011.PubMed/NCBI
|
|
25
|
Zuker M: Mfold web server for nucleic acid
folding and hybridization prediction. Nucleic Acids Res.
31:3406–3415. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Parr C, Sanders AJ, Davies G, et al:
Matriptase-2 inhibits breast tumor growth and invasion and
correlates with favorable prognosis for breast cancer patients.
Clin Cancer Res. 13:3568–3576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Parr C, Watkins G, Mansel RE and Jiang WG:
The hepatocyte growth factor regulatory factors in human breast
cancer. Clin Cancer Res. 10:202–211. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang WG, Hiscox SE, Parr C, et al:
Antagonistic effect of NK4, a novel hepatocyte growth factor
variant, on in vitro angiogenesis of human vascular endothelial
cells. Clin Cancer Res. 5:3695–3703. 1999.
|
|
29
|
Sanders AJ, Ye L, Mason MD and Jiang WG:
The impact of EPLINalpha (Epithelial protein lost in neoplasm) on
endothelial cells, angiogenesis and tumorigenesis. Angiogenesis.
13:317–326. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
David L, Feige JJ and Bailly S: Emerging
role of bone morphogenetic proteins in angiogenesis. Cytokine
Growth Factor Rev. 20:203–212. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cai J, Pardali E, Sanchez-Duffhues G and
ten Dijke P: BMP signaling in vascular diseases. FEBS Lett.
586:1993–2002. 2012. View Article : Google Scholar
|
|
32
|
Ye L, Lewis-Russell JM, Davies G, Sanders
AJ, Kynaston H and Jiang WG: Hepatocyte growth factor up-regulates
the expression of the bone morphogenetic protein (BMP) receptors,
BMPR-IB and BMPR-II, in human prostate cancer cells. Int J Oncol.
30:521–529. 2007.PubMed/NCBI
|
|
33
|
Ye L, Lewis-Russell JM, Sanders AJ,
Kynaston H and Jiang WG: HGF/SF up-regulates the expression of bone
morphogenetic protein 7 in prostate cancer cells. Urol Oncol.
26:190–197. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kanomata K, Kokabu S, Nojima J, Fukuda T
and Katagiri T: DRAGON, a GPI-anchored membrane protein, inhibits
BMP signaling in C2C12 myoblasts. Genes Cells. 14:695–702. 2009.
View Article : Google Scholar : PubMed/NCBI
|