Introduction
Lung cancer is the most common cancer and
responsible for 19.4% of cancer death with 87% mortality rate
worldwide. A third of patients with lung cancer are Chinese and the
fatality is as high as 91.3% in China (1). Deregulated cell proliferation and
failure of cell apoptosis are critical events to propel neoplastic
progression (2). Besides the
internal straightforward linear accumulation of oncogenic
mutations, potential proliferation and apoptosis-inhibitory signals
responding to external pressure account for evolution of tumor.
Hypoxia is a characteristic driver of solid carcinoma. During the
process of tumor formation and rapid growth, oxygen diffusion
limitation and poor blood perfusion lead to hypoxia (3). To adapt to low oxygen level,
anomalous proliferation and reduced apoptosis which result in lower
supply of oxygen have been developed in hypoxic tumors (4). The vicious circle of tumor
progression has long been a major focus for cancer treatment.
However, previous studies showed an inconsistent role of hypoxia in
A549 cells (5,6).
Hypoxia-inducible factor 1 (HIF-1) α plays a central
role in this process as a master regulator of oxygen homeostasis.
Clinical data indicate that HIF-1α expression is either positively
associated with tumor stage in brain tumor (7) or correlates with longer median
survival time among patients with non-small cell lung cancer
(8). Consistently, in vitro
studies present evidence that HIF-1α has a contradictory effect on
cell proliferation and apoptosis in lung cancer A549 cells
(9,10). It is proposed that HIF-1α level is
associated with tissue-and cell-special response to hypoxia
(11). Von Hippel-Lindau tumor
suppressor protein (pVHL), which is often defective or inactive in
cancer, mediated the ubiquitination and degradation of HIF-1α in
normoxia (12). In hypoxia, P53 is
revealed to have directly negative effect on the stability of
HIF-1α or indirectly negative impact via pVHL (13). A recent study showed that P53
regulates LPA-stimulated HIF-1α expression through binding to the
promoter of HIF-1α competed with Krüppel-like factors 5 (KLF5) in
colon cancer cells (14). Since
hypoxia exerts synergistic effect with LPA on HIF-1α expression and
cell survival (15), KLF5 may be
potentially involved in the regulation of HIF-1α following hypoxia.
KLF5 as a zinc finger transcription factor, is known to be
regulated by hypoxia in tumors (16). The overexpression of KLF5 is
reported to be associated with better patient survival in non-small
lung cancer (17) or shorter
overall survival in breast cancer (18). In view of this fact, it is likely
that the conflicting response to hypoxia of HIF-1α and KLF5 may
rely on the types and stages of tumors.
In this study, we assessed the effect of HIF-1α and
KLF5 on cell survival in A549 cells exposed to hypoxia, and a
hypothesis was made that there may be interaction between HIF-1α
and KLF5 involved in the adaptation to hypoxia. We further explored
the function regulation of HIF-1α by KLF5 to advance knowledge on
cancer remission.
Materials and methods
Cell lines and culture
Human non-small cell lung cancer cell line A549 was
obtained from the American Type Culture Collection (ATCC, Manassas,
VA, USA). All cells were cultured in RPMI-1640 supplemented with
10% (vol/vol) FBS (Gibco, Grand Island, NY, USA) and 1%
penicillin-streptomycin. Cells were maintained at 37°C under
normoxic condition in a 5% CO2, 95% ambient air
incubator (Hera cell 150, Heraeus, Langenese, Germany) or hypoxic
condition in a modular incubator (Galaxy R, RS Bitotech, Alloa, UK)
flushed with a gas mixture consisting of 1% O2, 5%
CO2, and 94% N2.
Cell viability assay
Approximately 2,000 cells were seeded in each well
of 96-well plate (Corning, Acton, MA, USA). After 72-h treatment,
10 μl Cell Counting Kit-8 (CCK-8) solution was added and cells were
incubated at 37°C for 4 h following the manufacturer’s instructions
(Dojindo Laboratories, Tokyo, Japan). Optical density (OD) values
(measuring wavelength at 450 nm, reference wavelength at 630 nm)
were obtained using an ELx800 Universal Microplate Reader (Bio-Tek
Instruments, Inc., Winooski, VT, USA).
Colony formation
Cells (100) were allocated in each well of 6-well
culture cluster (Corning). After attachment to plates, A549 cells
with indicated treatment were exposed to 20% O2 or 1%
O2 for 14 days. Colonies were washed three times with
cold PBS, fixed with 4% (vol/vol) paraformaldehyde for 20 min and
then stained with 0.2% (w/v) crystal violet for 10 min. All
colonies visible with the naked eye (>50 cells) were
counted.
Analysis of cell proliferation by CFSE
dilution
Cells for labeling were diluted to
1–5×106 per ml in FACS buffer (PBS/0.1% BSA) and stained
with an equal volume of carboxyfluorescein diacetate succinimidyl
ester (CFSE) at a final working concentration of 10 μM for 10 min
at 37°C. The staining was quenched by the addition of 5-fold
volumes of complete RPMI-1640 medium and incubating for 5 min at
37°C. Cells were pelleted and re-suspended in PRMI medium three
times. Then cells were plated in 6-well plates at 1×105
cells per well under normoxic or hypoxic condition for 72 h.
Harvested cells were measured using flow cytometer (BD Biosciences,
San Jose, CA, USA) with 488 nm excitation and analyzed with BD
FACSDiva 6.1.
Analysis of cell apoptosis by Annexin
V/PI assay
Cells apoptosis were measured using the Annexin
V/propidium iodide (PI) Detection kit (Beyotime, Shanghai, China)
by flow cytometry according to the manufacturer’s protocol. A549
cells were plated in the 6-well plates and treated with indicated
transfection and O2 supply. The floating cells were
collected and the adherent cells were harvested with trypsin
(without EDTA). All cells were pooled together, washed with
ice-cold PBS, and re-suspended in 400 μl binding buffer. Then the
cells were incubated in dark for 15 min at room temperature (RT) in
the presence of 5 μl Annexin V-FITC and 5 μl of PI. The stained
cells were analyzed by flow cytometry (BD Biosciences, San Jose,
CA, USA) to identify the early apoptotic (Annexin
V+/PI−) and late apoptotic (Annexin
V+/PI+) cells.
RNA isolation and real-time PCR
Total RNA was extracted using TRIzol (Takara,
Dalian, China) from A549 cells. RNA (0.5 μg) was converted into
cDNA in a final volume of 10 μl using the cDNA RT–PCR kit (Takara,
Dalian, China). Then cDNA was used as a template for quantitating
gene expression using SYBR Green real-time PCR kit (Takara) in an
ABI PRISM Fast 7500 system (Applied Biosystems, Foster City, CA,
USA) according to the manufacturer’s instructions. The
amplification conditions were preheat at 95°C for 30 sec, 40 cycles
of 95°C for 5 sec, 60°C for 34 sec followed by 95°C for 15 sec and
60°C for 1 min. Primers were as follows: β-actin
F-5′-agcgagcatcccccaaagtt-3′, R-5′-gggcacgaaggctcatcatt-3′; HIF-1α
F-5′-catctccatctcctacccaca-3′, R-5′-cttttcctgctctgtttggtg-3′; KLF5
F-5′-ccaagtcagtttcttccacaac-3′, R-5′-gtttctccaaatcggggttact-3′;
cyclin B1 F-5′-ctggataatggtgaatggacac-3′,
R-5′-cgatgtggcatacttgttcttg-3′; BIRC
F-5′-caccgcatctctacattcaaga-3′, R-5′-caagtctggctcgttctcagt-3′;
caspase-3 F-5′-atcacagcaaaaggagcagttt-3′,
R-5′-acaccactgtctgtctcaatgc-3′.
Transfection of siRNA
Small interfering RNA (siRNA) constructed with
HIF-1α or KLF5 and the non-targeting negative control siRNA were
synthesized by Ruibo Biotechnology Co. (Guangzhou, China). The
target sequences were the following: HIF-1α sense
5′-ugcucuuugugguuggaucua-3′, antisense 5′-uagauccaaccacaaagagca-3′;
KLF5 sense 5′-aagcucaccugaggacuca-3′, antisense
5′-ugaguccucaggugagcuu-3′. A549 cells (2×105 cells/well)
were seeded in the 6-well plates and were transfected with siRNA at
a final concentration of 50 nM using Lipofectamine 2000
(Invitrogen, Carlsbad, CA, USA) following the manufacturer’s
protocol. Cells were harvested after 24-h incubation for RNA
isolation and protein were extracted at 72 h after
transfection.
Immunoblotting and
co-immunoprecipitation
Whole cells in 6-well plates were washed with cold
PBS and isolated by RIPA lysis buffer (1% Triton X-100, 1%
deoxycholate, 0.1% SDS) for 30 min on ice. The lysates were
centrifuged at 12,000 rpm for 10 min, boiled with an addition of 5X
SDS sample buffer and stored at −20°C. Protein samples (50 μg) and
prestained protein marker (SM 0671, Fermentas, USA) were
electrophoresed in 10% SDS-PAGE at 100 V for 2 h and transferred to
a 0.22-μm pore size polyvinylidene fluoride (PVDF) membrane
(Immobilon, Millipore) at 200 mA for 2 h using Trans-Blot
Electrophoretic Transfer Cell (Bio-Rad Laboratories, Richmond, CA,
USA). The membrane was blocked and incubated with the appropriate
primary antibody: anti-HIF antibody (Novus Biologicals, NB100-134,
1:1,000), anti-KLF5 antibody (Santa Cruz, sc-22797, 1:200),
anti-GAPDH antibody (Proteintech, 10494-1-AP, 1:3,000), anti-cyclin
B1 (Proteintech, 55004-1-AP, 1:1,000), anti-survivin (Proteintech,
10508-1-AP, 1:1,000), anticaspase- 3 (Proteintech, 19677-1-AP,
1:1,000). After washing by Tris-buffered saline (TBS) containing
0.1% Tween-20, blots were incubated with secondary antibody
conjugated to horseradish peroxidase (HRP) (Proteintech, SA00001-1,
SA00001-2, 1:4,000) for 2 h at RT. The signals were dectcted by
enhanced chemiluminescent ECL kit (Pierce, Rockford, IL, USA) for 2
min and captured by Kodak X-ray film. For co-immunoprecipitation
(co-IP) analyses, cells were lysed in NP-40 lysis buffer (1%
NP-40). Protein samples (800 μg) were incubated with the
appropriate primary antibody (1–2 μg) and agarose beads (A/G plus,
Santa Cruz Biotechnologies) overnight at 4°C, followed by
immunoblotting (as described above).
Statistical analysis
Statistical analyses were carried out using GraphPad
Prism (version 5.0). The Student’s t-test was used to make a
statistical comparison between groups, two paired. For multi-group
analysis of variances, one-way ANOVA with Dennett’s multiple
comparison test and two-way ANOVA with Bonferroni post-test were
performed. All experiments were repeated at least four times and
the results are expressed as the mean ± SEM. P<0.05 was
considered to be statistically significant.
Results
Hypoxia promotes proliferation of A549
cells and inhibits apoptosis
Uncontrollable survival is a pivotal characteristic
in the process of tumor development. In order to identify the
effect of hypoxia on lung cancer cells, we performed CCK-8 test to
measure cell viability, colony forming assay for cell clonality,
CFSE dilution assay for cell proliferation and Annexin V/PI assay
for cell apoptosis in A549 cells exposed to 20% or 1%
O2. According to CCK-8 test results, hypoxic cell
viability evaluated by mitochondrial activity was significantly
higher than normoxic cells (Fig.
1A). In colony forming assay, colony numbers of cells under
hypoxic condition for 14 days increased 3-fold compared with cells
in normoxia (Fig. 1B and C). CFSE
dilution assay showed that hypoxia significantly increased cell
proliferation after a 3-day treatment (Fig. 1D and E). Percentages of apoptotic
cells were detected by Annexin V and PI staining and hypoxia
reduced both the early stage and the late stage apoptotic cells
(Fig. 1F and G).
Hypoxia upregulates the expression of
HIF-1α and KLF5
To investigate the role of HIF-1α and KLF5 in lung
cancer, we tested the expression of HIF-1α and KLF5 in A549 cells
exposed to hypoxia for different times by real-time PCR and
immunoblotting. HIF-1α mRNA levels were increased in response to
hypoxia within 0.5 h, peaked at 1 h and returned to basal levels
after 8 h of stimulation. Similarly, the mRNA levels of KLF5 were
elevated after 0.5 h and peaked at 2 h, then returned to basal
levels 8 h later (Fig. 2A). As
expected, HIF-1α and KLF5 protein levels were rapidly elevated
exposed to 1% O2 after 0.5 h and continued to rise
steadily up to 4 h (Fig. 2B and
C).
Effect of KLF5 and HIF-1α on cell
proliferation and apoptosis
To better characterize the role of HIF-1α and KLF5
in cell survival following hypoxia, we measured cell viability,
clonality, proliferation and apoptosis in cells treated with siRNA
targeting HIF-1α or KLF5. Cell viability with a significant
increase in hypoxia were decreased to normoxic level by transient
silencing of HIF-1α or KLF5. However, downregulation of HIF-1α and
KLF5 had no impact on cell viability in normoxia (Fig. 3A). Consistently, HIF-1α and KLF5
inhibition resisted cell survival and accelerated cell apoptosis in
hypoxia and did not affect normoxic cells (Fig. 3B–G).
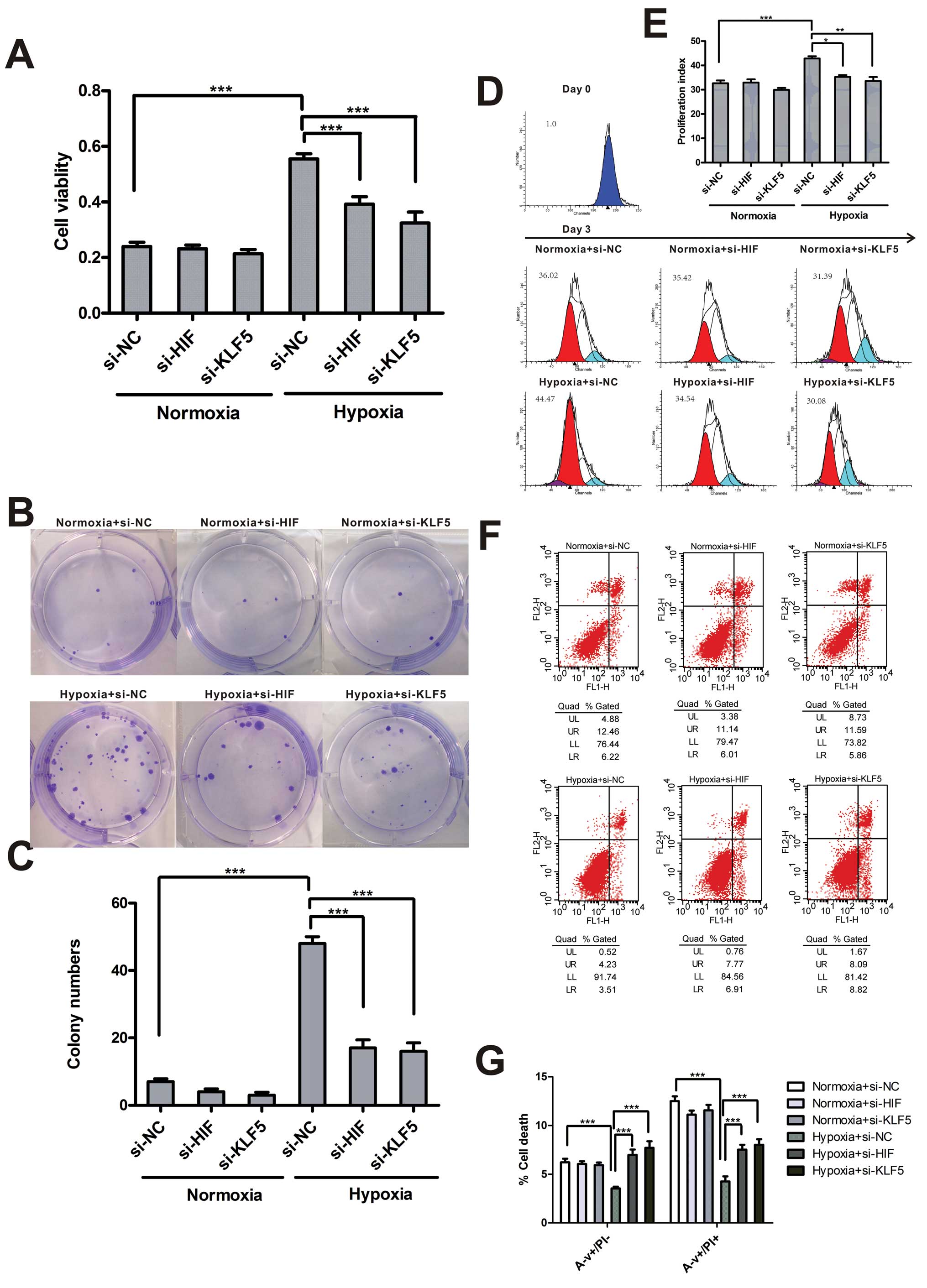 | Figure 3KLF5 and HIF-1α mediate proliferation
and apoptosis of hypoxic A549 cells. (A) CCK-8 test was conducted
in A549 cells transfected with negative control siRNA, HIF-1α-siRNA
or KLF5-siRNA in normoxia or hypoxia for 3 days. (B) Representative
colonies are shown for indicated cells exposed to different
O2 concentrations. (C) The number of colonies is shown.
(D) Analysis of indicated cells labeled CFSE following exposure to
20% O2 or 1% O2 for 3 days. (E) Proliferation
indices were calculated for each treatment group. (F) Annexin V/PI
assay was done to determine apoptosis of transfected cells exposed
to 20% O2 or 1% O2 for 3 days. (G) Percentage
of cell death containing early apoptosis, and later stage apoptosis
in transfected cells under different O2 supply
condition. Data (A, C, E and G) are shown as mean ± SEM; n=4.
*P<0.05, **P<0.01,
***P<0.001 (two-way ANOVA with Bonferroni post-test).
si-NC, negative control siRNA; si-HIF, HIF-1α siRNA; si-KLF5, KLF5
siRNA; N, normoxia; H, hypoxia. |
Regulation of downstream target genes and
proteins
We assessed whether KLF5 or HIF-1α knockout affected
the expression of relevant target genes and proteins. The results
showed that KLF5 and HIF-1α inhibition significantly decreased the
expression of cyclin B1, survivin and increased caspase-3
expression in mRNA and protein levels in hypoxia (Fig. 4), which were identified as the key
regulators of cell survival and apoptosis (19,20).
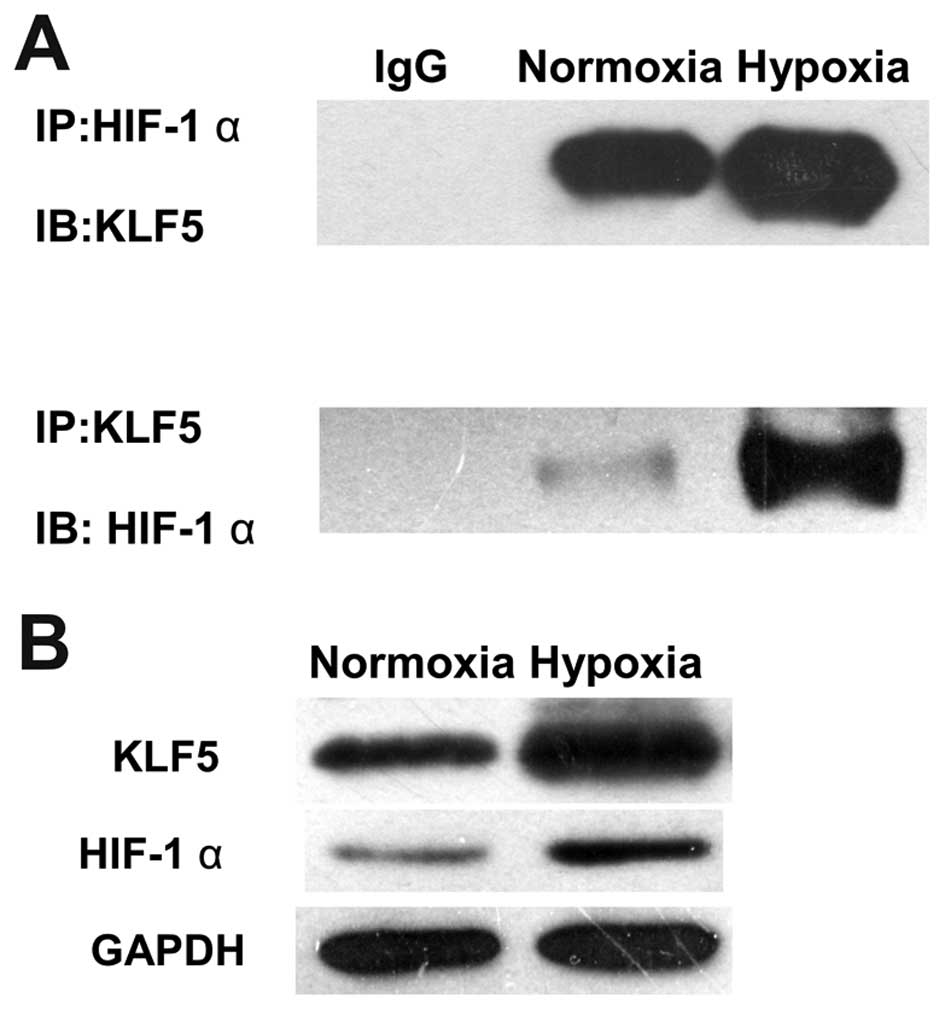
HIF-1α interacted with KLF5
Since HIF-1α and KLF5 both are hypoxia-regulated
transcriptional activators and involved in tumor progression, co-IP
assay was performed in A549 cells exposed to 20% or 1%
O2 to confirm the direct correlation of HIF-1α and KLF5.
KLF5 was found to co-immunoprecipitate with HIF-1α and vice versa.
The amount of co-immunoprecipitated KLF5 was increased in hypoxia
compared with normoxia (Fig. 5A).
Immunoblots described the same amounts of input and hypoxia-induced
increase in HIF-1α and KLF5 (Fig.
5B).
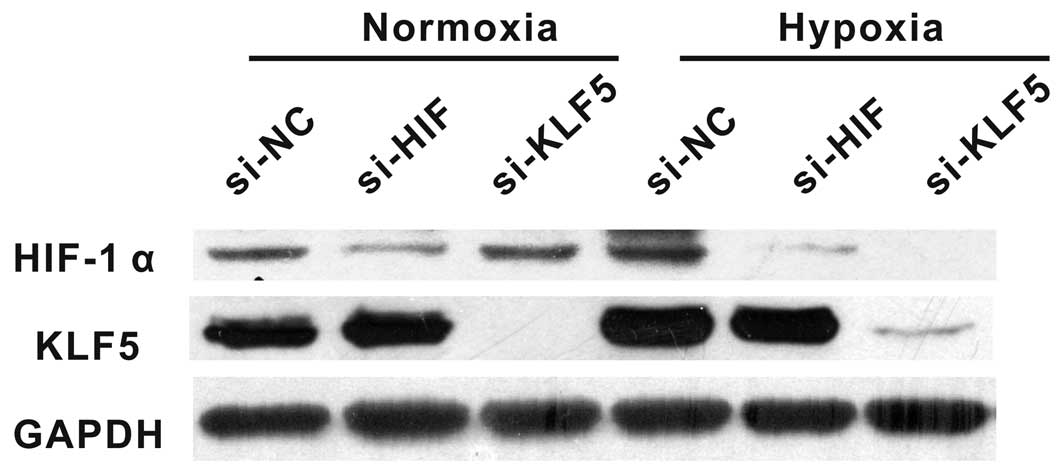
Hypoxia-induced HIF-1α overexpression is
KLF5-dependent
To evaluate the functional interaction of HIF-1α
with KLF5 in hypoxia, HIF-1α and KLF5 expression was measured in
A549 cells exposed to 20% or 1% O2 with the presence of
KLF5 or HIF-1α siRNA. Suppressing KLF5 by siRNA significantly
reduced HIF-1α expression compared with negative controls under
hypoxic condition while HIF-1α expression did not change in
normoxia. Likewise, treating cells with HIF-1α siRNA did not affect
the expression of KLF5 either in hypoxia or normoxia (Fig. 6).
Discussion
Hypoxia is described to be an important stimulating
factor to sustain an aggressive growth phenotype in cancer cells.
Multiple biological processes have been developed in hypoxic
tumors, including activation of glycolysis and angiogenesis,
pro-survival and invasiveness of cancer cells, metastasis and
resistance to chemotherapy and radiation, which lead to poorer
prognosis (21–23). The role of hypoxia in alveolar
epithelial cells remains ambiguous as a pro-apoptotic factor or
proliferation promoter. Therefore, we assessed the effect of
hypoxia on A549 cells in several ways and found that prolonged
hypoxia (1% O2 for 3 days) promoted cell survival and
inhibited cell apoptosis. Hypoxic levels and duration account for
the different responses of cultured cells. Anoxia
(O2<0.1%) and hypoxia persisting for too long induce
eventually cell death (24).
However, under low oxygen tension (1% O2), adaptive
response is developed to resist cell death and promote cell
survival, which is largely mediated by HIF-1, NF-κB and p53
(25). Compared with acute
hypoxia, chronic and sustained hypoxia addressed in our study is
more similar to the slowly progressive natural history of
tumor.
HIF-1α plays an important role in the adaptation to
hypoxia, which is underscored by the association with prognosis.
Plenty of HIF-1α inhibitors, including chemical inhibitors, protein
and nucleic acid agents, have been studied for targeted cancer
therapy (26). KLF5 is
predominantly present in epithelial tissues characterized by active
proliferation and exerts pleiotropic effects on the regulation of
tumor progression, cell proliferation and apoptosis by binding to
similar GC-rich promoter of target genes (27). In our experiments, expression of
HIF-1α and KLF5 was upregulated time-dependently in A549 cells
exposed to 1% O2. Interestingly, peak time of HIF-1α
protein expression was identical with KLF5. Hypoxia-induced
increase of cell viability, clonality, proliferation and decrease
of cell apoptosis were abrogated by siRNA targeting HIF-1α or KLF5,
implying their contribution to hypoxic microenvironment.
Cell proliferation is dependent on the response to
mitogenic signals to enter the cell cycle. There is evidence that
KLF5 mediates the activation of cyclin B1/Cdc2 complex to
accelerate mitosis (28).
Apoptosis is controlled by caspase family (29) and caspase-3 plays a key role in
both the extrinsic and the intrinsic apoptosis pathways (30). Survivin, as a direct inhibitor of
caspase-3, is implicated in promoting tumorigenesis and resisting
chemotherapy and radiation-induced apoptosis in non-small cell lung
cancer (31). Recent studies show
that KLF5 is involved in the upregulation of survivin via direct
interaction with p53 (32), and
knockout of KLF5 induces caspase-3 cleavage through pERK/MKP-1
signaling. HIF-1α is revealed to induce survivin expression at the
transcriptional level in prostate cells (33) and bind to the promoter of caspase-3
(34). As expected, cyclin B1 and
survivin were downregulated and caspase-3 was increased by HIF-1α
or KLF5 ablation. Therefore, our experiments clearly demonstrated
that KLF5 and HIF-1α played a vital role in the regulation of cell
survival and cell apoptosis via cyclin B1, survivin and caspase-3
in hypoxic lung cancer cells.
The expression and activity of HIF-1α are regulated
in tumor development by multiple signaling pathways and proteins,
including PKC/P13K/AKT/mTOR (35–37),
Ras/MEK/ERK (38), MAPK (39,40),
EGFR and IGF1R autocrine signaling (41), PTEN (37,42,43).
Under hypoxic condition, stabilized HIF-1α protein dimerizes with
HIF-1β in the nucleus and the complex binds to a core DNA hypoxia
response element (HRE) (44),
activating the expression of numerous tumor promoter genes,
including GLUT1 (10), VEGF
(45), NOS (46), IGF-2 (47) and ID2 (48). The increase of HIF-1α mRNA and
protein levels in our study supported the view that hypoxia-induced
HIF-1α accumulation originate from de novo gene
transcription and the protein stabilization (49). An array of proteins participate in
the regulation of HIF-1α degradation via binding to HIF-1α,
including ERRα (50), STAT3
(51), Hsp90 (52) and VHL (53).
In our study, KLF5 formed a complex with HIF-1α and
the interaction between them was oxygen-dependent. Furthermore,
silencing of KLF5 markedly inhibited HIF-1α production while HIF-1α
inhibition did not affect KLF5 expression, suggesting a possible
role of KLF5 in preventing the degradation of HIF-1α following
hypoxia. HIF-1α is recently proposed by Mori et al to
regulate KLF5 expression by modulating its interaction with KLF5 in
pancreatic cancer cell line (54).
Mori et al showed that HIF-1α co-immunoprecipitates with
KLF5 and vice versa, independent of hypoxia. The expression of
HIF-1α is not affected by the downregulation of KLF5. Because of
the particularly overexpression of HIF-1α in pancreatic cancer
cells, the functional regulation of KLF5 and HIF-1α are performed
in normoxia. Therefore, the differences between Mori et al
and our study may be accounted for by the types of cancer and the
duration of hypoxia. The new finding substantiated the direct
interaction of KLF5 with HIF-1α and the upregulation of HIF-1α by
KLF5 in the adaptation to hypoxia supported tumor growth, providing
a promising therapeutic target for cancer therapy.
KLF5 was strongly overexpressed in hypoxic A549
cells and mediated hypoxia-induced upregulation of HIF-1α.
Considering the multiple regulations of cell viability, clonality,
proliferation and apoptosis in hypoxia, KLF5 may be an attractive
target for therapy of lung cancer. Thus, treatment protocols using
KLF5 inhibitors require further investigation.
Acknowledgements
This study was supported by grants from Natural
Science Foundation of China (81270106), Ministry of Science and
Technology of China (2012BAI05B00) and Ministry of Public Health
(201002008).
References
|
1
|
Estimated Incidence, Mortality and
Prevalence Worldwide in 2012. //globocaniarc.fr/Pages/fact_sheets_cancer.aspx.
Accessed Mar 27, 2014
|
|
2
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wilson WR and Hay MP: Targeting hypoxia in
cancer therapy. Nature reviews Cancer. 11:393–410. 2011. View Article : Google Scholar
|
|
4
|
Brahimi-Horn MC, Chiche J and Pouyssegur
J: Hypoxia and cancer. J Mol Med. 85:1301–1307. 2007. View Article : Google Scholar
|
|
5
|
Zhang H, Okamoto M, Panzhinskiy E, Zawada
WM and Das M: PKC delta/midkine pathway drives hypoxia-induced
proliferation and differentiation of human lung epithelial cells.
Am J Physiol Cell Physiol. 306:C648–C658. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vaporidi K, Tsatsanis C, Georgopoulos D
and Tsichlis PN: Effects of hypoxia and hypercapnia on surfactant
protein expression proliferation and apoptosis in A549 alveolar
epithelial cells. Life Sci. 78:284–293. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zagzag D, Zhong H, Scalzitti JM, Laughner
E, Simons JW and Semenza GL: Expression of hypoxia-inducible factor
1alpha in brain tumors: association with angiogenesis, invasion,
and progression. Cancer. 88:2606–2618. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Volm M and Koomagi R: Hypoxia-inducible
factor (HIF-1) and its relationship to apoptosis and proliferation
in lung cancer. Anticancer Res. 20:1527–1533. 2000.PubMed/NCBI
|
|
9
|
Wang Q, Li LH, Gao GD, et al: HIF-1alpha
up-regulates NDRG1 expression through binding to NDRG1 promoter,
leading to proliferation of lung cancer A549 cells. Mol Biol Rep.
40:3723–3729. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Luo F, Liu X, Yan N, et al:
Hypoxia-inducible transcription factor-1alpha promotes
hypoxia-induced A549 apoptosis via a mechanism that involves the
glycolysis pathway. BMC Cancer. 6:262006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chi JT, Wang Z, Nuyten DS, et al: Gene
expression programs in response to hypoxia: cell type specificity
and prognostic significance in human cancers. PLoS Med. 3:e472006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim WY and Kaelin WG: Role of VHL gene
mutation in human cancer. J Clin Oncol. 22:4991–5004. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sermeus A and Michiels C: Reciprocal
influence of the p53 and the hypoxic pathways. Cell Death Dis.
2:e1642011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee SJ, No YR, Dang DT, et al: Regulation
of hypoxia-inducible factor 1alpha (HIF-1alpha) by lysophosphatidic
acid is dependent on interplay between p53 and Kruppel-like factor
5. J Biol Chem. 288:25244–25253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Park SY, Jeong KJ, Lee J, et al: Hypoxia
enhances LPA-induced HIF-1alpha and VEGF expression: their
inhibition by resveratrol. Cancer Lett. 258:63–69. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schumpelick VH-PB and Schackert HK:
Chirurgisches Forum und DGAV Forum. Springer; Berlin: 2009
|
|
17
|
Meyer SE, Hasenstein JR, Baktula A, et al:
Kruppel-like factor 5 is not required for K-RasG12D lung
tumorigenesis, but represses ABCG2 expression and is associated
with better disease-specific survival. Am J Pathol. 177:1503–1513.
2010. View Article : Google Scholar
|
|
18
|
Tong D, Czerwenka K, Heinze G, et al:
Expression of KLF5 is a prognostic factor for disease-free survival
and overall survival in patients with breast cancer. Clin Cancer
Res. 12:2442–2448. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Budihardjo I, Oliver H, Lutter M, Luo X
and Wang X: Biochemical pathways of caspase activation during
apoptosis. Annu Rev Cell Dev Biol. 15:269–290. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ambrosini G, Adida C and Altieri DC: A
novel anti-apoptosis gene, survivin, expressed in cancer and
lymphoma. Nat Med. 3:917–921. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Harris AL: Hypoxia - a key regulatory
factor in tumour growth. Nature reviews Cancer. 2:38–47. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Graves EE, Maity A and Le QT: The tumor
microenvironment in non-small-cell lung cancer. Semin Rad Oncol.
20:156–163. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cui J, Mao X, Olman V, Hastings PJ and Xu
Y: Hypoxia and miscoupling between reduced energy efficiency and
signaling to cell proliferation drive cancer to grow increasingly
faster. J Mol Cell Biol. 4:174–176. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bruick RK: Oxygen sensing in the hypoxic
response pathway: regulation of the hypoxia-inducible transcription
factor. Genes Dev. 17:2614–2623. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lenihan CR and Taylor CT: The impact of
hypoxia on cell death pathways. Biochem Soc Trans. 41:657–663.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu Y, Liu J and Huang H: Recent agents
targeting HIF-1alpha for cancer therapy. J Cell Biochem.
114:498–509. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Diakiw SM, D’Andrea RJ and Brown AL: The
double life of KLF5: opposing roles in regulation of
gene-expression, cellular function, and transformation. IUBMB Life.
65:999–1011. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nandan MO, Chanchevalap S, Dalton WB and
Yang VW: Kruppel-like factor 5 promotes mitosis by activating the
cyclin B1/Cdc2 complex during oncogenic Ras-mediated
transformation. FEBS Lett. 579:4757–4762. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Reed JC: Dysregulation of apoptosis in
cancer. J Clin Oncol. 17:2941–2953. 1999.PubMed/NCBI
|
|
30
|
Ghavami S, Hashemi M, Ande SR, et al:
Apoptosis and cancer: mutations within caspase genes. J Med Genet.
46:497–510. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Krepela E, Dankova P, Moravcikova E, et
al: Increased expression of inhibitor of apoptosis proteins,
survivin and XIAP, in non-small cell lung carcinoma. Int J Oncol.
35:1449–1462. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu N, Gu L, Findley HW, et al: KLF5
interacts with p53 in regulating survivin expression in acute
lymphoblastic leukemia. J Biol Chem. 281:14711–14718. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yun YJ, Li SH, Cho YS, Park JW and Chun
YS: Survivin mediates prostate cell protection by HIF-1alpha
against zinc toxicity. Prostate. 70:1179–1188. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Van Hoecke M, Prigent-Tessier AS, Garnier
PE, et al: Evidence of HIF-1 functional binding activity to
caspase-3 promoter after photothrombotic cerebral ischemia. Mol
Cell Neurosci. 34:40–47. 2007.PubMed/NCBI
|
|
35
|
Majumder PK, Febbo PG, Bikoff R, et al:
mTOR inhibition reverses Akt-dependent prostate intraepithelial
neoplasia through regulation of apoptotic and HIF-1-dependent
pathways. Nat Med. 10:594–601. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhong H, Chiles K, Feldser D, et al:
Modulation of hypoxia-inducible factor 1alpha expression by the
epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP
pathway in human prostate cancer cells: implications for tumor
angiogenesis and therapeutics. Cancer Res. 60:1541–1545. 2000.
|
|
37
|
Park JH, Lee JY, Shin DH, Jang KS, Kim HJ
and Kong G: Loss of Mel-18 induces tumor angiogenesis through
enhancing the activity and expression of HIF-1alpha mediated by the
PTEN/PI3K/Akt pathway. Oncogene. 30:4578–4589. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang M, Ma Q, Hu H, et al: Stem cell
factor/c-kit signaling enhances invasion of pancreatic cancer cells
via HIF-1alpha under normoxic condition. Cancer Lett. 303:108–117.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sodhi A, Montaner S, Patel V, et al: The
Kaposi’s sarcoma-associated herpes virus G protein-coupled receptor
up-regulates vascular endothelial growth factor expression and
secretion through mitogen-activated protein kinase and p38 pathways
acting on hypoxia-inducible factor 1alpha. Cancer Res.
60:4873–4880. 2000.
|
|
40
|
Fukuda R, Hirota K, Fan F, Jung YD, Ellis
LM and Semenza GL: Insulin-like growth factor 1 induces
hypoxia-inducible factor 1-mediated vascular endothelial growth
factor expression, which is dependent on MAP kinase and
phosphatidylinositol 3-kinase signaling in colon cancer cells. J
Biol Chem. 277:38205–38211. 2002. View Article : Google Scholar
|
|
41
|
Semenza GL: Targeting HIF-1 for cancer
therapy. Nat Rev Cancer. 3:721–732. 2003. View Article : Google Scholar
|
|
42
|
Zundel W, Schindler C, Haas-Kogan D, et
al: Loss of PTEN facilitates HIF-1-mediated gene expression. Genes
Dev. 14:391–396. 2000.PubMed/NCBI
|
|
43
|
Liu R, Zheng HQ, Zhou Z, Dong JT and Chen
C: KLF5 promotes breast cell survival partially through fibroblast
growth factor-binding protein 1-pERK-mediated dual specificity
MKP-1 protein phosphorylation and stabilization. J Biol Chem.
284:16791–16798. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Schofield CJ and Ratcliffe PJ: Oxygen
sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 5:343–354.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Forsythe JA, Jiang BH, Iyer NV, et al:
Activation of vascular endothelial growth factor gene transcription
by hypoxia-inducible factor 1. Mol Cell Biol. 16:4604–4613.
1996.PubMed/NCBI
|
|
46
|
Melillo G, Musso T, Sica A, Taylor LS, Cox
GW and Varesio L: A hypoxia-responsive element mediates a novel
pathway of activation of the inducible nitric oxide synthase
promoter. J Exp Med. 182:1683–1693. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Feldser D, Agani F, Iyer NV, Pak B,
Ferreira G and Semenza GL: Reciprocal positive regulation of
hypoxia-inducible factor 1alpha and insulin-like growth factor 2.
Cancer Res. 59:3915–3918. 1999.PubMed/NCBI
|
|
48
|
Lofstedt T, Jogi A, Sigvardsson M, et al:
Induction of ID2 expression by hypoxia-inducible factor-1: a role
in dedifferentiation of hypoxic neuroblastoma cells. J Biol Chem.
279:39223–39231. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yee Koh M, Spivak-Kroizman TR and Powis G:
HIF-1 regulation: not so easy come, easy go. Trends Biochem Sci.
33:526–534. 2008.PubMed/NCBI
|
|
50
|
Zou C, Yu S, Xu Z, et al: ERRalpha
augments HIF-1 signalling by directly interacting with HIF-1alpha
in normoxic and hypoxic prostate cancer cells. J Pathol. 233:61–73.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jung JE, Kim HS, Lee CS, et al: STAT3
inhibits the degradation of HIF-1alpha by pVHL-mediated
ubiquitination. Exp Mol Med. 40:479–485. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Minet E, Mottet D, Michel G, et al:
Hypoxia-induced activation of HIF-1: role of HIF-1alpha-Hsp90
interaction. FEBS Lett. 460:251–256. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ohh M, Park CW, Ivan M, et al:
Ubiquitination of hypoxia-inducible factor requires direct binding
to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol.
2:423–427. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mori A, Moser C, Lang SA, et al:
Upregulation of Kruppel-like factor 5 in pancreatic cancer is
promoted by interleukin-1beta signaling and hypoxia-inducible
factor-1alpha. Mol Cancer Res. 7:1390–1398. 2009. View Article : Google Scholar : PubMed/NCBI
|