Introduction
Osteosarcoma (OS) is the most common primary
malignant neoplasm in adolescents with an annual estimated
worldwide incidence of 4 million, with a peak incidence at the age
of 15–19 years (1). Osteosarcoma
is associated with abnormal differentiation caused by genetic and
epigenetic changes. Advances in osteosarcoma therapy have enhanced
patient outcomes. The most effective regimens currently include
neoadjuvant and adjuvant chemotherapy coupled with local control
that usually consists of limb-sparing surgery (2). Unfortunately, the cure rate is still
very poor due to pulmonary metastases (3). Therefore, the identification of
effector molecules and signaling pathways underlying resistance to
chemotherapy and malignancy is vital for osteosarcoma treatment.
Studies have investigated the genes associated with metastasis of
osteosarcoma, and microRNAs (miRNAs) have become a new research
hotspot in gene therapy.
microRNAs (miRNAs) are a class of 22–25 nucleotide
RNA molecules that negatively regulate gene expression in animals
and plants (4,5). Since the discovery of the role of
miRNAs in Caenorhabditis elegans development (6), a frequent disregulation of miRNAs has
been observed in diverse cancers, including synovial sarcoma, colon
cancer (7), breast cancer
(8), glioma (9), glioblastoma (10), hepatocellular carcinoma (11), lung (12) and gastric cancer (13). Some of these miRNA expression
profiles showed downregulation in tumors compared with normal
tissue (14), like miR-127 in
human bladder cancers (15) and
microRNA-34a in OS (16). However,
other miRNAs are upregulated in tumors, such as miR-150 in gastric
cancer (17) and miR-17-92 cluster
in renal cell carcinoma (18). The
alterations in miRNA expression may play a crucial role in the
initiation and progression of the above cancers (19), functioning as a novel class of
oncogenes and tumor suppressors (20,21).
Thus, miRNAs play an essential role in basic physiologic processes,
such as development, differentiation, proliferation and apoptosis
(22). However, their biological
function remains largely unknown.
miR-194 is specifically expressed in the human
gastrointestinal tract and is induced during intestinal epithelial
cell differentiation (23). The
regulatory role of miR-194 was first studied in normal and
malignant cells of the gastrointestinal tract (24). Overexpression of miR-194 in
gastrointestinal cancer cells suppresses cell migration, invasion
and metastasis (24). miR-194
functions as a tumor suppressor gene by downregulating targets such
as SSH2, HBEGF, IGF1R, CDH2
(N-cadherin) and TLN2 (23–27).
Hepatocyte nuclear factor (HNF) also induces miR-194 expression
during intestinal epithelial cell differentiation (23). In colon cancer tissue, miR-194 was
downregulated relative to normal mucosa (28). Low expression of miR-194 has been
associated with large tumor size and advanced stage in gastric
cancer (29). In endometrial
cancer cells, miR-194 has been reported to inhibit self-renewal
factor BMI-1, reduce cell invasion and inhibit
epithelial-mesenchymal transition (EMT) (30). The mutations of p53 tumor
suppressor gene, which directly regulates the expression of
miR-194, were found in 20–60% of sporadic OS (31). The reports suggested that miR-194
may function as a tumor suppressor in OS. However, the effects of
miR-194 in osteosarcoma have not been completely elucidated.
Therefore, it is of great significance to further study the
function and mechanism of miR-194 in osteosarcoma.
We carried out in vitro and in vivo
experiments to evaluate the effects of miR-194 and its possible
direct targets, IGF1R and CDH2, in tumor growth and
metastasis of SOSP-9607 and U2-OS cells. We also predicted its
putative target genes, which are correlated with tumor growth and
metastasis, using bioinformatics analysis. We report for the first
time that overexpression of miR-194 inhibited growth and metastasis
of osteosarcoma. In addition, miR-194 specifically downregulated
the expression of IGF1R and CDH2. miR-194 gene
therapy may prove to be a promising therapy for tumorsuppression in
osteosarcoma.
Materials and methods
Ethics statement
All research involving human tissue samples and
animals was approved by the Ethics Review Committee of Fourth
Military Medical University, Xi’an, Shaanxi, China (approval
ID:2013106) and written informed consent was obtained from all
participating patients.
Human tissue samples
A total of 107 pairs of human osteosarcoma tissue
samples were obtained from patients who underwent surgical
resection at the Tangdu Hospital of Fourth Military Medical
University between 2007 and 2010 and were diagnosed with
osteosarcoma based on histopathological evaluation. The biopsies
were immediately snap-frozen in liquid nitrogen after resection and
stored at -80°C. One section of each sample was stained with
hematoxylin-eosin (H&E) for histopathological evaluation. The
clinical stage of these osteosarcoma patients was classified
according to the sixth edition of the tumor-node-metastases (TNM)
classification of the International Union Against Cancer
(UICC).
All 107 osteosarcoma patients were studied in a
follow-up. The median follow-up was 42 months (range, 5–68 months).
During the follow-up period, 62 patients (57.9%) died of disease.
Distant metastases developed in 52 patients at a mean of 12.7
months (range, 3–41 months) after the original diagnosis. Of these
patients, 13 had bone metastases and 43 had lung metastases (4
patients had both bone and lung metastases).
Cell culture
Human osteosarcoma cell lines SOSP-9607 were
established and reserved in our laboratory as previously described
(32). Human osteosarcoma cell
lines U2-OS were purchased from the American Type Culture
Collection (ATCC, Manassas, VA, USA) and used within 6 months of
the purchase. SOSP-9607 cells were maintained in RPMI-1640 medium
(HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum
(FBS; HyClone), 2.0 mM l-glutamine, 100 U/ml penicillin and 100
μg/ml streptomycin, and incubated at 37°C in a humidified incubator
supplemented with 5% CO2 and 95% air. U2-OS cells were
maintained in the same conditions, except DMEM medium was used.
Cell line authentication was performed by the Orthopedics Oncology
Institute of Chinese PLA according to the UKCCCR guidelines every
2–3 months, including mycoplasma test by PCR and measurement of
cell proliferation by counting.
Generation of stable cell lines
Recombinant lentiviruses containing overexpression
of miRNA-194, for knocking down miRNA-194 and miRNA control were
purchased from Shanghai Genechem Co., Ltd. (Shanghai, China). The
precusor sequence of miR-194 was used for overexpression as
follows: AUGGUGUUAUCAAGUGUAACAGCAACUCCAUGUGG
ACUGUGUACCAAUUUCCAGUGGAGAUGCUGUUACU UUUGAUGGUUACCAA. The reverse
complementary sequence of miR-194 was used for the knock-down as
follows: TC CACATGGAGTTGCTGTTACA. Besides the multiple clone sites
of lentivirus expression vectors, there also was a GFP reporter
driven by an independent promoter (SV40 promoter) to indicate the
infection rate of the virus timely.
To generate the stable cell line, 1×104
cells were transfected with 5×105 transducing units of
lentiviruses. The supernatant was removed after 24 h and replaced
with complete culture medium. Infection efficiency was confirmed by
RT-PCR 96 h after infection and the cells were selected with 1
μg/ml puromycin for 2 weeks.
Reverse transcription and quantitative
real-time PCR
Total RNA containing miRNA and mRNA was extracted
from cells with TRIzol® reagent (Invitrogen, Carlsbad,
CA, USA), or from formalin- or paraformalin-fixed,
paraffin-embedded (FFPE) tissues with RecoverAll™ Total Nucleic
Acid Isolation kit (Ambion, Foster City, CA, USA; cat no. AM1975),
according to the manufacturer’s instructions. The RNA was
quantified by absorbance at 260 nm and transcribed into cDNA using
BioRT Two-Step RT-PCR kit (Bioer Technology, Inc., Hangzhou,
China). To evaluate IGF-1R and N-cadherin expression levels, 1 μg
of total RNA was used for reverse transcription with iScript cDNA
Synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA), according
to the manufacturer’s instructions. The sequences of the forward
and reverse primers for IGF1R were 5′-CGACATTGAGGAGGTCAC AGA-3′ and
5′-TGGGCACGAAGATGGAGTT-3′. The sequences of the forward and reverse
primers for N-cadherin were 5′-GTCAGCAGAAGTTGAAGAAATAGTG-3′ and
5′-GCAAGTTGATTGGAGGGATG-3′.
The sequences of the forward and reverse primers for
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were
5′-TGGGTGTGAACCATGAGAAGT-3′ and 5′-TGAGTCC TTCCACGATACCAA-3′. The
expression level of GAPDH was used as a control. To evaluate
hsa-mir-194 levels, the sequences of primers for miR-194:
5′-ACACTCCAGCTGGG TGTAACAGCAACTCCAT-3′ were used. U6 was used as a
control.
Apoptosis, proliferation and cell cycle
assays
Cultured cells were grown in 6-, 24- and 96-well
plates. Apoptosis and cell cycle were measured using flow
cytometry. The procedures were performed as previously described
(33). Cell viability was examined
using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide (MTT) assay. MTT was performed at 24, 48, 72, 96, 120 and
144 h. The absorbance at 492 nm was measured after incubation with
20 μl of MTT for 4 h. The curve of cell proliferation was then
drawn and the proliferation efficiency was examined. There were
6-wells in each group, the experiments were repeated three times
independently and the results were given as means ± SD. The plate
clone formation assay was performed as previously described
(34). Clones with >50 cells
were counted with an ordinary optical microscope and the clone
formation rate was calculated with the following formula: Plate
clone formation efficiency = (number of clones/number of cells
inoculated) × 100%.
Transwell cell migration and matrigel
invasion assays
The invasive potential of cells was measured in 6.5
mm Transwell with 8.0-mm pore polycarbonate membrane insert (cat.
3422; Corning, NY), according to the manufacturer’s instructions.
The filter of the top chamber was coated with 50 μl of diluted
matrigel and incubated at 37°C for 2 h. The lower chambers were
filled with 600 μl of RPMI-1640 medium containing 5% fetal bovine
serum (FBS) as chemoattractant. The suspension of 5,000 cells in
100 μl migration medium was added into each top chamber. After the
cells were incubated for 16 h, the non-invading cells that remained
on the upper surface were removed with a cotton swab. The invasive
cells in the lower surface of the membrane insert were fixed with
4% paraformaldehyde for 30 min, permeabilized with 0.2% Triton
X-100 at room temperature for 15 min and then stained with 0.1%
crystal violet for 5 min. The number of cells in the lower surface,
which had invaded through the membrane, was counted under a light
microscope in five random fields at a magnification of ×100. The
experiments were repeated three times independently and results
were expressed as means ± SD.
The procedure for transwell migration assays was the
same as the transwell invasion assay except that the filter of top
chamber was not coated with matrigel.
Wound healing migration assay
When the transfected and untransfected SOSP-9607 and
U2-OS cells were grown to confluence, a scratch in the cell
monolayer was made using a micropipette tip. Following incubation
of the cells under standard conditions for 24 h, the plates were
washed twice with fresh medium and images were captured at
different times. The migration potential was estimated by counting
the cells that migrated from the wound edge. The cell migration
rate was obtained by counting three fields per area and represented
as the average of six independent experiments over multiple
days.
Animal studies
Four-week-old female nude mice (BALB/c, nu/nu;
Experimental Animal Centre of the Fourth Military Medical
University in China), 17–22 g in weight, were maintained under
specific pathogen-free conditions with 12-h light/12-h dark cycles
at 26–28°C and 50–65% humidity. Animal feed and underpad, which
were purchased from the Experimental Animal Center, Fourth Military
Medical University, were autoclaved and vacuum packed. The water
was adjusted to a pH value of 2.8 and autoclaved before use.
Animal experiments were performed to evaluate
orthotopic tumor growth and spontaneous pulmonary metastasis
properties of osteosarcoma cells in vivo. Briefly, 4 groups
SOSP-9607 cells (overexpression of miRNA-194, for knocking down
miRNA-194, miRNA control and untransfected cells) suspension of
100,000 cells in 100 μl were injected into the proximal tibia of
each anesthetized nude mouse (n=10 animals per group). Every 7 days
post-inoculation, the length and width of individual orthotopic
tumor were measured with calipers, and the volume (mm3)
was calculated according to the formula: 1/2 × length ×
width2 (35). The curve
of orthotopic tumor growth was depicted 42 days after inoculation
mouse lungs and orthotopic tumors were harvested and weighed.
miR-194 expression levels in the orthotopic tumors were tested
using real-time RT-PCR, and the number of pulmonary metastatic
tumor nodules was counted under a low-power dissecting
stereomicroscope. Finally, mouse lungs were fixed with 10%
neutral-buffered formalin, embedded in paraffin, sectioned at 6 μm
and stained with H&E. The pulmonary metastases were imaged
under a light microscope at magnifications of ×40, ×100, 2×00 and
×400.
Protein extraction and western blot
analysis
Protein extracts were prepared through a modified
RIPA buffer with 0.5% sodium dodecyl sulfate (SDS) in the presence
of a proteinase inhibitor cocktail (Complete Mini; Roche
Diagnostics, Basel, Switzerland) and then were performed as
previously described (36).
Luciferase reporter assay
To validate IGF1R and N-cadherin as
target genes of miR-194 in osteosarcoma cells, luciferase assay was
performed as previously described (33).
Target prediction
Bioinformatics analysis was carried out using
specific programs: Pictar (http://pictar.mdc-berlin.de/), miRanda (http://www.microrna.org) and TargetScan (http://www.targetscan.org/).
Immunohistochemistry
The dilution of CDH2 and IGF1R antibody used for
immunohistochemistry was 1:100. Immunohistochemistry was carried
out as previously described (37).
The final scores of CDH2 and IGF1R expression were calculated as
previously described (38) and
classified as follows: 0–4, low; 5–9, high.
Statistical analysis
All values in the present study were expressed as
the means ± SD, and all error bars represent the standard deviation
of the mean. Student’s t test, one-way analysis of variance and
repeated measures data of ANOVA were used to determine
significance. Patient survival and their differences were
determined using the log-rank test. Cox regression (proportional
hazards model) was used for multivariate analysis of prognostic
factors. All statistical tests were two-sided. p<0.05 was
considered statistically significant. Statistical analyses were
performed using the SPSS 17.0 software (SPSS, Inc., Chicago, IL,
USA).
Results
Significant difference between F4 and
control F5M2 cells
F4 and F5M2 were the sublines originated from
SOSP-9607 using limited dilution method (32,39).
F5M2 cells show stronger proliferation and invasion than F4 cells,
which is useful in studies on metastatic mechanism of osteosarcoma
(32). In the present study, we
evaluated the expression of miR-194 using quantitative real-time
PCR. The results showed that miR-194 expression was significantly
lower in F5M2 cells compared with F4 cells (Fig. 1A; p<0.001). The results
suggested that miR-194 might play a vital role in the metastatic
processes.
Generation of stable cell lines
After transfection and selection of cells, the
experiments with SOSP-9607 cells and U2-OS cells were divided into
four groups including a blank group (untransfected cells), a
control group (cells transfected with the control lentivirus), an
OE group (overexpression of miRNA-194) and a KD group (knocked down
miRNA-194). These GFP-labeled oligonucleotides were detected using
fluorescence microscopy (Olympus, Tokyo, Japan) (Fig. 1B).
miR-194 expression levels in four groups were
measured using microscopy and stem-loop real-time RT-PCR. The
results (Fig. 1C and D) showed
that the level of miR-194 was significantly higher in the OE group
and lower in KD group compared with control and blank groups,
respectively. However, there were no significant differences
between the control and blank groups. These results indicated that
miR-194 recombinant lentiviruses could regulate miR-194 expression
effectively in both SOSP-9607 and U2-OS cells. These strategies
were then used as the basis of the remaining experiments.
miR-194 inhibits proliferation of
osteosarcoma in vitro
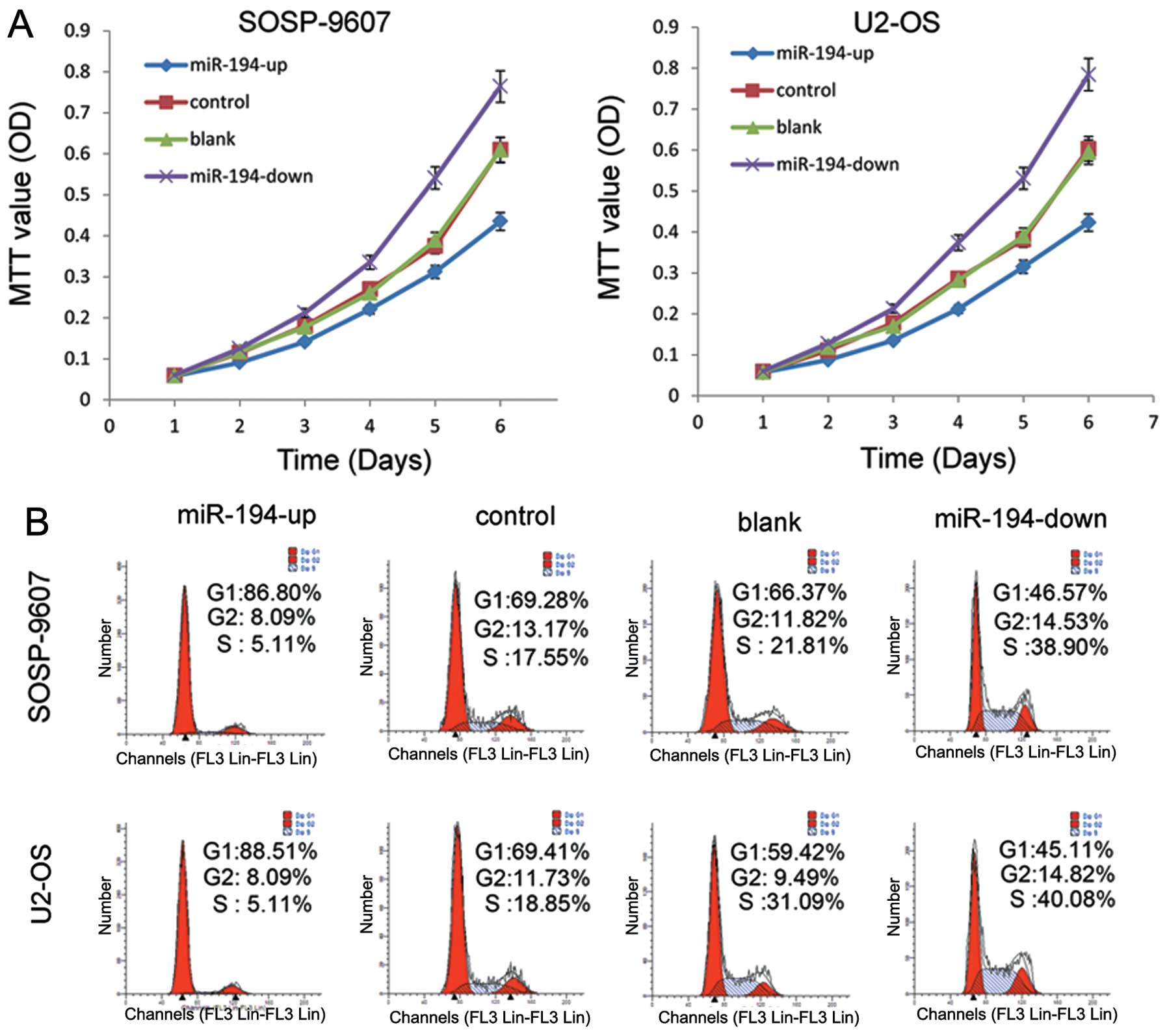
The results of MTT assay showed that SOSP-9607 cells
in OE groups exhibited a significant decline in proliferation
capacity compared with the other three groups (p<0.001), which
were negatively correlated with the exogenous miR-194 level
(Fig. 2A). In contrast, cells in
the KD group showed significantly enhanced proliferation
(p<0.001). No statistical difference was found between the blank
groups and control groups (p=0.541). We also tested U2-OS cells
(Fig. 2A). The results were
similar to the stably transfected SOSP-9607 cells.
Cell cycle distribution was detected by flow
cytometry. The results showed that more cells were in the G0/G1
phase in the OE group compared with the G0/G1 phase in KD group in
SOSP-9607 and U2-OS cells (Fig.
2B). These data demonstrated that miR-194 could inhibit the
proliferation in both SOSP-9607 and U2-OS cells.
miR-194 induces apoptosis
Apoptosis in the SOSP-9607 cell line was detected
using flow cytometry. SOSP-9607 cells in the upregulated groups
showed significantly increased spontaneous apoptosis compared with
the other three groups (p<0.001), with the cells in the
downregulated groups showing significantly decreased spontaneous
apoptosis. The cells in untransfected groups did not produce
noticeable changes compared with cells in the control groups
(p=0.147); (Fig. 3A and C).
Similar results were obtained in U2-OS cell lines (Fig. 3B and D). These results showed that
miR-194 could induce apoptosis in both SOSP-9607 and U2-OS
cells.
Effect of miR-194 on clone formation in
SOSP-9607 and U2-OS cells
The clone formation efficiency of SOSP-9607 cells
was as follows: OE (12.87±2.66%), control (49.00±4.80%), blank
(50.13±4.71%) and KD (77.93±3.30%) groups, respectively, after 21
days of culture (Fig. 4A and C).
No significant differences existed between the blank and control
SOSP-9607 cells (p=0.736). The clone formation efficiency of U2-OS
cells was: OE (9.07±2.93%), control (23.00±5.79%), blank
(22.8±3.41%) and KD (54.87±6.07%) groups, respectively (Fig. 4B and D). No significant difference
was observed between the blank and control cells (p=0.96).
Statistical analysis showed that miR-194 inhibits the clonogenicity
of osteosarcoma in vitro (P<0.001).
miR-194 inhibits migration and invasion
of osteosarcoma in vitro
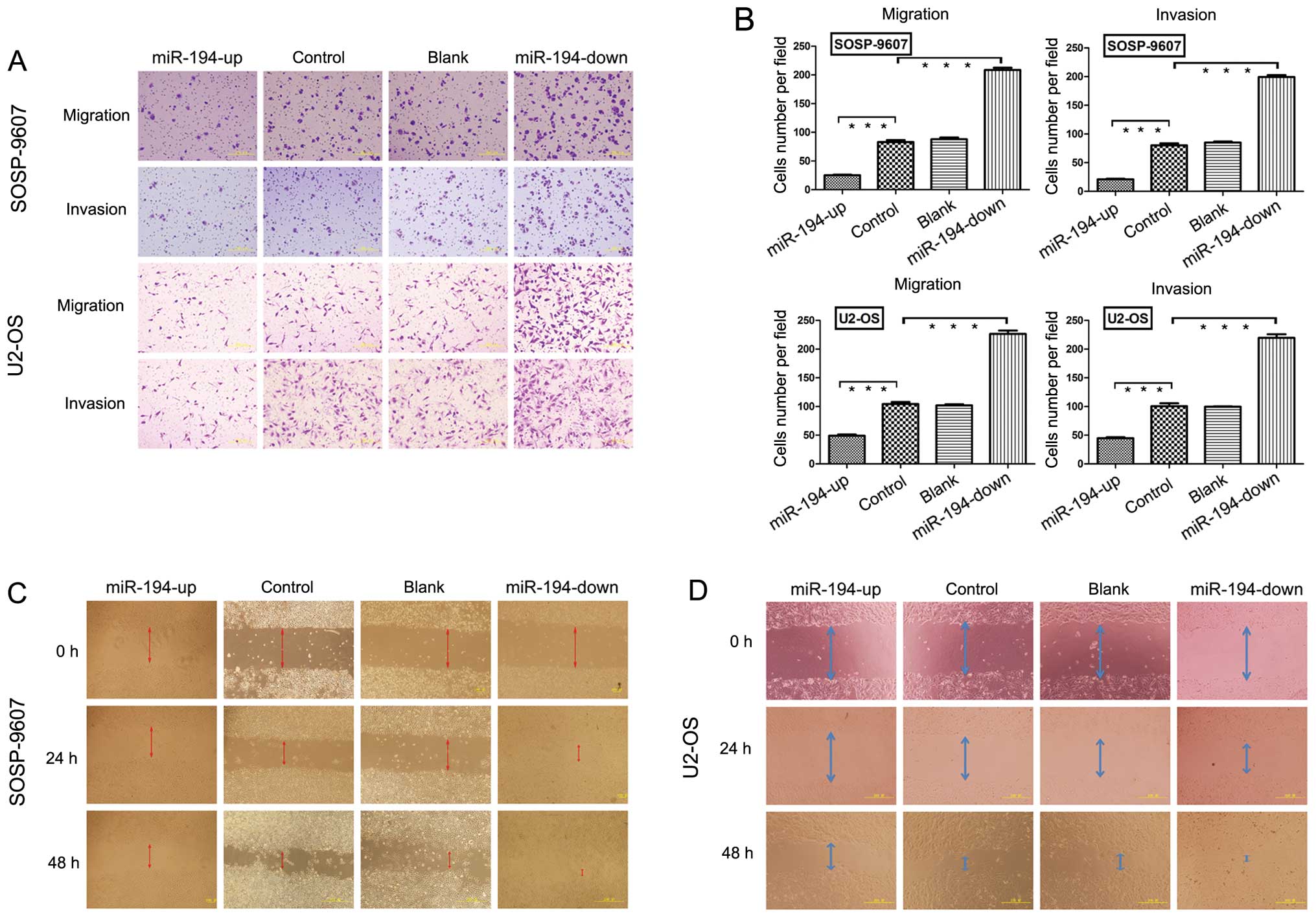
Results in the transwell migration assay showed
significantly lower numbers of OE SOSP-9607 cells (25.00±2.54;
p<0.001) compared with blank (88.00±6.59), control (83.00±7.51)
and KD SOSP-9607 cells (208.60±9.04) (Fig. 5A and B). No significant difference
was seen between the blank and control SOSP-9607 cells (p=0.266).
In the invasion assay, the OE SOSP-9607 cells (21.00±2.23;
p<0.001) passing through the matrigel were significantly lower
than the blank (80.00±8.30), control (85.00±4.12) and KD SOSP-9607
cells (199.20±7.72) (Fig. 5A and
B). No significant difference existed between the blank and
control SOSP-9607 cells (p=0.216) (Fig. 5B). Similar results were obtained in
U2-OS cell lines (Fig. 5A and B),
which strongly indicated that miR-194 had an important role in
reducing the migration and invasion of osteosarcoma in
vitro.
The wound healing assay showed that cells in OE
groups exhibited a significant decrease in migration rate compared
to the other three groups. KD groups of SOSP-9607 cells (or U2-OS)
nearly closed the wound 48 h after incubation, but not the other
three groups (Fig. 5C and D).
miR-194 inhibits tumor growth and
metastasis of osteosarcoma in vivo
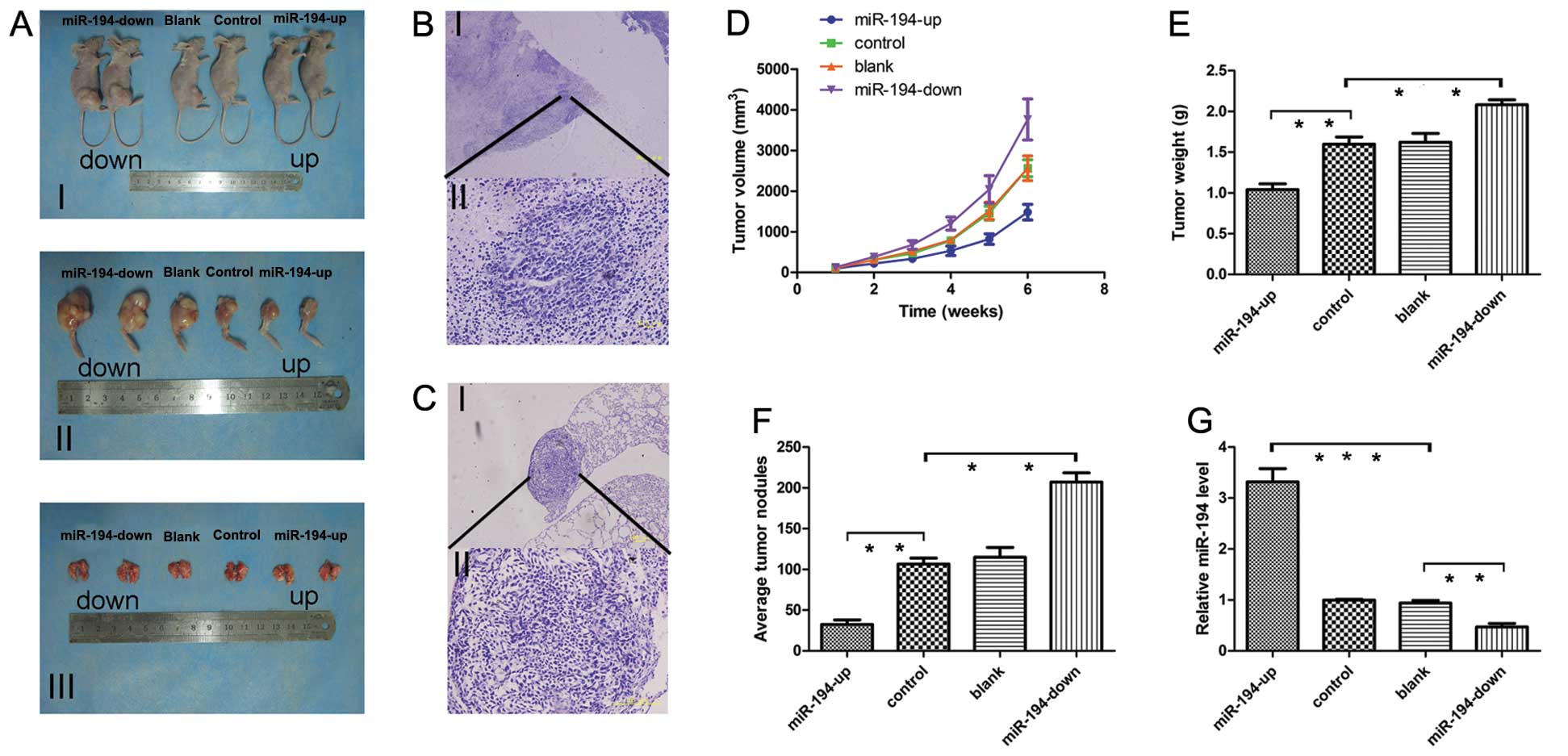
Four groups of stable cells (OE, control, blank and
KD SOSP-9607) were injected into proximal tibia of young nude mice.
To evaluate tumor growth, the length (L) and width (W) of
orthotopic tumor were measured every 7 days post-inoculation. The
volume of tumor was calculated according to the formula: Volume =
1/2 × L × W2, and the growth curve of orthotopic tumor
obtained. Progressive solid tumors were seen in all mice. By
contrast, cells in OE groups produced much smaller tumors, while KD
group generated the biggest size (Fig.
6A and D; P<0.05). The mice were sacrificed 42 days
post-inoculation. The mean tumor weight ± SD of orthotopic tumors
were as follows: OE group 1.04±0.159 g, control group 1.598±0.198
g, blank group 1.622±0.240 g and KD group 2.082±0.134 g (Fig. 6A and E). No significant difference
was observed between the blank and control cells (p=0.842). The
number of metastatic nodes was dramatically reduced in the nude
mice in OE group compared with the other groups (Fig. 6A and F). Tumor and metastases were
confirmed based on histopathological evaluation (Fig. 6B and C). Orthotopic tumors in the
OE group expressed higher miR-194 levels compared with other groups
(Fig. 6G) indicating that
exogenous miR-194 significantly inhibited the tumor growth and
metastasis in vivo.
Expression of miR-194 in osteosarcoma and
corresponding non-cancerous tissues
miR-194 expression was decreased in 59 of 107
(55.14%) tumor samples compared with their non-malignant
counterparts by real-time PCR (Fig.
7A). U6 was used as a control. However, no statistically
significant difference was observed between the cancer tissues
(mean ± SD, 3.6467±7.44944) and matched non-tumor adjacent tissues
(NATs) (mean ± SD, 5.3679±15.09357) (p=0.291) (Fig. 7B).
Downregulation of miR-194 is associated
with advanced clinicopathological features of osteosarcoma
The median miR-194 expression level in 107 patients
with osteosarcoma was 3.647. Patients were divided into two groups
according to their expression levels of miR-194, using its median
as a cut-off: high miR-194 expression group (n=41) and low miR-194
expression group (n=66). As shown in Table I, we found statistically
significant relationships between miR-194 expression and age
(p=0.0015), clinical stage (p=0.019), distant metastasis (p=0.0251)
and patient mortality (p=0.0065). No significant difference was
observed between the expression of miR-194 and the patient gender
(p=0.4038) and tumor size (p=0.6264).
 | Table IRelationship between expression of
miR-194, N-cadherin and IGF1R and clinicopathological factors in
107 osteosarcoma patients. |
Table I
Relationship between expression of
miR-194, N-cadherin and IGF1R and clinicopathological factors in
107 osteosarcoma patients.
| | miR-194
expression | | N-cadherin
expression | | IGF1R
expression | |
|---|
| |
| |
| |
| |
|---|
|
Characteristics | No. | Low no. | High no. | p-value | Low no. | High no. | p-value | Low no. | High no. | p-value |
|---|
| Gender | | | | 0.130 | | | 0.8110 | | | 0.4753 |
| Male | 62 | 42 | 20 | | 29 | 33 | | 26 | 36 | |
| Female | 45 | 24 | 21 | | 20 | 25 | | 22 | 23 | |
| Age (years) | | | | 0.0052a | | | 0.0957 | | | 0.5904 |
| ≥18 | 35 | 15 | 20 | | 12 | 23 | | 17 | 18 | |
| <18 | 72 | 51 | 21 | | 37 | 35 | | 31 | 41 | |
| Tumor size
(cm2) | | | | 0.1098 | | | 0.0769 | | | 0.0172a |
| ≥50 | 47 | 25 | 22 | | 17 | 30 | | 15 | 32 | |
| <50 | 60 | 41 | 19 | | 32 | 28 | | 33 | 27 | |
| Clinical stage | | | | 0.0034a | | | 0.0234a | | | 0.1218 |
| IIA | 32 | 13 | 19 | | 20 | 12 | | 18 | 14 | |
| IIB/III | 75 | 53 | 22 | | 29 | 46 | | 30 | 45 | |
| Distant
metastasis | | | | 0.0058a | | | 0.0081a | | | 0.0139a |
| Yes | 52 | 39 | 13 | | 17 | 35 | | 17 | 35 | |
| No | 55 | 27 | 28 | | 32 | 23 | | 31 | 24 | |
| Status | | | | 0.0065a | | | 0.0037a | | | 0.0221a |
| Survival | 45 | 21 | 24 | | 28 | 17 | | 26 | 19 | |
| Death | 62 | 45 | 17 | | 21 | 41 | | 22 | 40 | |
The median of miR-194 expression levels in all 99
paraformalin-fixed, paraffin-embedded (FFPE) tissues with
osteosarcoma was 5.74. The FFPE tissues were also divided: high
miR-194 expression group (n=21) and low miR-194 expression group
(n=78). As shown in Table II, we
found statistically significant relationships between miR-194
expression and age (p=0.037), tumor size (p=0.041), clinical stage
(p=0.039), distant metastasis (p=0.044) and patient mortality
(p=0.013). No significant difference was observed between the
expression of miR-194 and the patient gender (p=0.749). These
results revealed that loss of miR-194 was associated with some
clinicopathological features of OS.
 | Table IIRelationship between expression of
miR-194 and clinicopathological factors in 99 osteosarcoma
formalin- or paraformalin-fixed, paraffin-embedded (FFPE)
tissues. |
Table II
Relationship between expression of
miR-194 and clinicopathological factors in 99 osteosarcoma
formalin- or paraformalin-fixed, paraffin-embedded (FFPE)
tissues.
| miR194
expression |
|---|
|
|
|---|
|
Characteristics | No. of cases | Mean ± SD | p-value |
|---|
| Gender | | | 0.749 |
| Male | 55 |
5.9304±13.23676 | |
| Female | 44 | 4.6715±6.96565 | |
| Age (years) | | | 0.037a |
| ≥18 | 41 |
11.5227±15.43802 | |
| <18 | 54 | 1.4141±2.63859 | |
| Tumor size
(cm2) | | | 0.041a |
| ≥50 | 36 | 1.3000±3.38195 | |
| <50 | 63 |
7.7371±12.95206 | |
| Clinical stage | | | 0.039a |
| IIA | 27 |
12.9419±15.69269 | |
| IIB/III | 72 | 1.6235±4.33862 | |
| Distant
metastasis | | | 0.044a |
| Yes | 48 | 1.6254±4.66794 | |
| No | 51 |
10.5140±14.59265 | |
| Status | | | 0.013a |
| Survival | 47 |
9.4371±13.60451 | |
| Death | 52 | 0.5538±0.58681 | |
Downregulation of miR-194 confers poor
prognosis in patients with osteosarcoma
Patients with high miR-194 expression survived
significantly longer compared with low miR-194 expression based on
the log rank test (Fig. 7C;
p=0.0007). Similar results were obtained with FFPE tissues
(Fig. 7D; p=0.0004). These results
revealed that downregulation of miR-194 was associated with poor
prognosis of OS.
To identify whether miR-194 was an independent
prognostic covariate for osteosarcoma, we performed a multivariate
Cox proportional hazards analysis. In the final multivariate Cox
regression model, low levels of miR-194 expression in osteosarcoma
(p=0.001, relative risk = 0.390) and distant metastasis (p=0.001,
relative risk =2.386) were associated with a poor prognosis in
terms of overall survival, independent of other clinical covariates
(Table III). Similar results
were obtained in FFPE tissues (Table
III; p=0.023, relative risk = 0.371).
 | Table IIIMultivariate cox regression analysis
of prognostic variables in osteosarcoma and osteosarcoma FFPE
tissues. |
Table III
Multivariate cox regression analysis
of prognostic variables in osteosarcoma and osteosarcoma FFPE
tissues.
| Variables | B | P-value | Wald | Relative risk | 95% confidence
interval |
|---|
| 107 osteosarcoma
tissues | miR-194
expression | −0.943 | 0.001a | 11.813 | 0.390 | 0.228–0.667 |
| Age | −0.020 | 0.937 | 0.006 | 0.980 | 0.595–1.614 |
| Clinical stage | −0.261 | 0.297 | 0.771 | 0.771 | 0.431–1.378 |
| Distant
metastasis | 0.870 | 0.001a | 10.493 | 2.386 | 1.410–4.038 |
| Tumor size
(cm2) | 0.470 | 0.091 | 2.851 | 1.599 | 0.927–2.759 |
| 99 FFPE
tissues | miR-194
expression | −0.991 | 0.023a | 5.193 | 0.371 | 0.158–0.871 |
| Age | 0.036 | 0.893 | 0.018 | 1.037 | 0.612–1.756 |
| Clinical stage | −0.300 | 0.321 | 0.985 | 0.741 | 0.409–1.340 |
| Distant
metastasis | 0.898 | 0.005a | 7.913 | 2.455 | 1.313–4.592 |
| Tumor size
(cm2) | 0.308 | 0.260 | 1.269 | 1.360 | 0.796–2.324 |
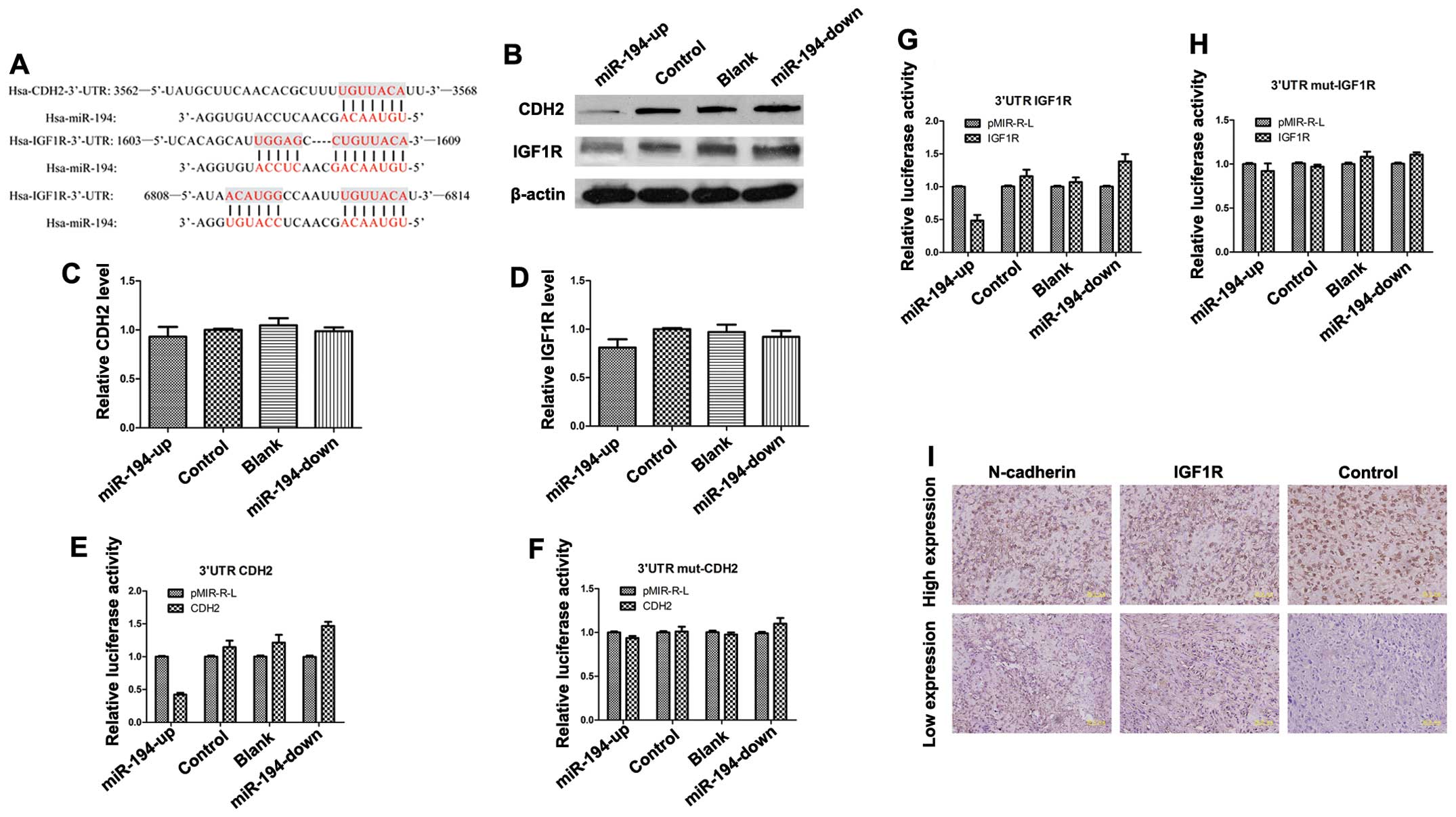
CDH2 and IGF1R are potential targets of
miR-194
We examined the potential targets of miR-194 by
searching the PicTar miRanda and TargetScan databases. We
identified a conserved domain within the 3′-UTR of CDH2
(N-cadherin) and IGF1R with a potential miR-194 binding
site (Fig. 8A). We examined the
expression of CDH2 and IGF1R on mRNA and protein
levels in OE, blank, control and KD SOSP-9607 cells using real-time
PCR and western blot analysis. The results showed that miR-194 had
no effect on CDH2 and IGF1R mRNA levels (Fig. 8C and D). However, the level of
endogenous N-cadherin protein in OE SOSP-9607 cells was
reduced compared with the other three cells normalized to an
endogenous reference β-actin protein (Fig. 8B). Overexpression of
N-cadherin protein was also found in KD SOSP-9607 cells
compared with the other three groups of cells (Fig. 8B). Western blot analysis
demonstrated a significant decrease in endogenous IGF1R
levels in OE group compared with the other three groups (Fig. 8B). The results indicate that
miR-194 may target CDH2 and IGF1R.
We assessed the interaction of miR-194 with
luciferase reporter assay in SOSP-9607 cells using a pMIR-REPORT™
Luciferase vector containing the 3′-UTR of CDH2 or a control
pMIR-REPORT™ Luciferase vector containing the same 3′-UTR with
mutated miR-194 seed nucleotides. Renilla luciferase vector
was used for normalization. The miR-194-up cells significantly
repressed the luciferase activity of the vector with the wild-type
CDH2 3′-UTR, whereas mutation of the seed sequence abolished
this repression (Fig. 8E and F).
Similar results of the IGF1R were also obtained (Fig. 8G and H).
Expression of IGF1R and N-cadherin
proteins were both inversely correlated with miR-194 expression and
regulated the migration and invasion of osteosarcoma cells
We examined IGF1R and N-cadherin
protein expression in 107 patients with osteosarcoma using
real-time quantitative PCR. Of the 41 osteosarcoma cases with
elevated miR-194, 25 (61.0%) showed low levels of
N-cadherin. High levels of N-cadherin were present in
42 of 66 (63.6%) cases with downregulated miR-194 (p<0.05). Of
the 41 osteosarcoma cases with elevated miR-194, 29 (70.7%) had low
levels of IGF1R. High levels of IGF1R were seen in 47
of 66 (71.2%) cases with downregulated miR-194 (p<0.001)
(Table IV). These findings
suggest that expression of the IGF1R and N-cadherin
proteins were inversely correlated with miR-194 expression in
osteosarcoma.
 | Table IVInverse correlation of expression of
miR-194 and N-cadherin and IGF1R in osteosarcoma (using real-time
quantitative PCR) and osteosarcoma FFPE tissues (using
immunohistochemistry analysis). |
Table IV
Inverse correlation of expression of
miR-194 and N-cadherin and IGF1R in osteosarcoma (using real-time
quantitative PCR) and osteosarcoma FFPE tissues (using
immunohistochemistry analysis).
| Group | High miR-194, n
(%) | Low miR-194, n
(%) | In all |
|---|
| 107 osteosarcoma
tissues | High
N-cadherin | 16 (39.0) | 42 (63.6) | 58 |
| Low N-cadherin | 25 (61.0) | 24 (36.4) | 49 |
| In all | 41 | 66 | 107 |
| High IGF1R | 12 (29.3) | 47 (71.2) | 59 |
| Low IGF1R | 29 (70.7) | 19 (28.8) | 48 |
| In all | 41 | 66 | 107 |
| 99 FFPE
tissues | High
N-cadherin | 5 (23.8) | 51 (65.4) | 56 |
| Low N-cadherin | 16 (76.2) | 27 (34.6) | 43 |
| In all | 21 | 78 | 99 |
| High IGF1R | 6 (28.6) | 62 (79.5) | 68 |
| Low IGF1R | 15 (71.4) | 16 (20.5) | 31 |
| In all | 21 | 78 | 99 |
We then examined IGF1R and N-cadherin
protein expression in 99 paraffin specimens of osteosarcoma using
immunohistochemistry analysis. Representative images of
IGF1R and N-cadherin are shown (Fig. 8I) and analyzed (Table IV). Of the 21 osteosarcoma cases
with elevated miR-194, 16 (76.2%) showed low levels of
N-cadherin. High levels of N-cadherin were present in
51 of 78 (65.4%) cases with downregulated miR-194 (p<0.01). Of
the 21 osteosarcoma cases with elevated miR-194, 15 (71.4%) had low
levels of IGF1R. High levels of IGF1R were seen in 62 of 78
(79.5%) cases with downregulated miR-194 (p<0.001). These
findings suggest that expression of the IGF1R and
N-cadherin proteins were inversely correlated with miR-194
expression in osteosarcoma.
The expression of N-cadherin was associated
with clinical stage (p=0.0354), distant metastasis (p=0.0271) and
survival (p=0.0014), while the expression of IGF1R was
associated with tumor size (p=0.0101), distant metastasis
(p=0.0259) and survival (p=0.0253) (Table I).
Discussion
Osteosarcoma is the most frequent primary solid
malignancy of bone, which is defined by the presence of malignant
mesenchymal cells which produce osteoid and/or immature bone
(40). Approximately 20% of
patients present with lung metastases at initial diagnosis and in
40% of patients metastases occur at a later stage. Eighty percent
of all metastases arise in the lungs, most commonly in the
periphery of the lungs, and exhibit resistance to conventional
chemotherapy (41).
Uncontrolled cell proliferation and aggressive tumor
cell metastasis are two essential steps during cancer progression.
Therefore, in the present study, we investigated the effects of
miR-194 on tumor growth and metastasis of osteosarcoma. We showed
that overexpressed miR-194 significantly suppressed proliferation,
migration and invasion of SOSP-9607 and U2-OS cells in
vitro. Our mouse model showed that miR-194 also significantly
inhibited orthotopic tumor growth and lung metastasis in
vivo. We have performed the largest study to date that assessed
the expression levels of miR-194 in osteosarcoma by real-time PCR.
However, no significant difference of miR-194 expression was found
in 107 cancerous and adjacent non-cancerous tissue pairs. Tissue
specificity might be one of the reasons which was associated with
the differences in miR-194 expression, as observed in cancer miRNA
signatures across different organs (42). Furthermore, different osteosarcoma
cell lines show different expression profiles of invasion, motility
and colony formation, with different mRNAs and miRNA expression
(43). We observed that decreased
expression of miR-194 was correlated with cancer progression and
poor prognosis in osteosarcoma patients, independent of other
clinicopathologic factors. Therefore, upregulated miR-194 was very
effective in inhibiting tumor growth and metastasis indicating that
miR-194 functions as a tumor suppressor gene and as a potential
therapeutic target.
Generally, metastatic models are conducted in nude
mice by injecting human osteosarcoma cells either intravenously or
subcutaneously (32). However,
these models are not clinically relevant since osteosarcoma does
not occur spontaneously (44). In
the present study, we selected a spontaneous metastatic model
involving orthotopic transplantation of osteosarcoma cells
resulting in spontaneous pulmonary metastases (32). The microenvironment of tibia in
nude mice resembled the tumor progression and metastases
development clinically.
Cadherins have a role in Ca2+-dependent
cell-cell interaction (45) as
well as acting as metastasis promoting or suppressing proteins in
different cancers (46,47). Insulin and insulin-like growth
factor receptor (IGFR)-mediated molecular pathways are important
effectors of neoplastic transformation in non-small cell lung
cancer (48) and squamous cell
carcinoma of the head and neck (49). IGFIR has a major role in
cancer cell proliferation and survival, and confers resistance to
cytotoxic, hormonal and targeted therapies (50). Our results indicate that miR-194
interacted with N-cadherin and IGF-IR and negatively
regulated their expression at the translational level, which also
indicated that miR-194 may suppress tumor growth and metastasis in
osteosarcoma cells by down-regulating N-cadherin and
IGF-IR.
Unlike siRNAs, which silence the expression of a
single gene, miRNAs mainly silence the expression of multiple genes
simultaneously. It is estimated that an average miRNA may have more
than 100 targets (51). It is
crucial to identify additional target genes that mediate the
miR-194-induced regulation of tumor metastasis. Predicting and
identifying the miR-194-targeting genes provides an experimental
basis for further research on regulatory mechanism of miR-194. By
using TargetScan 5.1 and PicTar, we predicted putative genes of
miR-194, and obtained several putative targets correlating with
tumor growth or metastasis, such as QKI, KIAA1239,
EPHA5, NACC2, MCTS1 and SAMD4A. In
general, the discovery of miRNA and their functions, has introduced
a new dimension to our existing knowledge of signaling molecules
and pathways for more precise therapeutic targeting. Further
investigation is required for characterization of miR-194 and other
miRNAs as prognostic and/or diagnostic markers in human
osteosarcoma.
In conclusion, the results demonstrate that miR-194
affected the growth and metastasis of osteosarcoma cells both in
vitro and in vivo. Overexpression of miR-194
downregulated the expression of N-cadherin and IGF-IR protein,
suggesting that miR-194 functions as tumor suppressor probably by
downregulating N-cadherin and IGF-IR in osteosarcoma.
Downregulation of miR-194 may be associated with tumor
aggressiveness and tumor metastasis of osteosarcoma, suggesting
that miR-194 may be an independent prognostic marker for
osteosarcoma. Other putative miR-194 target genes that are
potentially associated with the growth and metastasis of
osteosarcoma cells should be investigated. Finally, miR-194 may
prove to be a promising gene therapeutic agent. It could be
informative to confirm the putative target genes and further
investigate the underlying molecular mechanisms of miR-194 as a
tumor suppressor gene in osteosarcoma.
Acknowledgements
We would like to thank Chengkui Cai, Qiong Ma,
Guangyi Zhao, Yanhua Wen, Yunyan Liu, Lei Jin, Yinglong Zhang,
Shiju Yan, Cong Sun, Xin Wang and Chuan Dong for their excellent
technical assistance and helpful discussions.
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: data
from the Surveillance, Epidemiology, and End Results Program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marina N, Gebhardt M, Teot L and Gorlick
R: Biology and therapeutic advances for pediatric osteosarcoma.
Oncologist. 9:422–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Picci P: Osteosarcoma (osteogenic
sarcoma). Orphanet J Rare Dis. 2:62007. View Article : Google Scholar
|
|
4
|
Ambros V: microRNAs: tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
|
|
7
|
Sarver AL, Li L and Subramanian S:
MicroRNA miR-183 functions as an oncogene by targeting the
transcription factor EGR1 and promoting tumor cell migration.
Cancer Res. 70:9570–9580. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gong C, Yao Y, Wang Y, et al:
Up-regulation of miR-21 mediates resistance to trastuzumab therapy
for breast cancer. J Biol Chem. 286:19127–19137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mei J, Bachoo R and Zhang CL:
MicroRNA-146a inhibits glioma development by targeting Notch1. Mol
Cell Biol. 31:3584–3592. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Suh SS, Yoo JY, Nuovo GJ, et al:
MicroRNAs/TP53 feedback circuitry in glioblastoma multiforme. Proc
Natl Acad Sci USA. 109:5316–5321. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qi P, Cheng SQ, Wang H, Li N, Chen YF and
Gao CF: Serum microRNAs as biomarkers for hepatocellular carcinoma
in Chinese patients with chronic hepatitis B virus infection. PLoS
One. 6:e284862011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dang X, Ma A, Yang L, et al: MicroRNA-26a
regulates tumorigenic properties of EZH2 in human lung carcinoma
cells. Cancer Genet. 205:113–123. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Suzuki H, Yamamoto E, Nojima M, et al:
Methylation-associated silencing of microRNA-34b/c in gastric
cancer and its involvement in an epigenetic field defect.
Carcinogenesis. 31:2066–2073. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saito Y, Liang G, Egger G, et al: Specific
activation of microRNA-127 with downregulation of the
proto-oncogene BCL6 by chromatin-modifying drugs in human cancer
cells. Cancer Cell. 9:435–443. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yan K, Gao J, Yang T, et al: MicroRNA-34a
inhibits the proliferation and metastasis of osteosarcoma cells
both in vitro and in vivo. PLoS One. 7:e337782012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu Q, Jin H, Yang Z, et al: MiR-150
promotes gastric cancer proliferation by negatively regulating the
pro-apoptotic gene EGR2. Biochem Biophys Res Commun. 392:340–345.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chow TF, Mankaruos M, Scorilas A, et al:
The miR-17–92 cluster is over expressed in and has an oncogenic
effect on renal cell carcinoma. J Urol. 183:743–751. 2010.
|
|
19
|
Deng S, Calin GA, Croce CM, Coukos G and
Zhang L: Mechanisms of microRNA deregulation in human cancer. Cell
Cycle. 7:2643–2646. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Slaby O, Svoboda M, Fabian P, et al:
Altered expression of miR-21, miR-31, miR-143 and miR-145 is
related to clinicopathologic features of colorectal cancer.
Oncology. 72:397–402. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hino K, Tsuchiya K, Fukao T, et al:
Inducible expression of microRNA-194 is regulated by HNF-1α during
intestinal epithelial cell differentiation. RNA. 14:1433–1442.
2008.PubMed/NCBI
|
|
24
|
Meng Z, Fu X, Chen X, et al: miR-194 is a
marker of hepatic epithelial cells and suppresses metastasis of
liver cancer cells in mice. Hepatology. 52:2148–2157. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Krutzfeldt J, Rosch N, Hausser J,
Manoharan M, Zavolan M and Stoffel M: MicroRNA-194 is a target of
transcription factor 1 (Tcf1, HNF1α) in adult liver and controls
expression of frizzled-6. Hepatology. 55:98–107. 2012.PubMed/NCBI
|
|
26
|
Le XF, Almeida MI, Mao W, et al:
Modulation of MicroRNA-194 and cell migration by HER2-targeting
trastuzumab in breast cancer. PLoS One. 7:e411702012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hino K, Fukao T and Watanabe M: Regulatory
interaction of HNF1-α to microRNA-194 gene during intestinal
epithelial cell differentiation. Nucleic Acids Symp Ser (Oxf).
51:415–416. 2007.
|
|
28
|
Braun CJ, Zhang X, Savelyeva I, et al:
p53-Responsive micrornas 192 and 215 are capable of inducing cell
cycle arrest. Cancer Res. 68:10094–10104. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song Y, Zhao F, Wang Z, et al: Inverse
association between miR-194 expression and tumor invasion in
gastric cancer. Ann Surg Oncol. 19(Suppl 3): S509–S517. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dong P, Kaneuchi M, Watari H, et al:
MicroRNA-194 inhibits epithelial to mesenchymal transition of
endometrial cancer cells by targeting oncogene BMI-1. Mol Cancer.
10:992011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pichiorri F, Suh SS, Rocci A, et al:
Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs
the p53/ MDM2 autoregulatory loop in multiple myeloma development.
Cancer Cell. 18:367–381. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen X, Yang TT, Wang W, et al:
Establishment and characterization of human osteosarcoma cell lines
with different pulmonary metastatic potentials. Cytotechnology.
61:37–44. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao G, Cai C, Yang T, et al: MicroRNA-221
induces cell survival and cisplatin resistance through PI3K/Akt
pathway in human osteosarcoma. PLoS One. 8:e539062013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun D, Yang K, Zheng G, Li Z and Cao Y:
Study on effect of peptide-conjugated near-infrared fluorescent
quantum dots on the clone formation, proliferation, apoptosis, and
tumorigenicity ability of human buccal squamous cell carcinoma cell
line BcaCD885. Int J Nanomed. 5:401–405. 2010. View Article : Google Scholar
|
|
35
|
Naito S, von Eschenbach AC, Giavazzi R and
Fidler IJ: Growth and metastasis of tumor cells isolated from a
human renal cell carcinoma implanted into different organs of nude
mice. Cancer Res. 46:4109–4115. 1986.PubMed/NCBI
|
|
36
|
Pan Z, Zhao W, Zhang X, et al: Scutellarin
alleviates interstitial fibrosis and cardiac dysfunction of infarct
rats by inhibiting TGFβ1 expression and activation of p38-MAPK and
ERK1/2. Br J Pharmacol. 162:688–700. 2011.PubMed/NCBI
|
|
37
|
Osaki M, Takeshita F, Sugimoto Y, et al:
MicroRNA-143 regulates human osteosarcoma metastasis by regulating
matrix metalloprotease-13 expression. Mol Ther. 19:1123–1130. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kong W, He L, Coppola M, et al:
MicroRNA-155 regulates cell survival, growth, and chemosensitivity
by targeting FOXO3a in breast cancer. J Biol Chem. 285:17869–17879.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Grenman R, Burk D, Virolainen E, et al:
Clonogenic cell assay for anchorage-dependent squamous carcinoma
cell lines using limiting dilution. Int J Cancer. 44:131–136. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Arndt CA, Rose PS, Folpe AL and Laack NN:
Common musculoskeletal tumors of childhood and adolescence. Mayo
Clin Proc. 87:475–487. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bacci G, Rocca M, Salone M, et al: High
grade osteosarcoma of the extremities with lung metastases at
presentation: treatment with neoadjuvant chemotherapy and
simultaneous resection of primary and metastatic lesions. J Surg
Oncol. 98:415–420. 2008. View Article : Google Scholar
|
|
42
|
Baffa R, Fassan M, Volinia S, et al:
MicroRNA expression profiling of human metastatic cancers
identifies cancer gene targets. J Pathol. 219:214–221. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lauvrak SU, Munthe E, Kresse SH, et al:
Functional characterisation of osteosarcoma cell lines and
identification of mRNAs and miRNAs associated with aggressive
cancer phenotypes. Br J Cancer. 109:2228–2236. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Miretti S, Roato I, Taulli R, et al: A
mouse model of pulmonary metastasis from spontaneous osteosarcoma
monitored in vivo by Luciferase imaging. PLoS One. 3:e18282008.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Angst BD, Marcozzi C and Magee AI: The
cadherin superfamily: diversity in form and function. J Cell Sci.
114:629–641. 2001.PubMed/NCBI
|
|
46
|
Stemmler MP: Cadherins in development and
cancer. Mol Biosyst. 4:835–850. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Derycke LD and Bracke ME: N-cadherin in
the spotlight of cell-cell adhesion, differentiation,
embryogenesis, invasion and signalling. Int J Dev Biol. 48:463–476.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dziadziuszko R, Merrick DT, Witta SE, et
al: Insulin-like growth factor receptor 1 (IGF1R) gene copy number
is associated with survival in operable non-small-cell lung cancer:
a comparison between IGF1R fluorescent in situ hybridization,
protein expression, and mRNA expression. J Clin Oncol.
28:2174–2180. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Meyer F, Samson E, Douville P, Duchesne T,
Liu G and Bairati I: Serum prognostic markers in head and neck
cancer. Clin Cancer Res. 16:1008–1015. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Munagala R, Aqil F and Gupta RC: Promising
molecular targeted therapies in breast cancer. Indian J Pharmacol.
43:236–245. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: tools for microRNA genomics. Nucleic Acids
Res. 36:D154–D158. 2008. View Article : Google Scholar : PubMed/NCBI
|