Introduction
Osteosarcoma is the most common primary bone
malignancy in children and young adults. Osteosarcoma occurs in the
long bones of the limbs, particularly in the distal femur and
proximal tibia. Osteosarcoma is a locally aggressive tumor and
tends to produce early distant metastases, particularly to the
lung. Before 1970, amputation was the only treatment for
osteosarcoma patients and 80% patients died of metastatic disease
(1). Since the 1970s, the
combination of limb-sparing surgery and conventional chemotherapy
agents, including methotrexate (MTX), cisplatin (CDDP), and
doxorubicin, has been used to treat osteosarcoma. However, the
5-year patient survival has plateaued at ~60–70% (2).
Tumors are organized into a hierarchy of
heterogeneous cell populations. Recurrence and metastatic potential
may be due to a subpopulation of cells with stem cell-like
characteristics, such as cancer stem cells (CSCs) or
tumor-initiating cells (TICs), which maintain the capacity to
regenerate entire tumors (3).
Targeting the TICs in osteosarcoma may be a promising avenue to
explore for the development of new therapies for this devastating
disease.
Increasing evidence of the existence of TICs in
patients with osteosarcoma has been reported. Identification of
osteosarcoma TICs has been performed using CD133 (4,5),
side populations (6,7), PKH26 (8), ALDH1 (9,10),
and the promoter reporter assays of hTERT (11) and Oct3/4 (12). TIC-enriched osteosarcoma cell
populations exhibit capacity for self-renewal, multilineage
differentiation, tumorigenicity, and chemo- and radioresistance.
Furthermore, TICs are usually quiescent, with a low protein
turnover, decreased metabolism, and downregulation of proteasome
activity. The ubiquitin-proteasome system is the major
non-lysosomal system for the degradation of intracellular proteins.
Recently, cancer cells with low proteasome activity have been
identified as TICs in patients with breast cancer, glioma (13), pancreatic cancer (14), and esophageal cancer (15) using a fluorescence marker system
for the level of proteasome activity. However, no study has
reported the identification of TICs in human osteosarcoma cell
lines on the basis of low proteasome activity.
Here we showed that a small subpopulation of
osteosarcoma cells with low proteasome activity had TIC-like
properties. Human osteosarcoma cell lines were transfected with a
retroviral vector that monitored proteasome activity using a
fluorescent protein (ZsGreen). We isolated a fraction of cells with
low proteasome activity from these human osteosarcoma cell lines
and identified these cells to have tumor-initiating capacity.
Materials and methods
Cell culture
Human osteosarcoma cell lines U2-OS and MG-63 were
purchased from the American Type Culture Collection (Manassas, VA,
USA) and cultured in Dulbecco’s modified Eagle’s medium
(Invitrogen) supplemented with 10% fetal bovine serum (Gibco),
penicillin, and streptomycin (Sigma). All cells were grown in a
humidified incubator at 37°C with 5% CO2.
Generation of stable cell lines
expressing ZsGreen-cODC fusion proteins using retroviral
transduction
The retroviral expression vector
pQCXIN-ZsGreen-cODC, containing green fluorescence ZsGreen-labeled
degron ODC, was kindly provided by Dr Shinji Tanaka. The retroviral
vector was transfected into platinum A (Plat-A) to generate a
retrovirus. The vector was transfected into Plat-A retroviral
packaging cells using FuGENE 6 (Promega); the virus collected from
the supernatant of Plat-A was used to infect osteosarcoma cells.
The stable transfectants were selected with Geneticin (Invitrogen)
and the accumulation of ZsGreen-labeled degron ODC protein was
monitored by fluorescence microscopy and flow cytometry. To
determine if this was a stable transfection, the cells were exposed
to proteasome inhibitor MG-132 (Wako, Japan) for 12 h.
Time-lapse analysis
After FACS, ZsGreen+ cells were
separately plated at a density of 104 cells in 35-mm
dishes in Dulbecco’s modified Eagle’s medium (Invitrogen)
supplemented with 10% fetal bovine serum (Gibco) and Pen/Strep
(Sigma). After incubation in 5% CO2 at 37°C overnight,
cell attachment was confirmed. Image analysis was performed using a
FV1200 confocal microscope (Olympus).
Flow cytometry
We used a FACS Aria II (BD Biosciences, San Jose,
CA, USA) for cell sorting. Osteosarcoma cells were washed with
phosphate-buffered saline (PBS) and enzymatically dissociated with
0.05% trypsin-EDTA (Invitrogen). Cells were gently triturated and
filtered through cell strainer caps (35-μm mesh) to obtain a single
cell suspension (~1×107 cells/ml). The presence of
ZsGreen allows the selection of cells with low proteasome activity
using flow cytometry with standard fluorescein isothiocyanate
(FITC) for cell sorting.
Measurement of proteasome activity
Using Proteasome-Glo™ cell-based assays (Promega),
the chymotrypsin-like, trypsin-like, and caspase-like activities
were measured according to the manufacturer’s protocol.
Proteasome-Glo™ buffer was mixed with luciferin detection reagent,
and the substrate was added to the mixture and incubated at room
temperature for 1 h. An equal volume of proteasome-Glo™ reagent was
added to the samples and further incubated for 15 min. The
luminogenic substrate containing the Suc-LLVY sequence was
recognized by the proteasome. Following the proteasome cleavage,
the substrate for luciferase (aminoluciferin) was released,
allowing the luciferase reaction to proceed and produce light. The
luminescence was measured using a luminometer (Thermo Fisher
Scientific, Waltham, MA, USA).
Sphere formation assay
ZsGreen+ and ZsGreen− cells
were sorted by FACS, plated separately at a density of 10,000 cells
on low-attachment 6-well plates (Costar, Corning, NY, USA), and
incubated in serum-free Dulbecco’s modified Eagle’s medium/F12
medium (Invitrogen) supplemented with b-FGF, EGF (Sigma), and N2
(Wako). After 14 days, spheres with diameters >100 μm were
counted.
Tumorigenicity assay
Our animal studies were approved by the Animal
Experiments Committee of Osaka University (Suita, Japan). Following
FACS, the portions containing 1×103, 1×104
and 1×105 cells were mixed with BD Matrigel
(Becton-Dickinson, Franklin Lakes, NJ, USA) at a 1:1 ratio and were
subcutaneously injected into 4- to 5-week-old female NOD/SCID mice
(Charles River, Japan). The animals were anesthetized with
isofluorane and maintained under sterile airflow conditions during
the experiments.
Cell proliferation assay
Proliferation assays for the U2-OS and MG-63 cells
treated with MTX and CDDP (both purchased from Wako, Japan) were
performed to test chemosensitivity. Briefly, a total of
2×103 of both ZsGreen+ and
ZsGreen− cells were sorted, individually seeded into
96-well plates (Corning), and treated with varying concentrations
of the drugs for 72 h. The assay was performed using a commercially
available cell counting kit-8 (Dojindo, Japan). The remaining
viable cell count was determined by measuring the absorbance at 450
nm using an Enspire plate reader (Perkin-Elmer).
Clonogenic survival assay
The appropriate number of ZsGreen+ and
ZsGreen− cells were sorted, individually seeded in 6-cm
dishes, and exposed to radiation at 0, 2, 4, 6, and 8 Gy by Cs-137
gamma irradiation generated using a Gammacell 40 Exactor (MDS
Nordion, Ottawa, Canada). After incubation for 14 days, the
colonies were fixed and stained with crystal violet. Colonies
containing >50 cells were counted as survivors. At least three
parallel samples were scored in three to five repetitions conducted
for each irradiation condition.
Statistical analysis
Data are expressed as means ± SDs. Statistically
significant differences were determined by two-way ANOVA analysis
and Student’s t-test, where appropriate, and were defined as
p<0.05.
Results
Establishment of osteosarcoma cell lines
transfected with proteasome sensor vector
To monitor proteasome activity, two osteosarcoma
cell lines, U2-OS and MG-63, were stably transfected with
retroviral vector pQCXIN-ZsGreen-cODC. After geneticin selection
and single cell cloning, we established two osteosarcoma cell lines
transfected with a proteasome sensor vector. We divided these cells
into ZsGreen+ and ZsGreen− groups according
to their fluorescence intensity. We could easily identify
ZsGreen+ cells (Fig.
1). The FACS analysis showed that the fractions of
ZsGreen+ cells were 1.7% in U2-OS and 5.1% in MG-63
(Fig. 2A). To validate the
proteasome activity monitoring, we performed proteasome inhibition
of these established cell lines with MG-132. Fig. 2A shows that proteasome inhibition
dramatically increased the fraction of ZsGreen+ cells
from 1.7 to 93.7% in U2-OS and from 5.1 to 37.8% in MG-63.
Furthermore, we directly measured proteasome activity, such as
trypsin-like, chymotrypsin-like, and caspase-like activities, using
Proteasome Glo; this showed that ZsGreen+ cells had
lower proteasome activity compared with ZsGreen− cells
in MG-63 (Fig. 2B), but not in
U2-OS (data not shown).
ZsGreen+ cells regenerate
ZsGreen+ and ZsGreen− cells
TICs have self-renewal and multilineage
differentiation capacity. To test this, we separately cultured
ZsGreen+ and ZsGreen− cells after cell
sorting for U2-OS. Time-lapse imaging showed asymmetric divisions
of the ZsGreen+ cells into ZsGreen+ and
ZsGreen− cells (Fig.
3A), whereas the ZsGreen− cells did not divide into
ZsGreen+ cells. Similarly, the FACS analysis showed that
the ZsGreen+ cells of U2-OS regenerated into
ZsGreen+ and ZsGreen− cells, whereas the
ZsGreen− cells divided into only ZsGreen−
cells (Fig. 3B). These findings
indicate that the ZsGreen+ population had the capacity
for self-renewal and multilineage differentiation. However, the
ZsGreen+ and ZsGreen− cells of MG-63 divided
into both ZsGreen+ and ZsGreen− cells, which
indicated that both the populations had the capacity for
differentiation.
Sphere-forming capacity
To evaluate self-renewal, we evaluated the
sphere-forming capacity of ZsGreen+ and
ZsGreen− cells. A total of 10,000 ZsGreen+
and ZsGreen− cells were sorted and cultured in
serum-free conditions. The frequencies of sphere formation for
ZsGreen+ and ZsGreen− were 0.16 and 0.013%
(p<0.001) in U2-OS and 0.23 and 0.097% (p=0.027) in MG-63 cells
(Fig. 4A and B), respectively. The
ZsGreen+ cells showed a high frequency of sphere
formation compared with the ZsGreen− cells.
Tumorigenicity assay
TICs are primarily characterized by the ability to
form tumors in NOD/SCID mice. To determine whether
ZsGreen+ cells were more tumorigenic than
ZsGreen− cells in vivo, we injected both the
populations separately into NOD/SCID mice. After 3 months, no
tumors were observed (Table
I).
 | Table ITumorigenesis of ZsGreen+
and ZsGreen− cells of U2-OS in NOD/SCID mice. |
Table I
Tumorigenesis of ZsGreen+
and ZsGreen− cells of U2-OS in NOD/SCID mice.
| No. of cells per
injection |
|---|
|
|
|---|
|
1×103 |
1×104 |
1×105 |
|---|
| ZsGreen+
cells | 0/6 | 0/6 | 0/6 |
| ZsGreen−
cells | 0/6 | 0/6 | 0/6 |
Treatment resistance
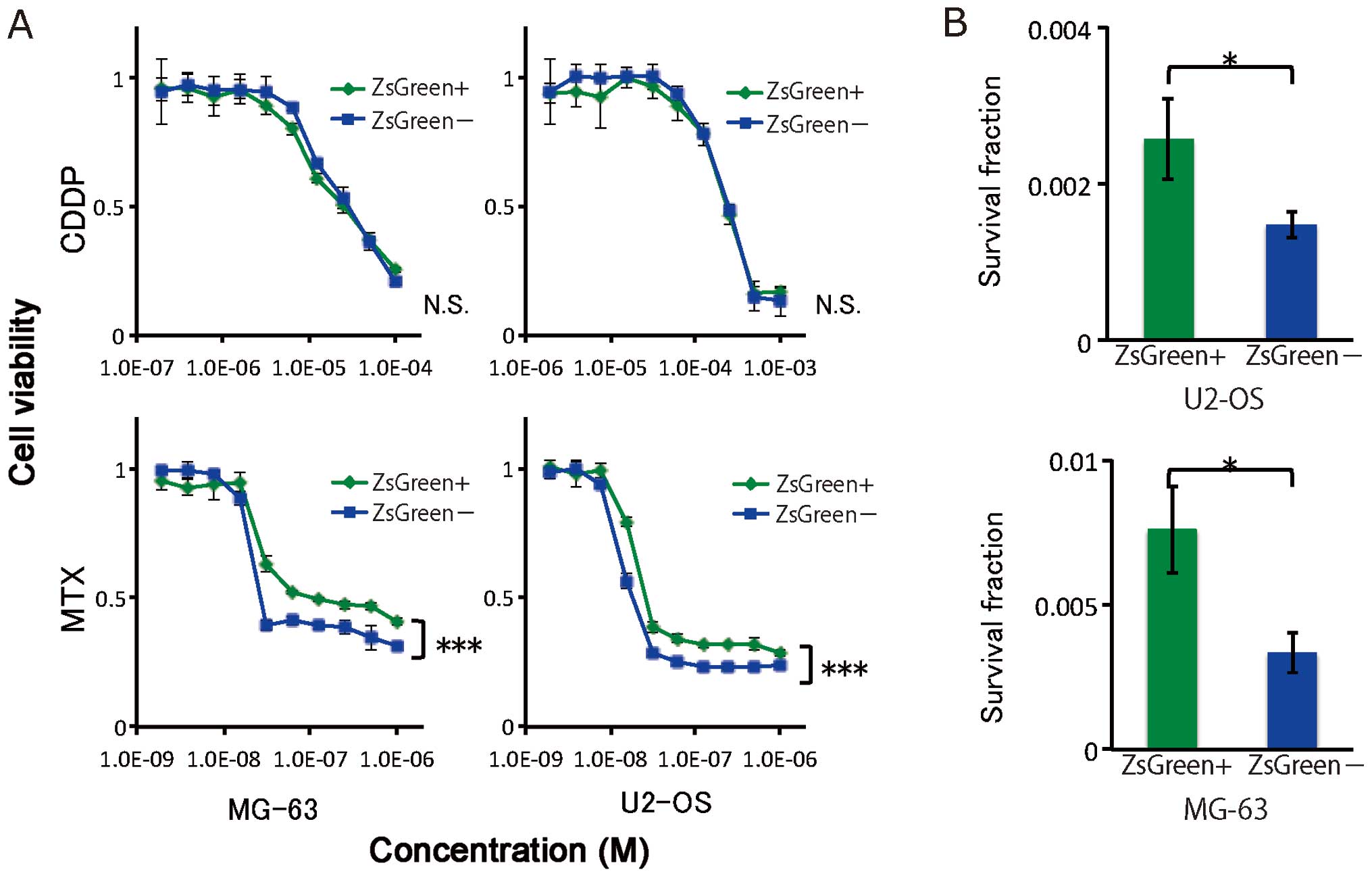
To test chemoresistance, ZsGreen+ and
ZsGreen− cells from two osteosarcoma cell lines were
exposed to MTX and CDDP. The difference in chemoresistance between
the ZsGreen+ and ZsGreen− cells was assessed
using the cell proliferation assay. Of the two cell lines, the
ZsGreen+ cells exhibited significant resistance to MTX,
but not to CDDP (Fig. 5A).
Next, to evaluate radioresistance, we performed
clonogenic survival assays for ZsGreen+ and
ZsGreen− cells of the two cell lines. As shown in
Fig. 5B, ZsGreen+ cells
were more radioresistant than ZsGreen− cells in both
cell lines. These results indicated that the ZsGreen+
cells were resistant to chemotherapy and radiotherapy.
Discussion
Our findings indicate that low proteasome activity
can serve as a marker to identify a subpopulation of TIC cells in
osteosarcoma. The ZsGreen+ cells displayed several
features that are typically observed in TICs, including
self-renewal, generation of differentiated progeny, and treatment
resistance. We observed that the fractions of ZsGreen+
cells were 1.7 and 5.1% in the U2-OS and MG-63 cells, respectively.
Previous reports have shown that the fractions of
ZsGreen+ cells were <4% in U87MG (13) and 0.5% in Panc-1 (14). The proteasome activity of
ZsGreen+ cells are lower than ZsGreen− cells
in MG-63, whereas no difference was found in U2-OS; this is likely
because this proteasome activity assay can detect partial
proteasome activity. The sphere-forming capacity in established
osteosarcoma cell lines has been previously demonstrated (4,8,9,11,12).
Interestingly, Honoki et al showed that the frequency of
sphere formation was 0.25% in ALDH1-positive MG-63 cells (9). Moreover, primary cultures of
osteosarcoma tended to have enhanced sphere-forming capacities
(5,8,16).
Although sphere-forming capacity may be a good in
vitro assay to study osteosarcoma TICs, the gold standard to
identify TICs is the formation of tumors after limiting dilution
transplantation in immunodeficient mice. There was no tumor
formation when we performed limiting dilution transplantation in
NOD/SCID mice. Wang et al also showed that the TIC-like
populations (high ALDH activity) of the osteosarcoma cell lines did
not form tumors, but they did show enhanced tumor formation in
populations with high ALDH activity obtained from a xenograft
(10). Similar to the observation
in this study, the population with low proteasome activity in
osteosarcoma cells in vivo may be more tumorigenic. We could
not inoculate a larger number of cells, such as 106
cells, because of the extensive long time required for the sorting
process.
We studied treatment resistance of the
ZsGreen+ cells and found that they were chemo- and
radioresistant. The mechanism by which treatment resistance occurs
in TICs has not been elucidated. Di Fiore et al manufactured
novel TIC-enriched cell lines such as 3AB-OS that were irreversibly
selected from MG-63 cells after long exposure with 3-aminobenzamide
(3AB) and expressed higher levels of the drug resistance marker
ABCG2 (17). Furthermore, Honoki
et al showed that the high ALDH1 population of MG-63
enhanced the resistance to CDDP and doxorubicin (9). Fujiawa et al recently found
that miR-133a expression was correlated with chemo-resistance
(18).
In conclusion, osteosarcoma cells with low
proteasome activity had TIC properties, including sphere formation
and chemo- and radioresistance. This result may lead to new
approaches for the development of a more specific therapy and for
an improvement in prognosis.
Acknowledgements
We thank Dr Shinji Tanaka at Department of
Hepatobiliary and Pancreatic Surgery, Graduate School of Medicine,
Tokyo Medical and Dental University for pQCXIN-ZsGreen-cODC
plasmid. This study was supported in part by a Grant-in-Aid for
Scientific Research and a grant from the Platform for Drug
Discovery, Informatics, and Structural Life Science, from the
Ministry of Education, Culture, Sports, Science and Technology; a
Grant-in-Aid from the Third Comprehensive 10-Year Strategy for
Cancer Control, Ministry of Health, Labour and Welfare, Japan.
H.I., M.K., N.N., and J.K. received partial support from Chugai
Co., Ltd., Yakult Honsha Co., Ltd., Merck Co., Ltd. and Taiho
Therapeutic Co., Ltd., through institutional endowments.
References
|
1
|
Marcove RC, Miké V, Hajek JV, Levin AG and
Hutter RV: Osteogenic sarcoma under the age of twenty-one. A review
of one hundred and forty-five operative cases. J Bone Joint Surg
Am. 52:411–423. 1970.PubMed/NCBI
|
|
2
|
Gill J, Ahluwalia MK, Geller D and Gorlick
R: New targets and approaches in osteosarcoma. Pharmacol Ther.
137:89–99. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meacham CE and Morrison SJ: Tumour
heterogeneity and cancer cell plasticity. Nature. 501:328–337.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tirino V, Desiderio V, d’Aquino R, et al:
Detection and characterization of CD133+ cancer stem
cells in human solid tumors. PLoS One. 3:e34692008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tirino V, Desiderio V, Paino F, et al:
Human primary bone sarcomas contain CD133+ cancer stem
cells displaying high tumorigenicity in vivo. FASEB J.
25:2022–2030. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Murase M, Kano M, Tsukahara T, et al: Side
population cells have the characteristics of cancer stem-like
cells/cancer-initiating cells in bone sarcomas. Br J Cancer.
101:1425–1432. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang M, Yan M, Zhang R, Li J and Luo Z:
Side population cells isolated from human osteosarcoma are enriched
with tumor-initiating cells. Cancer Sci. 102:1774–1781. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rainusso N, Man T-K, Lau CC, et al:
Identification and gene expression profiling of tumor-initiating
cells isolated from human osteosarcoma cell lines in an orthotopic
mouse model. Cancer Biol Ther. 12:278–287. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Honoki K, Fujii H, Kubo A, Kido A, Mori T,
Tanaka Y and Tsujiuchi T: Possible involvement of stem-like
populations with elevated ALDH1 in sarcomas for chemotherapeutic
drug resistance. Oncol Rep. 24:501–505. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang L, Park P, Zhang H, La Marca F and
Lin C-Y: Prospective identification of tumorigenic osteosarcoma
cancer stem cells in OS99-1 cells based on high aldehyde
dehydrogenase activity. Int J Cancer. 128:294–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu L, Liu S, Zhang C, et al: Enrichment of
human osteosarcoma stem cells based on hTERT transcriptional
activity. Oncotarget. 4:2326–2338. 2013.PubMed/NCBI
|
|
12
|
Levings PP, McGarry SV, Currie TP, et al:
Expression of an exogenous human Oct-4 promoter identifies
tumor-initiating cells in osteosarcoma. Cancer Res. 69:5648–5655.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vlashi E, Kim K, Lagadec C, et al: In vivo
imaging, tracking, and targeting of cancer stem cells. J Natl
Cancer Inst. 101:350–359. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Adikrisna R, Tanaka S, Muramatsu S, et al:
Identification of pancreatic cancer stem cells and selective
toxicity of chemotherapeutic agents. Gastroenterology. 143:234–245.
e72012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kano Y, Konno M, Kawamoto K, et al: Novel
drug discovery system for cancer stem cells in human squamous cell
carcinoma of the esophagus. Oncol Rep. 31:1133–1138.
2014.PubMed/NCBI
|
|
16
|
Ying M, Liu G, Shimada H, et al: Human
osteosarcoma CD49f(−)CD133(+) cells: impaired in osteogenic fate
while gain of tumorigenicity. Oncogene. 32:4252–4263. 2013.
|
|
17
|
Di Fiore R, Santulli A, Ferrante RD, et
al: Identification and expansion of human osteosarcoma-cancer-stem
cells by long-term 3-aminobenzamide treatment. J Cell Physiol.
219:301–313. 2009.PubMed/NCBI
|
|
18
|
Fujiwara T, Katsuda T, Hagiwara K, et al:
Clinical relevance and therapeutic significance of microRNA-133a
expression profiles and functions in malignant
osteosarcoma-initiating cells. Stem Cells. 32:959–973. 2014.
View Article : Google Scholar : PubMed/NCBI
|