Introduction
Renal cell carcinoma (RCC) has the highest fatality
rate in urinary tract malignant tumors and accounts for 2–3% of all
cancers worldwide, >102,000 RCC patients die annually of the
malignancy, the main type of RCC is classified as clear cell renal
cell carcinoma (ccRCC) (1,2). Due to RCC resistance to chemotherapy
and radiotherapy, the mortality of patients with advanced RCC is
very high. The localized RCC mainly relied on the surgical
treatment, while therapy options of advanced RCC are very limited
(3). Recent reports pointed out
that the molecular target drugs (e.g., sunitinib, and sorafenib)
were more effective in metastatic RCC (mRCC) and improved the
survival rate of patients with mRCC, but the long-term effect of
these drugs were limited to mRCC (4,5). The
molecular characteristics of RCC are complex and its mechanism
needs clarification (6).
Epigenetic alteration, especially aberrant
hypermethylation of the promoter region within a CpG island is
involved in silencing of transcription of classical tumor
suppressor genes (TSGs) in cancers and it was considered to be one
of the earliest and most frequent alterations in cancer (7), which had two main specific
mechanisms: one is that DNA methylation may inhibit gene expression
directly by blocking binding to DNA of factors required for optimal
transcription (8). The other is
that methylation affects gene expression directly by interfering
with transcription factor binding, and/or indirectly by recruiting
histone deacetylases through methyl-DNA-binding proteins (9), however, the gene methylation is
reversible, unlike mutation or loss of heterozigosity (LOH).
Therefore, we should search for novel TSGs by way of promoter CpG
methylation so as to reveal the epigenetic mechanism of
carcinogenesis and also identify potential tumor biomarkers for
early detection of RCC (10). At
present, many candidate TSGs silenced by DNA methylation
modification also have been reported in RCC, such as RASSF1
(11), DLC1 (12), and LRRC3B (13).
The large tumor suppressor 1 (LATS1) gene has
been identified as a TSG in Drosophila and encodes a
putative serine/threonine kinase at the earliest time, which is a
member of the nuclear Dbf2-related (NDR) family (14). LATS1 gene is located at
chromosome 6q25.1 and its open reading frame is 3,393 bp encoding a
1130-amino acid polypeptide with molecular weight of 126.87 kDa,
which is highly similar to LATS2 in structure and function
(15). Recently LATS1 has been
identified as a key factor of the Hippo signaling pathway that
plays pivotal roles in various biological processes such as cell
proliferation, genetic stability, cell migration, cell metastasis,
tumorigenesis, organ size control, stem cell differentiation and
renewal, drug resistance, spindle formation, actin polymerization
of modulation (16–19). However, studies have shown that the
expression of LATS1 was reduced or in deficient in a wide variety
of tumors, including gliomas (15), cervical cancer (20), gastric cancer (21), skin cancer (22), and metastatic prostate cancer
(23). LATS1 knockout mice
spontaneously developed non-metastatic soft tissue sarcomas and
metastatic ovarian stromal cell tumors (24). Therefore, LATS1 has been considered
as a TSG, but its role in human cancer is unclear (25). Recently, LATS1 downregulated by
promoter hypermethylation has been reported in various human tumors
including colorectal cancer (CRC) (26), head and neck squamous cell
carcinoma (HNSCC) (27), soft
tissue sarcoma (28), astrocytoma
(29), and breast cancer (30), which indicated that LATS1 promoter
hypermethylation was related to tumorigenesis. Thus, LATS1 may be a
potential target gene for cancer gene therapy. However, whether
LATS1 is subjected to epigenetic silencing and its function in RCC
is unclear.
In this study, we hypothesized that LATS1 is
epigenetically downregulated and functions as a TSG in RCC. To test
this point of view, we first dectected the expression of LATS1 in
RCC tissues by immunohistochemistry and reverse transcription
polymerase chain reaction (RT-PCR) and analyzed its relationship
with the clinicopathologic characteristics of RCC, then selected
786-O cells with low expression of LATS1 mRNA, which promoter
methylation was examined by bisulfite sequence-PCR (BSP), we
subsequently investigated the effects of LATS1 gene
demethylation and overexpression on the biological function and
Yes-associated protein (YAP) in human RCC 786-O cells. The ultimate
goal of this report is to determine whether LATS1 can be
used as a potential target gene for RCC diagnosis and therapy.
Materials and methods
Patients and tissue specimens
The study was approved by the local Ethics
Committees and informed written consent was obtained. Permission
for the use of tissue, it was obtained from the First Affiliated
Hospital of Chongqing Medical University from March to December
2012. Eligible patients were diagnosed to have clear cell carcinoma
of the kidney by pathological methods. The total number of patients
was 30, 15 male and 15 female, aged 36–77 with an average age of
63. We chose the RCC tissues and matched normal kidney tissues (4
cm away from the cancer tissues) undergone radical nephrectomy.
Each specimen was divided into two equal parts, one was drawn into
a Haoe frozen tube after being treated by diethylpyrocarbonate
(DEPC) water, liquid nitrogen frozen condensate, cryopreserved at
−80°C for RT-PCR experiments; the other was placed in 10% formalin
solution for the immunohistochemistry experiment.
Immunohistochemisty
The tissues were fixed with 10% formaldehyde
(ZSGB-BIO, Beijing, China) embedded in paraffin, sectioned into 4
μm thick slices and used for staining. In brief, anti-LATS1 (1:100)
(Santa Cruz Biotechnology, Inc., Dallas, TX, USA) antibodies were
applied to the paraffin section, after deparaffinization, antigen
reconditioning and serum (Gibco-BRL, Carlsbad, CA, USA) was
blocked. Then incubated in 37°C water bath for 2 h and the
secondary antibody and streptomycin antibiotics-peroxidase was
applied on the section. Visualization was performed using DAB
chromogen. Sections were restained with hematoxylin (Shanghai
BlueGene Biotech Co., Ltd., Shanghai, China), dehydrated, and
mounted in neutral gum, and analyzed using a bright field
microscope. The tissues were scored according to positive areas and
staining intensity. The percentage of positive areas was graded as
0 (≤5%), 1 (6–25%), 2 (26–50%), 3 (≥51%), and the staining
intensity was graded as 0–2 (i.e., 0, negative; 1, weak; 2,
strong). The two grades were multiplied and tissues were assigned
to one of three levels: 0 was negative, 1–4 was weak positive, 5–6
was strong positive.
Cell culture and 5-Aza
786-O and HEK-293 were purchased from American Type
Culture Collection (ATCC) (Rockville, MD, USA), 786-O cells were
maintained in RPMI-1640 containing 10% Gibco fetal bovine serum
(FBS) (Gibco-BRL) with 1% antibiotics. HEK-293 cells were grown in
DMEM/HG supplemented with FBS and antibiotics. All cells were
maintained at 37°C in a humidified incubator with 5%
CO2. Cells were treated with 5-Aza-2′-deoxycytidine
(5-Aza) (Sigma-Aldrich, St. Louis, MO, USA) 1 μM for 4 days.
Culture medium and 5-Aza were replaced daily.
RNA isolation and RT-PCR
Total RNA was isolated from RCC cells or tissues
using RNAiso Plus (Takara Bio, Inc., Osaka, Japan). The total RNA
quality was detected by UV spectrophotometer (Bio-Rad, Hercules,
CA, USA), 1 μg RNA was reverse to synthesis the single-stranded
cDNA using PrimeScript RT reagent kit and then amplified with
Premix Taq Q3 2.0 kit (both from Takara Bio, Inc.) according to the
manufacturer’s instructions. The primers used are as follows. LATS1
forward, 5′-CCACCCTACCCAAAACATCTG-3′ and reverse, 5′-CGC
TGCTGATGAGATTTGAGTAC-3′; YAP forward, 5′-TGA
ACAAACGTCCAGCAAGATAC-3′ and reverse, 5′-CAGCCC CCAAAATGAACAGTAG-3′.
GAPDH was used as internal control. GAPDH forward,
5′-ACCACCATGGAGAAGGC TGG-3′ and reverse,
5′-CTCAGTGTAGCCCAGGATGC-3′. PCR conditions were as follows: 94°C
for 5 min, followed by 30 cycles of 94°C for 30 sec, 56°C for 60
sec, and 72°C for 60 sec. The PCR products were run on a 2% agarose
gel. The relative expressions of LATS1 and YAP were quantitatively
measured using densitometry by Quantity One software (Bio-Rad).
Western blot analysis
Cells were scraped and lysates were prepared in 80
μl of radioimmunoprecipitation assay (RIPA) lysis buffer containing
1% phenylmethanesulfonyl fluoride (PMSF), and protein concentration
was determined by protein quantitated kit (all from Beyotime,
Shanghai, China). After mixing with loading buffer and
denaturalizing by boiling for 10 min, 50 μg of protein was loaded
and separated on 6% (for LATS1) or 10% (for YAP and β-actin) sodium
dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) at 80
V. The proteins were transferred onto polyvinylidene difluoride
(PVDF) membranes (Millipore, Billerica, MA, USA) at 250 mA. The
membranes were blocked by 5% non-fat dry milk in TBS containing
0.05% Tween-20 (TBST) for 2 h at room temperature, primary
antibodies against LATS1 (Santa Cruz Biotechnology, Inc.) at 1:150,
YAP (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) at 1:200,
and β-actin (Beyotime) at 1:500 were applied overnight at 4°C,
membranes were washed with TBST, and then incubated with
horseradish peroxidase-conjugated goat anti-rabbit secondary
antibody (Bioworld Technology Co., Ltd., Jiangsu, China) for 1 h at
37°C. The proteins of interest were visualized with the enhanced
ECL detection system (Beyotime). The densitometry of band was
quantified by Quantity One software (Bio-Rad).
BSP
Cells were collected and washed with PBS. BSPs were
carried out as described previously (31), DNA was extracted from the cells
using TIANamp Genomic DNA kit [Tiangen Biotech (Beijing) Co., Ltd.,
Beijing, China] according to the manufacturer’s protocol,
bisulfite-treated DNA was PCR amplified using primers BSP forward,
5′-AGAAGAAAGTTT TGGATTTATTAAAT-3′ and reverse, 5′-CATTTATAAATT
AACTTCTAAAATAC-3′. The PCR products were electrophoresed and
purified using Spin-X tubes, and then cloned into the pUC-T vector
(both from CWbiotech, Beijing, China), with 10 colonies randomly
chosen and sequenced.
Lentiviral vectors and infection
To overexpress LATS1 in 786-O cells,
LATS1-expressing lentiviruses were generated using the GV218 system
(Shanghai GeneChem Co., Shanghai, China) containing the full-length
coding region (from GAGGAT CCCCGGGTACCGGTCGCCACCATGAAGAGGAGTGAA
AAGCCAGAAGG to TCACCATGGTGGCGACCGGAA CATATACTAGATCGCGATTTTTAATC) of
LATS1. The recombinant virus was purified, and titrated by qPCR,
and its infectivity was detected after transduction at increasing
multiplicity of infection (MOI). The experiment was divided into
three groups: control group, mock virus and lentiviral-LATS1.
Flow cytometry analysis (FCM)
To evaluate the cell apoptosis and cell cycle, the
cells were digested and adjusted in density of 1×106/ml,
washed two times with PBS and add 1 ml PBS to beat it after
centrifugal, cells were stained with Annexin V-FITC (Nanjing KeyGen
Biotech Co., Ltd., Nanjing, China) and propidium iodide (PI) (BD
Biosciences, Shanghai, China) for 30 min at room temperature and
determined by FCM (Becton-Dickinson, Franklin Lakes, NJ, USA) to
detect the cell apoptosis. According to the aforementioned method,
the cells were centrifugated, fixed with 70% ethanol, and incubated
for 10 min with 2 mg/ml RNase (Sigma-Aldrich), the cellular DNA was
then stained with 50 ng/ml PI for 30 min at room temperature in
darkness, and the cell cycle was analyzed by FCM.
Cell counting kit-8 (CCK-8)
The cells were seeded in 96-well plates. After
demethylation or infection, the cell proliferation was determined
by using a CCK-8 (Nanjing KeyGen Biotech Co., Ltd.) following
manufacturer’s instructions. The optical density (OD) was measured
with a microplate reader (SpectraMax M2; Molecular Devices,
Sunnyvale, CA, USA) at 450 nm wavelength, then the cell
proliferation inhibition rate (IR) was calculated.
Statistical analysis
Statistical analyses were performed with SPSS,
version 19.0 (SPSS, Inc., Chicago, IL, USA). Data are shown as mean
values ± standard deviation (SD). Differences between the two
independent groups were analyzed by the Student’s t-test. The
χ2 test was used to calculate differences in the
patients’ age, gender, tumor stage, clinical stage and pathological
grade. P<0.05 was considered significant.
Results
Expression of LATS1 in RCC tissues and
cell lines
We examined the expression of LATS1 in 30 pairs of
RCC tissues and matched normal kidney tissues by RT-PCR and
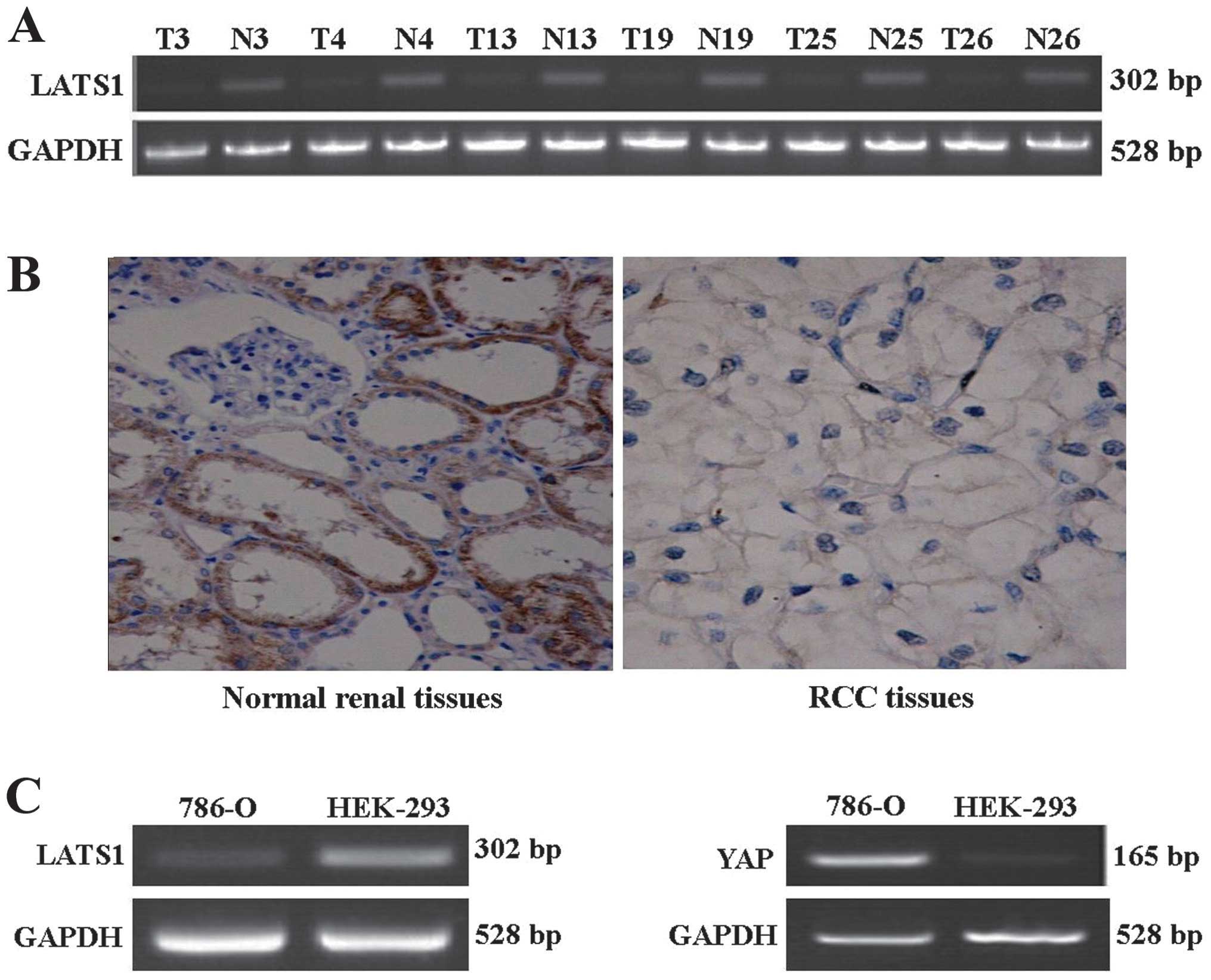
immunohistochemistry. RT-PCR results (Fig. 1A) showed that the expression of
LATS1 mRNA was significantly decreased (P<0.05) in RCC tissues,
while the expression of LATS1 mRNA in normal kidney tissues was
increased.
We examined the expression of LATS1 mRNA in 786-O
and HEK-293 cell line by RT-PCR. As shown in Fig. 1C, compared with HEK-293, the
expression of LATS1 mRNA was significantly decreased (P<0.05) in
786-O.
Immunohistochemistry revealed that the expression
rate of LATS1 in the RCC tissue was 46.7% (14/30) and negative or
weakly positive, while the expression rate of LATS1 in the normal
kidney tissues was 76.7% (23/30) and strongly positive (Fig. 1B). LATS1 mainly accumulated in
cytoplasm of kidney tubules and presented as brown-yellow or brown
particles.
LATS1 expression is related with
clinicopathologic characteristics of RCC
The relationships between LATS1 expression with the
clinicopathologic characteristics in RCC was analyzed. We did not
find a significant correlation of the expression of LATS1 with
patients’ gender, age, tumor size and renal vein metastasis in RCC
(P>0.05). However, we observed that the expression of LATS1 was
related to the clinical stage and pathological grade in RCC
(Table I).
 | Table IThe correlation of LATS1 protein
expression with clinicopathologic characteristics in RCC
(χ2 test). |
Table I
The correlation of LATS1 protein
expression with clinicopathologic characteristics in RCC
(χ2 test).
| | LATS1
expression | | |
|---|
| |
| | |
|---|
| Group | No. | (+) | (−) | Positive (%) | P |
|---|
| Gender | | | | | 0.464 |
| Male | 15 | 6 | 9 | 40 | |
| Female | 15 | 8 | 7 | 53.3 | |
| Age (years) | | | | | 0.232 |
| <60 | 18 | 10 | 8 | 55.6 | |
| ≥60 | 12 | 4 | 8 | 33.3 | |
| Renal vein
metastasis | | | | | 0.171 |
| Yes | 2 | 0 | 2 | 0 | |
| No | 28 | 14 | 14 | 50 | |
| Size of RCC
(cm) | | | | | 0.295 |
| >5 | 8 | 5 | 3 | 62.5 | |
| ≤5 | 22 | 9 | 13 | 40.9 | |
| Pathological
grading | | | | | 0.024 |
| Well | 8 | 6 | 2 | 75 | |
| Moderate | 13 | 7 | 6 | 53.8 | |
| Poor | 9 | 1 | 8 | 11.1 | |
| Clinical
staging | | | | | 0.024 |
| I–II | 17 | 11 | 6 | 64.7 | |
| III–IV | 13 | 3 | 10 | 23.1 | |
Methylation status of the LATS1 promoter
region
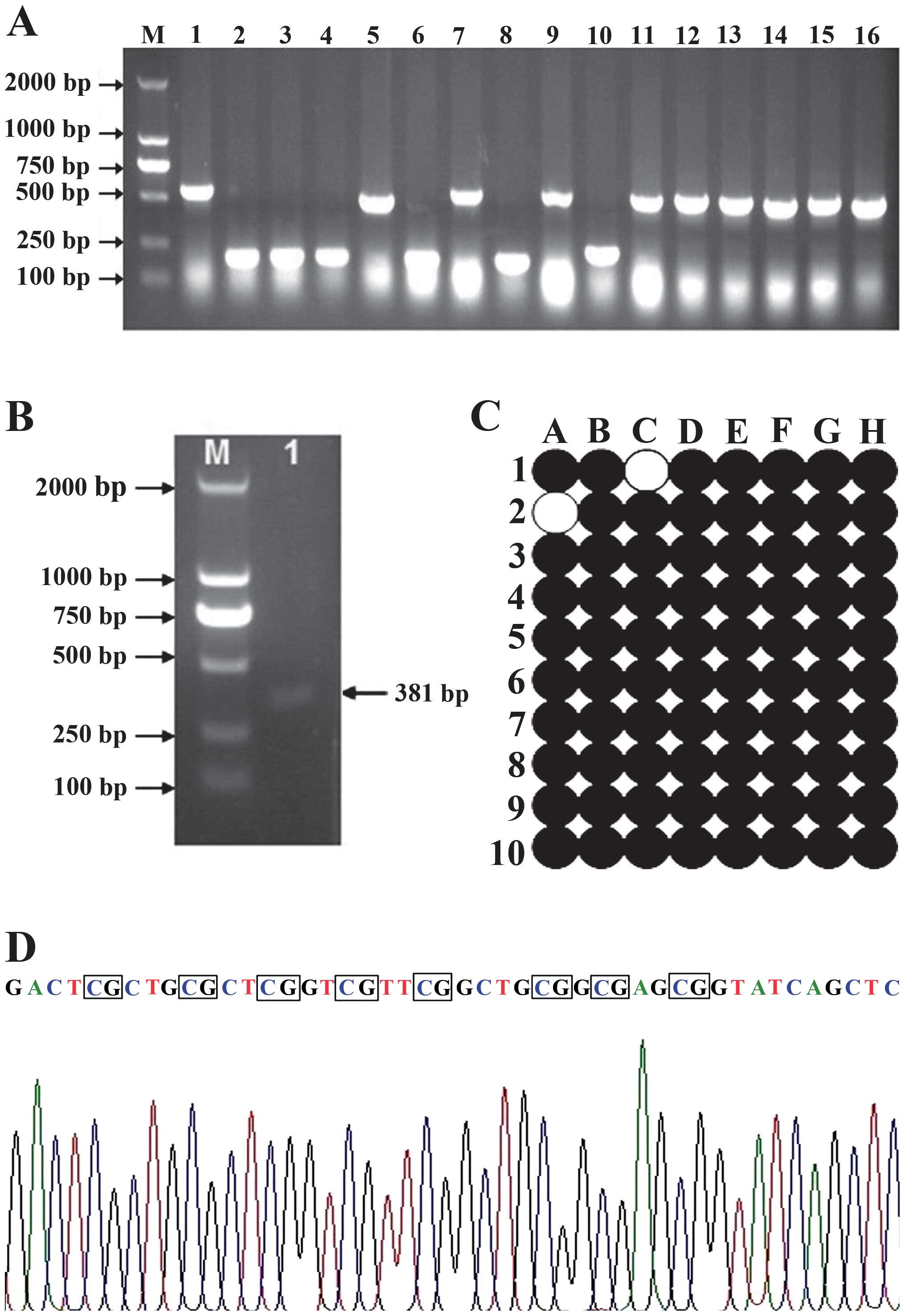
The LATS1 CpG island is located in chromosome 5′ UTR
of 6q24–25 (32). We selected
786-O with low expression of LATS1 mRNA, and the methylation status
at eight CpG sites of the LATS1 CpG islands from −600 to 500 bp was
characterized by BSP. This analysis indicated that the CpG islands
were densely methylated in 786-O cells. The methylation rate
accounted for 97.5% (Fig. 2).
Restoration of LATS1 expression and
downregulation of YAP expression by treatment with 5-Aza
demethylation
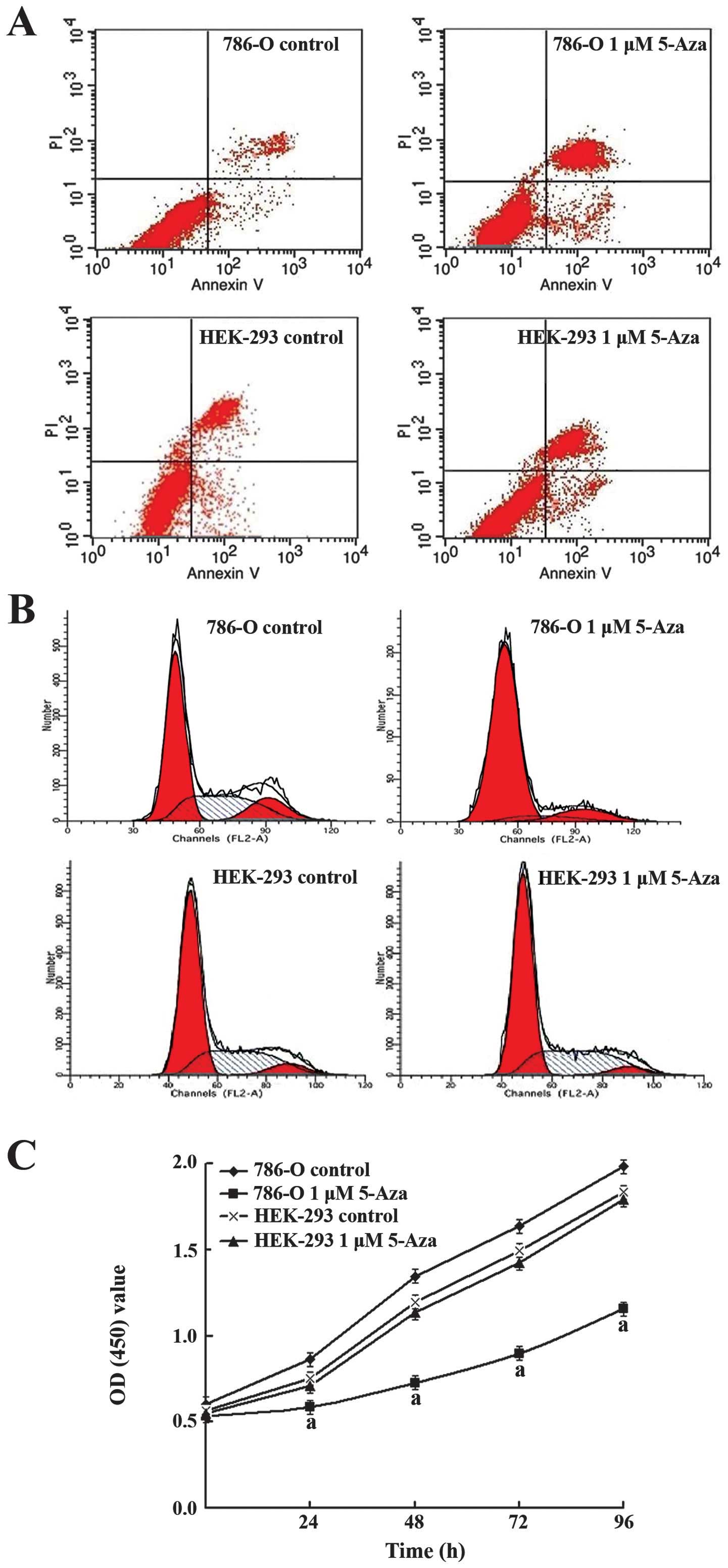
To test whether methylation directly induced LATS1
silencing, we demethylated the LATS1 gene in 786-O cells and
HEK-293 cells with 5-Aza, an inhibitor of DNA methyltransferases
(DNMTs). As shown in Fig. 3, after
treating with 1 μM 5-Aza for 96 h, the mRNA and protein of LATS1
were restored, while the mRNA and protein of YAP were downregulated
in 786-O cells, but, there was no obvious change in HEK-293 cells.
These results indicate that methylation directly mediates the
transcriptional silencing of LATS1 in RCC.
Biological function is affected by LATS1
demethylation with 5-Aza
To investigate the effects of LATS1 gene
demethylation on the biological function in human RCC cell line
786-O. According to the above, in the treatment of cells with 1 μM
5-Aza for 96 h, the FCM revealed that the apoptosis rate of 786-O
cells in experiment group (27.73±2.85)% was significantly higher
than that of control group (7.54±1.71)% (P<0.05), while the
apoptosis rate of HEK-293 had no obvious difference in the
experiment group (16.16±0.94)% from its control (15.77±0.98)%
(P>0.05). This result showed that demethylation of LATS1 induced
apoptosis of 786-O cells (Fig.
4A).
In order to ascertain the cell cycle, the FCM showed
that G1 stage (82.12±3.01)% of 786-O cells in experiment group was
significantly higher than that of control group (57.43±1.13)%
(P<0.05), while G1 stage (61.14±1.05)% of HEK-293 cells in
experiment group had no obvious difference from its control group
(60.35±0.94)% (P>0.05). These results indicate that
demethylation of LATS1 induced 786-O cell cycle to arrest in G1
stage (Fig. 4B).
To investigate cell proliferation, CCK-8 showed that
the OD value (1.16±0.01) of 786-O cells in the experimental group
was obviously lower than that of the control group (1.98±0.01)
(P<0.05) after treatment with 1 μM 5-Aza for 96 h. The cell
proliferation IR was 32, 46, 45, and 41%, respectively, after 24,
48, 72, and 96 h of LATS1 demethylation, while the HEK-293 cells
were not obviously inhibited. This result indicated that cell
proliferation was markedly inhibited by LATS1 demethylation with
5-Aza in 786-O cells (Fig.
4C).
Lentiviral vectors mediated LATS1
overexpression and downregulated YAP
To test the effect of LATS1 on YAP in RCC cells,
786-O cells were infected with lentiviral-LATS1 (Shanghai GeneChem
Co.) at MOI 60 for each viral vector after 96 h. Proportion of
infected 786-O cells was detected by FCM. Results showed that the
transfection efficiency in control group, mock virus group and
lentiviral-LATS1 group was 0.06, 95.96 and 81.69% (Fig. 5A), respectively. Expression of
green fluorescent protein (GFP) after transfection was detected by
fluorescence microscopy (Fig. 5B).
The expression of LATS1 and YAP was detected at both mRNA and
protein levels by RT-PCR and western blot analysis (Fig. 6). Compared with control group and
mock virus group, the data show that the expression of LATS1 mRNA
and protein in lentiviral-LATS1 group was dramatically increased
(P<0.05), but the expression of YAP mRNA and protein was clearly
decreased (P<0.05).
 | Figure 5Expression of green fluorescent
protein (GFP) in 786-O cells. (A) The transfection efficiency was
assayed by flow cytometry analysis (FCM). M1, untransfected 786-O
cells; M2, transfected 786-O cells. Control group represents
untransfected 786-O cells; Mock virus group represents the empty
vector transfected 786-O cells; Transfection efficiency was 95.96%
(M2); Lentiviral-large tumor suppressor 1 (LATS1) group represents
lentiviral-LATS1 transfected 786-O cells. Transfection efficiency
was 81.69% (M2). (B) Expression of GFP after transfection was
detected by fluorescence microscopy. a1, untransfected 786-O cells
under optical microscope; a2, untransfected 786-O cells under
fluorescence microscope; a3, the overlap figure of a1 and a2; b1,
the empty vector transfected 786-O cells under optical microscope;
b2, the empty vector transfected 786-O cells under fluorescence
microscope; b3, the overlap figure of b1 and b2; c1, 786-O cells
transfected with lentiviral-LATS1 under optical microscope; c2,
786-O cells transfected with lentiviral-LATS1 under fluorescence
microscope; c3, the overlap figure of c1 and c2. |
Effect of LATS1 overexpression on
biological function
To test the functions of LATS1 in RCC cell 786-O,
apoptosis in 786-O cells transduced by lentiviral-LATS1 after 96 h
was determined by FCM (Fig. 7A),
the percentage of apoptotic cells in the groups was: control group
(8.40±1.11)%, mock virus (8.12±1.01)%, lentiviral-LATS1
(22.76±1.09)%. Based on our data, it suggested that 786-O cells
transduced by lentiviral-LATS1 induced cell apoptosis.
To investigate the effect of LATS1-mediation on the
RCC cell cycle, the transfected 786-O cells were assayed by FCM.
The results showed increasing numbers of cells arrested in G1
stage. The frequency of cells in G1 was (58.20±1.27)% in the
control group, (58.12±1.06)% in mock virus group, and (79.06±1.43)%
in the lentiviral-LATS1 group. These results suggested LATS1 caused
G1 stage arrest (P<0.05) (Fig.
7B).
We used the CCK-8 assay to determine the cell
proliferation of 786-O cells which were transduced by
lentiviral-LATS1. During the first 3 days, we found the OD value
had no significant difference (P>0.05) in the three groups, but
starting from the fourth day, the lentiviral-LATS1 group
(1.512±0.019) was strikingly lower than the control group
(1.808±0.02) (P<0.05) and mock virus group (1.763±0.014)
(P<0.05), and the cell proliferation IR was 4.71, 5.43, 3.70,
16.37, and 22.85%, respectively, after transfecting cell for 24,
48, 72, 96, and 120 h by lentiviral-LATS1, but the mock virus group
and control group were not obviously inhibited. The results
presented in Fig. 7C show that
cell proliferation was markedly inhibited by the LATS1
gene.
Discussion
The Hippo signaling pathway has been shown to be
involved in tumorigenesis, and the core components of this pathway
include MST1/2, WW45, LATS1/2, MOB1, and YAP, and they interact
with each other (33). In mammals,
when the pathway is activated by cell-cell contact or high cell
density (34), a core kinase
cascade causes a series of reactions (25), MST1/2 kinase interacts with and
phosphorylates WW45, an adaptor protein. Together, this complex
phosphorylates and activates LATS1/2, which, together with its
co-factor MOB1, phosphorylates YAP. Once phosphorylated, YAP is
sequestered or degraded in the cytoplasm, which inhibits the
expression of proliferation related genes cyclin D1 and
cyclin E (35). If any one
of the core components mentioned above changes, it will lead to the
unlimited growth in tissues and organs, and eventually trigger
tumors. Recent research shows, that Hippo signaling pathway, in
addition to the core components found above, can interact with
KIBRA (36) and FRMD6 (37). Although these factors may belong to
the Hippo signaling pathway, how LATS1 is negatively regulated is
largely unknown. Research over the past decades has revealed that
LATS1 is a central part of a complex signal transduction cascade in
multicellular eukaryotes (38) and
a regulator in cellular homeostasis (39). Absence of LATS1 would lead to the
formation of a variety of cancers, including gliomas (15), cervical cancer (20), gastric cancer (21), skin cancer (22), metastatic prostate cancer (23), ovarian stromal cell tumors
(24). However, the expression of
LATS1 in RCC is unclear.
In this study, we first demonstrated that LATS1 was
silenced in RCC tissues and found LATS1 mainly accumulated in
cytoplasm of kidney tubules by immunohistochemistry. This suggested
RCC is likely to begin in the renal tubules. Furthermore, the
expression of LATS1 protein was related with clinicopathologic
characteristics of RCC, and the expression of LATS1 was related to
the tumor clinical stage and pathological grade. We subsequently
measured the expression of LATS1 mRNA in human RCC tissues and
matched normal kidney tissues, and found that the expression of
LATS1 mRNA was significantly decreased in RCC tissues. We further
validated the downregulation of LATS1 mRNA in RCC cells by RT-PCR.
The results suggested that the decreased expression of LATS1
contributes to RCC progression and may play a role of TSG in
tumorigenesis of RCC.
Though studies have shown that LATS1 inactivation
was caused by other mechanisms, including LATS1 regulated by
ubiquitination regulatory factors (40), integrin-linked kinase (ILK)
(41), protease-activated
receptors (PARs) (42), G
protein-coupled receptor (GPCR) signaling pathway (43), LATS1 was supposed to be inactivated
in three major mechanisms: LOH, gene mutations, and
hypermethylation of its promoter region (32). Despite the relatively high
frequency of LOH at the locus containing LATS1 reported in breast
cancers (44,45), only one specimen with reduced LATS1
expression was demonstrated with an allelic LOH and somatic
mutation of LATS1 was not detected in 25 breast cancers by
RT-PCR-SSCP (46). Takahashi et
al (30) reported that
methylation frequency of LATS1 was 56.7% in breast cancer, which
indicated that hypermethylation of the promoter regions of LATS1 is
likely to play an important role in the downregulation of mRNA
levels in breast cancers. These results showed LATS1 was unlikely
to be inactivated in such a classic way as a combination of LOH and
somatic mutation, but was more likely to be induced by
hypermethylation. Therefore, we selected the 786-O cells which
decreased LATS1, and analyzed promoter methylation of LATS1 at
eight CpG sites from −600 to 500 bp by BSP. We found its promoter
was densely methylated, the methylation rate accounted for 97.5%.
Other similar results were also reported, Wierzbicki et al
(26) found that the promoter
regions of LATS1 was hypermethylated as high as 57% in 44 CRCs, and
they concluded that decreased expression of LATS1 in CRC was
associated with promoter hypermethylation, but not microsatellite
instability (MSI) status. Steinmann et al (27) analyzed the promoter methylation of
LATS1 in 54 HNSCC specimens, and found that its hypermethylation
accounted for 24%, and that a trend of increased LATS1 methylation
in more advanced tumor stages and less differentiated HNSCC was
observed. Jiang et al (29)
found that the promoter of LATS1 was hypermethylated as high as
63.66% in 88 astrocytomas, indicating that LATS1 may be a useful
target for astrocytoma therapy. However, the above studies only
reported LATS1 methylation in tumors, did not investigate how LATS1
demethylation affected tumor cells. Therefore, we used 1 μM 5-Aza
to process 786-O cells for 4 days, and we found LATS1 demethylation
could restore its expression and downregulate YAP. It demonstrated
that the inactivation of LATS1 was not caused by a genetic
alteration, such as mutation, but by a reversible epigenetic
mechanism.
We explored the effect of LATS1 overexpression on
YAP in 786-O cells. 786-O cells were infected with lentiviral-LATS1
at MOI 60 after 96 h, and the transfection efficiency was 81.69% by
FCM. GFP expression in 786-O cells was high and the exogenous
expression of LATS1 strongly downregulated YAP. Its proposed
mechanism may be that YAP was phosphorylated at S127 by LATS1 and
likely directly interacted by YAP WW domain and LATS1 PPXY motif to
activate YAP HXRXXS motif to phosphorylate, which results in YAP
binding to 14-3-3 protein and cytoplasmic sequestration (47), YAP might be also phosphorylated by
LATS1 kinases at S381, which caused casein kinase 1 (CK1)
phosphorylation in succession and recruited ubiquitin factors E3 to
degrade YAP in the cytoplasm (48).
The dynamic balance between cell proliferation and
apoptosis maintains the normal size of the tissues and organs and
the homeostasis of the organisms. However, tumorigenesis which is
often related to cell apoptosis is restrained (49). We measured effect of LATS1
overexpression and demethylation on cell apoptosis and
proliferation. The results showed the percentage of apoptotic cells
in lentiviral-LATS1 groups was higher than other groups and the
cell proliferation was inhibited clearly in a time-dependent manner
by lentiviral-LATS1. We also found the apoptosis rate of 786-O
cells in experiment group was significantly higher than that of
control group, and the 786-O cells proliferation was obviously
inhibited with 1 μM 5-Aza treatment for 96 h, our data for the
first time suggested LATS1 overexpression and demethylation
obviously induced cell apoptosis and inhibited proliferation in
786-O cells. The mechanism may be through the upregulating
pro-apoptosis protein p53 and Bax (50) or enhancing the stability of p53
(51) to induce cell apoptosis and
inhibit proliferation.
Regulation of cell cycle is a refined biological
process and depends on a series of cell engine molecules which form
a complex molecular signal network system, but any one of the
molecules or signals which is abnormal will result in disorder of
cell cycle regulation and lead to tumorigenesis. LATS1 is a member
of the subfamily of protein kinases including Dbf2, Orb6, Cot-1,
NDR, and Kpm, which are involved in cell cycle regulation (52), so we investigated the cell cycle by
FCM, and found that exogenous expression of LATS1 and demethylation
strongly induced cell cycle arrested in G1 stage. Consistent with
our notion, Li et al (53)
found 3,3′-diindolylmethane (DIM) was able to upregulate expression
of LATS1 and induce cell arrest in G1 stage in human gastric cancer
cell lines (SNU-1 and SNU-484). Its mechanism is probably that the
DIM through MST1/2-LATS1-MOB1 complex promotes an active Hippo
signaling pathway and favors YAP phosphorylation. Our studies were
different from these research results. Yang et al (50) and Xia et al (54) reported that overexpression of LATS1
caused G2/M arrest through the inhibition of CDC2 kinase activity
in breast cancer cells. Its potential reason may derive from the
fact that cell cycle was detected in different tumor cells selected
or in different microenvironments of tumor cells.
In conclusion, our research strongly indicates that
LATS1 is a TSG in RCC and associated with tumor clinical stage and
pathological grade. We are the first to report on that LATS1 is
silenced at least in part through promoter hypermethylation in RCC
cells. Demethylation of LATS1 promoter by 5-Aza was able to restore
the expression by itself, to downregulate YAP, inhibit cell
proliferation, induce cell apoptosis and cell cycle arrest in G1
stage. Moreover, overexpression of LATS1 by lentivirus mediation
was also able to downregulate YAP, inhibit cell proliferation,
induce cell apoptosis and cell cycle arrest in G1 stage. Thus, it
would be worth further investigating the possible use of LATS1
methylation as a target for future molecular therapy and
diagnosis.
Acknowledgements
This study was supported by the National Natural
Science Foundation of P.R. China (30972999) and Science and Project
of Chongqing Municipal Health Bureau (2013-2-082).
References
|
1
|
Junker K, Ficarra V, Kwon ED, Leibovich
BC, Thompson RH and Oosterwijk E: Potential role of genetic markers
in the management of kidney cancer. Eur Urol. 63:333–340. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: the impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Linehan WM and Rathmell WK: Kidney cancer.
Urol Oncol. 30:948–951. 2012. View Article : Google Scholar
|
|
4
|
Motzer RJ, Hutson TE, Tomczak P, et al:
Overall survival and updated results for sunitinib compared with
interferon alfa in patients with metastatic renal cell carcinoma. J
Clin Oncol. 27:3584–3590. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Escudier B, Eisen T, Stadler WM, et al:
Sorafenib in advanced clear-cell renal-cell carcinoma. New Engl J
Med. 356:125–134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cancer Genome Atlas Research Network.
Comprehensive molecular characterization of clear cell renal cell
carcinoma. Nature. 499:43–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ricketts CJ, Morris MR, Gentle D, et al:
Genome-wide CpG island methylation analysis implicates novel genes
in the pathogenesis of renal cell carcinoma. Epigenetics.
7:278–290. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Watt F and Molloy PL: Cytosine methylation
prevents binding to DNA of a HeLa cell transcription factor
required for optimal expression of the adenovirus major late
promote. Genes Dev. 2:1136–1143. 1988. View Article : Google Scholar
|
|
9
|
Zhu WG, Srinivasan K, Dai Z, et al:
Methylator of adjacent CpG sites affects Sp1/Sp3 binding and
activity in the p21(Cip1) promoter. Mol Cell Biol. 23:4056–4065.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Morris MR, Ricketts C, Gentle D, et al:
Identification of candidate tumour suppressor genes frequently
methylated in renal cell carcinoma. Oncogene. 29:2104–2117. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Reu FJ, Leaman DW, Maitra RR, et al:
Expression of RASSF1A, an epigenetically silenced tumor suppressor,
overcomes resistance to apoptosis induction by interferons. Cancer
Res. 66:2785–2793. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Q, Ying J, Zhang K, et al: Aberrant
methylation of the 8p22 tumor suppressor gene DLC1 in renal
cell carcinoma. Cancer Lett. 249:220–226. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kondratov AG, Stoliar LA, Kvasha SM, et
al: Methylation pattern of the putative tumor-suppressor gene
LRRC3B promoter in clear cell renal cell carcinomas. Mol Med
Rep. 5:509–512. 2012.PubMed/NCBI
|
|
14
|
Justice RW, Zilian O, Woods DF, Noll M and
Bryant PJ: The Drosophila tumor suppressor gene warts
encodes a homolog of human myotonic dystrophy kinase and is
required for the control of cell shape and proliferation. Genes
Dev. 9:534–546. 1995.
|
|
15
|
Ji T, Liu D, Shao W, Yang W, Wu H and Bian
X: Decreased expression of LATS1 is correlated with the progression
and prognosis of glioma. J Exp Clin Cancer Res. 31:672012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao B, Tumaneng K and Guan KL: The Hippo
pathway in organ size control, tissue regeneration and stem cell
self-renewal. Nat Cell Biol. 13:877–883. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu T, Bachman J and Lai ZC: Evidence for a
tumor suppressor role for the large tumor suppressor genes
LATS1 and LATS2 in human cancer. Genetics.
195:1193–1196. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Visser-Grieve S, Zhou Z, She YM, Huang H,
Cyr TD, Xu T and Yang X: LATS1 tumor suppressor is a novel
actin-binding protein and negative regulator of actin
polymerization. Cell Res. 21:1513–1516. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yabuta N, Mukai S, Okamoto A, et al:
N-terminal truncation of Lats1 causes abnormal cell growth control
and chromosomal instability. J Cell Sci. 126:508–520. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Visser S and Yang X: Identification of
LATS transcriptional targets in HeLa cells using whole human genome
oligonucleotide microarray. Gene. 449:22–29. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou GX, Li XY, Zhang Q, et al: Effects of
the hippo signaling pathway in human gastric cancer. Asian Pac J
Cancer Prev. 14:5199–5205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nishio M, Hamada K, Kawahara K, et al:
Cancer susceptibility and embryonic lethality in Mob1a/1b
double-mutant mice. J Clin Invest. 122:4505–4518. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao B, Li L, Wang L, Wang CY, Yu J and
Guan KL: Cell detachment activates the Hippo pathway via
cytoskeleton re organization to induce anoikis. Genes Dev.
26:54–68. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
St John MA, Tao W, Fei X, et al: Mice
deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours
and pituitary dysfunction. Nat Genet. 21:182–186. 1999.PubMed/NCBI
|
|
25
|
Yu FX and Guan KL: The Hippo pathway:
regulators and regulations. Genes Dev. 27:355–371. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wierzbicki PM, Adrych K, Kartanowicz D, et
al: Underexpression of LATS1 TSG in colorectal cancer is associated
with promoter hypermethylation. World J Gastroenterol.
19:4363–4373. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Steinmann K, Sandner A, Schagdarsurengin U
and Dammann RH: Frequent promoter hypermethylation of tumor-related
genes in head and neck squamous cell carcinoma. Oncol Rep.
22:1519–1526. 2009.PubMed/NCBI
|
|
28
|
Seidel C, Schagdarsurengin U, Blümke K, et
al: Frequent hypermethylation of MST1 and MST2 in soft tissue
sarcoma. Mol Carcinog. 46:865–871. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang Z, Li X, Hu J, Zhou W, Jiang Y, Li G
and Lu D: Promoter hypermethylation-mediated down-regulation of
LATS1 and LATS2 in human astrocytoma. Neurosci Res. 56:450–458.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Takahashi Y, Miyoshi Y, Takahata C,
Irahara N, Taguchi T, Tamaki Y and Noguchi S: Down-regulation of
LATS1 and LATS2 mRNA expression by promoter hypermethylation and
its association with biologically aggressive phenotype in human
breast cancers. Clin Cancer Res. 11:1380–1385. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tao Q, Huang H, Geiman TM, Lim CY, Fu L,
Qiu GH and Robertson KD: Defective de novo methylation of viral and
cellular DNA sequences in ICF syndrome cells. Hum Mol Genet.
11:2091–2102. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hisaoka M, Tanaka A and Hashimoto H:
Molecular alterations of h-warts/LATS1 tumor suppressor in human
soft tissue sarcoma. Lab Invest. 82:1427–1435. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nishio M, Otsubo K, Maehama T, Mimori K
and Suzuki A: Capturing the mammalian Hippo: elucidating its role
in cancer. Cancer Sci. 104:1271–1277. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bao Y, Hata Y, Ikeda M and Withanage K:
Mammalian Hippo pathway: from development to cancer and beyond. J
Biochem. 149:361–379. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Schlegelmilch K, Mohseni M, Kirak O, et
al: Yap1 acts downstream of α-catenin to control epidermal
proliferation. Cell. 144:782–795. 2011.
|
|
36
|
Moleirinho S, Chang N, Sims AH, et al:
KIBRA exhibits MST-independent functional regulation of the Hippo
signaling pathway in mammals. Oncogene. 32:1821–1830. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Angus L, Moleirinho S, Herron L, et al:
Willin/FRMD6 expression activates the Hippo signaling pathway
kinases in mammals and antagonizes oncogenic YAP. Oncogene.
31:238–250. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Badouel C and McNeill H: SnapShot: the
hippo signaling pathway. Cell. 145:484. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Visser S and Yang X: LATS tumor
suppressor. a new governor of cellular homeostasis. Cell Cycle.
9:3892–3903. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Salah Z, Cohen S, Itzhaki E and Aqeilan
RI: NEDD4 E3 ligase inhibits the activity of the Hippo pathway by
targeting LATS1 for degradation. Cell Cycle. 12:3817–3823. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Serrano I, McDonald PC, Lock F, Muller WJ
and Dedhar S: Inactivation of the Hippo tumour suppressor pathway
by integrin-linked kinase. Nat Commun. 4:29762013. View Article : Google Scholar
|
|
42
|
Mo JS, Yu FX, Gong R, Brown JH and Guan
KL: Regulation of the Hippo-YAP pathway by protease-activated
receptors (PARs). Genes Dev. 26:2138–2143. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu FX, Zhao B, Panupinthu N, et al:
Regulation of the Hippo-YAP pathway by G-protein-coupled receptor
signaling. Cell. 150:780–791. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Noviello C, Courjal F and Theillet C: Loss
of heterozygosity on the long arm of chromosome 6 in breast cancer:
possibly four regions of deletion. Clin Cancer Res. 2:1601–1606.
1996.PubMed/NCBI
|
|
45
|
Lee EY, To H, Shew JY, Bookstein R, Scully
P and Lee WH: Inactivation of the retinoblastoma susceptibility
gene in human breast cancers. Science. 241:218–221. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Morinaga N, Shitara Y, Yanagita Y, et al:
Molecular analysis of the h-warts/LATS1 gene in human breast
cancer. Int J Oncol. 17:1125–1129. 2000.
|
|
47
|
Hong W and Guan KL: The YAP and TAZ
transcription co-activators: key downstream effectors of the
mammalian Hippo pathway. Semin Cell Dev Biol. 23:785–793. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hergovich A: Regulation and functions of
mammalian LATS/NDR kinases: looking beyond canonical Hippo
signalling. Cell Biosci. 3:322013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Valis K, Prochazka L, Boura E, et al:
Hippo/Mst1 stimulates transcription of the proapoptotic mediator
NOXA in a FoxO1-dependent manner. Cancer Res. 71:946–954. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang X, Li DM, Chen W and Xu T: Human
homologue of Drosophila lats, LATS1, negatively regulate
growth by inducing G(2)/M arrest or apoptosis. Oncogene.
20:6516–6523. 2001.PubMed/NCBI
|
|
51
|
Matallanas D, Romano D, Al-Mulla F, et al:
Mutant K-Ras activation of the proapoptotic MST2 pathway is
antagonized by wild-type K-Ras. Mol Cell. 44:893–906. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hori T, Takaori-Kondo A, Kamikubo Y and
Uchiyama T: Molecular cloning of a novel human protein kinase, kpm,
that is homologous to warts/lats, a Drosophila tumor
suppressor. Oncogene. 19:3101–3109. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li XJ, Park ES, Park MH and Kim SM:
3,3′-Diindolylmethane suppresses the growth of gastric cancer cells
via activation of the Hippo signaling pathway. Oncol Rep.
30:2419–2426. 2013.
|
|
54
|
Xia H, Qi H, Li Y, et al: LATS1 tumor
suppressor regulates G2/M transition and apoptosis. Oncogene.
21:1233–1241. 2002. View Article : Google Scholar : PubMed/NCBI
|