Introduction
Autophagy is an evolutionarily conserved, multistep
cellular response to multiple forms of stress such as deprivation
of nutrition and oxygen. Depending on the circumstances, autophagy
either inhibits or promotes cell growth (1). To enable cell survival under
conditions of stress, autophagy contributes to genomic stability by
eliminating damaged organelles and protein aggregates that produce
reactive oxygen species which cause DNA damage (2). In cancer cells, autophagy is often
caused by hypoxia (3), and leads
to resistance to radiotherapy and chemotherapy (4,5).
By virtue of rapid cell proliferation, overwhelming
growth, increased oxygen consumption, and insufficient blood
supply, intratumoral hypoxia is one of the most important
characteristics of solid tumors (6,7).
Cellular response to hypoxia is partially orchestrated by the
activation of hypoxia-inducible factors (HIFs). During hypoxia, the
HIF-1α protein is protected from degradation, following which it
translocates to the nucleus and binds to constitutively expressed
HIF-1β, thereby activating a mass of HIF target genes involved in
proliferation, energy metabolism, erythropoiesis, and angiogenesis
in both normal tissues and cancer cells (8,9).
Autophagy is also involved in a HIF-1α-mediated cell survival
mechanism (10–12).
MicroRNAs are a group of small non-coding RNAs,
usually 21–24 nucleotides in length. They are involved in
posttranscriptional regulation of genes by pairing with target
mRNAs to induce cleavage or translational silencing (13). A number of miRNAs can be up- or
downregulated by hypoxia; specifically, microRNA-210 is extremely
sensitive to oxygen deprivation. In a hypoxic environment, the
expression of miR-210 is upregulated by HIF-1α (9,14,15).
Bcl-2, a potential target of miR-210 (9), forms a complex with Beclin-1, a Bcl-2
homology 3 (BH3) domain-only protein that plays important roles in
autophagosome formation, autolysosome fusion (16), and promotion of autophagy. The
interaction of Bcl-2 and Beclin-1 inhibits autophagy (17). Therefore, we hypothesized that
miRNA-210/Bcl-2 may potentially play a role in autophagy under
hypoxia.
In the present study, we investigated the mechanism
of hypoxia on autophagy and radiosensitivity in colon cancer
cells.
Materials and methods
Cell culture
Human colon cancer cell lines SW480 and SW620 (Cell
Bank, Shanghai Institutes for Biological Sciences, China) were
cultured in L-15 culture media (Sigma, CA, USA) supplemented with
10% fetal bovine serum (Biowest, Loire Valley, France) at 37°C with
free gas exchange with atmospheric air. Hypoxia treatment was
performed using a tri-gas incubator (37°C, 5% CO2, 93%
N2 and 2% O2; YCP-50S, Changsha Huaxi
Electronic Technology Co., Ltd., Hunan, China) for different
periods.
Irradiation
X-ray irradiation (RT) was performed at a dose rate
of 3.68 Gy/min using the Small Animal Radiation Research Platform
(GulmayMedical, ND, USA) with a total dose of 4 Gy determined
according to our preliminary research.
RNA isolation and quantitative real-time
polymerase chain reaction (qRT-PCR)
Cells were grown to a density of 1×107
and digested with trypsin. Total RNA was isolated using the TRIzol
reagent (RNAprep Pure Cell/Bacteria kit, Tiangen, Beijing, China),
following the manufacturer’s instructions. RNA (500 ng) was used as
the template for reverse transcription to obtain cDNA using
PrimeScript™ RT Reagent kit (Perfect Real-Time, Takara, Japan).
QRT-PCR analysis was performed using the following sequence of the
reverse transcription primer of miR-210: 5′-GTCGTATCCAGTGCAGGG
TCCGAGGTATTCGCACTGGATACGACTCAGCC-3′. The following primers were
used for quantitative PCR: miR-210 primers: sense,
5′-CTGTGCGTGTGACAG-3′, and antisense, 5′-GTGCAGGGTCCGAGGT-3′; U6
primers: sense, 5′-CTCG CTTCGGCAGCACA-3′, and antisense,
5′-AACGCTTCACG AATTTGCGT-3′; HIF-1α primer, sense, 5′-CGTTCCTTCGA
TCAGTTGTC-3′, and antisense, 5′-TCAGTGGTGGCAGTG GTAGT-3′. The
expression of HIF-1α and miR-210 was evaluated using LightCycler
480 SYBR Green I Master (Roche, Basel, Switzerland).
Western blotting
Cells were lysed with RIPA lysis buffer (Roche) and
total protein was quantified using Pierce BCA protein assay kit
(Thermo Fisher, MA, USA). For analysis, an equal amount of protein
extracts was separated by SDS-PAGE on a 10% (w/v) polyacrylamide
gel, followed by electrotransfer onto a BioTrace NT Membrane (Pall,
NY, USA). The blots were blocked for 1 h with blocking buffer 5%
(w/v) fat-free milk, 0.1% (v/v) Tween-20 in PBS. The following
antibodies were used: anti-HIF-1α rabbit monoclonal antibody
(Epitomics, CA, USA), anti-LC3 mouse monoclonal antibody (Medical
& Biological Laboratories Co., Ltd., Japan), anti-Beclin-1
rabbit monoclonal antibody (Cell Signaling Technology, MA, USA),
and anti-Bcl-2 rabbit monoclonal antibody (Cell Signaling
Technology), anti-ATG12 rabbit monoclonal antibody (Cell Signaling
Technology), anti-GAPDH mouse monoclonal antibody (Cell Signaling
Technology) and anti-β-actin mouse monoclonal antibody (Cell
Signaling Technology). After incubation with horseradish
peroxidase-conjugated secondary anti-rabbit (Millipore, MA, USA) or
anti-mouse antibodies (GeneTex, TX, USA), the protein bands were
detected using the ECL blotting detection reagent (Thermo Fisher,
MA, USA), imaged and quantified using Chemioscope Mini system
(ChemiQ 4800, Bioshine, Shanghai, China).
Transfection of HIF-1α and miRNA-210
siRNA
The cells were plated for 24 h before transfection
and then transfected with miR-210 siRNA or HIF-1α siRNA using
X-tremeGene siRNA reagent (Roche). The siRNAs were synthesized by
Shanghai GenePharma Co. and contained the following sequences:
miR-210 siRNA sense, 5′-UCAGCCGCUGUCAC ACGCACAG-3′; HIF-1α siRNA
sense, 5′-CCACCACUGA UGAAUUAAATT-3′, and antisense, 5′-UUUAAUUCAUCA
GUGGUGGTT-3′.
Autophagy detection
The cells were digested with trypsin and washed
twice with phosphate-buffered saline (PBS, pH 7.4) (Hyclone, UT,
USA), centrifuged for 5 min at 400 g and resuspended in a binding
buffer. The autophagosomes were marked using Cyto-ID Autophagy
Detection kit (Enzo, NY, USA). The marked cells were photographed
using fluorescence microscopy (Eclipse Ti-S, Nikon, Japan) and the
fluorescence intensity was detected by flow cytometry (FC500 MPL,
Beckman Coulter, CA, USA).
Apoptosis detection
The cells were digested and washed twice with
phosphate-buffered saline (PBS, pH 7.4; Hyclone), centrifuged for 5
min at 400 g and resuspended in a binding buffer. The cells were
then stained using Annexin V-FITC Apoptosis Detection kit (BD
Biosciences, NJ, USA), followed by the addition of propidium iodide
(BD Biosciences). The samples were analyzed by flow cytometry
(FC500 MPL, Beckman Coulter).
Colony assay
Irradiated cells were counted, diluted in L-15
culture medium, and reseeded in flasks, followed by incubation for
14 days. After this period, the cells were fixed with
polyoxymethylene (Sinopharm Chemical Reagent Co., Ltd., China) and
stained with crystal violet. The stained cells were viewed under
microscope (Eclipse Ti-S, Nikon) and colonies with >50 cells
were calculated. Each experiment included at least 3–6 replicates
and all the experiments were repeated 2–3 times.
Statistical analysis
All the data are presented as mean ± standard
deviation of more than 3 independent experiments. Statistical
analysis was performed using Student’s t-test with SPSS, P<0.05
was considered to be statistically significant.
Results
Effect of hypoxia on the expression of
HIF-1α and miR-210
In the cell lines SW480 and SW620, the relative
expression of miR-210 increased significantly after >12 h of
hypoxic treatment, compared to the normoxic treatment (Fig. 1A, P<0.05). The mRNA expression
of HIF-1α remained unchanged under hypoxic treatment compared to
normoxic treatment in the two cell lines (Fig. 1B, P>0.05). HIF-1α protein was
increased after 12 h of hypoxic treatment in both cell lines
(Fig. 1C).
Relationship between HIF-1α and miR-210
after hypoxic treatment
Under hypoxic conditions, the protein expression of
HIF-1α decreased after transfection with HIF-1α siRNA in both the
cell lines (Fig. 2A). The
expression of miR-210 also decreased significantly from 24 h after
transfection with HIF-1α siRNA under hypoxic treatment (Fig. 2B, P<0.05). In addition, HIF-1α
protein was decreased after miR-210 expression was downregulated
(Fig. 2A), suggesting that the
expression of miR-210 may have a positive feedback effect on the
expression of HIF-1α.
Relationship between the expression of
miR-210, Bcl-2, and autophagy under hypoxic condition
Compared to normoxic treatment, hypoxic conditions
led to a significant increase in autophagy. However, autophagy
induced by hypoxia was attenuated significantly after miRNA-210
siRNA transfections (Fig. 3,
P<0.05).
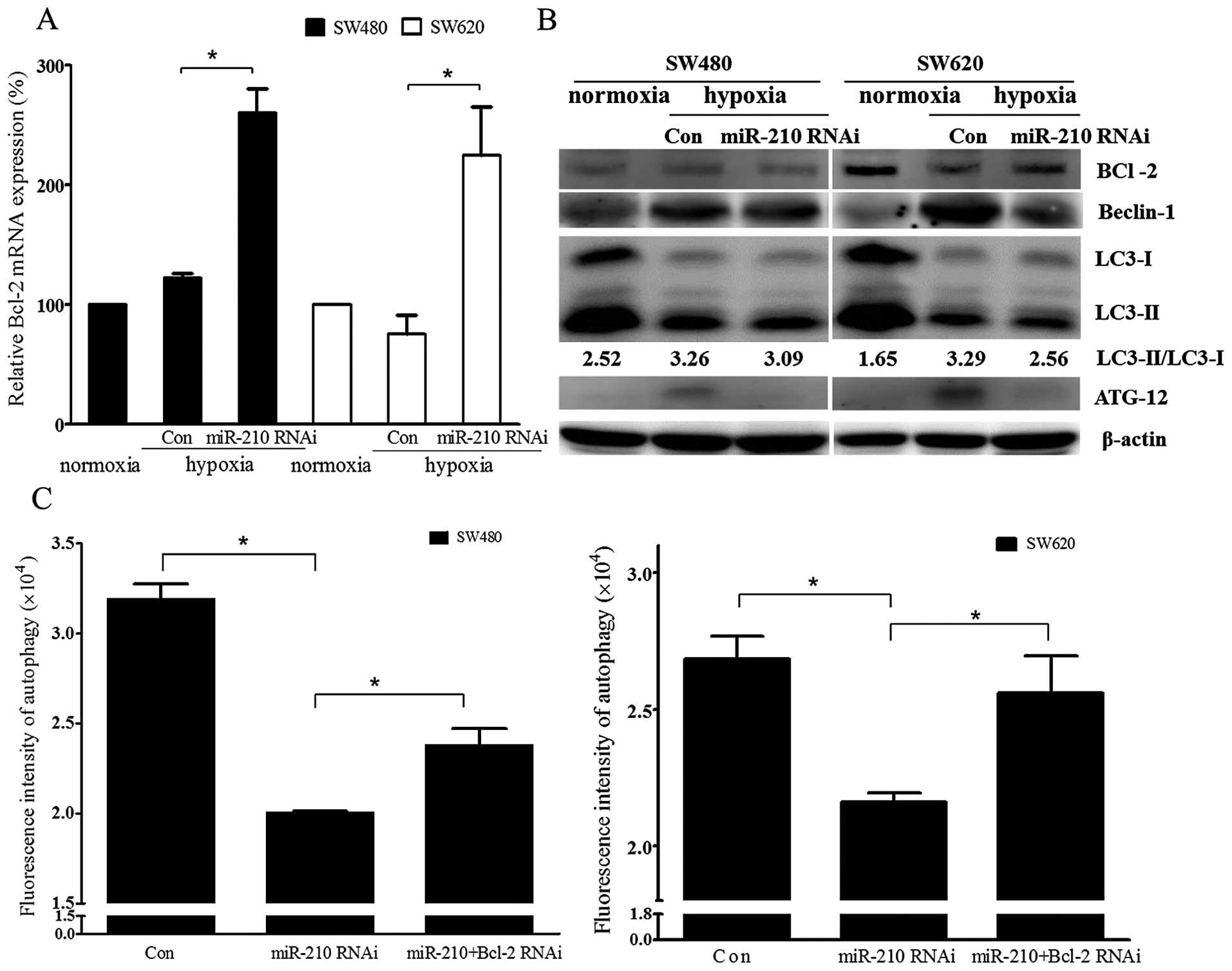
Under hypoxic condition, Bcl-2 mRNA expression was
significantly increased after miRNA-210 was downregulated by siRNA
in both the cell lines (Fig. 4A,
P<0.05). Bcl-2 protein expression was decreased significantly in
SW620 cells after hypoxic treatment. However, the change of bcl-2
protein expression after hypoxic treatment in SW480 cells was not
found. Autophagy-related proteins including Beclin-1, ATG12 and
LC3II as well as the ratio of LC3II/LC3I were decreased after
miRNA-210 siRNA treatment under hypoxic treatment compared with
hypoxic treatment only group (Fig.
4B). Although autophagy induced by the hypoxic treatment was
attenuated by the downregulation of miRNA-210, ablation of both
miR-210 and Bcl-2 by siRNAs remarkably rescued this attenuation in
both SW480 and SW620 cells (Fig.
4C, P<0.05).
Effect of miR-210 and Bcl-2 on
radiosensitivity of colon cancer cells under hypoxic condition
Under hypoxic conditions, apoptosis induced by X-ray
irradiation (RT) increased after miR-210 siRNA transfections,
compared with RT alone (Fig. 5,
P<0.01 for SW480 and P<0.05 for SW620). However, there was no
difference in apoptosis between miR-210 siRNA-transfected group and
either the miRNA-210 or the Bcl-2siRNA-transfected group. On the
other hand, cell colony formation assay showed that the survival of
cells was decreased when miRNA-210 expression was downregulated
under hypoxic conditions after RT, while ablation of both miR-210
and Bcl-2 increased the colony formation activity (Fig. 6, P<0.05). These findings
demonstrate that the inhibition of miR-210 leads to increased
radiosensitivity by upregulating Bcl-2 expression under hypoxic
conditions, which was not related to apoptosis induced by
radiation.
Discussion
Autophagy is an evolutionarily conserved, cellular
degradation pathway that eliminates damaged or superfluous proteins
and organelles (18,19). Cytosolic material is sequestered in
autophagosomes, delivered to lysosomes for degradation, and
recycled to sustain cell viability (20). Previous studies demonstrate that
autophagy can cause resistance to radio- and chemotherapy in
breast, prostate, head and neck, and ovarian cancers (5,21–23).
Hypoxia is a well-known inducer of autophagy
(24), one of the most remarkable
features of solid tumors. Under these conditions, HIF-1α plays a
central role in various cellular responses (25); particularly, it promotes cell
proliferation and survival through MYC and PI3K/AKT (26,27);
increases angiogenesis through VEGF and PDGF (28); and influences cell metabolism by
targeting GLUT1 (29).
Hypoxia-induced autophagy is also involved in a HIF-1α-mediated
general mechanism of cell survival. It has been identified that
BNIP3 and BNIP3L are the downstream targets of HIF-1α-mediated
autophagy (10).
In addition, miR-210 acts as downstream of HIF-1α,
which is consistently upregulated in multiple cancer cell lines
under hypoxic conditions (30–32).
It promotes migration and invasion and is proposed as an
independent prognostic marker for the overall survival of colon
cancer patients (33). Chio et
al reported that miR-210 could mediate hypoxia-induced neural
apoptosis by targeting Bcl-2, and discovered a miR-210-specific
binding element in the 3′-UTR of the Bcl-2 mRNA (34).
The anti-apoptotic protein Bcl-2 inhibits apoptosis
by binding to Bax or Bak (35).
Previous studies demonstrate that the overexpression of Bcl-2
induces resistance to chemo- and radiotherapy in cancer patients
(36). Bcl-2 plays a dual role as
an anti-apoptotic, anti-autophagic protein, and is closely related
to ROS levels (37). BH3 agonists
can specifically interrupt the binding of Bcl-2/XL and Bak/Bax by
competing for the BH3 domain. Hepatocellular carcinoma cells with
increased levels of Bcl-2 exhibit resistance to BH3 agonists by
activating the ROS-JNK-autophagy pathway (38). Silencing of Bcl-2 by siRNA induces
autophagic cell death, but not apoptosis in MCF-7 breast cancer
cells with high level of Bcl-2 expression (39). Similarly, we did not observe any
effect of Bcl-2 silencing on apoptosis induced by radiation
treatment in colon cancer cells. The crosstalk between apoptosis
and autophagy is known to control cell survival and death (40), primarily by JNK1-mediated
phosphorylation of Bcl-2. Under conditions of nutrient starvation,
low level of Bcl-2 phosphorylation initially occurs and promotes
survival by activating autophagy; with prolonged starvation, higher
levels of Bcl-2 phosphorylation promote apoptosis (41). This indicates that Bcl-2 acts as a
switch for the regulation of apoptosis and autophagy.
In this study, we demonstrated that autophagy is
enhanced by hypoxia, and negatively influences radiosensitivity in
two colon cancer cell lines. We demonstrated that HIF-1α causes
hypoxia-induced overexpression of miR-210, while the downregulation
of miR-210 has a negative effect on the expression of HIF-1α.
Furthermore, downregulation of Bcl-2 was discovered under hypoxic
environment. Beclin-1 is known to regulate the initial steps of
autophagy. Beclin-1 possesses a BH-3 domain that binds Bcl-2 and
other Bcl-2 homologues such as Bcl-XL and Mcl-1 (42). The association of Bcl-2 and
Beclin-1 during normal growth conditions may prevent inappropriate
activation of autophagy while disruption of this interaction may
induce autophagy (17,41,43).
In conclusion, our study demonstrates that hypoxia
induces HIF-1α and its downstream target miR-210, which inhibits
the expression of Bcl-2 and enhances autophagy, thereby
contributing to radioresistance in colon cancer cells. However, a
comprehensive analysis of the interactions between HIF-1α, miR-210,
and Bcl-2 remains to be done. Further studies in other cancer cells
and animal models are necessary to understand the crosstalk between
apoptosis and autophagy. A better understanding of the mechanism of
hypoxia-induced autophagy may be of potential value for improving
the effectiveness of radiotherapy.
Acknowledgements
This study was supported by the Scientific Research
Foundation for the Returned Overseas Chinese Scholars (no. N130204)
from China State Education Ministry, the National Natural Science
Foundation of China (nos. 81202148 and 31370838), the Foundation of
Fudan University 985 Project (985IIIYPT06), the Shanghai Pujiang
Program (no. 13PJ1401600), the Foundation of Shanghai Committee of
Science and Technology of China (no. 12DZ2260100).
References
|
1
|
Avivar-Valderas A, Bobrovnikova-Marjon E,
Alan Diehl J, Bardeesy N, Debnath J and Aguirre-Ghiso JA:
Regulation of autophagy during ECM detachment is linked to a
selective inhibition of mTORC1 by PERK. Oncogene. 32:4932–4940.
2013. View Article : Google Scholar :
|
|
2
|
Liu B, Wen X and Cheng Y: Survival or
death: disequilibrating the oncogenic and tumor suppressive
autophagy in cancer. Cell Death Dis. 4:e8922013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Goda N and Kanai M: Hypoxia-inducible
factors and their roles in energy metabolism. Int J Hematol.
95:457–463. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chaachouay H, Ohneseit P, Toulany M,
Kehlbach R, Multhoff G and Rodemann HP: Autophagy contributes to
resistance of tumor cells to ionizing radiation. Radiother Oncol.
99:287–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He WS, Dai XF, Jin M, Liu CW and Rent JH:
Hypoxia-induced autophagy confers resistance of breast cancer cells
to ionizing radiation. Oncol Res. 20:251–258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peitzsch C, Perrin R, Hill RP, Dubrovska A
and Kurth I: Hypoxia as a biomarker for radioresistant cancer stem
cells. Int J Radiat Biol. 90:636–652. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin SC, Liao WL, Lee JC and Tsai SJ:
Hypoxia-regulated gene network in drug resistance and cancer
progression. Exp Biol Med (Maywood). 239:779–792. 2014. View Article : Google Scholar
|
|
8
|
Semenza GL: Hypoxia and cancer. Cancer
Metastasis Rev. 26:223–224. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Majmundar AJ, Wong WJ and Simon MC:
Hypoxia-inducible factors and the response to hypoxic stress. Mol
Cell. 40:294–309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bellot G, Garcia-Medina R, Gounon P, et
al: Hypoxia-induced autophagy is mediated through hypoxia-inducible
factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol
Cell Biol. 29:2570–2581. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang H, Bosch-Marce M, Shimoda LA, et al:
Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic
response to hypoxia. J Biol Chem. 283:10892–10903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Saito S and Nakashima A: The role of
autophagy in extravillous trophoblast function under hypoxia
(Review). Placenta. 34(Suppl): S79–S84. 2013. View Article : Google Scholar
|
|
13
|
Janga SC and Vallabhaneni S: MicroRNAs as
post-transcriptional machines and their interplay with cellular
networks. Adv Exp Med Biol. 722:59–74. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Camps C, Buffa FM, Colella S, et al:
hsa-miR-210 is induced by hypoxia and is an independent prognostic
factor in breast cancer. Clin Cancer Res. 14:1340–1348. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fasanaro P, D’Alessandra Y, Di Stefano V,
et al: MicroRNA-210 modulates endothelial cell response to hypoxia
and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol
Chem. 283:15878–15883. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Maejima Y, Kyoi S, Zhai P, et al: Mst1
inhibits autophagy by promoting the interaction between Beclin1 and
Bcl-2. Nat Med. 19:1478–1488. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pattingre S, Tassa A, Qu X, et al: Bcl-2
antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell.
122:927–939. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mathew R, Karantza-Wadsworth V and White
E: Role of autophagy in cancer. Nat Rev Cancer. 7:961–967. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu S, Chu Y, Yang Y, et al: Inhibition of
macrophage autophagy induced by Salmonella enterica serovar typhi
plasmid. Front Biosci. 19:490–503. 2014. View Article : Google Scholar
|
|
20
|
Nakatogawa H, Suzuki K, Kamada Y and
Ohsumi Y: Dynamics and diversity in autophagy mechanisms: lessons
from yeast. Nat Rev Mol Cell Biol. 10:458–467. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Apel A, Herr I, Schwarz H, Rodemann HP and
Mayer A: Blocked autophagy sensitizes resistant carcinoma cells to
radiation therapy. Cancer Res. 68:1485–1494. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sannigrahi MK, Singh V, Sharma R, Panda NK
and Khullar M: Role of autophagy in head and neck cancer and
therapeutic resistance. Oral Dis. June 3–2014.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang J and Wu GS: Role of autophagy in
cisplatin resistance in ovarian cancer cells. J Biol Chem.
289:17163–17173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Klionsky DJ and Emr SD: Autophagy as a
regulated pathway of cellular degradation. Science. 290:1717–1721.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ruan K, Song G and Ouyang G: Role of
hypoxia in the hallmarks of human cancer. J Cell Biochem.
107:1053–1062. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Koshiji M, Kageyama Y, Pete EA, Horikawa
I, Barrett JC and Huang LE: HIF-1alpha induces cell cycle arrest by
functionally counteracting Myc. EMBO J. 23:1949–1956. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Agani F and Jiang BH: Oxygen-independent
regulation of HIF-1: novel involvement of PI3K/AKT/mTOR pathway in
cancer. Curr Cancer Drug Targets. 13:245–251. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Clara CA, Marie SK, de Almeida JR, et al:
Angiogenesis and expression of PDGF-C, VEGF, CD105 and HIF-1alpha
in human glioblastoma. Neuropathology. 34:343–352. 2014.PubMed/NCBI
|
|
29
|
Liu Y, Li YM, Tian RF, et al: The
expression and significance of HIF-1alpha and GLUT-3 in glioma.
Brain Res. 1304:149–154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Devlin C, Greco S, Martelli F and Ivan M:
miR-210: More than a silent player in hypoxia. IUBMB Life.
63:94–100. 2011.PubMed/NCBI
|
|
31
|
Chan YC, Banerjee J, Choi SY and Sen CK:
miR-210: the master hypoxamir. Microcirculation. 19:215–223. 2012.
View Article : Google Scholar :
|
|
32
|
Grosso S, Doyen J, Parks SK, et al:
MiR-210 promotes a hypoxic phenotype and increases radioresistance
in human lung cancer cell lines. Cell Death Dis. 4:e5442013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qu A, Du L, Yang Y, et al:
Hypoxia-inducible MiR-210 is an independent prognostic factor and
contributes to metastasis in colorectal cancer. PLoS One.
9:e909522014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chio CC, Lin JW, Cheng HA, et al:
MicroRNA-210 targets antiapoptotic Bcl-2 expression and mediates
hypoxia-induced apoptosis of neuroblastoma cells. Arch Toxicol.
87:459–468. 2013. View Article : Google Scholar
|
|
35
|
Zhou F, Yang Y and Xing D: Bcl-2 and
Bcl-xL play important roles in the crosstalk between autophagy and
apoptosis. FEBS J. 278:403–413. 2011. View Article : Google Scholar
|
|
36
|
Kitada S, Takayama S, De Riel K, Tanaka S
and Reed JC: Reversal of chemoresistance of lymphoma cells by
antisense-mediated reduction of bcl-2 gene expression. Antisense
Res Dev. 4:71–79. 1994.PubMed/NCBI
|
|
37
|
Kaminskyy VO and Zhivotovsky B: Free
radicals in cross talk between autophagy and apoptosis. Antioxid
Redox Signal. 21:86–102. 2014. View Article : Google Scholar
|
|
38
|
Ni Z, Wang B, Dai X, et al: HCC cells with
high levels of Bcl-2 are resistant to ABT-737 via activation of the
ROS-JNK-autophagy pathway. Free Radic Biol Med. 70:194–203. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Akar U, Chaves-Reyez A, Barria M, et al:
Silencing of Bcl-2 expression by small interfering RNA induces
autophagic cell death in MCF-7 breast cancer cells. Autophagy.
4:669–679. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mukhopadhyay S, Panda PK, Sinha N, Das DN
and Bhutia SK: Autophagy and apoptosis: where do they meet?
Apoptosis. 19:555–566. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wei Y, Sinha S and Levine B: Dual role of
JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis
regulation. Autophagy. 4:949–951. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Oberstein A, Jeffrey PD and Shi Y: Crystal
structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a
novel BH3-only protein. J Biol Chem. 282:13123–13132. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Maiuri MC, Criollo A, Tasdemir E, et al:
BH3-only proteins and BH3 mimetics induce autophagy by
competitively disrupting the interaction between Beclin 1 and
Bcl-2/Bcl-X(L). Autophagy. 3:374–376. 2007. View Article : Google Scholar : PubMed/NCBI
|