Introduction
Non-coding RNAs (ncRNAs) are found in the genome of
humans, mouse and other animals. However, the functions of ncRNAs
are only partially understood. ncRNAs are mainly classified into
housekeeping or regulatory ncRNAs (1–3).
Based on transcript size, regulatory ncRNAs can be further grouped
into 2 subclasses: small ncRNAs (20–200 nt) and long ncRNAs
(lncRNAs, >200 nt). microRNAs (miRNAs) have been the most
extensively investigated of the small ncRNAs, and estimates suggest
that >1,000 miRNAs regulate up to 30% of all protein-encoding
genes (4–7). Characterization of the functional and
clinical significance of some ncRNAs has shown that they are key
factors in gene regulation and influence normal and cancer cell
phenotypes (4,8–10).
Recent data have demonstrated that >3,000 human
long intervening non-coding RNAs (lincRNAs) and most long ncRNAs
are associated with DNA-binding proteins such as
chromatin-modifying complexes (11) and epigenetically regulate the
expression of multiple genes (12,13).
Transcription of lncRNAs has been shown to modulate gene activity
in response to external oncogenic stimuli and DNA damage (14). This finding indicates the potential
involvement of lncRNAs in the pathogenesis of human diseases, most
notably in cancer (15). HOX
transcript antisense intergenic RNA (HOTAIR) is a 2158-bp
lncRNA that was identified from a custom tiling array of the
HOXC gene cluster. Interaction of HOTAIR with the
polycomb repressive complex 2 (PRC2), which is composed of EZH2,
SUZ12 and EED, leads to the trimethylation of histone H3 lysine 27
and establishment of the repressive H3K27me3 chromatin mark
(11). HOTAIR has been
shown to inhibit tumor suppressor genes such as HOXD10,
PGR, and the proto-cadherin gene family in breast cancer
cells (16). HOTAIR is a
negative prognostic factor for breast, liver, colon, pancreatic and
cervical cancer (17–19). Furthermore, increased HOTAIR
expression has been correlated with enhanced breast and colon
cancer metastasis. Although HOTAIR has been shown to play a
critical role in the progression of breast, liver, colon and
pancreatic cancers, little is known about the molecular mechanisms
in cervical cancer.
Cervical cancer is the third most common cancer and
the fourth leading cause of cancer death in women worldwide
(20). Widespread implementation
of Pap smear screening programs in recent years has decreased the
incidence and mortality of cervical cancer in many countries
(21). Despite this, cervical
cancer continues to be a major public health problem (21). Cancer cell motility and invasion
play a crucial role in the mortality of cervical cancer patients
(22). Therefore, major research
efforts have focused on the identification of tumor-specific
markers for predicting the biological behavior of cervical cancers.
Several miRNAs, including miR-214, miR-143, miR-375, miR-23b and
miR-20, have been shown to modulate cervical cancer cell motility
and invasion; these may represent potential prognostic markers for
predicting the aggressiveness of cervical cancer (23–27).
Increased understanding of the molecular mechanisms underlying
cervical carcinogenesis and progression is required to identify
reliable prognostic markers associated with tumor
aggressiveness.
In the present study, we determined the expression
and clinical significance of HOTAIR in cervical cancer. We
found that HOTAIR was highly expressed in cervical cancer
and was associated with disease recurrence. Furthermore,
HOTAIR knockdown inhibited proliferation, migration and
invasion of human cervical cancer cell lines. Also, we examined the
molecular events that occur downstream of HOTAIR involvement
in cervical cancer migration and invasion. These findings provide
novel insights into the role of HOTAIR in the metastatic
progression of cervical cancer.
Materials and methods
Human tissues
Cervical cancer samples were obtained from 111
female patients who underwent surgery at Yonsei Severance Hospital,
Yonsei University, between 2007 and 2012. Specimens from patients
with newly diagnosed invasive [FIGO (International Federation of
Gynecology and Obstetrics) stage IA-IVB] cervical cancer who had
not received prior treatment were included in the study. Forty
samples of normal cervix from patients undergoing simple
hysterectomy because of uterine leiomyomata were obtained as
controls. Specimens from patients with concomitant gynecological
cancer were excluded from the study. All specimens were immediately
frozen in liquid nitrogen and stored at −80°C until RNA extraction.
The study was conducted according to the principles in the
Declaration of Helsinki and was approved by the ethical committee
of Yonsei Severance Hospital. Informed consent was obtained from
all patients. The clinical information is summarized in Table I.
 | Table IAssociation between HOTAIR
expression and clinicopathologic factors in cervical cancer
(n=111). |
Table I
Association between HOTAIR
expression and clinicopathologic factors in cervical cancer
(n=111).
| | HOTAIR
expression | |
|---|
| |
| |
|---|
| n (%) | Low | High | P-valuea |
|---|
| Age (mean ±
SD) | 111 | 50.4±2.51 | 50.8±1.29 | 0.8809 |
| Stage | | | | 0.7671 |
| I | 43 (38.74) | 10 | 33 | |
| II | 56 (50.45) | 10 | 46 | |
| III–IV | 12 (10.81) | 2 | 10 | |
| Cell type | | | | 0.2334 |
| SCC | 78 (70.27) | 17 | 61 | |
| Adeno | 24 (21.62) | 2 | 22 | |
| Mixed | 3 (2.7) | 1 | 2 | |
| Other | 6 (5.41) | 2 | 4 | |
| Tumor size
(cm) | | | | 0.8839 |
| <4 | 66 (60) | 14 | 52 | |
| ≥4 | 44 (40) | 8 | 36 | |
| Lymphatic
invasion | | | | |
| Yes | 58 (52.25) | 10 | 48 | 0.6351 |
| No | 53 (47.75) | 12 | 41 | |
| Lymph node
metastasis | | | | 0.0437 |
| Yes | 35 (31.53) | 3 | 32 | |
| No | 76 (68.47) | 19 | 57 | |
Cell culture
SiHa (squamous cervical carcinoma), HeLa (epitheloid
cervical carcinoma) and Caski (epidermoid cervical carcinoma
established from a metastasis in the small bowel mesentery) human
cervical cancer cell lines obtained from the American Type Culture
Collection (ATCC, Rockville, MD, USA). SiHa and HeLa cells were
cultured in Dulbecco’s modified Eagle’s medium, and Caski cells
were cultured in RPMI-1640 medium (Gibco-BRL, Gaithersburg, MD,
USA). The human keratinocyte cell line HaCaT was cultured in
RPMI-1640 medium. The culture media were supplemented with 10%
(vol/vol) fetal bovine serum and penicillin/streptomycin. The cell
lines were maintained at 37°C in a humidified atmosphere of 5%
CO2 and 95% air. Cells with a passage number <20 were
used in all experiments.
Quantitative real-time polymerase chain
reaction (qRT-PCR)
Total RNA was extracted from cancerous/non-cancerous
specimens or cell lines using TRIzol® reagent
(Invitrogen, Carlsbad, CA, USA), and 2 μg of total RNA was reverse
transcribed into first-strand cDNA by using a reverse transcription
reagent kit (Invitrogen) according to the manufacturer’s protocol.
qRT-PCR was performed using the SYBR® Green real-time
PCR kit (Toyobo, Co., Ltd., Osaka, Japan) in a 20-μl reaction
volume, which contained 10 μl of SYBR-Green Master PCR Mix, 5 pmole
each of forward and reverse primers, 1 μl of diluted cDNA template,
and appropriate amounts of sterile distilled water. Conditions for
the amplification of genes were as follows: initial denaturation at
95°C for 3 min; 40 cycles of denaturation at 95°C for 15 sec,
annealing at 60°C for 60 sec, and elongation at 72°C for 60 sec;
and final elongation at 72°C for 5 min. qRT-PCR was performed on
the ABI StepOnePlus Real-Time PCR system (Applied Biosystems,
Foster City, CA, USA). All quantifications were performed with
U6 as the internal standard. The PCR primer sequences were
as follows: HOTAIR, 5′-GGTAGAAAAAGCAACCACGAAGC-3′ (sense)
and 5′-ACATAAACCTCTGTCTGTGAG TGCC-3′ (antisense); E-cadherin,
5′-ATTCTGATTCTGC TGCTCTTG-3′ (sense) and
5′-AGTAGTCATAGTCCTGGTCCT-3′ (antisense); β-catenin,
5′-TGCAGTTCGCCTTCACTATG-3′ (sense) and 5′-ACTAGTCGTGGAATGGCACC-3′
(antisense); vimentin, 5′-TGGATTCACTCCCTCTGGTT-3′ (sense) and
5′-GGTCATCGTGATGCTGAGAA-3′ (antisense); snail,
5′-GAGGCGGTGGCAGACTAG-3′ (sense) and 5′-GACACATCGGTCAGACCAG-3′
(antisense); twist, 5′-CGGGAGTCCGCAGTCTTA-3′ (sense) and
5′-TGAATCTTGCTCAGCTTGTC-3′ (antisense); and U6,
5′-CTCGCTTCGGCAGCACA-3′ (sense) and 5′-AACGCTTCAGGAATTTGCGT-3′
(antisense). Relative gene expression was analyzed using the
2−ΔΔCT method, and the results were expressed as extent
of change with respect to control values. qRT-PCR experiments were
replicated at least 3 times.
Small interfering RNA (siRNA)
transfection
HOTAIR siRNA (siHOTAIR-1 and siHOTAIR-2) and
negative control siRNA (siNC) were purchased from Bioneer (Daejeon,
Korea). Cells (5×104 cells/well) were seeded into 6-well
plates and were transfected with 10 nM siRNA in phosphate-buffered
saline (PBS) using the G-Fectin kit (Genolution Pharmaceuticals
Inc., Seoul, Korea) according to the manufacturer’s protocol. These
siRNA-transfected cells were used in the in vitro assays 48
h post-transfection. The target sequences for HOTAIR siRNAs
were as follows: siRNA-1, 5′-UUUUCUACCAGGUCGGUAC-3′ and siRNA-2,
5′-AAUUCUUAAAUUGGGCUGG-3′.
Plasmid constructs and the generation of
stable cell line
The human HOTAIR transcript variant 3 cDNA
was amplified by PCR and was inserted into the pLenti6/V5-D-TOPO
vector according to ViraPower™ Lentiviral Expression systems
(Invitrogen). Briefly, plasmid was transfected into the 293FT cell
line and then lentivirus was infected in desired cell line.
Selection of HOTAIR stable transfected cells was performed
in medium containing blasticidin (Invitrogen).
Cell proliferation assay
Cell proliferation was evaluated using the Cell
Counting Kit-8 (CCK-8) assay (Dojindo Laboratories, Kumamoto,
Japan). Cells (2×103 cells/well) were seeded into
96-well flat-bottomed plates in 100 μl of complete medium. The
cells were incubated overnight to allow for cell attachment and
recovery and were then transfected with siNC or siHOTAIR for 24,
48, 72 and 96 h. CCK-8 solution (10 μl) was added to each well, and
the cells were incubated for an additional 2 h. Absorbance was
measured at 450 nm using a microplate reader. Three independent
experiments were performed in triplicate.
Matrigel invasion assay
The Matrigel invasion assay was performed using the
BD BioCoat Matrigel Invasion Chamber (pore size: 8 mm, 24-well; BD
Biosciences, Bedford, MA, USA) according to the manufacturer’s
protocol. siHOTAIR-transfected cells and siNC-transfected cells
(5×104 cell/plate) were plated in the upper chamber in
serum-free medium, and complete medium was added to the bottom
chamber. The Matrigel invasion chamber was incubated for 48 h at
37°C under 5% CO2. Non-invading cells were removed from
the upper chamber using cotton-tipped swabs. Cells that had invaded
through the pores onto the lower side of the filter were stained
(Diff-Quik; Sysmex, Kobe, Japan), and these were counted using a
hemocytometer. The number of invaded siHOTAIR-transfected cells was
expressed as fold-change relative to siNC-transfected cells, which
was set at 1. The assay was replicated at least 3 times.
Wound healing migration assay
Cells transfected with siNC or siHOTAIR
(5×105 cells/well) were seeded into 6-well culture
plates with serum-containing medium and were cultured until the
cell density reached ~90% confluence. The serum-containing medium
was removed, and cells were serum starved for 24 h. When the cell
density reached ~100% confluence, an artificial homogeneous wound
was created by scratching the monolayer with a sterile 200-μl
pipette tip. After scratching, the cells were washed with
serum-free medium. Images of cells migrating into the wound were
captured at 0, 24 and 48 h using a microscope. The assay was
performed in triplicate.
Western blot analysis
Cells were transfected with siNC or siHOTAIR for 48
h, washed with ice-cold 0.01 M PBS (pH 7.2), and lysed in lysis
buffer [50 mM Tris-HCl (pH 7.4), 150 mM saline, 1% Nonidet P-40,
and 0.1% sodium dodecyl sulfate (SDS)] supplemented with protease
inhibitors. Protein concentrations were determined using Bio-Rad
protein assay reagent according to the Bradford method (Bio-Rad
Laboratories, Hercules, CA, USA). Samples were boiled for 5 min,
subjected to 10% SDS-PAGE, and transferred electrophoretically to
polyvinylidene difluoride membranes (Millipore, Billerica, MA,
USA). Membranes were blocked with 5% non-fat dried milk in 1X
Tris-buffered saline containing 0.1% Tween-20 (TBST; pH 7.6) at
room temperature for 1 h and were then incubated with primary
antibody at 4°C overnight under constant agitation. The primary
antibodies used included: rabbit anti-human VEGF (1:500 dilution;
Abcam, Cambridge, MA, USA), rabbit anti-human MMP-9 (1:1,000
dilution; Cell Signaling Technology, Beverly, MA, USA), rabbit
anti-human E-cadherin (1:1,000 dilution; Cell Signaling
Technology), rabbit anti-human β-catenin (1:1,000 dilution; Cell
Signaling Technology), mouse anti-human Vimentin (1:1,000 dilution;
Sigma, St. Louis, MO, USA), mouse anti-human Snail (1:1,000
dilution; Cell Signaling Technology), rabbit anti-human Twist
(1:1,000 dilution; Abcam), and mouse anti-human β-actin antibody
(1:5,000 dilution; Sigma). Membranes were washed 3 times with 1X
TBST, incubated with a horseradish peroxidase-conjugated
anti-rabbit secondary antibody (1:2,000 dilution; Abcam) or
anti-mouse secondary antibody (1:2,000 dilution; Abcam) for 1 h at
room temperature under constant agitation, and then washed 3 times
with 1X TBST. Proteins were visualized using an enhanced
chemiluminescence system (ECL™; Amersham, Little Chalfont, UK), and
band intensities were quantified using the Luminescent image
analyzer (LAS 4000 mini; Fujifilm, Uppsala, Sweden).
Statistical analysis
SPSS software (standard version 20.0; IBM) was used
for all statistical analyses. Data are expressed as the mean ±
standard deviation (SD). The association between HOTAIR
expression and clinicopathological characteristics was assessed
using the Pearson’s χ2 test, Student’s t-test, and
Fisher’s exact test. Overall survival was analyzed by the
Kaplan-Meier method, and the differences between groups were
estimated by the log-rank test. Multivariate survival analysis was
performed for the significant parameters in the univariate analysis
using the stepwise Cox regression model analysis. All statistical
tests were two-sided, and P<0.05 was considered to indicate a
statistically significant result.
Results
Association between HOTAIR expression and
clinicopathologic factors in cervical cancer
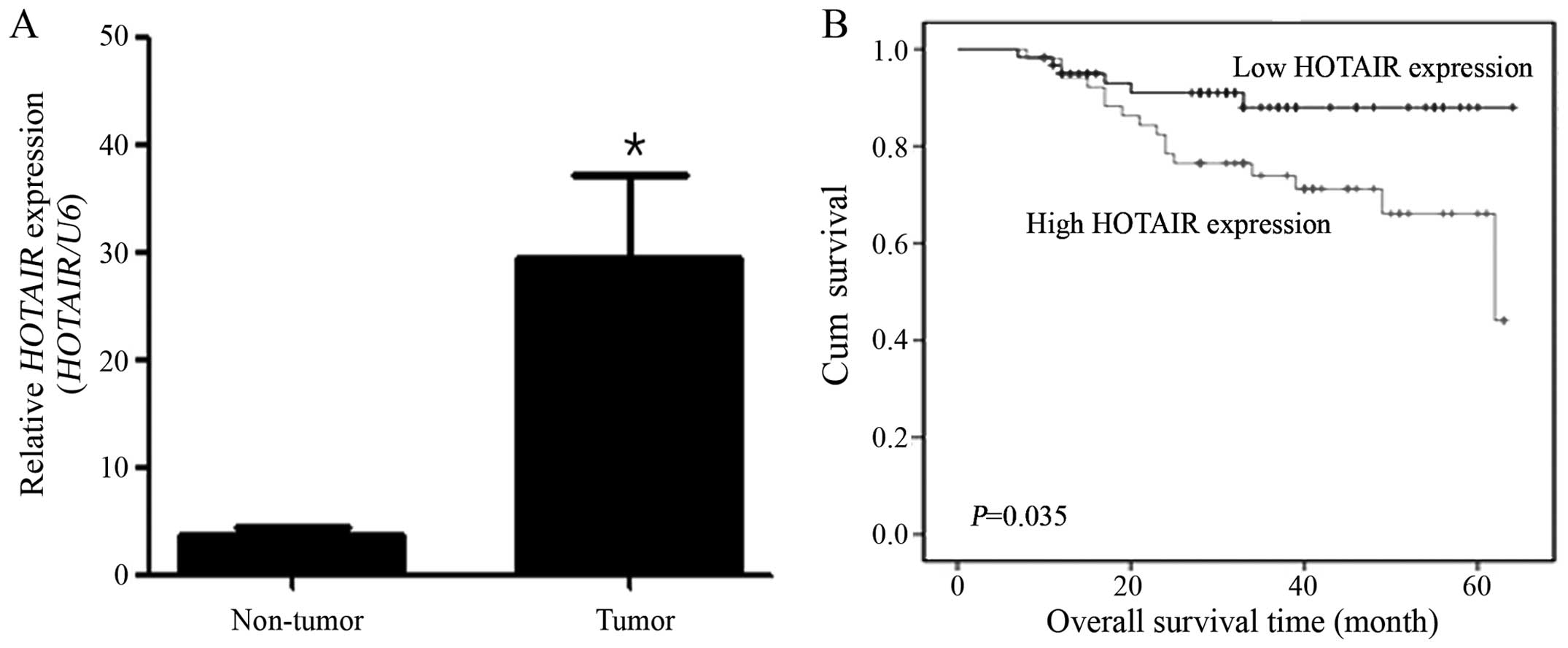
The expression of HOTAIR lncRNA was
determined in cervical cancer tissues (n=111) and corresponding
normal tissues (n=40) using qRT-PCR. HOTAIR expression in
cervical cancer tissues was >30-fold that in non-cancerous
tissues (Fig. 1A), suggesting that
the expression of HOTAIR is upregulated in cervical cancer.
To evaluate the prognostic value of HOTAIR for predicting
clinical outcome in cervical cancer, HOTAIR expression
levels were determined in an independent panel consisting of 111
cervical cancer patients with extensive clinical follow-up
(Table I). The patients were
divided into low (n=22) and high (n=89) HOTAIR expression
groups, and clinicopathologic features were compared between the
two groups. Age, stage, cell type and lymphatic invasion were not
significantly different between the low and high HOTAIR
expression groups. In contrast, HOTAIR expression was
correlated with lymph node metastasis (P=0.0437). Multivariate Cox
regression model analysis was performed to further evaluate the
prognostic significance of HOTAIR expression and
clinicopathologic characteristics on recurrence (Table II). HOTAIR expression was a
significant prognostic indicator for recurrence in cervical cancer
patients (relative risk=5.281; P=0.0493). As shown in Fig. 1B, HOTAIR expression levels
were correlated with overall survival HOTAIR (log-rank test;
P=0.035). These data suggest that HOTAIR expression
represent an independent prognostic factor for survival and that
the overexpression of HOTAIR might play an important role in
the program of cervical cancer.
 | Table IIMultivariate analysis for recurrence
in cervical cancer patients. |
Table II
Multivariate analysis for recurrence
in cervical cancer patients.
| Recurrence |
|---|
|
|
|---|
| Factor | HR | 95% CI | P-value |
|---|
| HOTAIR (Low
vs. high) | 5.281 | 1.005–27.742 | 0.0493 |
| Age | 0.949 | 0.907–0.993 | 0.024 |
| Stage (I vs.
II) | 0.484 | 0.148–1.582 | 0.2298 |
| Stage (I vs.
III–IV) | 2.428 | 0.484–12.168 | 0.2807 |
| Cell type (SCC vs.
adeno) | 2.288 | 0.768–6.819 | 0.1375 |
| Cell type (SCC vs.
mixed) | 44.548 | 8.469–234.335 | <0.001 |
| Cell type (SCC vs.
other) | 4.607 | 0.906–23.411 | 0.0655 |
| Tumor size (<4
vs. ≥4 cm) | 1.651 | 0.529–5.152 | 0.3876 |
| Lymphatic invasion
(Yes vs. no) | 0.974 | 0.391–2.426 | 0.9543 |
| Lymph node
metastasis (Yes vs. no) | 0.824 | 0.265–2.561 | 0.7384 |
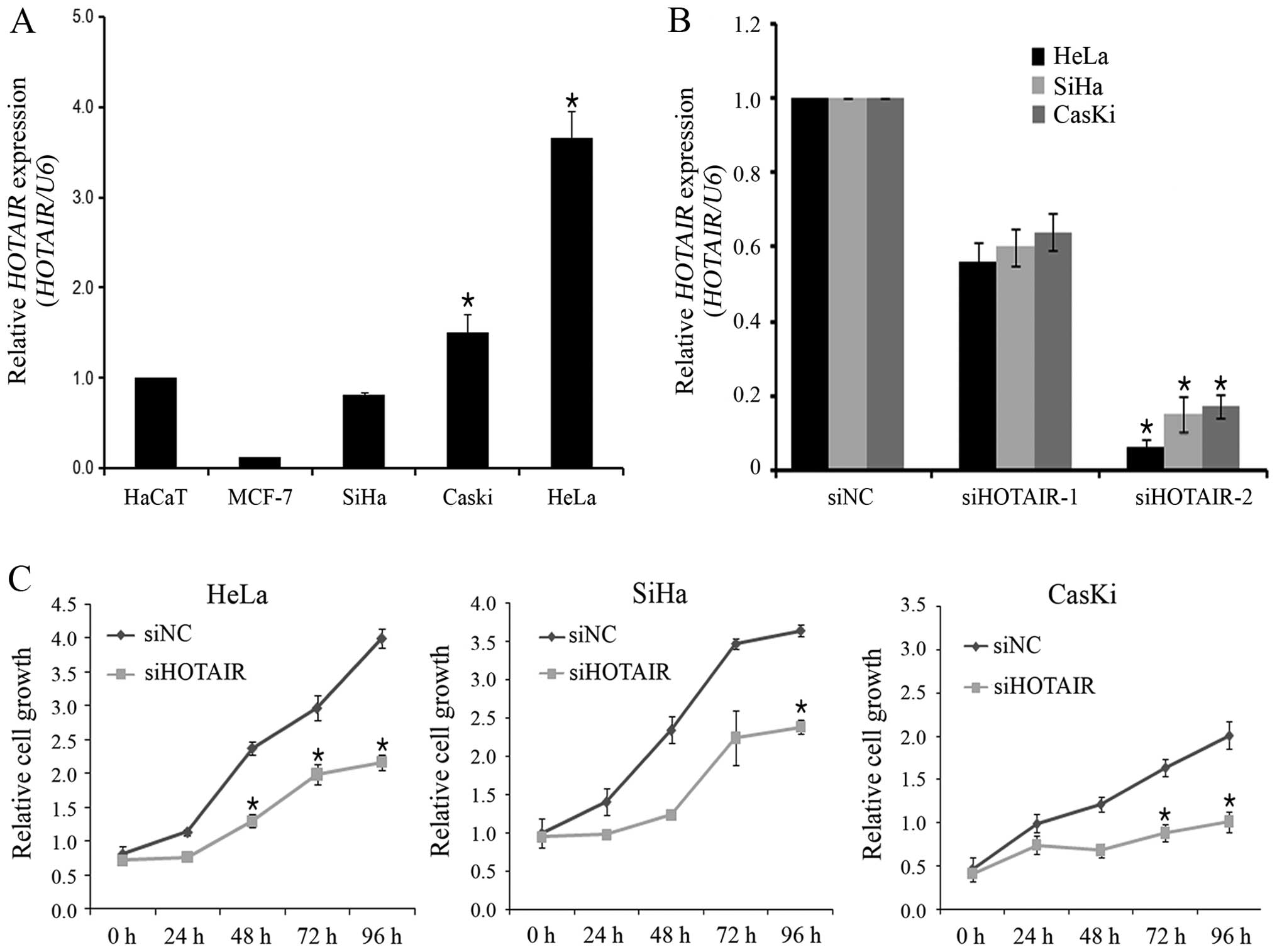
HOTAIR knockdown decreases cell
proliferation in cervical cancer cells
To determine the functional role of HOTAIR in
cervical cancer, siRNA was used to downregulate HOTAIR
expression. For this, HOTAIR expression in SiHa, Caski and
HeLa cervical cancer cell lines was first determined using qRT-PCR.
As shown in Fig. 2A, HOTAIR
expression levels were higher in HeLa cells than in SiHa and Caski
cells. Therefore, HeLa cells were used for siRNA-mediated knockdown
of HOTAIR expression. The knockdown efficiency of the 2
HOTAIR-specific siRNAs (siHOTAIR-1 and siHOTAIR-2) was
evaluated, and siHOTAIR-2 was found to have higher silencing
efficiency than siHOTAIR-1 did (Fig.
2B). Therefore, siHOTAIR-2 was selected for use in the
subsequent in vitro biological assays. To determine the role
of HOTAIR in cervical cancer cell growth,
siHOTAIR-transfected cells were used in the CCK-8 assay.
siRNA-mediated knockdown of HOTAIR decreased cell
proliferation by 30% at 96 h post-transfection in HeLa cells
(Fig. 2C). Also, HOTAIR
siRNA inhibited cell proliferation in SiHa and Caski cells. This
finding indicates that HOTAIR is involved in the
proliferation of cervical cancer cells.
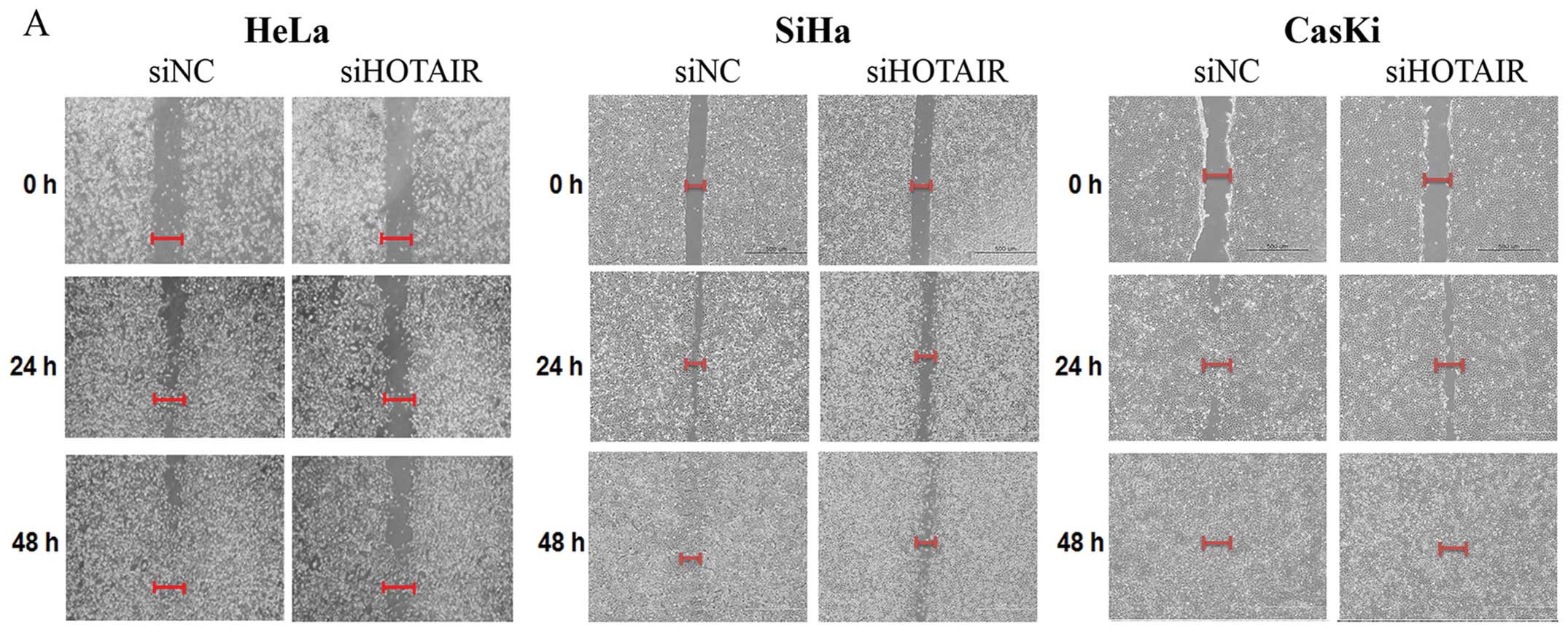
HOTAIR promotes cervical cancer cell
migration and invasion
To investigate the effect of HOTAIR on
migration and invasion, siHOTAIR-transfected cells were used in
wound healing and Matrigel invasion assays, respectively. The width
of the wound closure was larger in siHOTAIR-transfected cells than
in siNC-transfected of HeLa, SiHa and Caski cells (Fig. 3A). Therefore, downregulation of
HOTAIR decreased the migration of cervical cancer cells. We
also tested whether HOTAIR knockdown has an inhibitory
effect on HeLa cell invasion. Knockdown of HOTAIR inhibited
HeLa cell invasion >80% (Fig.
3B). To further assess the role of HOTAIR in the
pathogenesis of cervical cancer, SiHa cell lines stably expressing
ectopic HOTAIR were established (Fig. 3C). Consistent with the previous
results, stable HOTAIR overexpression in SiHa cells resulted
in a significantly increase the invasion ability of SiHa cells
(Fig. 3D). Collectively, these
results indicate that HOTAIR has an important role in the
migratory and invasive phenotype of cervical cancer cells.
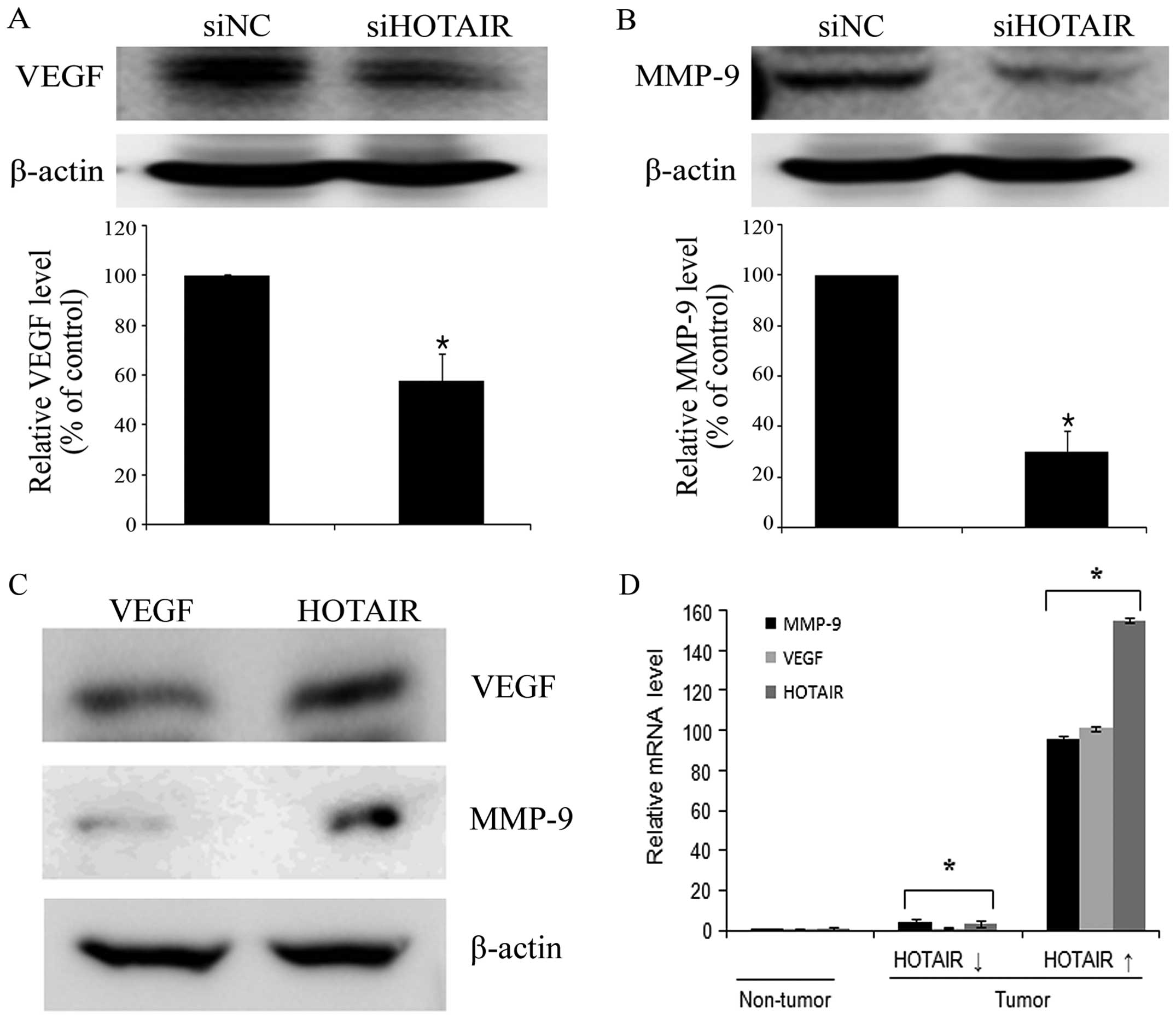
HOTAIR upregulates VEGF and MMP-9
expression in cervical cancer cells
VEGF and MMP-9 play an important role in tumor
progression by promoting migration and invasion (28,29).
Therefore, the effect of HOTAIR on the expression levels of
these proteins was determined in HeLa cells. VEGF and MMP-9 protein
expressions were significantly lower in siHOTAIR-transfected cells
than in siNC-transfected cells (Fig.
4A and B). In contrast, HOTAIR overexpression in SiHa
cells promoted VEGF and MMP-9 protein expression (Fig. 4C). In addition, the high expression
level of HOTAIR in cervical cancer tissues associated with
upregulation of VEGF and MMP-9 expression levels compared with the
low expression groups (Fig. 4D).
Taken together, our findings suggest that HOTAIR may promote
cervical cancer cell migration and invasion through the
upregulation of VEGF and MMP-9 expression.
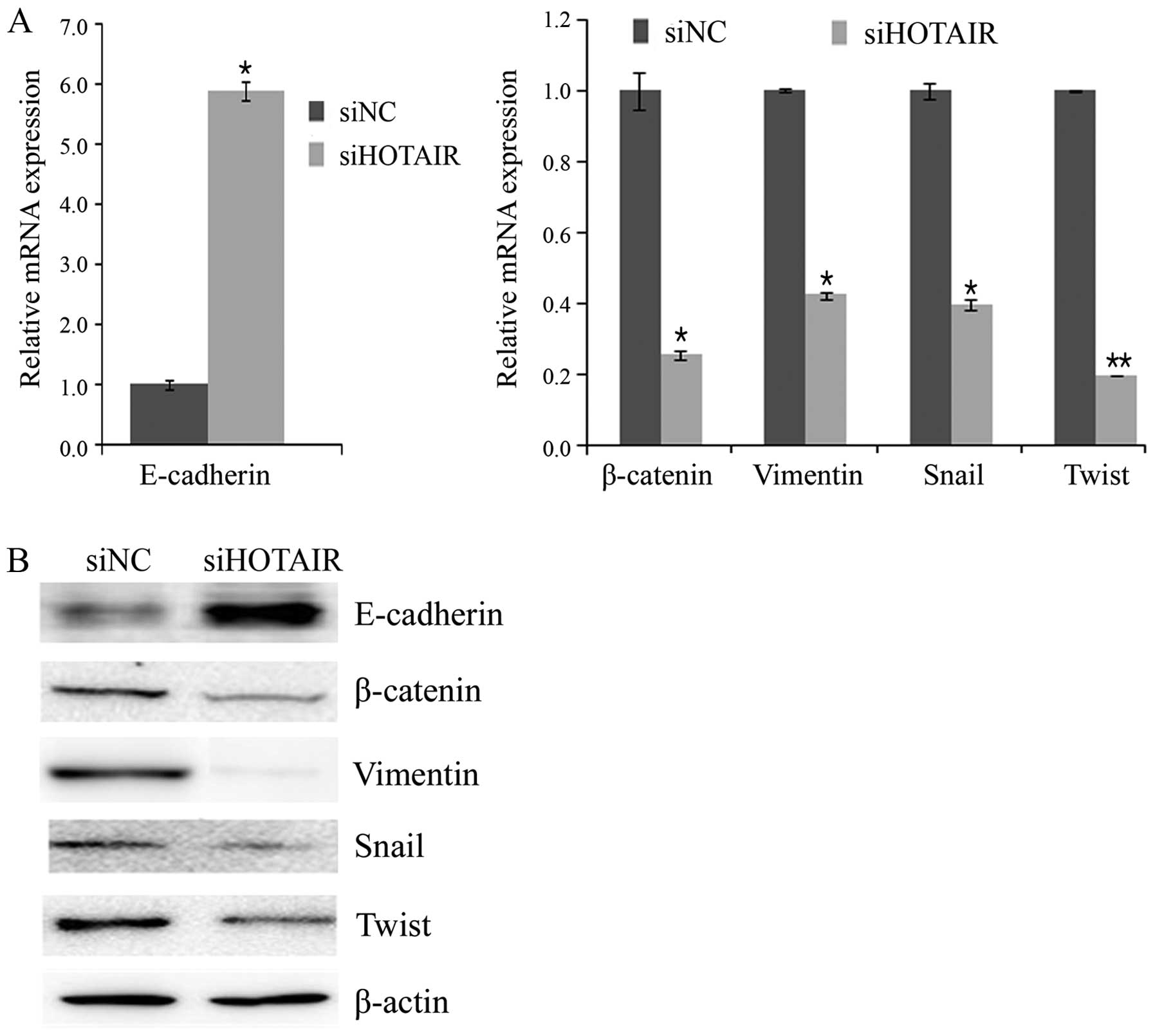
Inhibition of HOTAIR reversed EMT-related
genes in cervical cancer cells
Because the EMT is important in cell migration and
invasion, we also investigated whether direct inhibition of
HOTAIR could reverse EMT-related markers in HeLa cells using
real-time RT-PCR and western blot assays following HOTAIR
knockdown. As anticipated, the siHOTAIR resulted in an increase in
the expression of E-cadherin and a decrease in the expression of
β-catenin and vimentin (Fig. 5).
Next, we assessed the effect of HOTAIR knockdown on the
expression of following transcription factors known to promote EMT:
Snail and Twist. siHOTAIR-transfected cells expressed lower level
of snail and twist compared with the siNC-transfected cells
(Fig. 5). Collectively, the
dysregulation of the expression of EMT-related genes partially
explains the involvement of HOTAIR in cervical cancer cell
migration and invasion.
Discussion
In the present study, we found that HOTAIR
expression was higher in cervical cancer tissues than in
corresponding non-cancerous tissues and that it was associated with
recurrence in cervical cancer patients. Knockdown of HOTAIR
expression decreased cell growth, migration and invasion in
cervical cancer cells. The pro-metastatic effects of HOTAIR
are likely partially mediated by the regulation of the expression
of a number of genes involved in cell migration, invasion and EMT,
including VEGF, MMP-9, E-cadherin, β-catenin, Vimentin, Snail and
Twist. Together, our findings suggest that HOTAIR may
represent a potential biomarker and therapeutic target for cervical
cancer.
Although the functional role of small regulatory
ncRNAs such as miRNAs in human cancers is now well established,
little is known about the regulatory roles of lncRNAs and their
relevance to human disease. LncRNAs are transcripts of at least 200
nucleotides without protein-coding potential. Like their
protein-coding counterparts, many lncRNAs are capped, spliced and
polyadenylated (30). Recent data
have shown the tissue-specific expression patterns for lncRNAs.
Nevertheless, the growing catalog of functionally characterized
lncRNAs reveals that these transcripts are important in different
physiological processes (31,32),
and therefore, altered expression of lncRNAs may promote cancer
development and progression (33).
Recently, the lncRNA HOTAIR was associated with metastatic
progression in human breast cancer, hepatocellular carcinoma,
cervical and pancreatic cancer (16–19).
In the present study, HOTAIR expression was associated with
disease recurrence in cervical cancer patients and increased the
proliferation, migration, and invasion of cervical cancer cells
in vitro. Recent reports have shown that lncRNAs are crucial
for the regulation of chromatin structure, gene expression and
translational control (34,35).
However, the detailed functional impact and clinical significance
of lncRNA-mediated changes in chromatin and gene expression remain
to be elucidated. HOTAIR recruits PRC2 to specific target
genes in the genome, which leads to H3K27 trimethylation and
epigenetic silencing of metastatic suppressor genes (16). Therefore, modifications of
DNA-binding proteins by HOTAIR regulates global gene
expression. Kogo et al (18) showed that HOTAIR expression
was closely correlated with PRC2 occupancy in colorectal cancer
patients. Furthermore, in a recent study, HOTAIR-mediated
chromatin changes promoted breast cancer metastasis (16). The fact that HOTAIR drives
genome-wide chromatin reprogramming suggests that long-range
regulation by lncRNAs may be a widespread mechanism. This is
supported by a study showing that >20% of tested lncRNAs are
bound by PRC2 and other chromatin modifiers (13). These findings provoke questions
regarding the initial triggers for HOTAIR overexpression and
whether understanding of lncRNA mechanics may have clinical
relevance.
The recurrence rate after radical surgery in stage
I-II cervical cancer is ~15–30%, and the prognosis of recurrent
patients is suboptimal (36).
Therefore, identification of reliable biomarkers for predicting
recurrence is needed to improve the prognosis of cervical cancer
patients. Pelvic lymph node metastasis is the most important
postoperative risk factor for recurrence or failure to survive, and
thus, cervical cancer patients with metastasis in the pelvic lymph
nodes require adjuvant therapy (21,37,38).
In the present study, we showed that high HOTAIR expression
was correlated with lymph node metastasis and recurrence in
cervical cancer. Therefore, analysis of HOTAIR expression in
cervical cancer patients may predict the risk of recurrence and,
therefore, help guide treatment decisions. Despite the prognostic
significance of HOTAIR for tumor recurrence, the results of
the present study should be viewed cautiously because of the
relatively small sample size. Larger prospective studies are needed
to confirm our findings.
HOTAIR has been shown to increase the
invasion of many types of cancer cells including pancreatic,
breast, colon, and liver cancer cells (16–18).
In the present study, we found that downregulation of HOTAIR
expression decreased cervical cancer cell proliferation, migration
and invasion. Therefore, HOTAIR exerts pro-oncogenic
activities in cervical cancer and may promote a more aggressive and
metastatic phenotype. MMPs play a crucial role in cancer cell
invasion and metastasis. MMP-9, which degrades basement membrane
collagen, has been shown to promote tumor cell invasion and
metastasis and decrease survival in many types of cancer (29,39).
It has been generally accepted that tumor angiogenesis plays a
critical role in tumor growth, invasion and metastasis. Among the
angiogenic factors, VEGF has been shown to have a pivotal role in
tumor angiogenesis (40).
Knockdown of HOTAIR was associated with reduced expression
of VEGF and MMP-9 in BEL7402 hepatocellular carcinoma cells
(41). Furthermore, HOTAIR
knockdown inhibited proliferation, migration, and invasion through
modulation of the extracellular matrix. We also found that
downregulation of HOTAIR decreased the expression of VEGF
and MMP-9. Taken together, our findings demonstrate that
HOTAIR accelerates the aggressiveness of cervical cancer
cells through the upregulation of VEGF and MMP-9.
The functional importance of HOTAIR for the
activation of invasion indicates that further studies should
identify the role of HOTAIR in EMT process (15). It has been demonstrated that
knockdown of HOTAIR could reverse EMT process in gastric
cancer cells (42). These findings
prompted us to determine whether HOTAIR promotes cervical
cancer metastasis by regulating the expression of EMT-related
genes. As expected, our data suggest that HOTAIR knockdown
was dysregulated the expression of EMT-related genes (E-cadherin,
β-catenin, Vimentin, Snail and Twist), implying that these genes
participate in HOTAIR-induced cervical cancer
metastasis.
In conclusion, our results suggest that
HOTAIR is associated with recurrence in cervical cancer.
Moreover, HOTAIR may promote cervical cancer progression by
inducing cell migration and invasion through the upregulation of
VEGF, MMP-9 and expression of EMT-related genes. Thus,
HOTAIR may represent a potential therapeutic target and a
prognostic marker for cervical cancer.
Acknowledgements
The present research was supported by the Basic
Science Research Program through the National Research Foundation
of Korea (NRF) funded by the Ministry of Education, Science and
Technology (NRF-2012R1A1A2004523; NRF-2012R1A1A2040271) and the
faculty research grant from the Yonsei University College of
Medicine for 2014 (6-2014-0025). We thank Dae Ryong Kang and Myeong
Hwa Kang for their excellent technical assistance.
Abbreviations:
|
lncRNAs
|
long non-coding RNAs
|
|
HOTAIR
|
Hox transcript antisense intergenic
RNA
|
|
VEGF
|
vascular endothelial growth factor
|
|
MMP-9
|
matrix metalloproteinase-9
|
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
miRNAs
|
microRNAs
|
References
|
1
|
Guttman M, Amit I, Garber M, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bernstein E and Allis CD: RNA meets
chromatin. Genes Dev. 19:1635–1655. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mattick JS: The genetic signatures of
noncoding RNAs. PLoS Genet. 5:e10004592009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Borchert GM, Lanier W and Davidson BL: RNA
polymerase III transcribes human microRNAs. Nat Struct Mol Biol.
13:1097–1101. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pillai RS, Bhattacharyya SN and Filipowicz
W: Repression of protein synthesis by miRNAs: how many mechanisms?
Trends Cell Biol. 17:118–126. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Berezikov E and Plasterk RH: Camels and
zebrafish, viruses and cancer: a microRNA update. Hum Mol Genet.
14:R183–R190. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Prasanth KV and Spector DL: Eukaryotic
regulatory RNAs: an answer to the ‘genome complexity’ conundrum.
Genes Dev. 21:11–42. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Perez DS, Hoage TR, Pritchett JR, et al:
Long, abundantly expressed non-coding transcripts are altered in
cancer. Hum Mol Genet. 17:642–655. 2008. View Article : Google Scholar
|
|
10
|
Guttman M, Donaghey J, Carey BW, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rinn JL, Kertesz M, Wang JK, et al:
Functional demarcation of active and silent chromatin domains in
human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khalil AM, Guttman M, Huarte M, et al:
Many human large intergenic noncoding RNAs associate with
chromatin-modifying complexes and affect gene expression. Proc Natl
Acad Sci USA. 106:11667–11672. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hung T, Wang Y, Lin MF, et al: Extensive
and coordinated transcription of noncoding RNAs within cell-cycle
promoters. Nat Genet. 43:621–629. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gupta RA, Shah N, Wang KC, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim K, Jutooru I, Chadalapaka G, et al:
HOTAIR is a negative prognostic factor and exhibits pro-oncogenic
activity in pancreatic cancer. Oncogene. 32:1616–1625. 2013.
View Article : Google Scholar
|
|
18
|
Kogo R, Shimamura T, Mimori K, et al: Long
noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang L, Liao LM, Liu AW, et al:
Overexpression of long noncoding RNA HOTAIR predicts a poor
prognosis in patients with cervical cancer. Arch Gynecol Obstet.
290:717–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kodama J, Seki N, Masahiro S, et al:
Prognostic factors in stage IB–IIB cervical adenocarcinoma patients
treated with radical hysterectomy and pelvic lymphadenectomy. J
Surg Oncol. 101:413–417. 2010.PubMed/NCBI
|
|
22
|
Noordhuis MG, Fehrmann RS, Wisman GB, et
al: Involvement of the TGF-beta and beta-catenin pathways in pelvic
lymph node metastasis in early-stage cervical cancer. Clin Cancer
Res. 17:1317–1330. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kang HW, Wang F, Wei Q, et al: miR-20a
promotes migration and invasion by regulating TNKS2 in human
cervical cancer cells. FEBS Lett. 586:897–904. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qiang R, Wang F, Shi LY, et al: Plexin-B1
is a target of miR-214 in cervical cancer and promotes the growth
and invasion of HeLa cells. Int J Biochem Cell Biol. 43:632–641.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu L, Yu X, Guo X, et al: miR-143 is
downregulated in cervical cancer and promotes apoptosis and
inhibits tumor formation by targeting Bcl-2. Mol Med Rep.
5:753–760. 2012.
|
|
26
|
Wang F, Li Y, Zhou J, et al: miR-375 is
down-regulated in squamous cervical cancer and inhibits cell
migration and invasion via targeting transcription factor SP1. Am J
Pathol. 179:2580–2588. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Au Yeung CL, Tsang TY, Yau PL and Kwok TT:
Human papillomavirus type 16 E6 induces cervical cancer cell
migration through the p53/microRNA-23b/urokinase-type plasminogen
activator pathway. Oncogene. 30:2401–2410. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim KJ, Li B, Winer J, et al: Inhibition
of vascular endothelial growth factor-induced angiogenesis
suppresses tumour growth in vivo. Nature. 362:841–844. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Curran S and Murray GI: Matrix
metalloproteinases: molecular aspects of their roles in tumour
invasion and metastasis. Eur J Cancer. 36:1621–1630. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carninci P, Kasukawa T, Katayama S, et al:
The transcriptional landscape of the mammalian genome. Science.
309:1559–1563. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dinger ME, Amaral PP, Mercer TR, et al:
Long noncoding RNAs in mouse embryonic stem cell pluripotency and
differentiation. Genome Res. 18:1433–1445. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pang KC, Dinger ME, Mercer TR, et al:
Genome-wide identification of long noncoding RNAs in
CD8+ T cells. J Immunol. 182:7738–7748. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hall PA and Russell SH: New perspectives
on neoplasia and the RNA world. Hematol Oncol. 23:49–53. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Umlauf D, Fraser P and Nagano T: The role
of long non-coding RNAs in chromatin structure and gene regulation:
variations on a theme. Biol Chem. 389:323–331. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Morris KV and Vogt PK: Long antisense
non-coding RNAs and their role in transcription and oncogenesis.
Cell Cycle. 9:2544–2547. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim MK, Jo H, Kong HJ, et al:
Postoperative nomogram predicting risk of recurrence after radical
hysterectomy for early-stage cervical cancer. Int J Gynecol Cancer.
20:1581–1586. 2010.
|
|
37
|
Soliman PT, Frumovitz M, Sun CC, et al:
Radical hysterectomy: a comparison of surgical approaches after
adoption of robotic surgery in gynecologic oncology. Gynecol Oncol.
123:333–336. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Biewenga P, van der Velden J, Mol BW, et
al: Prognostic model for survival in patients with early stage
cervical cancer. Cancer. 117:768–776. 2011. View Article : Google Scholar
|
|
39
|
Wiercinska E, Naber HP, Pardali E, van der
Pluijm G, van Dam H and ten Dijke P: The TGF-beta/Smad pathway
induces breast cancer cell invasion through the up-regulation of
matrix metalloproteinase 2 and 9 in a spheroid invasion model
system. Breast Cancer Res Treat. 128:657–666. 2011. View Article : Google Scholar
|
|
40
|
Burger RA: Role of vascular endothelial
growth factor inhibitors in the treatment of gynecologic
malignancies. J Gynecol Oncol. 21:3–11. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Geng YJ, Xie SL, Li Q, Ma J and Wang GY:
Large intervening non-coding RNA HOTAIR is associated with
hepatocellular carcinoma progression. J Int Med Res. 39:2119–2128.
2011. View Article : Google Scholar
|
|
42
|
Xu ZY, Yu QM, Du YA, et al: Knockdown of
long non-coding RNA HOTAIR suppresses tumor invasion and reverses
epithelial-mesenchymal transition in gastric cancer. Int J Biol
Sci. 9:587–597. 2013. View Article : Google Scholar : PubMed/NCBI
|