Introduction
Lung cancer is a major cause of cancer-related
mortality worldwide, accounting for 18% (1.4 million) of cancer
deaths in 2008 (1). It is
traditionally classified into two major subtypes, small cell lung
cancer and non-small cell lung cancer (NSCLC), the latter of which
covers ~85% of newly diagnosed lung cancers, and is further
subdivided into two major histological subtypes, adenocarcinoma and
squamous cell carcinoma (SCC), which account for ~38 and 20% of all
lung cancers, respectively (2).
Although historically these histological subtypes did not
significantly affect treatment decisions (3), recent advances in NSCLC chemotherapy
have introduced treatment options that are subtype-dependent. For
example, a folate antimetabolite, pemetrexed, has been used in
first-line (4), second-line
(5), and maintenance (6) settings for patients with non-SCC.
Bevacizumab, an angiogenesis inhibitor, is considered inadequate
for SCC because it increases the risk of fatal hemoptysis and is
less effective (7,8). In addition, discovery of biomarkers
such as recurrent mutations in the epidermal growth factor receptor
(EGFR) kinase and a fusion gene of EML4-anaplastic lymphoma kinase
has led to a marked change in lung adenocarcinoma treatment
(9–12). However, these mutations occur only
in adenocarcinoma patients who have never smoked, but are not
present in SCC cases that are invariably associated with tobacco
smoking (13). In fact, targeted
agents developed for lung adenocarcinoma have been largely
ineffective against SCC (14),
therefore SCC is still treated by conventional platinum-based
chemotherapy with little improvement (3).
Recently, some studies have reported several genetic
changes related to SCC, such as amplification of TP63,
PIK3CA, PDGFRA, SOX2, or FGFR1 and
mutations in TP53, EGFR, PIK3CA,
NRE2L2, PTEN, and DDR2 (15,16).
However, they have not been effective in the clinical setting thus
far. For this reason, many researchers are exploring driver
mutations as well as targeted agents for SCC through gene
expression profiling and sequencing studies (16,17).
Therefore, we carried out a comprehensive gene expression analysis
to identify genes that are specifically and highly expressed in
lung SCC, and detected the family with sequence similarity 83,
member B (FAM83B) gene. FAM83B has been reported as an
important intermediary in EGFR/RAS signaling, and is highly
expressed at the mRNA level in several cancers, including breast,
cervix, bladder, lung, testis thyroid, and ovary cancer (18). However, detailed examination of its
protein expression and association with clinicopathological factors
in SCC patients has not been previously undertaken. Therefore, we
conducted western blotting and immunohistochemical analyses and
found that FAM83B protein was also increased in lung SCC compared
with lung adenocarcinoma or adjacent normal tissues, and that
high-expression levels of FAM83B were associated with a high
disease-free survival (DFS) rate.
Materials and methods
Ethics statement
This study was approved by the Ethics Committee of
Fukushima Medical University (Fukushima, Japan) (approval no.
1713). Written informed consent was obtained from all participants
involved. We obtained ethics approval from the Ethics Committees at
all Institutions where samples were analyzed.
Case selection
This study was conducted in a cohort of patients
with NSCLC who underwent pulmonary resection at Fukushima Medical
University Hospital between 2005 and 2011. The tumor samples of 215
patients (SCC 113 cases, adenocarcinoma 102 cases) were examined
for FAM83B expression. Tumor samples were selected from patients
who fulfilled all of the following criteria: i) patients suffering
from primary NSCLC with confirmed stage (T1–T3, pN0-pN2, and pM0);
ii) patients who underwent curative surgery but did not receive any
preoperative treatment; and iii) patients whose clinical follow-up
data were available. Follow-up information of at least 5 years was
available for this study. Use of all clinical materials was
approved by the Institutional Ethics Committee in Fukushima Medical
University. Formalin-fixed, paraffin-embedded samples from all
cases were used for immunohistochemistry (IHC). For the
comprehensive gene expression analysis, 64 normal lung samples, 60
cases of adenocarcinoma, and 20 cases of SCC were used. For western
blotting, three normal lung samples, 5 cases of adenocarcinoma, and
5 cases of SCC were used.
Comprehensive gene expression
analysis
A small fraction (7×7 mm) of each surgical specimen
was excised and frozen in liquid nitrogen. Frozen samples were
processed for total RNA extraction using ISOGEN (Nippon Gene Co.,
Ltd., Tokyo, Japan) and for purification of poly(A)++RNA
using a MicroPoly(A)Purist kit (Ambion, Austin, TX, USA). Human
common reference RNA was prepared by mixing equal amounts of
poly(A)+RNA extracted from 22 human cancer cell lines (A431, A549,
AKI, HBL-100, HeLa, HepG2, HL60, IMR-32, Jurket, K562, KP4, MKN7,
NK-92, Raji, RD, Saos-2, SK-N-MC, SW-13, T24, U251, U937, and Y79)
to reduce cell type-specific bias in expression (19).
Synthetic polynucleotides (80-mers) representing
31,797 species of human transcripts (MicroDiagnostic, Tokyo, Japan)
were arrayed using a custom arrayer. SuperScript II (Invitrogen
Life Technologies, Carlsbad, CA, USA) and Cyanine 5-dUTP
(Perkin-Elmer, Boston, MA, USA) was used to synthesize labeled cDNA
from 2 μg sample RNA, while Cyanine 3-dUTP (Perkin-Elmer)-labeled
cDNA was synthesized from 2 μg reference RNA. Hybridization was
performed with a Labeling and Hybridization kit (MicroDiagnostic).
Signals were measured with a GenePix 4000B Scanner (Axon
Instruments, Inc., Union city, CA, USA) and then processed into
primary expression ratios (ratio of the cyanine-5 intensity of each
sample to the cyanine-3 intensity of the human common reference
RNA). Each ratio was normalized by multiplication with the
normalization factors using GenePix Pro 3.0 software (Axon
Instruments, Inc.). The primary expression ratios were converted
into log2 values (designated log ratios). We assigned an
expression ratio of 1 (log ratio of 0) for spots that exhibited
fluorescence intensities under the detection limits, and we
included these in the signal calculation of the mean averages. The
data were processed using Microsoft Excel software (Microsoft,
Bellevue, WA, USA) and MDI gene expression analysis software
package (MicroDiagnostic). Data corresponding to FAM83B were
extracted, and statistical analysis of the Kruskal-Wallis test was
performed using GraphPad Prism ver. 6.0 (GraphPad Software, Inc.,
San Diego, CA, USA).
Immunoblotanalysis
Frozen tissues from tumor and non-tumor regions were
homogenized with RIPA buffer [150 mM sodium chloride, 1% NP-40, 1%
sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 25 mM
Tris-HCl (pH 7.6)] containing protease inhibitor cocktail (Roche
Diagnostics, Indianapolis, IN, USA) using a glass homogenizer
(Wheaton Dounce Tissue Grinder) on ice. The homogenate were
centrifuged at 10,000 × g at 4°C for 20 min to remove debris, and
the supernatants were mixed with an equal volume of a 20 mM
Tris-HCl (pH 6.8) buffer containing 2% SDS, 12% glucose, 2%
2-mercaptoethanol, 0.002% PBS, and BPB. The proteins contained in
the supernatants were separated by SDS-PAGE using 5–20% gradient
polyacrylamide gels (SuperSep Ace 5–20%; Wako Pure Chemical
Industries, Ltd., Osaka, Japan) according to the Laemmli method
(20). Separated proteins were
transferred to polyvinylidene difluoride membranes (Millipore,
Billerica, MA, USA), according to the method of Towbin et al
(21). The membranes were blocked
with 5% skim milk in T-PBS (0.137 M NaCl, 2.6 mM KCl, 1.8 mM
KH2PO4, 8.1 mM
Na2HPO4·12H2O, and 0.005%
Tween-20) and incubated with primary antibodies overnight at 4°C.
The membranes were incubated with anti-FAM83B (1:2,000, HPA031464;
Atlas Antibodies AB, Stockholm, Sweden) or anti-GAPDH (1:2,500, no.
2118; Cell Signaling Technology, Inc., Danvers, MA, USA) as primary
antibodies, and then with horseradish peroxidase-conjugated goat
anti-rabbit immunoglobulin G antibody (1:10,000; GE Healthcare Life
Sciences, Tokyo, Japan) as a secondary antibody. The signals were
detected by ImageQuant LAS 4000 using Prime Western Blotting
Detection Reagent (both from GE Healthcare Life Sciences).
IHC and quantitative analysis
Tissue specimens were fixed in formalin and embedded
in paraffin. Sections were autoclaved in 0.01 M citrate buffer (pH
6.0) for antigen retrieval. After blocking in 5% skim milk,
sections were incubated with a rabbit polyclonal anti-FAM83B
antibody (HPA031464; Atlas Antibodies AB) at a dilution of 1:100 at
4°C overnight. They were further incubated for 20 min at room
temperature with a biotinylated goat anti-rabbit IgG (1:400
dilution, Vectastain Elite ABC kit; Vector Laboratories, Inc.,
Burlingame, CA, USA), and then with avidin-biotin-HRP regent (1:200
dilution; Vectastain Elite ABC kit; Vector Laboratories, Inc.) for
30 min at room temperature. They were observed under a microscope
(BX50; Olympus, Tokyo, Japan) and positivity was judged when
>10% of the area was occupied with positive cells.
For quantitation of staining intensity, tissue
sections were immunohistochemically stained without nuclear counter
staining. For each specimen, five regions of 680×860 μm each were
randomly selected, and the images were captured with a microscope
(BX51) equipped with a 20× objective lens (UPlanSApo) and a CCD
camera (DP71) (all from Olympus). Images with no tissues sections
were also acquired as a background signal. All images were
converted to 256-level gray scale images and then inverted using
ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Mean intensity values were measured only in the tumor regions, from
which the background value was subtracted. According to the values,
patients were divided into two groups; values with equal or higher
than median were classified into a ‘high-expression group’, while
those less than the median into a ‘low-expression group’.
Statistical analysis
Associations of FAM83B expression levels with
clinical characteristics were evaluated using Pearson’s
χ2 test. DFS and overall survival (OS) in patients with
completely resected lung cancers were analyzed. DFS was measured
from the time of surgery to initial tumor relapse (local recurrence
or distant). OS was calculated from the time of surgery to death at
last follow-up date, and 95% confidence interval was evaluated by
survival analysis using the Kaplan-Meier method. Survival outcomes
for the high- versus the low-expression group were compared using
the log-rank test. Statistical significance was set at P<0.05
for all analyses. Multivariate analysis was performed using Cox
regression analysis with the following pre-specified variables:
gender, pathological stage, smoking, and FAM83B status. All
statistical analyses were performed using SPSS version 20.0
software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered
statistically significant.
Results
FAM83B expression in lung cancer and
normal tissue
We first extracted the expression ratios of FAM83B
from the comprehensive gene expression analysis data, and compared
these among 20 cases of SCC, 60 cases of adenocarcinoma, and 64
samples of adjacent normal tissue. As shown in Fig. 1A, FAM83B mRNA levels in SCC were
significantly higher than those in adenocarcinoma (P<0.0001) and
normal tissues (P<0.0001), but there was no significant
difference between levels in adenocarcinoma and normal tissues
(Fig. 1A). We next analyzed FAM83B
protein expression in five SCC cases, five adenocarcinoma cases,
and three normal tissue samples by western blotting. FAM83B protein
was detected as a signal with an apparent molecular mass of ~110
kDa. The FAM83B signal intensities in lung cancer were stronger
than that in adjacent normal tissue, and the SCC samples expressed
FAM83B at higher levels in comparison with the adenocarcinoma
samples (Fig. 1B).
 | Figure 1The family with sequence similarity 83
member B (FAM83B) expression in lung cancer and adjacent normal
tissue. (A) Expression ratios for FAM83B mRNA in clinical samples
of adenocarcinoma (Ad), squamous cell carcinoma (SCC), and adjacent
normal lung tissues (NC) were extracted from data obtained via a
comprehensive gene expression analysis, and plotted. Horizontal
bars indicate medians, *P<0.0001. (B) FAM83B
expression was examined by western blotting of samples from Ad (5
cases), SCC (5 cases), and NC (3 cases). GAPDH protein expression
was examined as an internal control. (C) Immunohistochemistry (IHC)
of FAM83B in Ad, SCC, and NC. Paraffin sections were stained using
an anti-FAM83B antibody, which was detected by DAB staining.
Representative images for each tissue are shown. Boxed regions in
the left column are magnified and shown in the right column. Note
that immunoreactivity for FAM83B was detected in the cytoplasm and
near plasma membranes (arrowheads) in SCC, but were hardly detected
in Ad and NC. Scale bars, 100 μm. |
Localization of FAM83B in lung cancer and
normal lung tissue
To evaluate FAM83B expression in paraffin-embedded
tissue samples that were retrospectively collected, we attempted
immunohistochemical analyses using anti-FAM83B polyclonal
antibodies. As shown in Fig. 1C,
while the FAM83B signal was only weakly or barely detected in
adjacent normal and lung adenocarcinoma tissues, some SCC tissues
showed highly intense signals. The staining patterns of FAM83B
differed in the same tissue sample as well as among SCC cases; it
was often observed along the cell surface and occasionally in the
cytoplasm, and in other cells it was localized both in the plasma
membranes and cytoplasm (Fig. 1C).
When positivity was examined, 94.7% of SCCs (107 out of 113 cases)
and 14.7% of adenocarcinoma (15 out of 102 cases) were positive,
and overall 56.7% (122 out of 215 cases) of lung cancers were
positive. In contrast, all corresponding adjacent normal lung
tissues were negative. Thus, sensitivity and specificity were
calculated as 94.5 and 85.3%, respectively.
Relationship between FAM83B and
clinicopathological variables of SCC
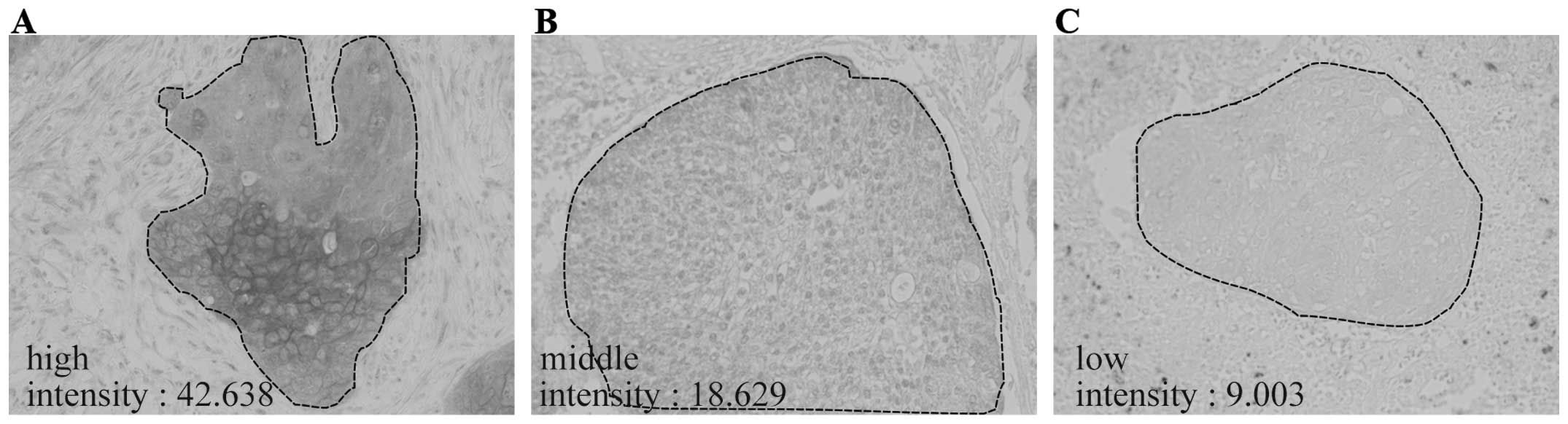
The immunoreactivities for FAM83B varied among SCC
tissues (Fig. 2A–C). Therefore, to
examine the relationship between FAM83B protein levels and
clinicopathological factors, FAM83B signal intensities were
quantified by image analysis. When 107 FAM83B-positive cases of
lung SCC were analyzed, the intensity values for FAM83B were
distributed from 3.37 to 63.79 with a median of 17.01 (Fig. 2D). Then, the 107 cases were divided
into a high-expression group (54 cases) and a low-expression group
(53 cases), and were subjected to an association analysis with
clinicopathological factors. Clinicopathological data for SCC
patients included in this analysis are summarized in Table I. Notably, the majority of patients
were male (88.8%), aged ≥65 years (81.3%), and smokers (Brinkman
index ≥600, 86.0%). Percentages of patients with pathologic stage I
and II+III were 57.0 and 43.0%, respectively. As a result, a
marginal relationship between FAM83B and vascular invasion was
found but this was not statistically significant (P=0.066), while
there was no significant association between FAM83B protein
expression and any of the factors examined including age, gender,
smoking history, pT factor, pN factor, p stage, tumor
differentiation, pleural invasion, or lymphatic vessel
invasion.
 | Table IRelationship between FAM83B expression
and clinicopathological parameters in SCC. |
Table I
Relationship between FAM83B expression
and clinicopathological parameters in SCC.
| | FAM83B
expression | |
|---|
| |
| |
|---|
| Characteristic | Total n=107 | FAM83B high n=54
(50.5%) | FAM83B low n=53
(49.5%) | P: high vs.
low |
|---|
| Gender |
| Female | 12 (11.2%) | 7 (58.3%) | 5 (41.7%) | 0.563 |
| Male | 95 (88.8%) | 47 (49.5%) | 48 (50.5%) | |
| Age (years) |
| <65 | 20 (18.7%) | 7 (35.0%) | 13 (65.0%) | 0.125 |
| ≥65 | 87 (81.3%) | 47 (54.0%) | 40 (46.0%) | |
| Smoking history
(BI) |
| <600 | 15 (14.0%) | 8 (53.3%) | 7 (46.7%) | 0.811 |
| ≥600 | 92 (86.0%) | 46 (50.0%) | 46 (50.0%) | |
| pT factor |
| T1 | 36 (33.6%) | 22 (61.1%) | 14 (38.9%) | 0.117 |
| T2+T3 | 71 (66.4%) | 32 (45.1%) | 39 (54.9%) | |
| pN factor |
| N0 | 76 (71.0%) | 41 (53.9%) | 35 (46.1%) | 0.260 |
| N1+N2 | 31 (29.0%) | 13 (41.9%) | 18 (58.1%) | |
| p-TNM stage |
| Stage I | 61 (57.0%) | 33 (54.1%) | 28 (45.9%) | 0.387 |
| Stage II/III | 46 (43.0%) | 21 (45.7%) | 25 (54.3%) | |
| Tumor
differentiation |
| Well/moderate | 80 (74.8%) | 43 (53.8%) | 37 (46.2%) | 0.242 |
| Poorly | 27 (25.2%) | 11 (40.7%) | 16 (59.3%) | |
| Pleural
invasion |
| Positive | 34 (31.8%) | 14 (41.2%) | 20 (58.8%) | 0.190 |
| Negative | 73 (68.2%) | 40 (54.8%) | 33 (45.2%) | |
| Lymphatic vessel
invasion |
| Positive | 46 (43.0%) | 20 (43.5%) | 26 (56.5%) | 0.209 |
| Negative | 61 (57.0%) | 34 (55.7%) | 27 (44.3%) | |
| Vascular
invasion |
| Positive | 53 (49.5%) | 22 (41.5%) | 31 (58.5%) | 0.066 |
| Negative | 54 (50.5%) | 32 (59.3%) | 22 (40.7%) | |
Survival outcomes according to FAM83B
expression
We drew Kaplan-Meier survival estimates for the OS
and DFS and compared the two groups using the log-rank test. In the
high-expression group, there was a significant extension in the DFS
(HR: 0.491, P=0.042; Fig. 2E)
while OS did not differ significantly (HR: 0.848, P=0.650; Fig. 2F). We also applied univariate
analysis to evaluate associations between DFS and several important
clinicopathological factors including age, gender, smoking history,
pT factor, pN factor, p stage, tumor differentiation, pleural
invasion, lymphatic vessel invasion, vascular invasion, or FAM83B
expression by the Cox proportional hazards model. pT factor, pN
factor, p stage, pleural invasion, lymphatic vessel invasion,
vascular invasion, and low protein expression of FAM83B were
significantly associated with DFS (Table II). In a multivariate statistical
analysis, however, the association did not reach statistical
significance (P=0.197) (Table
II), indicating that FAM83B expression could not be considered
as an independent prognostic factor.
 | Table IICox proportional hazards model
analysis of prognostic factors in patients with SCC. |
Table II
Cox proportional hazards model
analysis of prognostic factors in patients with SCC.
| Variables | Hazard ratio (95%
Cl) |
Unfavorable/favorable | P |
|---|
| Univariate
analysis |
| FAM83B | 0.489
(0.240–0.996) | Weak/strong | 0.049 |
| Age (years) | 1.088
(0.449–2.637) | ≥65/<65 | 0.852 |
| Gender | 1.077
(0.379–3.063) | Male/female | 0.889 |
| pT factor | 3.142
(1.293–7.638) | T2+T3/T1 | 0.012 |
| pN factor | 2.577
(1.288–5.156) | N1+N2/N0 | 0.007 |
| p stage | 2.309
(1.156–4.611) | II+III/I | 0.018 |
| Pleural
invasion | 4.167
(2.088–8.316) |
Positive/negative | 0.0001 |
| Lymphatic vessel
invasion | 3.482
(1.703–7.122) |
Positive/negative | 0.001 |
| Vascular
invasion | 2.302
(1.131–4.684) |
Positive/negative | 0.021 |
| Multivariate
analysis |
| FAM83B | 0.610
(0.288–1.294) | Weak/strong | 0.197 |
| Age (years) | 1.321
(0.519–3.362) | ≥65/<65 | 0.559 |
| Gender | 1.385
(0.467–4.108) | Male/female | 0.557 |
| pT factor | 1.685
(0.557–5.096) | T2+T3/T1 | 0.355 |
| pN factor | 1.767
(0.577–5.408) | N1+N2/N0 | 0.319 |
| p stage | 0.960
(0.331–2.784) | II+III/I | 0.94 |
| Pleural
invasion | 2.538
(1.084–5.944) |
Positive/negative | 0.032 |
| Lymphatic vessel
invasion | 2.453
(1.072–5.615) |
Positive/negative | 0.034 |
| Vascular
invasion | 0.955
(0.423–2.157) |
Positive/negative | 0.912 |
Discussion
It has recently been reported that FAM83B can act as
an important intermediary in aberrant EGFR/RAS signaling, and is
actually highly expressed in several cancer tissues, such as breast
and lung cancer (18). In the
present study, we intensively examined the expression of FAM83B in
lung cancer at both the mRNA and protein level, and found that it
was highly expressed in lung SCC rather than in lung adenocarcinoma
or adjacent non-cancer regions. Importantly, higher FAM83B
expression evaluated by IHC was associated with longer DFS.
Previously, Cipriano et al (18) investigated a microarray data set
obtained from Oncomine (https://www.oncomine.org/), and found that FAM83B
expression was associated with specific cancer subtypes, increased
tumor grade, and decreased OS. In the case of lung cancers, they
demonstrated that FAM83B expression was higher in SCC than
adenocarcinoma (P=0.00084), and that it was associated with
increasing T stage (P=0.016). Therefore, our data support their
conclusions by additionally examining FAM83B protein levels, and
further provide significant evidence that it is associated with
better prognosis of SCC.
Lung cancer is divided into two major subgroups,
small-cell lung cancer or NSCLC, by its clinical features and the
selection of treatment type. Thus, in the past, NSCLC was treated
according to a ‘uniform’ strategy. However, it has recently been
recognized that histological subtypes are also important for lung
cancer treatment, because several optional treatments have been
proposed as specific for non-SCC. Unfortunately, there is no
effective therapy for lung SCC. Therefore, accurate pathological
diagnosis, particularly to discriminate non-SCC from SCC in biopsy
samples, is an important step in its treatment. However, it has
recently been reported that ~20% of hematoxylin-eosin-stained
biopsy specimens from NSCLC fail to be appropriately diagnosed, and
are known as ‘not otherwise specified’ with poor prognosis
(22). Therefore, more effective
diagnostic markers for each tissue type of NSCLC are required.
To date, only a few markers for lung SCC have been
found, including cytokeratin 5/6 (CK5/6) and p63. CK5/6 is used as
a marker of SCC, and its immunohistochemical detection showed
61–100% sensitivity and 79–93% specificity for SCC (23,24).
p63 is a transcription factor belonging to the p53 family, and has
been clinically used as a diagnostic marker (25). However, problems occasionally
result because of its low specificity; it also shows positivity in
16–65% of lung adenocarcinomas (26). Recently, development of specific
antibodies against p40 (ΔNp63) together with immunohistochemical
evaluation of TTF-1 and p40 have made it possible to completely
discriminate lung adenocarcinoma and SCC (27). Immunohistochemical detection of
FAM83B in this study showed 94.5% sensitivity and 85.3% specificity
for SCC. This highly accurate result indicates that FAM83B could be
a reliable diagnostic marker for lung SCC. Further development of
the detection system would be helpful for accurate and rapid
diagnosis.
Prognosis of lung SCC is generally worse than that
of lung adenocarcinoma (28). It
is well known that lung SCC is associated with high smoking rates
and complications such as interstitial pneumonia and chronic
obstructive pulmonary disease resulting from smoking history, which
hampers optimal treatments of chemotherapy, including adjuvant
chemotherapy. Therefore, selection of appropriate treatments in
so-called ‘personalized therapy’ is more important in determining
the treatment strategy for lung SCC, where the information of
prognostic biomarkers with higher reliability is helpful. Our data
demonstrated that patients with high FAM83B expression tended to
exhibit longer DFS (P=0.042), indicating that FAM83B is a candidate
biomarker that can predict prognosis of SCC.
At present, we do not know the mechanism by which
high expression of FAM83B results in longer DFS. FAM83B protein has
an amino-terminal domain of unknown function (DUF1669), which is
conserved among FAM83 members and contains a putative phospholipase
D-like motif that is critical for FAM83B-mediated transformation
activity. It has previously been shown that FAM83B can associate
with CRAF, p85α and p110α subunits of PI3K, AKT, and EGFR (18,29,30),
and is also able to activate phospholipase D via interaction with
EGFR (29). These researchers
concluded that FAM83B is involved downstream of EGFR, mediating
both the MAPK and PI3K/AKT signaling pathways. Consequently,
increased expression of FAM83B resulted in the transformation of
human mammary epithelial cells (18,29,30).
This mechanistic model is consistent with the fact that FAM83B is
highly expressed in breast cancer, and that its expression level is
associated with its malignancy (18). However, in the case of lung SCC,
our results indicate that high expression of FAM83B would predict
better prognosis. Although it seems contradictory, there has been
another example. The transcription factor SOX2 has been identified
as an amplified lineage-survival oncogene in lung and esophageal
SCC, and its overexpression has been shown to be associated with
better prognosis (31–33). The most plausible explanation would
be that SOX2 expression might promote squamous differentiation
rather than malignant dedifferentiation. FAM83B may also be
involved in a similar mechanism, though further studies are
required.
Proteome analyses have identified FAM83B as a novel
interactor for APC and AXIN-1, both of which are components of a
destruction complex of β-catenin and thus regulate the WNT
signaling pathway (34). Such
additional signaling pathways together with EGFR pathways may be
differentially operated in different tissues and during the context
of differentiation. Taken together, our findings suggest that
although potential FAM83B-targeted therapy might be effective for
breast cancer, it would not necessarily be true for lung SCC. More
detailed analyses of fundamental signaling pathways using cell
lines derived from SCC would give us better conclusions for the
function of FAM83B in lung SCC.
Acknowledgements
This research was partially supported by grants for
translational research programs from New Energy and Industrial
Technology Development Organization (NEDO) (Tokyo, Japan) and
Fukushima Prefecture. Dr Y. Yanagisawa and Dr R. Honma are
employees of Nippon Gene Co., Ltd. We are extremely grateful to Ms.
Y. Kikuta for her technical assistance.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Travis WD: Pathology of lung cancer. Clin
Chest Med. 32:669–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schiller JH, Harrington D, Belani CP, et
al: Comparison of four chemotherapy regimens for advanced
non-small-cell lung cancer. N Engl J Med. 346:92–98. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Scagliotti GV, Parikh P, von Pawel J, et
al: Phase III study comparing cisplatin plus gemcitabine with
cisplatin plus pemetrexed in chemotherapy-naive patients with
advanced-stage non-small-cell lung cancer. J Clin Oncol.
26:3543–3551. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hanna N, Shepherd FA, Fossella FV, et al:
Randomized phase III trial of pemetrexed versus docetaxel in
patients with non-small-cell lung cancer previously treated with
chemotherapy. J Clin Oncol. 22:1589–1597. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ciuleanu T, Brodowicz T, Zielinski C, et
al: Maintenance pemetrexed plus best supportive care versus placebo
plus best supportive care for non-small-cell lung cancer: a
randomised, double-blind, phase 3 study. Lancet. 374:1432–1440.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sandler A, Gray R, Perry MC, et al:
Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell
lung cancer. N Engl J Med. 355:2542–2550. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hainsworth JD, Fang L, Huang JE, et al:
BRIDGE: an open-label phase II trial evaluating the safety of
bevacizumab + carboplatin/paclitaxel as first-line treatment for
patients with advanced, previously untreated, squamous non-small
cell lung cancer. J Thorac Oncol. 6:109–114. 2011. View Article : Google Scholar
|
|
9
|
Turner NC and Seckl MJ: A therapeutic
target for smoking-associated lung cancer. Sci Transl Med.
2:62ps562010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Soda M, Choi YL, Enomoto M, et al:
Identification of the transforming EML4-ALK fusion gene in
non-small-cell lung cancer. Nature. 448:561–566. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Paez JG, Janne PA, Lee JC, et al: EGFR
mutations in lung cancer: correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khuder SA: Effect of cigarette smoking on
major histological types of lung cancer: a meta-analysis. Lung
Cancer. 31:139–148. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rekhtman N, Paik PK, Arcila ME, et al:
Clarifying the spectrum of driver oncogene mutations in
biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS
and presence of PIK3CA/AKT1 mutations. Clin Cancer Res.
18:1167–1176. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim HS, Mitsudomi T, Soo RA and Cho BC:
Personalized therapy on the horizon for squamous cell carcinoma of
the lung. Lung Cancer. 80:249–255. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cancer Genome Atlas Research Network.
Comprehensive genomic characterization of squamous cell lung
cancers. Nature. 489:519–525. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gold KA, Wistuba II and Kim ES: New
strategies in squamous cell carcinoma of the lung: identification
of tumor drivers to personalize therapy. Clin Cancer Res.
18:3002–3007. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cipriano R, Graham J, Miskimen KL, et al:
FAM83B mediates EGFR- and RAS-driven oncogenic transformation. J
Clin Invest. 122:3197–3210. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Miura A, Honma R, Togashi T, et al:
Differential responses of normal human coronary artery endothelial
cells against multiple cytokines comparatively assessed by gene
expression profiles. FEBS Lett. 580:6871–6879. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Laemmli UK: Cleavage of structural
proteins during the assembly of the head of bacteriophage T4.
Nature. 227:680–685. 1970. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Towbin H, Staehelin T and Gordon J:
Electrophoretic transfer of proteins from polyacrylamide gels to
nitrocellulose sheets: procedure and some applications. Proc Natl
Acad Sci USA. 76:4350–4354. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ou SH and Zell JA: Carcinoma NOS is a
common histologic diagnosis and is increasing in proportion among
non-small cell lung cancer histologies. J Thorac Oncol.
4:1202–1211. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Noh S and Shim H: Optimal combination of
immunohistochemical markers for subclassification of non-small cell
lung carcinomas: a tissue microarray study of poorly differentiated
areas. Lung Cancer. 76:51–55. 2012. View Article : Google Scholar
|
|
24
|
Ocque R, Tochigi N, Ohori NP and Dacic S:
Usefulness of immu-nohistochemical and histochemical studies in the
classification of lung adenocarcinoma and squamous cell carcinoma
in cytologic specimens. Am J Clin Pathol. 136:81–87. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Conde E, Angulo B, Redondo P, et al: The
use of P63 immunohistochemistry for the identification of squamous
cell carcinoma of the lung. PLoS One. 5:e122092010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bishop JA, Teruya-Feldstein J, Westra WH,
Pelosi G, Travis WD and Rekhtman N: p40 (ΔNp63) is superior to p63
for the diagnosis of pulmonary squamous cell carcinoma. Mod Pathol.
25:405–415. 2012. View Article : Google Scholar
|
|
27
|
Pelosi G, Fabbri A, Bianchi F, et al:
ΔNp63 (p40) and thyroid transcription factor-1 immunoreactivity on
small biopsies or cellblocks for typing non-small cell lung cancer:
a novel two-hit, sparing-material approach. J Thorac Oncol.
7:281–290. 2012. View Article : Google Scholar
|
|
28
|
Kogure Y, Ando M, Saka H, et al: Histology
and smoking status predict survival of patients with advanced
non-small-cell lung cancer. Results of West Japan Oncology Group
(WJOG) Study 3906L. J Thorac Oncol. 8:753–758. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cipriano R, Bryson BL, Miskimen KL, et al:
Hyperactivation of EGFR and downstream effector phospholipase D1 by
oncogenic FAM83B. Oncogene. 33:3298–3306. 2014. View Article : Google Scholar :
|
|
30
|
Cipriano R, Miskimen KL, Bryson BL, Foy
CR, Bartel CA and Jackson MW: FAM83B-mediated activation of
PI3K/AKT and MAPK signaling cooperates to promote epithelial cell
transformation and resistance to targeted therapies. Oncotarget.
4:729–738. 2013.PubMed/NCBI
|
|
31
|
Bass AJ, Watanabe H, Mermel CH, et al:
SOX2 is an amplified lineage-survival oncogene in lung and
esophageal squamous cell carcinomas. Nat Genet. 41:1238–1242. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hussenet T, Dali S, Exinger J, et al: SOX2
is an oncogene activated by recurrent 3q26.3 amplifications in
human lung squamous cell carcinomas. PLoS One. 5:e89602010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wilbertz T, Wagner P, Petersen K, et al:
SOX2 gene amplification and protein overexpression are associated
with better outcome in squamous cell lung cancer. Mod Pathol.
24:944–953. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hilger M and Mann M: Triple SILAC to
determine stimulus specific interactions in the Wnt pathway. J
Proteome Res. 11:982–994. 2012. View Article : Google Scholar :
|