Introduction
Glioblastoma (GBM) is the most common and lethal
primary malignancy of the central nervous system. Even with
surgical resection and aggressive treatment with chemo- and
radiotherapy, the prognosis remains very poor. A wide variety of
novel therapeutic approaches have been developed and are currently
under study as potential treatments for GBM (1).
The anticancer drug doxorubicin (DOX) primarily
exhibits a wide spectrum of cytotoxic effects (2). Its planar three-ring structure
stabilizes the topoisomerase II-DNA cleavable complex by DNA
intercalation and enhances cleavage of DNA at both strands in a
topoisomerase II-dependent manner. DOX also reacts with cellular
formaldehyde to form DNA adducts (3). To effectively utilize the antitumor
function of DOX, there have been few studies focusing on the other
mechanisms of DOX (4).
Epigenetic lesions in DNA without mutations in the
coding regions have been shown to be common phenomena in the
pathogenesis of GBM, especially the methylation-mediated silencing
of tumor suppressor genes such as VHL, p16, E-cadherin, PTEN, p21
and RECK, MGMT, RASSF1A (5–8).
When DNA is methylated in the promoter region of genes where
transcription is initiated, genes are inactivated and silenced
(9). The cancer methylome is
highly disrupted, making DNA methylation an excellent target for
anticancer therapies. Several small synthetic and natural molecules
are thus able to reverse the DNA hypermethylation through
inhibition of DNA methyltransferase (DNMT). Over the last few
decades, an increasing number of DNMT inhibitors targeting DNA
methylation have been developed to increase efficacy with reduced
toxicity (10). Tumor suppressor
gene inactivation has previously been correlated with DNMT1
overexpression in various types of cancers (11). Knockdown of DNMT1 can repress tumor
suppressor genes (12). Previous
studies have clarified that DOX is likely to affect DNA methylation
by inhibiting catalytic activity of DNMT1 (13). We therefore hypothesized that DOX
might indirectly alter epigenetic regulation of gene
expression.
MicroRNAs (miRNAs) are small non-coding RNAs that
act through post-transcriptional silencing in critical regulatory
roles in multiple cellular functions (14). MicroRNAs represent an abundant
class of endogenously 18–25 nucleotide non-coding RNA molecules
which silence gene expression through a process of
post-transcriptional modification. As one of the first miRNAs
detected in the human genome, miR-21 has been validated to be
involved in many different types of human cancers. Through
targeting of PTEN, PDCD4, RECK and other signal transduction
pathways, miR-21 regulates the proliferation, apoptosis, and
invasion of hepatocellular cancer, colorectal cancer, breast cancer
(15–19). Thus, targeting miR-21 and
inhibiting its activity may be emerging as a promising therapeutic
option and offer a potential new mode of cancer therapy.
Currently, therapies which simultaneously administer
small molecular chemotherapeutic drug with gene medicine are common
and effective ways to treat cancer (20). Cheng et al demonstrated that
the folate-targeted co-delivery of Bcl-2 siRNA and DOX system
caused not only an obvious reduced expression of anti-apoptotic
Bcl-2 gene but also a remarkable elevated level of the
pro-apoptotic Bax gene, resulting in the significantly apoptosis in
tumor tissues (21). Based on
that, we hypothesized DOX and miR-21 inhibitor (miR-21i) could
regulate gene expression synergistically to inhibit tumor
cells.
In the above study, we detected the expression of
tumor suppressor genes E-cadherin, RECK, PTEN, p21, VHL and
miR-200a/b/429 and miR-181d to discuss the effect of DOX and
miR-21i on GBM suppression. In our present study, we discovered
that DOX caused not only downregulation of DNMT1 but also obviously
upregulation of PTEN and p21 genes as well as 4 non-coding miRNAs
in the transcription stage. At the same time, miR-21i can regulate
gene expression through post-transcriptional regulation. In
addition, we have shown that combining DOX and miR-21i enhanced
methylation associated tumor suppressor gene expression, this
synergetic effect took place at the transcriptional level and
post-transcriptional level. Furthermore, co-treatment with DOX and
miR-21i strengthened antitumor effect, resulting in reduced tumor
cell migration and cell invasion in vitro.
Materials and methods
Reagents, cell culture and
transfection
The antisense oligonucleotide sequence of
2′-O-methyl (2′-O-Me) miR-21 inhibitor was: 5′-GTC CAC TCT TGT CCT
CAA TG-3′. A scrambled inhibitor sequence (5′-AAG GCA AGC UGA CCC
UGA AGU-3′) was used as the negative control. They were chemically
synthesized by Shanghai Gene Pharma (Shanghai, China) and dissolved
in diethylpyrocarbonate (DEPC) water and frozen at −20°C. DOX
hydrochloride was purchased from Sigma-Aldrich. It dissolved in PBS
and diluted in serum-free medium.
Human glioma cell lines U87 (PTEN del/EGFR wt), U87
EGFRvIII (PTEN del/EGFR mut), LN229 (PTEN wt/EGFR wt) were obtained
from ATCC (American Type Culture Collection, Manassas, VA, USA).
The human GBM cell lines U87, U87 EGFRvIII, and LN229 were
maintained in Dulbecco’s modified Eagle’s medium (DMEM)
(Invitrogen) supplemented with 10% FBS (heat-inactivated fetal
bovine serum) (Hyclone) at 37°C, 5% CO2. The miR-21
inhibitor was transfected with Lipofectamine 3000 (Invitrogen,
Grand Island, NY, USA). Transfections with hsa-miR-21 inhibitor and
scrambled inhibitor were performed in serum-free medium 24 h after
plating. Cell transfection used Lipofectamine 3000 according to the
manufacturer’s instructions. For each 6-well, miRNA in 125 μl of
serum-free medium was mixed with 5 μl of Lipofectamine 3000 in 125
μl of the same medium and allowed to stand at room temperature for
5 min. The mixture was then added to cells and after 6 h the medium
was changed to complete medium.
Evaluation of DOX cytotoxicity
The cytotoxicity of DOX was evaluated by the 3-(4,
5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT)
assay. Five thousand cells/well were seeded in 96-well plates at
37°C for 24 h in 100 μl DMEM, which was supplemented with 10% FBS,
2 mM of glutamine (Sigma), 100 mg/ml of penicillin (Sigma) and 100
mg/ml of streptomycin (Sigma). Cancer cells were exposed to
different concentrations of DOX (0, 0.5, 1, 1.5, 2, 4, 8 and 10
μM/l) for 48 h. To assess cell viability, 20 μl of MTT (5 mg/ml)
was added to each well and the cells were incubated at 37°C for
another 4 h. The reaction was then stopped by dissolving the cells
in 200 μl of dimethyl sulfoxide (DMSO) with shaking for 15 min to
dissolve the formazan crystals formed by the living cells.
Quantification measurements (optical density) were obtained at a
wavelength of 490 nm using spectrophotometric analysis. Cells
without treatment were used as control.
Flow cytometric analysis of cellular
uptake of DOX
Cells (2×105) were cultured in a 6-well
plate at 37°C for 24 h in 2 ml DMEM. Cancer cells were then exposed
to different concentrations of DOX (0, 0.5, 1.5 and 8 μM/l) for 6
h. At the end of the incubation period, cells were trypsinized and
washed three times with PBS then fixed and resuspended in 75% ethyl
alcohol of the corresponding temperature. Uptake rates were
detected via flow cytometry (Becton-Dickinson, USA). Cells were
passed through a 37-μm nylon filter to ensure a single-cell
suspension. Laser excitation was at 488 nm and fluorescence was
detected at 575 nm. Files were collected of 20,000 gated events and
analyzed with the FACS software program.
Confocal fluorescence microscopy analysis
of DOX intra-cellular uptake and distribution
A confocal fluorescent microscopy was used to
compare the intracellular uptake of DOX (excitation/emission:
480/575 nm) and to investigate their cellular distribution. Cells
(2×105) were grown on glass cover slips in a 6-well
plate at 37°C for 24 h in 2 ml DMEM. Cancer cells were then exposed
to different concentrations of DOX (0, 0.5, 1.5, 8 μM/l) for 6 h.
At the end of the incubation period, the medium was removed and the
cells were washed three times with cold PBS and then fixed in 4%
paraformaldehyde for 30 min. The fixed cells were washed three
times with PBS at room temperature on a shaker. For nucleus
labeling, cells were incubated with Clear-Mount (aqueous)
containing DAPI (excitation/emission: 345/661 nm) for 10 min. The
fluorescent images of cells were analyzed using a laser scanning
confocal microscope.
Protein extraction and western blot
analysis
Glioma cells were treated with DOX/miR-21i alone or
compound respectively. Cells were harvested 48 h after
transfection. Each group of cells were then washed in cold PBS
three times and then solubilized in 1% Nonidet P-40 lysis buffer.
Homogenates were clarified by centrifugation at 12,000 rpm for 15
min at 4°C, and protein concentrations were determined with
Nanodrop spectrophotometer (Gene, USA). The protein contents of the
lysates (50 μg) were subjected to sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12, 10 and
8%, which were then transferred onto PVDF membranes (Millipore,
Billerica, MA, USA). The membranes were probed overnight with
primary antibodies against DNMT1 (1:1,000 dilution, Cell Signaling
Technology, USA), E-cadherin (1:1,000 dilution, Abcam, UK), RECK
(1:1,000 dilution, Santa Cruz, USA), PTEN (1:1,000 dilution, Santa
Cruz), VHL (1:1,000 dilution, Abcam), p21 (1:1,000 dilution, Cell
Signaling Technology) and GAPDH (1:1,000 dilution, Santa Cruz). The
membranes were subsequently washed three times with PBS to remove
excess primary antibodies, and incubated with appropriate
HRP-conjugated secondary antibodies (1:1,000 dilution, Beijing
Zhongshan Bio Corp., Beijing, China). GAPDH was selected as a
housekeeping gene.
RNA extraction and real-time PCR
Total RNA was extracted from cultured cells with
TRIzol reagent (Invitrogen) according to standard protocol. To
detect the concentration of total mRNA, a nanodrop
spectrophotometer (Gene) was utilized. Reverse transcription (RT)
was conducted with the Go Scsipt™ Reverse Transcription System
(Promega, USA). MJ-real time PCR (Bio-Rad, USA) was used to achieve
the amplification reaction and the protocol was carried out for 40
cycles at 95°C for 3 min, 95°C for 15 sec and 60°C for 30 sec. Both
RT and PCR primers were purchased from Gene Pharma. The relative
expression of miRNA/mRNA was evaluated via comparative CT
(threshold cycle) and was normalized to the expression of U6/GAPDH
RNA (Table I). All RT-PCR
reactions were performed in triplicate.
 | Table IRepresentative gene primers utilized
to perform quantitative PCR of mRNA. |
Table I
Representative gene primers utilized
to perform quantitative PCR of mRNA.
| Gene | Primer sequences
(5′-3′) |
|---|
| DNMT1 | Forward:
GGTGGAGAGTTATGACGAG
Reverse: TAGAATGCCTGATGGTCTG |
| E-cadherin | Forward:
TGATTCTCTGCTCGTGTT
Reverse: CGTTCAAGTAGTCATAGTCC |
| RECK | Forward:
GCTGTAGAAACCTTACTTACTG
Reverse: GCTATTGCTTTCCACATCTC |
| PTEN | Forward:
CTTCTACTGCCTCCAACAC
Reverse: AGACGAATAATCCTCCGAAC |
| VHL | Forward:
GTAGCGGTTGGTGACTTG
Reverse: CCCTGGTTTGTTCCTCTG |
| p21 | Forward:
CCCTTGTCCTTTCCCTTC
Reverse: GTGCCCTTCTTCTTGTGT |
| GAPDH | Forward:
CCGGGAAACTGTGGCGTGATGG
Reverse: AGGTGGAGGAGTGGGTGTCGCTGTT |
Wound healing assay
Prior to wounding, cell culture and transfection
conditions were optimized to ensure a homogeneous and viable cell
monolayer. One day prior to transfection, equal quantities of GBM
U87 and LN229 cells (2×105) were seeded in 6-well
plates. The U87 and LN229 cells were treated with PBS, scrambled
inhibitor, DOX, miR-21i or DOX/miR-21i, respectively. Cell
transfection used Lipofectamine 3000 according to the
manufacturer’s instructions. When cell confluence reached ~90% at
~24 h post-transfection, an artificial homogenous wound was made to
the monolayer using a sterile plastic 200 μl micropipette tip.
Following wounding, debris was removed by washing cells three times
with PBS. At different time-points, cells migrated into the wounded
area or cells with extended protrusions from the wound border were
photographed at ×200 magnification under a light microscope.
Invasion assays
Invasive capacities of human GBM U87 and LN229 cells
were tested via in vitro invasion assays (Becton-Dickinson
Bio-Coat Matrigel Invasion Chamber). The top chamber of a transwell
chamber was incubated with 60 μl Matrigel diluted with DMEM (1:2,
Matrigel: DMEM) at 37°C for 30 min. The Matrigel solidified and
acted as an extracellular membrane (ECM) for tumor cell invasion
analysis. The U87 and LN229 cells were treated with PBS, scrambled
inhibitor, DOX, miR-21i or DOX/miR-21i, respectively. Cell
transfection used Lipofectamine 3000 according to the
manufacturer’s instructions. After 24 h, each group of cells were
adjusted to 5×105/ml in DMEM and 100 μl of the
resuspended cell solution was added to the top chamber over the
Matrigel, with 100 μl of serum-free DMEM added up to 200 μl cell
solution. The cells were induced to invade toward a chemoattractant
filled with 500 μl of DMEM (with 10% FBS) which was placed into the
lower chambers of the wells. The transwell plate was assembled and
incubated at 37°C, in a 5% CO2 incubator. Following 24-h
incubation, the non-invading cells were removed from the upper
surfaces of the invasion membranes. Cells were stained by crystal
violet for 3 min, and washed with PBS to remove excess stain. The
chambers were gently scraped with a wet cotton swab. Images of each
well were captured by microscopic analysis with an Olympus Vanox.
All experiments were performed in triplicate.
Statistical analysis
All the experiments were carried out in triplicate
and data were analyzed using Windows SPSS software. Quantitative
values are expressed as means ± standard error, statistical
analyses were performed using t-test. Differences were considered
significant for p≤0.05. One-way ANOVA was used to test for
differences among at least 3 groups, and the least significant
difference post-hoc test was utilized to obtain individual p-values
followed by ANOVA. The t-test was utilized to determine differences
in each dual group comparison.
Results
Modes of cell death induced by DOX
Human GBM cells were used to identify and
characterize the various types of cell deaths induced by DOX. The
cytotoxicity of the DOX was evaluated by MTT assay in human glioma
cells. Human GBM cells LN229 (PTEN wt/EGFR wt), U87 (PTEN del/EGFR
wt) and U87 EGFRvIII (PTEN del/EGFR mut) were first infected with
different concentration gradient of DOX. We found that the three
kinds of cell lines produced different reactions to the tested
range of DOX concentrations (0.5–10 μM). As shown in Fig. 1A-a, LN229 cell line survival rates
decreased gradually with the increase of drug concentrations. U87
and U87 EGFRvIII cell line survival rates, however, decreased
radically in the low drug concentration (lower in 2 μM) but higher
doses of DOX did not result in significant apoptosis, which
appeared to be a relatively high platform on the survival curve
under the conditions of relatively high drug concentrations. The
survival rate was ~40%. The results reveal that PTEN or EGFR may
affect the biological activity of DOX in tumor cells and lead to
drug resistance. Several studies have shown that the signaling
pathway activated by the lipid kinase phosphoinositide 3-kinase
(PI3K) and the serine/threonine kinase, protein kinase B (PKB) or
Akt, play a more important role in chemoresistance including DOX
(22,23). Given that tumor suppressor gene
PTEN is a negative regulator of the PI3K pathway, the most
extensive evidence for the involvement of the PI3K pathway in human
cancer stems from studies of the PTEN. Loss of PTEN can be
sustained activation of this pathway. PI3K pathway activation
contributes to the effects chemoresistance (24). On the other hand, the increasing of
mutant EGFR receptor (EGFRvIII) expression lead to continuous
activation of EGFR. EGFRvIII signaling also activates the PI3K
pathway in glioblastoma cell lines (25,26).
Thus, loss of PTEN or EGFR mutations may play an important role in
glioblastoma cell chemoresistance to DOX. We believe this is worthy
of further study.
 | Figure 1Effects of DOX/miR-21i on cell
viability and characterization of DOX uptake by GBM cell lines. (A)
Effect of DOX/miR-21i on cell viability: (a) U87, U87-EGFRvIII and
LN229 GBM cell lines were treated with various concentrations of
DOX (0, 0.5, 1, 1.5, 2, 4, 8 or 10 μM) and after 48-h incubation an
MTT assay was performed. The viability of the untreated cells was
regarded as 100%. Combination of DOX and miR-21i in the U87 (b-1)
and U87-EGFRvIII (b-2) cell lines. (B) Cellular uptake of DOX
detected by flow cytometry. (C) Cellular distribution of DOX
detected by confocal microscopy. The two cell lines U87 and
U87-EGFRvIII were treated with DOX (0, 0.5, 1.5 and 8.0 μM) in a
6-h incubation at 37°C. Cells were counter-stained with DAPI (for
nuclei), DOX (red fluorescence). Error bars represent the mean ± SD
obtained from 3 independent experiments. |
miR-21i and DOX on proliferation of GBM
cells
Previous studies have clarified that the expression
of miR-21 was upregulated in human GBM. In order to examine whether
miR-21 could modulate the chemosensitivity to DOX in GBM cells, we
detected the expression of miRNAs in DOX-resistant cells. miR-21i
was transfected after treatment of DOX in U87 and U87 EGFRvIII cell
lines. Sensitivity to DOX was increased by the specific inhibition
of miR-21 of which the maximal inhibition differed for the two GBM
cell lines. The results showed that miR-21 downregulated
Dox-resistant U87 cells when treated with DOX at concentrations
ranging from 0.5 to 10 μM as measured by MTT assay (Fig. 1A-b), markedly enhanced cell death
was observed. This increased sensitivity to DOX was not, however,
observed in U87 EGFRvIII cells after downregulating miR-21 on high
DOX concentration, but rather simply delayed the emergence of
plateaus in high drug concentrations. These data suggest that the
synergistic effect appeared at low concentrations, and inhibition
of miR-21 could sensitize GBM cells to anticancer drug DOX.
Characterization of DOX uptake in tumor
cells
We examined cellular uptake of DOX at different
concentrations. Flow cytometry and confocal microscopy were
performed to observe cellular uptake and distribution of DOX. We
chose 0.5, 1.5 and 8 μM as the representative drug concentrations.
Flow cytometry revealed that the U87 cellular uptake of DOX
dramatically increased from 3.46 to 99.48%, as did the U87-EGFRvIII
cell line, changing from 3.10 to 98.65% (Fig. 1B). The highest DOX concentration
displayed the highest cellular uptake with 6-h incubation. These
results were further confirmed by confocal microscopy. The majority
of visible DOX fluorescence (red) was mainly in the nuclei.
Significantly higher intracellular DOX fluorescence intensity was
observed in the nucleus of the U87/U87-EGFRvIII cell lines with the
increase of DOX concentration (Fig.
1C), indicating an increased uptake of DOX by these cells.
Taken together, our data show that DOX is localized in the nucleus
and the amount in the nucleus gradually increased with the increase
of drug concentration. At the same time, it suggests that loss of
PTEN or EGFR mutations may be a reason for GBM cell chemoresistance
to DOX.
DOX may act as a demethylation drug
reactivating DNA methylation-silenced tumor suppressor genes
Anticancer drug DOX exhibits a wide spectrum of
cytotoxic effects primarily as a topoisomerase II α poison.
However, recent studies indicated that topoisomerase II may not be
the main target. Other cellular responses to doxorubicin have also
emerged, including the inhibition of the DNMT1 (13). DNMT1 is the primary enzyme
responsible for maintenance of DNA methylation on genomic DNA. To
assess whether DOX could lead to demethylation and introduction of
tumor suppressor genes through blocking DNMT1 expression, we first
analyzed the expression of DNMT1. After treating three GBM cells
with DOX for 48 h, western blot (Fig.
2A-a) and PCR (Fig. 2A-b)
analysis indicated that the protein and mRNA expression levels of
DNMT1 was clearly reduced. Reports have shown that epigenetic
lesions in DNA without mutations in the coding regions are a common
phenomenon in the tumorigenesis of GBM, especially the
methylation-mediated silencing of tumor suppressor genes such as
VHL, PTEN, E-cadherin, DAPK, MGMT, EMP3 and p21 (27). We then investigated whether DOX
could affect tumor suppressor encoding genes associated with
methylation such as E-cadherin, RECK, PTEN, VHL and p21. Western
blot analysis indicated that certain protein expression was
dramatically increased, including PTEN, and p21 (Fig. 2B). However, the expression levels
of E-cadherin, RECK, and VHL were not obviously changed. In order
to further confirm the role of tumor suppressor gene methylation
influenced by DOX, we performed Real time-PCR analyses to measure
the expression of mRNA. Similar to western blot analysis, the mRNA
expression of PTEN, and p21 was dramatically increased (Fig. 2C). However, the expression levels
of E-cadherin, RECK, and VHL were not markedly changed.
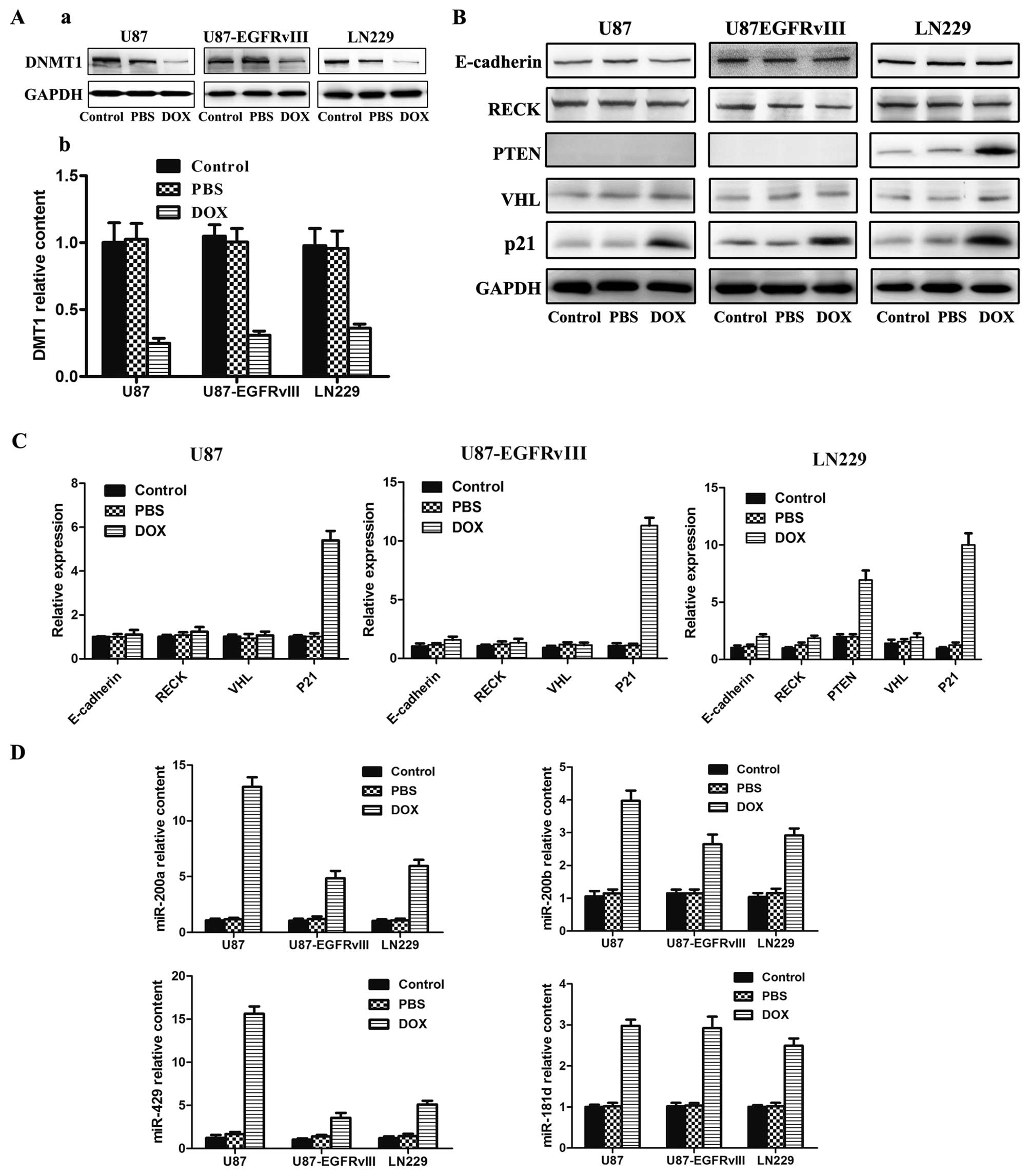 | Figure 2Effects of DOX on the functions of
U87, U87-EGFRvIII and LN229 cell lines. (A) DNMT1 relative
expression assayed using western blotting and RT-PCR. (B) Western
blot detection of E-cadherin, RECK, PTEN, VHL and p21 expression.
(C) RT-PCR analysis of E-cadherin, RECK, PTEN, VHL and p21 mRNA
expression. (D) RT-PCR analysis of 4 miRNAs expression in the
indicated cells. The cell lines were treated with DOX for 48 h at
the concentration of 0.4, 0.4 and 0.5 μM respectively. Western
blotting, GAPDH was used as the loading control. PCR, error bars
represent the mean ± SD obtained from 3 independent
experiments. |
After it was ascertained that DOX can affect
expression of some tumor suppressor genes, we investigated whether
DOX could regulate tumor suppressor non-coding genes. The number of
miRNAs with putative tumor suppressor functions undergoing promoter
CpG island hypermethylation in human cancer is one of the most
common causes of aberrant silencing. Recent reports have also
indicated the presence of hypermethylation-associated silencing of
some miR-200 family members in cancer cells, and most importantly,
the DNA methylation associated silencing of the miR-200 family
determines the evolving epithelial-mesenchymal transition
phenotypes (28). miR-181d was
downregulated in human GBM, and acts as a tumor suppressor in GBM
by targeting K-ras and Bcl-2 (29). Furthermore, our recent study
demonstrated that regulation of Wnt/β-catenin signaling pathway
affected GBM proliferation, and migration (30). We chose miRNAs which associated
with tumor invasion: miR-200a/b/429, and miR-181d. Forty-eight
hours later, the expression levels of miR-200a/b/429 and miR-181d
were increased in varying degrees (Fig. 2D). This indicated that DOX can
promote expression of tumor suppressor miRNAs.
Global changes in gene expression induced
by miR-21i in GBM cells
MiRNAs are small endogenous non-coding RNAs which
downregulate gene expression primarily by binding to the 3′UTR of
the target gene region (31).
MicroRNA-21 negatively regulates several targets, and thus impacts
tumorigenesis. Several targets of miR-21 have been experimentally
validated, including PTEN, VHL and RECK. Ectopic expression of
these targets may exert differing functional effects on
tumorigenesis. All three cell lines were infected with miR-21i for
48 h. We then similarly investigated tumor suppressor encoding
genes E-cadherin, RECK, PTEN, VHL, p21 and tumor suppressor
non-coding genes miR-200a/b/429, miR-181d. Western blotting of the
infected cells showed that the protein levels were increased for
E-cadherin, RECK, PTEN, VHL, but p21 was virtually unchanged
(Fig. 3A). Furthermore, PCR
results revealed that expression levels of tumor suppressor
miR-200a/b/429 and miR-181d were increased (Fig. 3B). These results provided evidence
that downregulation of miR-21 could play the role of inhibiting GBM
through upregulation tumor suppressor genes.
Co-treatment of DOX and miR-21 inhibitor
enhances tumor suppressor genes expression
Although effective, DOX can promote expression of
tumor suppressor genes and miRNA can regulate expression of tumor
suppressor genes, single transfection DOX or miR-21 inhibitor is
not very efficient in enhancing tumor-suppressor gene expression.
We suspect that the combination DOX and miR-21 inhibitor can
enhance the expression of tumor suppressor genes. To further
investigate the expression level of coding genes and non-coding
genes, three GBM cell types were simultaneously co-transfected with
DOX and miR-21i. Consistent with our hypothesized results, western
blotting revealed that protein levels of coding genes E-cadherin,
RECK, PTEN, VHL and p21 were markedly increased in the co-treatment
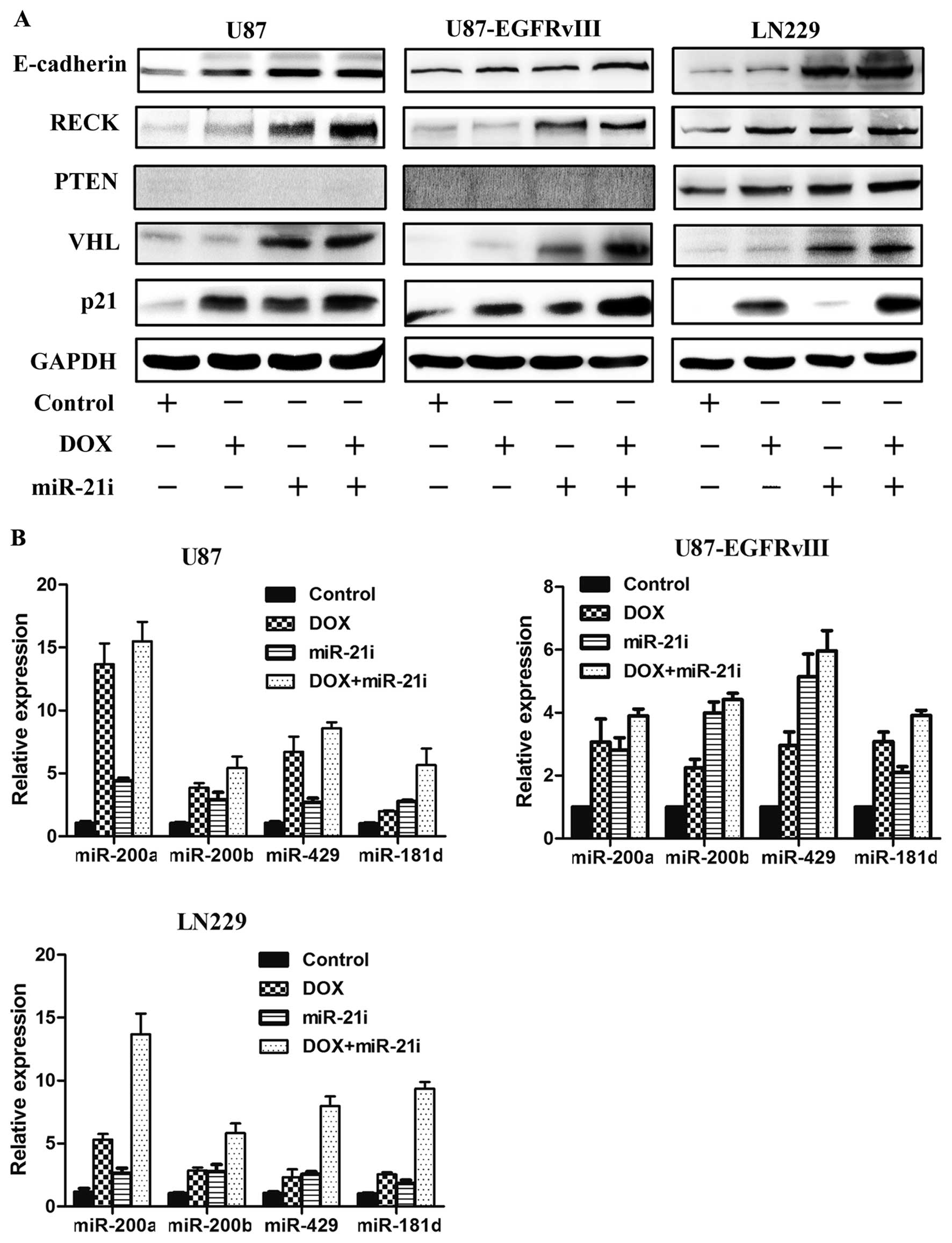
group as compared with the DOX or the miR-21i group alone (Fig. 4A). Furthermore, in the combined
treatment, a significant inducement of the miRNAs (miR-200a/b/429,
miR-181d) was observed with a greater level than that of DOX or
miR-21i treatment alone, as shown in Fig. 4B. In addition, the treatment with
DOX in combination with miR-21i induces a highly synergistic effect
that upregulates tumor suppressor genes in GBM cells.
Regulation of tumor cell activity by DOX
and miR-21i in vitro
Due to the aggressive growth characteristics of GBM,
we investigated the regulation of tumor cell activity by DOX and
miR-21i. Our previous study showed that elevated miR-200a inhibited
cell growth and invasion through the Wnt/β-catenin pathway in
vitro and in vivo (32,33).
We likewise know about miR-181d mediated suppression of
β-catenin/Wnt signaling (30). The
expression level of miR-200 family/miR-181d clearly influenced the
biological activity of GBM cells. We therefore investigated two
major biological activities of tumor cells migration, and invasion
potential. Migration and invasion potential are important
biological characteristics of malignant tumor cells. To examine the
cell migration in vitro, the scratch assay was employed, and
more decreased mobility was observed in the DOX and miR-21i group
(Fig. 5A). This result indicated
an inhibitory effect of DOX and miR-21i on the migration ability.
The number of U87 and LN229 cells invading through the Matrigel in
the DOX and miR-21i group was significantly decreased compared to
single DOX, miR-21i, control, PBS and scrambled inhibitor group
(Fig. 5B). In summary, our results
demonstrated that combination treatment with DOX and miR-21i
significantly reduced tumor cell migration and cell invasion
compared with DOX or the miR-21i treatment alone.
Discussion
The occurrence of GBM is associated not only with
genetic changes but also with epigenetic alterations such as
aberrations in DNA methylation patterns (27). Almost half of tumor-suppressor
genes have been shown to be transcriptionally silenced (34). Silencing by DNA hypermethylation in
GBM affects genes involved in key cellular functions such as the
cell cycle (p16 and p21), tumor suppression (VHL and PTEN), DNA
repair, and genome integrity (MGMT and MLH1) as well as tumor
invasion and apoptosis (CDH1 and RECK) (7,27,35,36).
For non-coding genes, miRNAs induce heritable changes in gene
expression without altering DNA sequences and thus contribute to
the epigenetic landscape. Therefore, CpG island promoter
hypermethylation is one of the causes of the silencing of tumor
suppressor miRNAs with a similar chromatin context to coding genes
(37). Because cytosine
methylation within the promoter regions of genes can cause
transcriptional silencing, demethylation may activate the
expression of genes that activate tumor suppressors in turn.
Evidence has proved that anticancer drugs with DOX can act as DNA
hypomethylating agents. According to Hanafy et al (4), total methylation percentage was
markedly reduced from 62.2 (control) to 36.7% by the action of DOX.
SP1049C, a Pluronic-based micellar formulation of DOX,
significantly increased gene promoter demethylation compared to
saline control P388 cancer stem cells in vivo (38). These results suggest that
anticancer drug DOX may act as DNA hypo-methylating agent.
DNMT1 is the best studied methyltransferase
responsible for maintaining DNA methylation patterns in genomic DNA
during DNA replication. High levels of DNMT1 expression have been
reported to transcriptionally silence tumor suppressor genes
(39,40). Since DNMT1 promotes methylation of
DNA and is a key factor in maintaining DNA methylation, some recent
research was committed to detect or downregulate DNMT1 (41,42).
Further studies revealed DOX can interact with DNA including the
formation of doxorubicin-DNA adducts, occur primarily at CpG
sequences, then inhibit the DNA methyltransferase DNMT1 (13). Inactivation of glutamic acid
decarboxylase 67 (GAD67) and reelin might be due to the aberrant
methylation of promoter-associated CpG islands. The doxorubicin
decreased levels of DNMT1 were previously reported exploited to
actively repress the GAD67 and reelin promoter eventually
significantly increasing expression of reelin and GAD67 (43). Furthermore, knockdown of DNMT1
expression caused an increase in chemosensitivity toward cisplatin
(39). Based on the ability of DOX
to intercalate DNA and DNMT1 it can be recruited to DNA damage
sites (44). We therefore
hypothesized that DOX might act as a potential demethylating agent
like 5-Aza-2′-deoxycytidine. In this study, E-cadherin, RECK, PTEN,
VHL, p21 coding genes were demonstrated to be silenced and
associated with the tumor invasion miRNAs (miR-200a/b/429, miR-181d
non-coding genes). In the present study, we showed for the first
time that the protein and mRNA expression of DNMT1 were reduced and
the expression of PTEN and p21 was increased after DOX exposure in
GBM cells. Rajendran et al (45) found that PTEN and p21 gene
promoters displayed hypermethylation in the glioma cell lines.
Downregulation of DNMT1 with DNMT inhibitor 5-azacytidine
consequentially increase the expression of PTEN and p21, as shown
through MS-PCR study. Furthermore, 4 miRNAs were similarly induced
after DOX treatment. Lujambio et al (46) showed that some miRNAs were
upregulated in a DNMT1 and DNMT3B double knockout in colon cancer
cell line model. Davalos et al (28) found that treatment with the DNMT
agent 5′-aza-2′-deoxycytidine in the miR-200 family hypermethylated
cancer cell lines with the DNA-demethylating agent
5′-aza-2′-deoxycytidine increase the expression of the miRNAs.
Although it remains to be confirmed in future experiments, DOX
could detect changes in promoter methylation of tumor suppressor
genes. These data indicated that DOX could block expression of
DNMT1 resulting in re-expression of certain silencing genes and
recovering the function of some tumor suppressor genes. Although
the precise mechanism for this remains unclear, here we propose a
potential mechanism of DOX in methylation. However, the expression
of E-cadherin, RECK and VHL gene mRNA were very weak. Single DOX
cannot effectively influence tumors. Neoplasms of high malignancy
and metastasis are still considered incurable. Therefore, there is
an urgent need to develop potential schemes for cancer
treatment.
Currently, miRNA-based gene therapy offers the
theoretical appeal of targeting multiple gene networks and has
garnered increasing attention (47). Previous studies have clarified that
the expression of miR-21 was upregulated in human GBM tissues as
well as in other cancers (48,49).
Previous studies have indicated that miR-21 was shown to regulate
the proliferation, apoptosis, and invasion of glioma by directly
targeting E-cadherin, PTEN, VHL and RECK (50–52).
In our study, we demonstrated that downregulation of miR-21 could
upregulate the expression of E-cadherin RECK, PTEN, VHL and a set
of miRNAs including miR-200a/b/429 and miR-181d in U87,
U87EGFRvIII, LN229 cells.
Regardless of whether DOX or miR-21i alone can
upregulate the tumor suppressor genes, DOX plays a role of
methylation in the stage of transcription regulation of gene
expression. Furthermore, miRNAs act as post-transcriptional gene
regulators. We therefore postulate a combination of drug and gene
through regulation of the transcription and post-transcription to
simultaneously regulate the expression of tumor suppressor genes.
Our previous studies showed that miR-21i enhanced GBM cells
sensitivity to Taxol, 5-FU and TMZ demonstrating miR-21 plays a
critical role in drug chemosensitivity (53–55).
In-depth studies still need to be performed to confirm the effects
of the combined miR-21 inhibitor and DOX. The results of western
blotting and PCR corresponded well with the fact that DOX and
miR-21 can enhance expression of tumor suppressor genes.
It is well known that the process of
epithelial-to-mesenchymal transition (EMT), characterized by loss
of intercellular adhesion and polarity, cytoskeletal reorganization
that enhances cell motility, and degradation of the basement
membrane has been associated with tumor progression and metastasis
(56). miR-200 family has been
shown to regulate EMT by downregulating the expression of the
transcriptional ZEB factors. Previous studies demonstrated that
miR-200a regulates EMT through direct targets of β-catenin and ZEB
in glioblastoma and gastric adenocarcinoma (32,33)
and miR-181d was believed to be a tumor suppressor in GBM directly
targeting Wnt/β-catenin signaling promoting tumor proliferation,
migration and invasion (30).
miR-21 can induce EMT during TGF-β in many tumor cell lines
(57). In addition, our previous
studies have verified that PTEN inhibits β-catenin activation via
downregulation of pAKT (58), so
the expression levels of PTEN may impact the EMT process via the
regulation of β-catenin. We further validated the upregulation of
RECK had a significant impact on tumor growth and invasion in
vitro and in vivo (51). In our study, we demonstrated that
co-treatment DOX and miR-21i can upregulate the expression of PTEN,
RECK, miR-200a/b/429 and miR-181d to suppress tumor migration and
invasion activity. The combination DOX and miR-21i may facilitate
the inhibition of EMT in GBM.
In the present study, we provide evidence that DOX
may be involved in epigenetic regulation of transcriptional
activity of tumor suppressor genes in GBM. Whether the analogue can
lead to hypomethylation of gene promoters wait further testing.
Here we hypothesized changes in DNMT1 protein and mRNA levels in
conjunction with DOX acting as a hypomethylation agent. We have
demonstrated that the combination of DOX and miR-21i can enhance
the expression of tumor suppressor genes compared to treatment
alone. Additionally, we have shown combination treatment improved
the cytotoxicity of DOX and decreased the migration, invasion
abilities of the tumor cells, and demonstrated synergistic effect
of anti-glioma in vitro.
In conclusion, DOX may promote expression of tumor
suppressors through demethylation to inhibit development of tumor
cells and miR-21 as microRNA in oncogenesis enhancing tumor cell
sensitivity to DOX so as to improve anticancer effect, which may
come into play at the post-transcriptional stage. Our study
unravels a new important element of DOX anticancer activity and
demonstrates its potential applications in demethylation. This
study provides novel insights into the mechanisms of
chemotherapeutic drug DOX with miRNA in regulation of tumor genes.
This will offer a strong rationale for therapeutic applications
involving cancer in the future. Further studies are under way to
investigate new delivery to co-deliver DOX and miRNA to overcome
blood-brain barrier (BBB) and enhance chemosensitivity.
Acknowledgements
This study was partially supported by the National
High Technology Research and Development Program 863 (2014AA021102
and 2012AA02A508), the China National Natural Scientific Fund
(81372703), and the Natural Science Foundation of Tianjin Municipal
Science and Technology Commission (12ZCDZSY17300).
References
|
1
|
Wilson TA, Karajannis MA and Harter DH:
Glioblastoma multiforme: state of the art and future therapeutics.
Surg Neurol Int. 5:642014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hande KR: Clinical applications of
anticancer drugs targeted to topoisomerase II. Biochim Biophys
Acta. 1400:173–184. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Swift LP, Rephaeli A, Nudelman A, Phillips
DR and Cutts SM: Doxorubicin-DNA adducts induce a non-topoisomerase
II-mediated form of cell death. Cancer Res. 66:4863–4871. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hanafy FM, Salem T, El-Aziz A, EL-Fiky B
and Shokair M: Influence of anticancer drugs on DNA methylation in
liver of female mice. Am J Mol Biol. 1:62–69. 2011. View Article : Google Scholar
|
|
5
|
Yu J, Zhang H, Gu J, et al: Methylation
profiles of thirty four promoter-CpG islands and concordant
methylation behaviours of sixteen genes that may contribute to
carcinogenesis of astrocytoma. BMC Cancer. 4:652004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Horiguchi K, Tomizawa Y, Tosaka M, et al:
Epigenetic inactivation of RASSF1A candidate tumor suppressor gene
at 3p21.3 in brain tumors. Oncogene. 22:7862–7865. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wiencke JK, Zheng S, Jelluma N, et al:
Methylation of the PTEN promoter defines low-grade gliomas and
secondary glioblastoma. Neuro Oncol. 9:271–279. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cankovic M, Mikkelsen T, Rosenblum ML and
Zarbo RJ: A simplified laboratory validated assay for MGMT promoter
hypermethylation analysis of glioma specimens from formalin-fixed
paraffin-embedded tissue. Lab Invest. 87:392–397. 2007.PubMed/NCBI
|
|
9
|
Bird A: DNA methylation patterns and
epigenetic memory. Genes Dev. 16:6–21. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vijayaraghavalu S and Labhasetwar V:
Efficacy of decitabine-loaded nanogels in overcoming cancer drug
resistance is mediated via sustained DNA methyltransferase 1
(DNMT1) depletion. Cancer Lett. 331:122–129. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Robert MF, Morin S, Beaulieu N, et al:
DNMT1 is required to maintain CpG methylation and aberrant gene
silencing in human cancer cells. Nat Genet. 33:61–65. 2003.
View Article : Google Scholar
|
|
12
|
Zhou W, Chen H, Hong X, Niu X and Lu Q:
Knockdown of DNA methyltransferase-1 inhibits proliferation and
derepresses tumor suppressor genes in myeloma cells. Oncol Lett.
8:2130–2134. 2014.PubMed/NCBI
|
|
13
|
Yokochi T and Robertson KD: Doxorubicin
inhibits DNMT1, resulting in conditional apoptosis. Mol Pharmacol.
66:1415–1420. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar :
|
|
15
|
Gaur AB, Holbeck SL, Colburn NH and Israel
MA: Downregulation of Pdcd4 by mir-21 facilitates glioblastoma
proliferation in vivo. Neuro Oncol. 13:580–590. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ziyan W, Shuhua Y, Xiufang W and Xiaoyun
L: MicroRNA-21 is involved in osteosarcoma cell invasion and
migration. Med Oncol. 28:1469–1474. 2011. View Article : Google Scholar
|
|
17
|
Kim N, Kim H, Jung I, Kim Y, Kim D and Han
YM: Expression profiles of miRNAs in human embryonic stem cells
during hepatocyte differentiation. Hepatol Res. 41:170–183. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gabriely G, Wurdinger T, Kesari S, et al:
MicroRNA 21 promotes glioma invasion by targeting matrix
metalloproteinase regulators. Mol Cell Biol. 28:5369–5380. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qiu LY and Bae YH: Self-assembled
polyethylenimine-graft-polyb(epsilon-caprolactone) micelles as
potential dual carriers of genes and anticancer drugs.
Biomaterials. 28:4132–4142. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheng D, Cao N, Chen J, Yu X and Shuai X:
Multifunctional nanocarrier mediated co-delivery of doxorubicin and
siRNA for synergistic enhancement of glioma apoptosis in rat.
Biomaterials. 33:1170–1179. 2012. View Article : Google Scholar
|
|
22
|
Jin W, Wu L, Liang K, Liu B, Lu Y and Fan
Z: Roles of the PI-3K and MEK pathways in Ras-mediated
chemoresistance in breast cancer cells. Br J Cancer. 89:185–191.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li B, Li J, Xu WW, et al: Suppression of
esophageal tumor growth and chemoresistance by directly targeting
the PI3K/AKT pathway. Oncotarget. 5:11576–11587. 2014.PubMed/NCBI
|
|
24
|
Oki E, Baba H, Tokunaga E, et al: Akt
phosphorylation associates with LOH of PTEN and leads to
chemoresistance for gastric cancer. Int J Cancer. 117:376–380.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Choe G, Horvath S, Cloughesy TF, et al:
Analysis of the phosphatidylinositol 3′-kinase signaling pathway in
glioblastoma patients in vivo. Cancer Res. 63:2742–2746.
2003.PubMed/NCBI
|
|
26
|
Huang PH, Mukasa A, Bonavia R, et al:
Quantitative analysis of EGFRvIII cellular signaling networks
reveals a combinatorial therapeutic strategy for glioblastoma. Proc
Natl Acad Sci USA. 104:12867–12872. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Martinez R and Esteller M: The DNA
methylome of glioblastoma multiforme. Neurobiol Dis. 39:40–46.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Davalos V, Moutinho C, Villanueva A, et
al: Dynamic epigenetic regulation of the microRNA-200 family
mediates epithelial and mesenchymal transitions in human
tumorigenesis. Oncogene. 31:2062–2074. 2012. View Article : Google Scholar :
|
|
29
|
Wang XF, Shi ZM, Wang XR, et al: MiR-181d
acts as a tumor suppressor in glioma by targeting K-ras and Bcl-2.
J Cancer Res Clin Oncol. 138:573–584. 2012. View Article : Google Scholar
|
|
30
|
Shi ZD, Qian XM, Zhang JX, et al: BASI, a
potent small molecular inhibitor, inhibits glioblastoma progression
by targeting microRNA-mediated beta-catenin signaling. CNS Neurosci
Ther. 20:830–839. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cong N, Du P, Zhang A, et al:
Downregulated microRNA-200a promotes EMT and tumor growth through
the wnt/beta-catenin pathway by targeting the E-cadherin repressors
ZEB1/ZEB2 in gastric adenocarcinoma. Oncol Rep. 29:1579–1587.
2013.PubMed/NCBI
|
|
33
|
Su J, Zhang A, Shi Z, et al: MicroRNA-200a
suppresses the Wnt/beta-catenin signaling pathway by interacting
with beta-catenin. Int J Oncol. 40:1162–1170. 2012.PubMed/NCBI
|
|
34
|
Kulis M and Esteller M: DNA methylation
and cancer. Adv Genet. 70:27–56. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Baylin SB and Herman JG: DNA
hypermethylation in tumorigenesis: epigenetics joins genetics.
Trends Genet. 16:168–174. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kato K, Long NK, Makita H, et al: Effects
of green tea poly-phenol on methylation status of RECK gene and
cancer cell invasion in oral squamous cell carcinoma cells. Br J
Cancer. 99:647–654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lopez-Serra P and Esteller M: DNA
methylation-associated silencing of tumor-suppressor microRNAs in
cancer. Oncogene. 31:1609–1622. 2012. View Article : Google Scholar :
|
|
38
|
Alakhova DY, Zhao Y, Li S and Kabanov AV:
Effect of doxorubicin/pluronic SP1049C on tumorigenicity,
aggressiveness, DNA methylation and stem cell markers in murine
leukemia. PLoS One. 8:e722382013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mutze K, Langer R, Schumacher F, et al:
DNA methyltransferase 1 as a predictive biomarker and potential
therapeutic target for chemotherapy in gastric cancer. Eur J
Cancer. 47:1817–1825. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Clements EG, Mohammad HP, Leadem BR, et
al: DNMT1 modulates gene expression without its catalytic activity
partially through its interactions with histone-modifying enzymes.
Nucleic Acids Res. 40:4334–4346. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nasonkin IO, Merbs SL, Lazo K, et al:
Conditional knockdown of DNA methyltransferase 1 reveals a key role
of retinal pigment epithelium integrity in photoreceptor outer
segment morphogenesis. Development. 140:1330–1341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xu M, Gao J, Du YQ, et al: Reduction of
pancreatic cancer cell viability and induction of apoptosis
mediated by siRNA targeting DNMT1 through suppression of total DNA
methyltransferase activity. Mol Med Rep. 3:699–704. 2010.
|
|
43
|
Kundakovic M, Chen Y, Costa E and Grayson
DR: DNA methyltransferase inhibitors coordinately induce expression
of the human reelin and glutamic acid decarboxylase 67 genes. Mol
Pharmacol. 71:644–653. 2007. View Article : Google Scholar
|
|
44
|
Mortusewicz O, Schermelleh L, Walter J,
Cardoso MC and Leonhardt H: Recruitment of DNA methyltransferase I
to DNA repair sites. Proc Natl Acad Sci USA. 102:8905–8909. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rajendran G, Shanmuganandam K, Bendre A,
Muzumdar D, Goel A and Shiras A: Epigenetic regulation of DNA
methyltransferases: DNMT1 and DNMT3B in gliomas. J Neurooncol.
104:483–494. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lujambio A, Ropero S, Ballestar E, et al:
Genetic unmasking of an epigenetically silenced microRNA in human
cancer cells. Cancer Res. 67:1424–1429. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bader AG, Brown D and Winkler M: The
promise of microRNA replacement therapy. Cancer Res. 70:7027–7030.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gao W, Shen H, Liu L, Xu J, Xu J and Shu
Y: MiR-21 over-expression in human primary squamous cell lung
carcinoma is associated with poor patient prognosis. J Cancer Res
Clin Oncol. 137:557–566. 2011. View Article : Google Scholar
|
|
49
|
Lakomy R, Sana J, Hankeova S, et al:
MiR-195, miR-196b, miR-181c,
miR-21expressionlevelsandO-6-methylguanine-DNA methyltransferase
methylation status are associated with clinical outcome in
glioblastoma patients. Cancer Sci. 102:2186–2190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shi Z, Zhang J, Qian X, et al: AC1MMYR2,
an inhibitor of dicer-mediated biogenesis of Oncomir miR-21,
reverses epithelial-mesenchymal transition and suppresses tumor
growth and progression. Cancer Res. 73:5519–5531. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Han L, Yue X, Zhou X, et al: MicroRNA-21
expression is regulated by beta-catenin/STAT3 pathway and promotes
glioma cell invasion by direct targeting RECK. CNS Neurosci Ther.
18:573–583. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang KL, Han L, Chen LY, et al: Blockage
of a miR-21/EGFR regulatory feedback loop augments anti-EGFR
therapy in glioblastomas. Cancer Lett. 342:139–149. 2014.
View Article : Google Scholar
|
|
53
|
Ren Y, Zhou X, Mei M, et al: MicroRNA-21
inhibitor sensitizes human glioblastoma cells U251 (PTEN-mutant)
and LN229 (PTEN-wild type) to taxol. BMC Cancer. 10:272010.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ren Y, Kang CS, Yuan XB, et al:
Co-delivery of as-miR-21 and 5-FU by poly(amidoamine) dendrimer
attenuates human glioma cell growth in vitro. J Biomater Sci Polym
Ed. 21:303–314. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Qian X, Ren Y, Shi Z, et al:
Sequence-dependent synergistic inhibition of human glioma cell
lines by combined temozolomide and miR-21 inhibitor gene therapy.
Mol Pharm. 9:2636–2645. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zavadil J, Narasimhan M, Blumenberg M and
Schneider RJ: Transforming growth factor-beta and microRNA: mRNA
regulatory networks in epithelial plasticity. Cells Tissues Organs.
185:157–161. 2007. View Article : Google Scholar
|
|
58
|
Han L, Yang Y, Yue X, et al: Inactivation
of PI3K/AKT signaling inhibits glioma cell growth through
modulation of beta-catenin-mediated transcription. Brain Res.
1366:9–17. 2010. View Article : Google Scholar : PubMed/NCBI
|