Introduction
The prognosis for lung cancer continues to remain
poor, and in stage I–III cases, this is largely due to drug
resistance leading to recurrence after adjuvant chemotherapy.
Energy-dependent rapid drug efflux and multidrug-resistant
molecules are known as factors involved in the resistance of
several types of cancer cells; however, drug retention does not
always correlate with its cytotoxicity, and therefore, there have
been many cases in which the mechanism of resistance remains
unknown. In addition, excluding specific gene mutations for
molecules such as epidermal growth factor receptor (EGFR), K-ras,
and EML/ALK4 in molecular-targeted therapy, markers to determine
drug sensitivity and to predict recurrence after chemotherapy are
yet to be identified. Therefore, it is urgent to seek universal
markers in which constitutive expression reflects drug resistance
in lung cancer.
Cancer cells have the capacity for self-renewal
through uncontrolled proliferation and dedifferentiation, similar
to embryonic stem (ES) cells (1).
Several molecules that are expressed during early embryonic
development are important in the maintenance of mouse ES cell
self-renewal (2–5). These molecules also generate and
maintain the ability of induced pluripotent stem (iPS) cells to
self-renew in mice and humans (6–8). Of
these molecules, Sall4 is a key factor in maintaining the
undifferentiated state and cell proliferation (9). Knockdown of Sall4 expression leads to
catastrophic ES-cell proliferation, and Sall4-knockout mice
do not survive until embryonic day 7 (9).
Sall4 is the mouse homolog of the Drosophila
homeotic gene spalt (sal) and is required for the
early development of the posterior head and anterior tail of
Drosophila (10).
Sal also regulates pattern formation and cell fate decisions
in the wing disc, trachea, and sensory organs. Mutations in the
human homolog SALL4 are known to cause Okihiro syndrome
(Duane-radial ray syndrome), characterized by limb deformities and
loss of eye movement (11,12). In some cases, anomalies of the
rectum, ear, heart, and kidney are also observed. The
SALL4/Sall4 gene is constitutively expressed in human
and mice CD34-positive hematopoietic stem cells (13). Interestingly, the overexpression of
Sall4 leads to leukemogenesis by increasing the number of
leukemic cells with markers for stem cells in 50% of transgenic
mice (13). In fact, SALL4
is overexpressed in various types of human hematopoietic
malignancies, such as acute myelocytic and lymphocytic leukaemia
(14,15).
Moreover, SALL4 upregulates the expression of the
oncogene Bmi-1 in human hematopoietic stem cells and
leukemic cells (16). Bmi-1
activates telomerase reverse transcriptase, thereby inducing
telomerase activity and leading to the transformation of human
non-cancerous epithelial cells (17). Bmi-1 also inhibits the function of
INK4a/ARF, usually by disturbing cyclin-dependent kinases 2, 4, and
6 (18), indicating that Bmi-1
expression leads to the progression of the cell cycle from G1 to
the S phase. Consequently, SALL4 expression may lead to
transformation of non-cancerous cells.
Previously, we found that SALL4 mRNA expression is
significantly higher in cancerous cells than in the non-cancerous
tissues of breast and lung cancer patients (19,20).
SALL4 is already highly expressed at the early-stage IA,
especially in lung cancer, suggesting that it has an essential role
in carcinogenesis (20).
SALL4 expression may characterize a feature of drug
resistance, which has been observed in the stem cell population;
evidence for this hypothesis has been demonstrated in a recent
study using leukemic cells (21).
However, the role of SALL4 in drug resistance has not yet
been reported in other cancers. Furthermore, the relationship
between SALL4 expression and prognosis, especially for
recurrence, remains unclear. Therefore, in this study, we examined
the effect of alteration of SALL4 expression on resistance
to anticancer drugs and analyzed the relationship between the
expression levels of SALL4 before chemotherapy and the
recurrence of lung cancer.
Materials and methods
Patients and tissue samples
Paraffin-embedded tissue samples from 31 lung
cancers (13 adenocarcinomas, 14 squamous cell carcinomas, and 4
small-cell lung cancers) were obtained after surgery. The tissue
samples were stained with hematoxylin/eosin and reviewed by
experienced pathologists. Clinicopathological factors and clinical
stages were evaluated on the basis of the tumor-node-metastasis
staging system.
Anticancer drugs
Cisplatin (CDDP), carboplatin (CBDCA), and
paclitaxel (PTX) were purchased from Sigma-Aldrich (St. Louis, MO,
USA). Each drug was dissolved in water or DMSO, and small aliquots
at high concentration were frozen at −40°C until use.
Cellculture
The human lung cancer cell line A549 was cultured in
Eagle’s minimal essential medium (MEM) and non-essential amino acid
solution (both from Sigma-Aldrich Japan K.K., Tokyo, Japan)
supplemented with 10% heat-inactivated fetal bovine serum (FBS)
(Invitrogen Life Technologies, Carlsbad, CA, USA). SBC-3 was
cultured in MEM (Sigma-Aldrich) supplemented with 10%
heat-inactivated FBS (Invitrogen Life Technologies). The cells were
grown at 37°C in a humidified atmosphere of 5% CO2.
RNA extraction and quantification of
SALL4 mRNA expression
Total RNA from the lung cancer samples in
formalin-fixed, paraffin-embedded tissues or from cancer cell lines
was extracted using an RNeasy FFPE isolation kit or RNeasy Plus
Mini kit (both from Qiagen, Tokyo, Japan) according to the
manufacturer’s instructions. The expression of SALL4 mRNA was
determined using quantitative reverse transcription-polymerase
chain reaction (RT-qPCR) according to the experimental procedure
described in our earlier report (19,20).
Western blotting
After the various treatments indicated in each
figure, the cells were harvested in a lysis buffer (50 mM Tris-HCl,
pH 8.0, 150 mM NaCl, 5 mM EDTA) containing a protease inhibitor
cocktail (Sigma-Aldrich), sonicated for 30 sec, and centrifuged at
15,000 rpm for 5 min. The supernatants were mixed with 1:2 volumes
of sample buffer (red loading buffer reagent; New England Biolabs,
Ipswich, MA, USA) and boiled for 2 min, and separated on a 4–20%
Tris-glycine gradient gel (Invitrogen Life Technologies) under
denaturing conditions. The proteins were electroblotted onto a
nitrocellulose membrane and reacted with antibody against SALL4
(ab29112; Abcam, Tokyo, Japan) or β-actin (monoclonal AC-15;
Sigma-Aldrich). Then, each protein was detected using ECL Prime
Western Blotting Detection Reagent (GE Healthcare Japan Corp.,
Tokyo, Japan), according to the manufacturer’s instructions. The
bands were visualized and imaged using ChemiDoc XRS Plus (Bio-Rad,
Hercules, CA, USA).
Microarray analysis
Total RNA from gene-transduced cells was prepared
and subjected to industrial analysis. Quality control was
performed, and global gene expression profiling was carried out by
Takara Bio, Inc. (Shiga, Japan) using the Agilent SurePrint G3
Human GE 8×60K Microarrays, as per the Agilent One-Color
Microarray-Based Gene Expression Analysis protocol (Agilent
Technologies, Inc., Santa Clara, CA, USA). The slides were scanned
using an Agilent Technologies Microarray Scanner, and the image
data were processed using Agilent Feature Extraction software,
version 10.10.1.1. The gene expression levels were compared after
global normalization. The data have been deposited at the National
Center for Biotechnology Information (NCBI) Gene Expression Omnibus
with the accession no. GSE56595.
Measurement of cell viability
Cells transduced with small inhibitory RNA (siRNA)
were plated in 96-well plates at a density of 500–1,000 cells/well
in the media supplemented with 10% FBS. The cells were allowed to
adhere for 24 h, and anticancer drugs were added after an
additional 24 h incubation. The culture plate was subjected to the
Cell Titer-Glo™ Luminescent Cell Viability assay (Promega Corp.,
Madison, WI, USA), according to the manufacturer’s instructions.
The level of ATP-derived luminescent signal, which correlates with
the number of viable cells, was measured using a Veritas™
Microplate Luminometer (Promega Corp.).
Cell cycle analysis
Cells plated onto 100-mm culture dishes (Costar™;
Corning, Inc., Corning, NY, USA) were trypsinized and washed with
FBS-free media and PBS. The cells were collected and fixed with 70%
methanol at 4°C. The cells were then treated with phosphate-citrate
buffer for 5 min. After centrifugation, the pellets were suspended
in 300 μl of 1% FBS-PBS and then treated with a final concentration
of 100 μg/ml RNase A and 10 μg/ml propidium iodide at room
temperature for 30 min. After staining, 20,000 cells/sample were
analyzed on a FACSCanto flow cytometer (Becton Dickinson Japan Co.,
Ltd., Tokyo, Japan).
SALL4 expression vector
The vector, inserted with the full-length
SALL4 gene (pCMV6-SALL4), was purchased from Origene
Technologies, Inc. (Rockville, MD, USA), and large-scale
preparation was performed using the competent cells. A control
vector (pCMV6-Mock) was constructed by digesting parental vector
using AsiSI (SfaAI) and MluI, resulting in a
lack of SALL4 sequence. Both the plasmids were transfected into
cells using a Nucleofector II device and a Cell Line Nucleofector
kit T (Lonza Japan, Ltd., Tokyo, Japan), according to the
manufacturer’s instructions.
Transduction of siRNA against SALL4
HP GenomeWide siRNA, designed to target the coding
region (exon 2) of SALL4 (GenBank accession no. NM-020436),
was purchased from Qiagen. Single-strand RNAs were annealed by
incubating each strand in the siRNA suspension buffer at 90°C for 1
min and then at 37°C for 1 h. Non-silencing control (NSC) RNA
(Qiagen) was used as a transduction control. The transduction of
siRNA was performed using the Nucleofector II device and the Cell
Line Nucleofector kit T (Lonza Japan, Ltd.), according to the
manufacturer’s instructions.
In brief, approximately 2–4×106 cells
were cultured under normal conditions to sub-confluency, and
1×106 cells were transduced with siRNA or NSC in a
cuvette. Next, 5×104 of these transduced cells were
plated on 3 ml of medium supplemented with 10% FBS in a 6-well
plate (Costar™; Corning, Inc.). After 24–48 h, the expression of
the silenced mRNA was quantified by TaqMan RT-PCR. The cells were
collected at different periods and subjected to the cell viability
assay and cell cycle analysis.
Statistical analysis
Data are presented as mean ± SD. Statistical
analysis was performed by paired Student’s t-test and the
Mann-Whitney rank sum test. P<0.05 was considered to be
significant.
Results
Effect of SALL4 siRNA transduction on
drug sensitivity
To determine the role of SALL4 expression as a
drug-resistant factor in cancer cells, we transduced SALL4
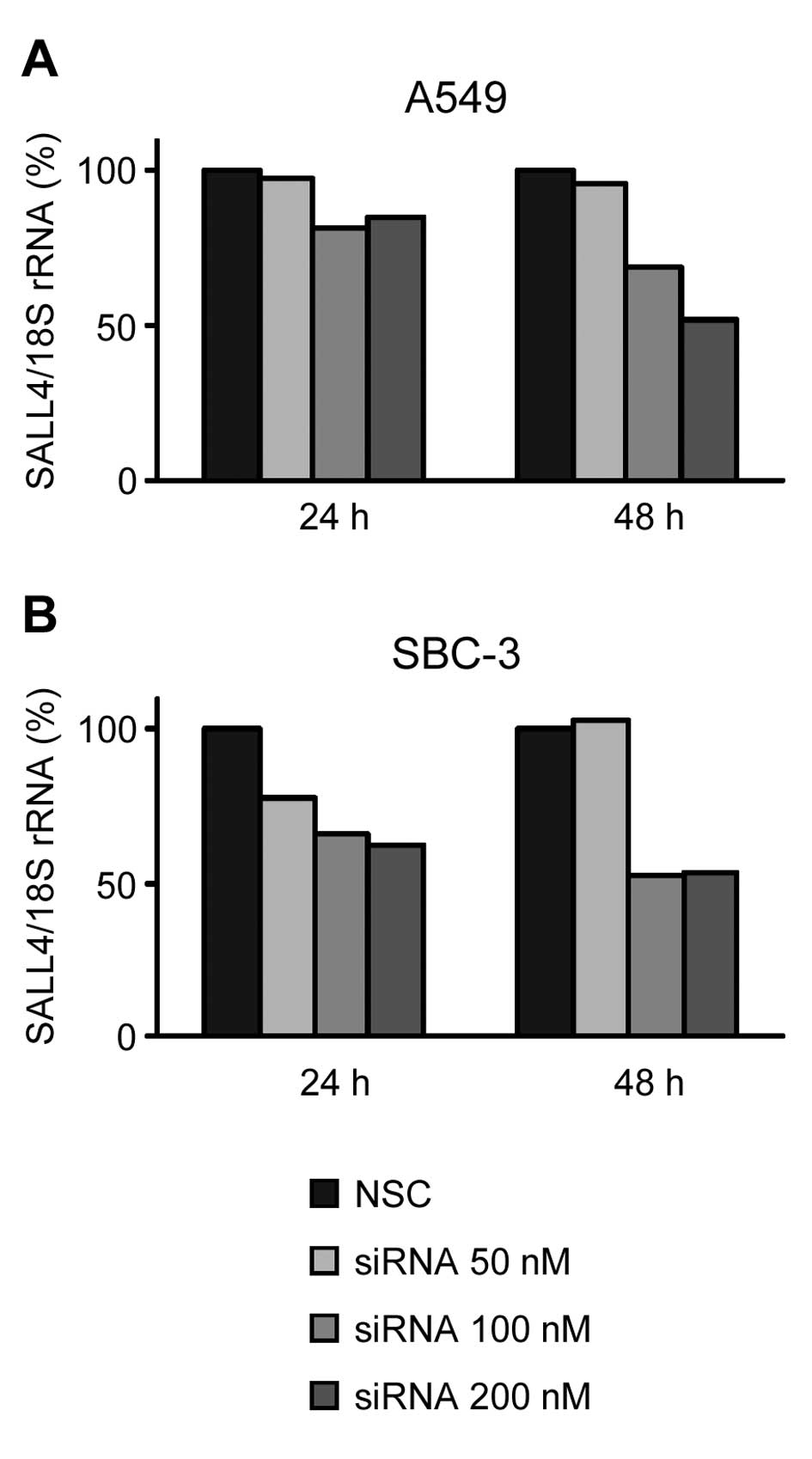
siRNA into lung cancer cells. In A549 and SBC-3 cells, the
transduction of siRNA decreased the SALL4 mRNA expression level to
~50% of the levels in NSC-transduced cells at 48 h after
transduction (Fig. 1A and B). To
use the cells with same extent of inhibitory effect on mRNA
expression, different siRNA concentrations (200 nM for A549 cells
and 100 nM for SBC-3 cells) were used. Next, we determined how the
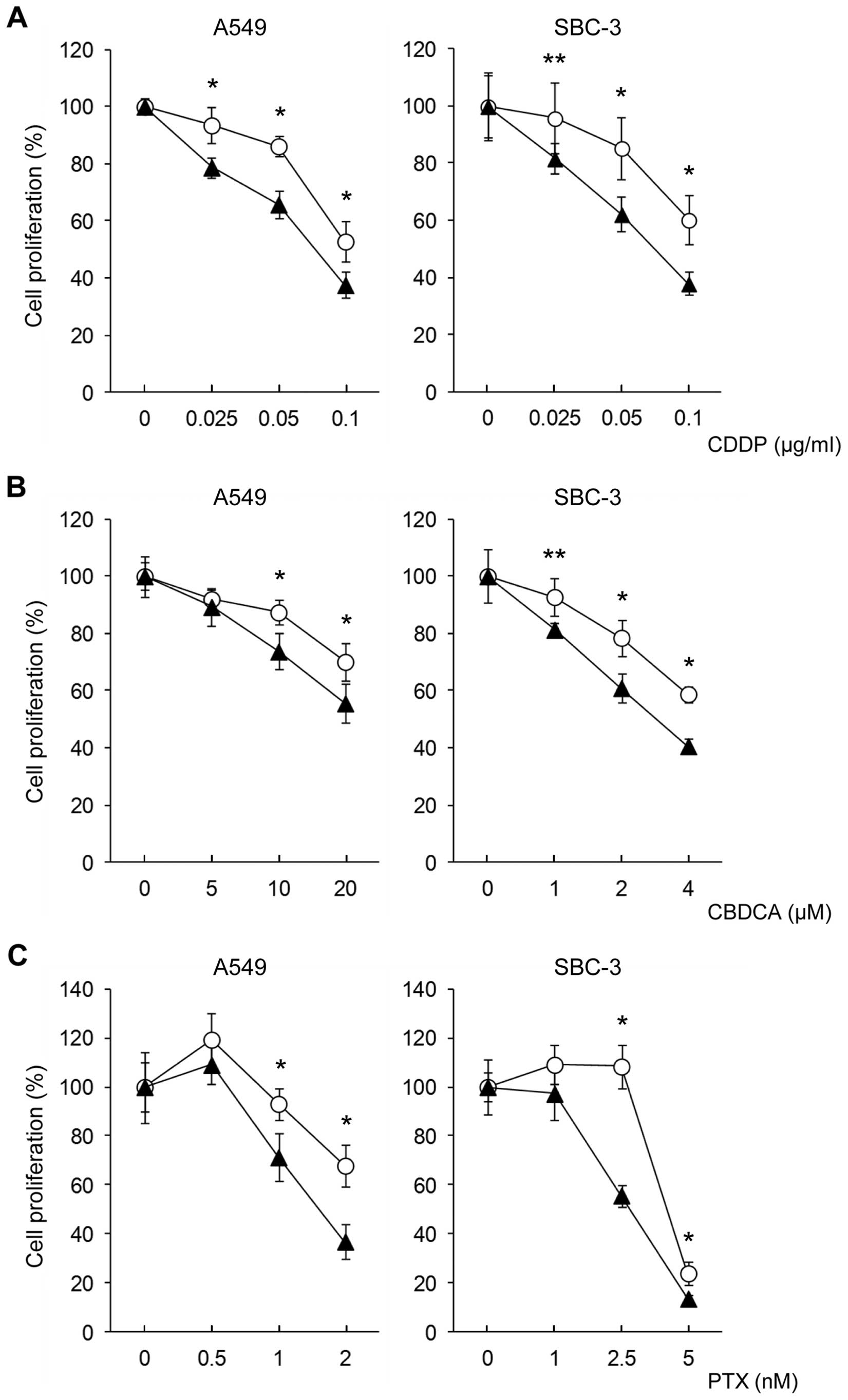
transduction of SALL4 siRNA affects drug sensitivity. We
observed a slight decrease in growth at 48 h after transduction of
NSC RNA and siRNA. After 48 h of siRNA transduction, the cells were
treated with each anticancer drug. After treatment with CDDP,
CBDCA, or PTX, the sensitivity of all anticancer drugs was
significantly increased in siRNA-transduced cells. Drug sensitivity
curves at day 4 after the treatment are shown in Fig. 2. The alteration of drug sensitivity
was observed to be most potent in the PTX-treated cells and was
also more potent in SBC-3 cells with relatively higher SALL4 mRNA
expression, confirmed before starting the experiments. After
treatment, the number of apparent apoptotic cells had not
increased, and arrested growth and relatively larger cells were
observed on day 4. In cell cycle analysis, the cell cycle pattern
was observed to be altered in the cells treated with anticancer
drugs after siRNA transduction; no increase in the sub-G1 fraction,
reflecting apoptotic cells, was observed in any of the treatments
(data not shown).
Microarray analysis of molecules
regulated by SALL4
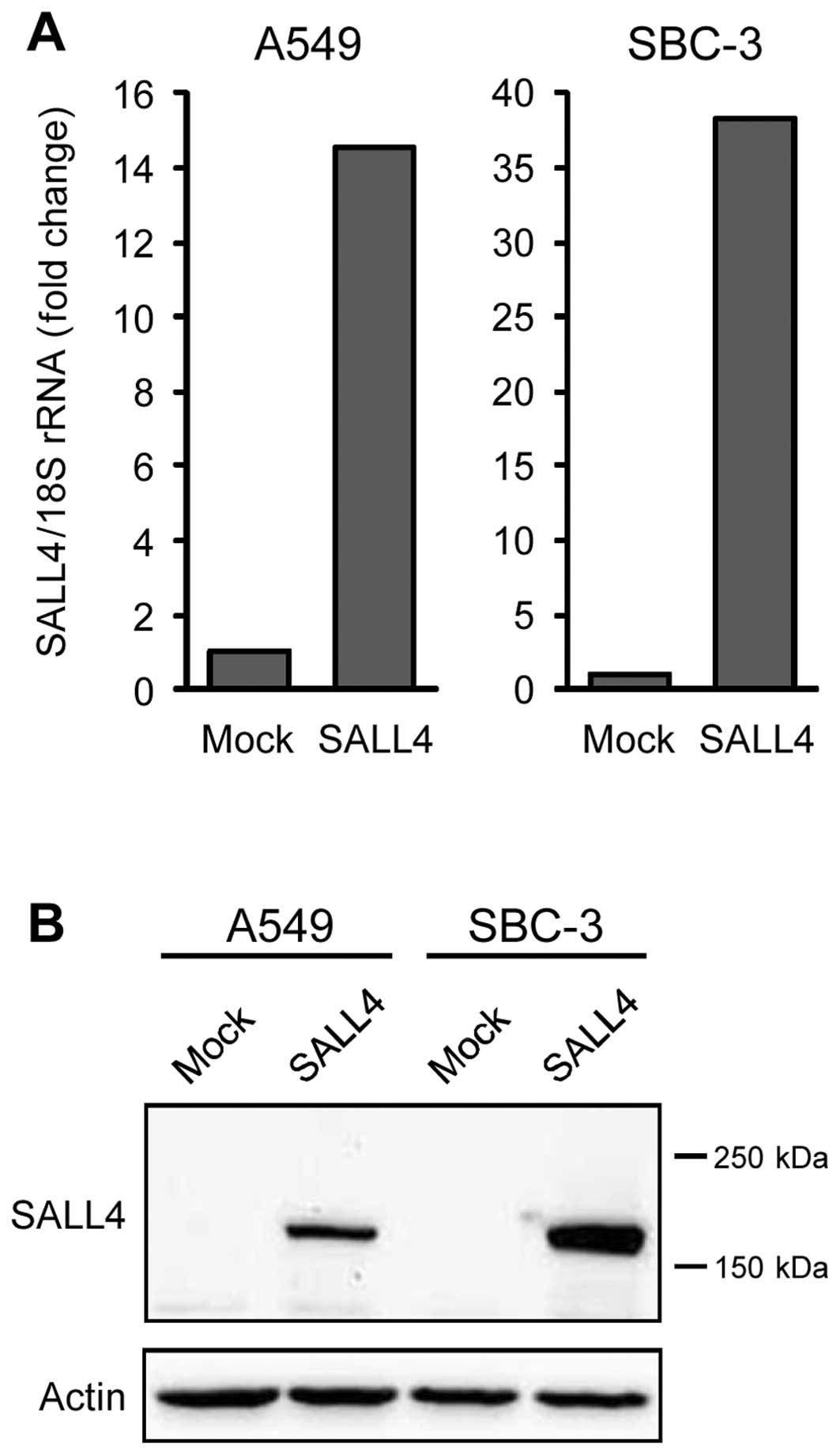
To examine the mechanism by which SALL4 regulates
drug sensitivity, ~65,000 molecules on a microarray chip were
analyzed using lung cancer cells transduced with a SALL4 expression
vector (pCMV6-SALL4). In these cells, SALL4 mRNA and protein
expression markedly increased at 24 h after transduction, unlike
that in the cells transduced with the control vector (pCMV6-Mock)
(Fig. 3A and B); these cells were
used for microarray analysis. Expression of numerous molecules was
altered in A549 and SBC-3 cells, and molecules showing >2-fold
change in expression in both cell types were taken to be candidate
molecules. Only four molecules, chorionic somatomammotropin
hormone-1/human placental lactogen (CSH-1/HPL), interleukin-6
(IL-6), transmembrane protein 229B (TMEM229B), and ameloblastin,
met this criterion (Table I).
 | Table IMolecules showing alteration of gene
expression >2-fold change both in A549 and SBC-3 cells by
microarray. |
Table I
Molecules showing alteration of gene
expression >2-fold change both in A549 and SBC-3 cells by
microarray.
| Molecules | Log2 ratio
(A549) | Log2 ratio
(SBC-3) | Major molecular
function |
|---|
| CSH-1/HPL | 4.81 (28.1)a | 8.22 (298.2) | Placental
development
Binding to mammary gland cell membrane |
| IL-6 | 1.43 (2.69) | 1.25 (2.38) | Growth of malignant
tumors
Transmission of survival signal via STAT3 |
| TMEM229B | 2.26 (4.78) | 2.61 (6.10) | Unknown |
| Ameloblastin | 2.26 (4.78) | 2.37 (5.17) | Enamel formation of
teeth |
Expression of SALL4 mRNA in lung
cancers
TaqMan RT-PCR was performed on cancerous tissue
samples obtained from patients with lung cancer. The patient
characteristics and clinical backgrounds are listed in Table II. First, the relationship between
SALL4 mRNA expression and the clinicopathological backgrounds were
analyzed (Table III). The mean
value was affected by one case showing extremely high expression;
however, gender, age, pathologic type, and pathologic stage showed
no statistically significant effect. On the contrary, in the group
showing recurrence of cancer after chemotherapy, the SALL4 mRNA
level was significantly higher than that in the group without
recurrence (Table III, p=0.031).
The mean SALL4 mRNA expression level in the group showing
recurrence (125.0±318.5) was 34-fold higher than that in the group
without recurrence (3.7±9.2) (Fig.
4). Even when four patients with small cell lung carcinoma were
excluded, the group with recurrence showed significantly higher
expression (139.8±341.4) compared to the expression (1.1±2.9) in
the group without recurrence (p<0.05). When the cut-off value
was set at the mean ± 2SD in cases without recurrence, cases
showing positive expression (8/15 cases) showed a shorter period
until recurrence (177±117 days) compared to that observed in the
seven negative cases (300±242 days). Even when two patients with
small cell lung carcinoma were excluded, the period until
recurrence was shorter (208±106 days) in the positive cases.
 | Table IIClinicopathologic backgrounds of the
patients with lung cancer. |
Table II
Clinicopathologic backgrounds of the
patients with lung cancer.
| R | No. | G | Age | Smoking | Stage | Type | Adjuvant
chemotherapy | Daysa | Daysb |
|---|
| (−) | 1 | M | 69 | Yes | IIIA | SCC | CDDP+PTX | N.A. | 874 |
| 2 | M | 64 | Yes | IIIA | SCC | CBDCA+PTX | N.A. | 746 |
| 3 | F | 71 | No | IIIA | Ad | CBDCA+PTX | N.A. | 802 |
| 4 | M | 56 | Yes | IIA | Ad | CBDCA+PTX | N.A. | 748 |
| 5 | M | 63 | Yes | IIB | SCC | CBDCA+PTX | N.A. | 660 |
| 6 | F | 66 | No | IA | Ad | UFT | N.A. | 562 |
| 7 | M | 65 | Yes | IA | SCLC | CDDP+CPT11 | N.A. | 1274 |
| 8 | M | 71 | Yes | IIA | SCLC | CPT11 | N.A. | 998 |
| 9 | M | 76 | Yes | IIIA | Ad | CBDCA+PEM | N.A. | 391 |
| 10 | M | 69 | Yes | IB | SCC | CBDCA+PTX | N.A. | 273 |
| 11 | M | 57 | Yes | IIIA | Ad | CBDCA+PEM | N.A. | 296 |
| 12 | M | 75 | Yes | IIIA | SCC | CBDCA+GEM | N.A. | 296 |
| 13 | F | 61 | No | IB | Ad | UFT | N.A. | 1372 |
| 14 | M | 71 | Yes | IIB | SCC | CBDCA+PTX | N.A. | 1405 |
| 15 | M | 66 | Yes | IIIA | SCC | CDDP+VNR | N.A. | 1645 |
| 16 | M | 71 | Unknown | IIB | SCC | CBDCA+PTX | N.A. | 1807 |
| (+) | 17 | F | 66 | Yes | IIIA | SCC |
CDDP/CBDCA+CPT11 | 275 | N.A. |
| 18 | F | 58 | Yes | IIA | Ad | CBDCA+PTX | 95 | N.A. |
| 19 | M | 67 | Yes | IB | SCLC | CBDCA+VP16 | 143 | N.A. |
| 20 | F | 54 | Yes | IIIA | Ad | CBDCA+PEM/PEM | 44 | N.A. |
| 21 | M | 67 | Yes | IIIA | SCC | CBDCA/PTX | 151 | N.A. |
| 22 | M | 68 | Yes | IIIA | SCLC | CBDCA+VP16 | 31 | N.A. |
| 23 | F | 58 | No | IIIB | Ad | CBDCA+PEM | 139 | N.A. |
| 24 | M | 62 | Yes | IIIA | SCC | CBDCA+DOC | 175 | N.A. |
| 25 | F | 74 | No | IIIA | Ad | CBDCA+PTX | 308 | N.A. |
| 26 | F | 51 | Yes | IIIA | Ad | CBDCA+PTX | 735 | N.A. |
| 27 | M | 59 | Yes | IIB | SCC | CDDP+VNR | 181 | N.A. |
| 28 | M | 72 | Yes | IIIA | SCC | CBDCA+PTX | 410 | N.A. |
| 29 | M | 59 | Yes | IA | SCC | CBDCA+PEM/VP16 | 411 | N.A. |
| 30 | F | 72 | Yes | IIB | Ad | CBDCA+PTX | 374 | N.A. |
| 31 | M | 49 | Yes | IIIA | Ad | CBDCA+PTX | 47 | N.A. |
 | Table IIIRelationship between SALL4 mRNA
expression and clinicopathologic backgrounds. |
Table III
Relationship between SALL4 mRNA
expression and clinicopathologic backgrounds.
| Factors | No. | Mean | SD | P-value |
|---|
| Gender |
| Male | 21 | 16.8 | 29.4 | 1.000 |
| Female | 10 | 158.1 | 391.9 | |
| Age |
| <60 years
old | 9 | 36.2 | 75.5 | 0.457 |
| ≥60 years old | 22 | 73.1 | 265.5 | |
| Pathologic
diagnosis |
| Ad | 13 | 25.1 | 64.1 | 0.207 |
| SCC | 14 | 107.5 | 332.0 | |
| SCLC | 4a | 25.8 | 12.1 | |
| Stage |
| I+II | 14 | 15.0 | 31.8 | 0.892 |
| III | 17 | 101.5 | 302.5 | |
| Prognosis |
| Desease-free | 16 | 3.7 | 9.2 | 0.031 |
| Reccurence | 15 | 125.0 | 318.5 | |
Discussion
We have previously reported that SALL4, a
gene essential for stem cell replication, showed an upregulated
expression in the cancerous cells than in the non-cancerous cells
in lung cancer patients (19,20).
However, its clinical significance, other than as a marker to
support the diagnosis of cancer, has not been determined. In the
present study, we report the first evidence that SALL4
expression could be a resistance factor against anticancer drugs in
lung cancer. The majority of recent studies investigating the
factors that determine drug sensitivity in lung cancer have focused
on the expression or mutation of target molecules such as EGFR,
K-ras, and EML4/ALK preceding molecular target therapy. However, no
markers that can be put to clinical use have yet been developed for
the factors regulating the sensitivity of conventional
chemotherapeutic drugs such as CDDP, CBDCA, and PTX. For example,
the anti-apoptotic molecule bcl-2 has been reported to produce
resistance to anticancer drugs when overexpressed in lung cancer
cells (22), leading to the
development of a bcl-2 inhibitor to increase drug sensitivity.
However, bcl-2 expression has not yet been sufficiently analyzed in
clinical samples, and therefore its significance as a prognostic
factor remains uncertain. Although the sample numbers are rather
small and a definitive conclusion cannot be reached, the results of
the present study clarify that SALL4 expression before
therapy tends to be higher in cases resulting in recurrence after
adjuvant chemotherapy. In addition, the results showed that the
period until recurrence is shorter in cases showing higher
SALL4 expression. These results indicate that measurement of
SALL4 expression may be useful to estimate the existence of
very small amounts of residual cancer cells, which cannot be
detected by conventional computed tomography (CT) or magnetic
resonance imaging (MRI), after surgery.
Comparison of recurring and non-recurring cases
showed no bias on the basis of the type of cancer drugs used; both
platinum drugs and taxol are widely used. Further, an in
vitro sensitivity test of SALL4 inhibition showed
altered sensitivity to all types of anticancer drugs. Taken
together with data from clinical samples, it appears that SALL4
might be a universal resistance factor against anticancer
drugs.
In the present study, in cases where SALL4
siRNA increased the sensitivity, the in vitro concentration
of anticancer drugs used was less than the concentration measurable
in the blood after administration of a clinical dose. This suggests
that SALL4 inhibition could augment cancer-cell sensitivity
to relatively lower concentrations of both platinum drugs and
taxol, of which dosage cannot be increased because of adverse
effects, such as gastrointestinal toxicity, renal toxicity, and
decreased platelet counts. In addition, both non-small cell
carcinoma of A549 cells and small cell SBC-3 cells showed increased
drug sensitivity, suggesting that SALL4 could be target molecules
for augmenting drug sensitivity, regardless of the cancer type.
A previous study has shown that the drug
transporters ABCG2 and ABCA3 can be induced by SALL4 in leukemic
cells (21). However, our
microarray analysis using SALL4-overexpressed lung cancer cells did
not reveal any alteration in the expression levels of either of
these mRNAs (data not shown). In this experiment, four molecules
showed elevated expression in both examined cell lines. In these
candidates, molecular function of CSH-1/HPL, TMEM229B, and
ameloblastin in cancer cells have not been previously reported.
Only IL-6 is known to transmit growth and survival signals via
STAT3 activation in lung cancer cells, and SALL4 expression is
reported to be upregulated by STAT3 (23–25),
suggesting that the STAT3 pathway may be involved in drug
resistance and that a positive-feedback loop may exist between
SALL4 and IL-6 via STAT3. Further study may prove this speculation
and clarify the molecular significance of CSH-1/HPL, TMEM229B, and
ameloblastin.
Molecules regulating stem cell replication are known
to show reciprocal augmentation of gene expression. SALL4 and
Nanog, a factor which maintains the undifferentiated state of stem
cells, have been reported to show reciprocally augmented gene
expression in mouse ES cells (26). In the preliminary experiment, we
recently found that SALL4 expression vector-transduced cells showed
an increase in Nanog mRNA expression (data not shown). The
molecular function of Nanog in cancer cells remains unclear, but it
is possible that SALL4 and Nanog cooperatively form a fundamental
feature for maintaining the undifferentiated state and promotion of
cell proliferation.
The results of the present study clarify that SALL4
acts as a constitutive resistance factor against anticancer drugs
and suggest that recurrence after adjuvant chemotherapy could be
predicted in lung cancer cases showing overexpression of
SALL4 before therapy. In addition, because both the
combination of SALL4 inhibition and anticancer drugs, and
siRNA alone can produce remarkable inhibition of cell proliferation
in some cell lines (20), SALL4
shows promise as a novel therapeutic target.
The involvement of SALL4 in acquired resistance
remains unclear. We preliminarily examined SALL4 expression
in cells cultured under conditions of step-wise increase of CDDP
concentration. SALL4 expression increased for several weeks
but then decreased to the level of constitutive expression. This
observation suggests the involvement of SALL4 in stress
response, but its significance as a factor for acquired resistance
remains to be investigated.
Acknowledgements
This study was supported by the grants from Japan
Society for the Promotion of Science (Tokyo, Japan).
References
|
1
|
Pardal R, Molofsky AV, He S and Morrison
SJ: Stem cell self-renewal and cancer cell proliferation are
regulated by common networks that balance the activation of
proto-oncogenes and tumor suppressors. Cold Spring Harb Symp Quant
Biol. 70:177–185. 2005. View Article : Google Scholar
|
|
2
|
Yasuda SY, Tsuneyoshi N, Sumi T, Hasegawa
K, Tada T, Nakatsuji N and Suemori H: NANOG maintains self-renewal
of primate ES cells in the absence of a feeder layer. Genes Cells.
11:1115–1123. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou Q, Chipperfield H, Melton DA and Wong
WH: A gene regulatory network in mouse embryonic stem cells. Proc
Natl Acad Sci USA. 104:16438–16443. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang J, Tam WL, Tong GQ, et al: Sall4
modulates embryonic stem cell pluripotency and early embryonic
development by the transcriptional regulation of Pou5f1. Nat Cell
Biol. 8:1114–1123. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang J, Chan YS, Loh YH, et al: A core
Klf circuitry regulates self-renewal of embryonic stem cells. Nat
Cell Biol. 10:353–360. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Okita K, Ichisaka T and Yamanaka S:
Generation of germline- competent induced pluripotent stem cells.
Nature. 448:313–317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tsubooka N, Ichisaka T, Okita K, Takahashi
K, Nakagawa M and Yamanaka S: Roles of Sall4 in the generation of
pluripotent stem cells from blastocysts and fibroblasts. Genes
Cells. 14:683–694. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu J, Vodyanik MA, Smuga-Otto K, et al:
Induced pluripotent stem cell lines derived from human somatic
cells. Science. 318:1917–1920. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sakaki-Yumoto M, Kobayashi C, Sato A, et
al: The murine homolog of SALL4, a causative gene in Okihiro
syndrome, is essential for embryonic stem cell proliferation, and
cooperates with Sall1 in anorectal, heart, brain and kidney
development. Development. 133:3005–3013. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jürgens G: Head and tail development of
the Drosophila embryo involves spalt, a novel homeotic gene. EMBO
J. 7:189–196. 1998.
|
|
11
|
Al-Baradie R, Yamada K, St Hilaire C, et
al: Duane radial ray syndrome (Okihiro syndrome) maps to 20q13 and
results from mutations in SALL4, a new member of the SAL family. Am
J Hum Genet. 71:1195–1199. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kohlhase J, Heinrich M, Liebers M, et al:
Cloning and expression analysis of SALL4, the murine homologue of
the gene mutated in Okihiro syndrome. Cytogenet Genome Res.
98:274–277. 2002. View Article : Google Scholar
|
|
13
|
Ma Y, Cui W, Yang J, et al: SALL4, a novel
oncogene, is constitutively expressed in human acute myeloid
leukemia (AML) and induces AML in transgenic mice. Blood.
108:2726–2735. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cui W, Kong NR, Ma Y, Amin HM, Lai R and
Chai L: Differential expression of the novel oncogene, SALL4, in
lymphoma, plasma cell myeloma, and acute lymphoblastic leukemia.
Mod Pathol. 19:1585–1592. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang J, Chai L, Gao C, et al: SALL4 is a
key regulator of survival and apoptosis in human leukemic cells.
Blood. 112:805–813. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang J, Chai L, Liu F, et al: Bmi-1 is a
target gene for SALL4 in hematopoietic and leukemic cells. Proc
Natl Acad Sci USA. 104:10494–10499. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dimri GP, Martinez JL, Jacobs JJ, et al:
The Bmi-1 oncogene induces telomerase activity and immortalizes
human mammary epithelial cells. Cancer Res. 62:4736–4745.
2002.PubMed/NCBI
|
|
18
|
Liu L, Andrews LG and Tollefsbol TO: Loss
of the human polycomb group protein BMI1 promotes cancer-specific
cell death. Oncogene. 25:4370–4375. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kobayashi D, Kuribayashi K, Tanaka M and
Watanabe N: SALL4 is essential for cancer cell proliferation and is
overex-pressed at early clinical stages in breast cancer. Int J
Oncol. 38:933–999. 2011.PubMed/NCBI
|
|
20
|
Kobayashi D, Kuribayashi K, Tanaka M and
Watanabe N: Overexpression of SALL4 in lung cancer and its
importance in cell proliferation. Oncol Rep. 26:965–970.
2011.PubMed/NCBI
|
|
21
|
Jeong HW, Cui W, Yang Y, Lu J, He J, Li A,
Song D, Guo Y, Liu BH and Chai L: SALL4, a stem cell factor,
affects the side population by regulation of the ATP-binding
cassette drug transport genes. PLoS One. 6:e183722011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Abou El Hassan MA, Mastenbroek DC,
Gerritsen WR, Giaccone G and Kruyt FA: Overexpression of Bcl2
abrogates chemo- and radiotherapy-induced sensitisation of NCI-H460
non-small-cell lung cancer cells to adenovirus-mediated expression
of full-length TRAIL. Br J Cancer. 91:171–177. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yi H, Cho HJ, Cho SM, Jo K, Park JA, Kim
NH, Amidon GL, Kim JS and Shin HC: Blockade of interleukin-6
receptor suppresses the proliferation of H460 lung cancer stem
cells. Int J Oncol. 41:310–316. 2012.PubMed/NCBI
|
|
24
|
Song L, Rawal B, Nemeth JA and Haura EB:
JAK1 activates STAT3 activity in non-small-cell lung cancer cells
and IL-6 neutralizing antibodies can suppress JAK1-STAT3 signaling.
Mol Cancer Ther. 10:481–494. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bard JD, Gelebart P, Amin HM, Young LC, Ma
Y and Lai R: Signal transducer and activator of transcription 3 is
a transcriptional factor regulating the gene expression of SALL4.
FASEB J. 23:1405–1414. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu Q, Chen X, Zhang J, et al: Sall4
interacts with Nanog and co-occupies Nanog genomic sites in
embryonic stem cells. J Biol Chem. 281:24090–24094. 2006.
View Article : Google Scholar : PubMed/NCBI
|