Introduction
Cervical cancer is a potentially preventable
disease; however, it is the third most commonly diagnosed cancer
and the fourth leading cause of cancer deaths in women worldwide,
accounting for 9% (529,800) of the new cancer cases and 8%
(275,100) of the cancer deaths among women in 2008 (1–3).
More than 85% of these cases and deaths occur in developing
countries, including China (1–3).
Cervical cancer is thought to develop through a multistep process
involving virus, tumor suppressor genes, proto-oncogenes and
immunological factors (4,5). It is known that human papillomavirus
(HPV) infection is necessary, but insufficient to cause malignancy
indicating the importance of other factors for malignant conversion
of high-grade HPV infection (6–9). Key
events that drive cancer are influenced by a multitude of factors
that still remain to be understood (10–12).
The etiology of cervical carcinoma remains poorly understood.
The FOS-like antigen-1 (Fra-1) is a member of the
FOS transcription factor family playing important roles in
transformation, proliferation, and metastasis (13–18).
Fra-1 is extensively phosphorylated in response to serum mitogens
or insulin in normal cell types, or in response to oncogenic RAS in
transformed thyroid lines (19–22).
In addition, the extent of Fra-1 phosphorylation is cell cycle
regulated, being further increased in the G2/M cell fraction
(13,23–25).
The results obtained from various studies show different
implications for Fra-1 according to tumor type. Fra-1
overexpression is predominantly associated with a large variety of
epithelial tumors, including thyroid, breast, lung, brain,
nasopharyngeal, esophageal, endometrial, prostate and colon
carcinomas, along with glioblastomas and mesotheliomas (26,27).
Fra-1 is downregulated in the tumorigenic cell lines CGL3 and HeLa
compared to the non-tumorigenic 444 cells. It inhibits the
tumorigenicity of cervical carcinoma cell lines (28). Fra-1 has tumor-suppressing function
upon micro-cell transfer in HPV-16- and HPV-18-positive cervical
carcinoma cells (29). Thus, it is
urgent to explore the relationship between Fra-1 and cervical
carcinoma.
Tumor suppressor p53 is the central component of a
system maintaining the genetic stability of animal and human
somatic cells (30–33). One of the important functions of
p53 is to recognize when DNA damage has occurred in a cell and
arrest the growth of that cell in the G1 period of the cell cycle
to allow for DNA repair or, if repair is not possible, to lead that
cell into cell-mediated death or suicide, called apoptosis
(32–35). The p53 gene plays the key
role in maintaining the genetic homogeneity of somatic cells and is
most often affected in cancer (32–37).
We examined the expression levels of Fra-1 and the
key molecules of p53 signaling pathway in cervical cancer tissues.
At the same time, the effects and possible mechanism of Fra-1 were
studied in a cervical cancer cell line.
Materials and methods
Cell culture
A human HeLa cervical cancer cell line was cultured
in DMEM supplemented with 10% fetal bovine serum (FBS) (Gibco by
Life Technologies™, Grand Island, NY, USA), 100 U/ml penicillin and
100 μg/ml streptomycin at 37°C in the presence of 5%
CO2.
Tumor samples
Twenty participants were recruited at the Third
Xiangya Hospital, Central South Uuniversity (Hunan, China). Consent
forms were obtained from individual patients, and experimental
protocols were approved by the Institutional Review Board of the
Third Xiangya Hospital. At the Third Xiangya Hospital, 20
participants were women with histologically confirmed cervical
cancer (Table I). All subjects
enrolled in the study were Chinese. Cervical cancer tissue and
corresponding non-tumor normal tissue were collected, and each
biopsy sample was divided into two sections, one was submitted to
routine histological diagnosis, and the remaining section was
evaluated by qPCR and western blotting.
 | Table ICharacteristics of cervical cancer
patients. |
Table I
Characteristics of cervical cancer
patients.
| Samples | Age (years) | HPV type | Histological
diagnose | Stagea |
|---|
| 1 | 43 | 33, 58 | Cervical poorly
differentiated squamous cell cancer | Ib1 |
| 2 | 39 | 16 | Cervical
intermediately differentiated squamous cell cancer | IIa1 |
| 3 | 42 | 16 | Cervical
intermediately differentiated squamous cell cancer | IIb |
| 4 | 45 | (−) | Cervical poorly
differentiated squamous cell cancer | Ib1 |
| 5 | 60 | 16 | Cervical
intermediately differentiated squamous cell cancer | IIb |
| 6 | 60 | 16 | Cervical
intermediately differentiated squamous cell cancer | IIb |
| 7 | 70 | 16 | Cervical
intermediately differentiated squamous cell cancer | IIa1 |
| 8 | 49 | (−) | Cervical
intermediately differentiated squamous cell cancer | IIa2 |
| 9 | 37 | 16, 58 | Cervical
intermediately differentiated squamous cell cancer | IIa1 |
| 10 | 44 | 16 | Cervical
intermediately differentiated squamous cell cancer | IIa2 |
| 11 | 46 | 52 | Cervical
intermediately differentiated squamous cell cancer | IIb |
| 12 | 42 | (−) | Cervical
intermediately differentiated squamous cell cancer | IIa2 |
| 13 | 43 | 45 | Cervical
intermediately differentiated squamous cell cancer | IIa2 |
| 14 | 61 | 16 | Cervical
intermediately differentiated squamous cell cancer | IIb |
| 15 | 36 | 59 | Cervical poorly
differentiated squamous cell cancer | Ib1 |
| 16 | 36 | 59 | Cervical poorly
differentiated squamous cell cancer | Ib1 |
| 17 | 57 | 16 | Cervical poorly
differentiated squamous cell cancer | Ib1 |
| 18 | 66 | 16, 33 | Cervical
intermediately differentiated squamous cell cancer | IIb |
| 19 | 43 | 18, 35 | Cervical poorly
differentiated squamous cell cancer | IIa1 |
| 20 | 43 | 45 | Cervical
intermediately differentiated squamous cell cancer | IIa2 |
RNA extraction and quantitative real-time
PCR
Total RNA was extracted from the biopsy samples with
RNeasy® kit (Qiagen, Carlsbad, CA, USA) according to the
manufacture’s instructions. The total RNA sample (1 μg) was used to
generate cDNA. Reverse transcription was carried out as described
previously (38–42). After the RT reaction, the PCR
reaction was preceded by 94°C for 5 min, then 30 cycles for Fra-1
of 94°C for 45 sec, 55°C for 45 sec, and 72°C for 1 min followed by
72°C for 7 min. All RT-PCR reactions were repeated at least three
times at different number of extension cycles to avoid false
results of the PCR. Glyceraldehyde 3-phosphate dehydrogenase
(GAPDH) was used as an endogenous control for normalization. The
sequences of the primers used for RT-PCR were as follows: Fra-1
forward, 5′-cgaaggccttgtgaacagat-3′ and reverse,
5′-cttctgcttctgcagctcct-3′; GAPDH forward,
5′-cgaccactttgtcaagctca-3′ and reverse, 5′-actgagtgtggcagggactc-3′.
Expression of mRNA was assessed by evaluating cycle threshold (CT)
values. The CT values were normalized with the expression levels of
GAPDH and the relative amount of mRNA specific to each of the
target genes was calculated using the 2−ΔΔCT method
(42,43).
Immunohistochemistry (IHC) and evaluation
of staining
IHC was done using the peroxidase-anti-peroxidase
technique following a microwave antigen retrieval procedure.
Antibody for Fra-1 was purchased from ImmunoWay
Biotechnology Co. (Newark, DE, USA). Antibody against Fra-1
(1:100) was overlaid on cervical cancer and corresponding non-tumor
normal tissue sections and incubated overnight at 4°C. Secondary
antibody incubation (Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA) was performed at room temperature for 30 min.
Sections were blindly evaluated by two investigators
in an effort to provide a consensus on staining patterns by light
microscopy (Olympus, Tokyo, Japan). Fra-1 staining was
assessed according to the methods described by Hara and Okayasu
(44) with minor modifications.
Each case was rated according to a score that added a scale of
intensity of staining to the area of staining. At least 10
high-power fields were chosen randomly, and >1,000 cells were
counted for each section. The intensity of staining was graded on
the following scale: 0, no staining; 1+, mild staining; 2+,
moderate staining; 3+, intense staining. The area of staining was
evaluated as follows: 0, no staining of cells in any microscopic
fields; 1+, <30% of tissue stained positive; 2+, 30–60% stained
positive; 3+, >60% stained positive. The minimum score when
summed (extension + intensity) was, therefore, 0, and the maximum,
6. A combined staining score (extension + intensity) of ≤ 2 was
considered to be a negative staining (low staining); 3–4, a
moderate staining; and 5–6, a strong staining.
Construction of pEGFP-N1-Fra-1 vector and
cell transfection
The pEGFP-N1-Fra-1 plasmid constructed to target
Fra-1 (RefSeq ID: NM_001300844.1) was obtained from Shanghai
Genechem Co., Ltd. (Shanghai, China). pEGFP-N1 plasmid (Shanghai
Genechem Co., Ltd.) was cut with EcoRI/BamHI and
ligated by T4 DNA ligase with gene encoding Fra-1, making the
Fra-1-pEGFP construct. The fusion sequences were verified by DNA
sequencing using ABI 3730. The empty pEGFP-N1 vector was used as a
negative control.
To establish a stable Fra-1-expressing cell line,
the plasmid pEGFP-N1/Fra-1 or control empty vector pEGFP-N1 was
transfected into HeLa cells, using Lipofectamine (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer’s
instructions, followed by G418 selection. The stable transfectants,
HeLa/Fra-1 and HeLa/vector, were isolated and the transcription of
Fra-1 protein was determined by western blot experiments.
Cell proliferation assay
The impact of Fra-1 on HeLa cell proliferation was
measured by MTT assay as described previously (34). Briefly, HeLa cells (HeLa,
HeLa/vector, and HeLa/Fra-1 cells) (104 cells/well) were
cultured in triplicate with 10% FCS DMEM in 96-well plates,
respectively. The cells were then exposed to 5 mg/ml MTT for 4 h.
The generated formazan was dissolved with dimethyl sulfoxide and
measured at 570 nm using an ELx800 Microplate Reader (BioTek
Instruments, Inc., Winooski, VT, USA).
The effect of Fra-1 to cervical cancer
cell apoptosis
Cell apoptosis was analyzed by flow cytometry
analysis using a MoFlo™ XDP High-Performance Cell Sorter (Beckman
Coulter, Miami, FL, USA) PI and Hoechst 33342 double staining
(Nanjing KeyGen Biotech., Co., Ltd., Jiangsu, China). Briefly, HeLa
cells (HeLa, HeLa/vector, and HeLa/Fra-1 cells) were seeded at a
density of 3×105 cells/well in 24-well culture plates.
Cells were collected in an Eppendorf tube 24 h and washed twice
with PBS by centrifugation. The supernatants were discarded. To
detect apoptosis, 500 μl PBS, 5 μl Hoechst 33342 and 5 μl PI were
added to each tube, and the contents of the tube were mixed in the
dark, at room temperature for 15 min, followed by FCM testing. The
data acquired were analyzed with Summit v5.2 software.
Western blotting
Proteins of the biopsy samples were prepared by
lysis buffer. The protein concentrations were determined using the
Bicinchoninic Acid Protein Assay method (Pierce Biotechnology,
Rockford, IL, USA). Extracts containing 50 μg of proteins were
separated in 10% SDS-PAGE gels and electroblotted onto
nitrocellulose membrances (HyClone Laboratories, Inc., Logan, UT,
USA). The membranes were blocked using Tris-buffered
saline/Tween-20 (25 mM Tris-HCl, 150 mM NaCl, pH 7.5, and 0.05%
Tween-20) containing 5% non-fat milk followed by overnight
incubation at 4°C with primary antibodies (rabbit anti-Fra-1
antibody, 1:300, ImmunoWay Biotechnology Co.; rabbit anti-MDM2
antibody, 1:200, and rabbit anti-p53 antibody, 1:200, Wuhan Boster
Biological Technology, Ltd., Hubei, China). After three washes,
secondary antibodies (anti-horseradish peroxidase antibodies,
1:2,000; Santa Cruz Biotechnology, Inc.) were added, and incubated
for 1 h. Then anti-GAPDH antibody (1:3,000; Santa Cruz
Biotechnology, Inc.) was used as a loading control.
Statistical analysis
Differences of non-parametric variables were
analyzed by the Fisher’s exact test using EPI software (EPI Info,
version 3.2.2, www.CDC.gov/epiinfo/). Differences of
the quantitative variables between groups were analyzed by
Student’s t-test using SPSS 13.0 program (SPSS, Inc., Chicago, IL,
USA). P<0.05 was considered statistically significant.
Results
Detection of mRNA expression levels of
Fra-1 gene in cervical cancer
To detect the mRNA expression levels of Fra-1
gene in cervical cancer and the adjacent non-cancerous tissues, we
chose 20 cervical cancer tissues and the adjacent non-cancerous
tissues to perform real-time quantitative RT-PCR of Fra-1
genes. Sample spreadsheet of data analysis was constructed by the
2−ΔΔCT method. The fold change in the expression of the
Fra-1 gene relative to the internal control gene
(GAPDH) was studied. The expression of Fra-1 gene was
downregulated in cervical cancer (Table II). Compared with the control
samples, the normalized Fra-1 gene expression in cervical
cancer was 0.32 times, 95% confidence interval (CI) was
0.22–0.48.
 | Table IIIdentification of the mRNA expression
level of Fra-1 in cervical cancer and adjacent non-cancerous
tissues by qPCR. |
Table II
Identification of the mRNA expression
level of Fra-1 in cervical cancer and adjacent non-cancerous
tissues by qPCR.
| Gene | Sample | No. | Fra-1 CT (mean ±
SD) | GAPDH CT (mean ±
SD) | ΔCT (mean ±
SD) | Δ ΔCT (mean ±
SD) | Folda |
|---|
| Fra-1 | Cervical
cancer | 20 | 32.70±1.37 | 19.08±0.79 | 13.62±0.51 | 1.61±0.56 | 0.32
(0.22–0.48) |
| Non-cancerous
tissues | 20 | 33.08±1.65 | 20.07±0.84 | 12.01±0.45 | 1.61±0.56 |
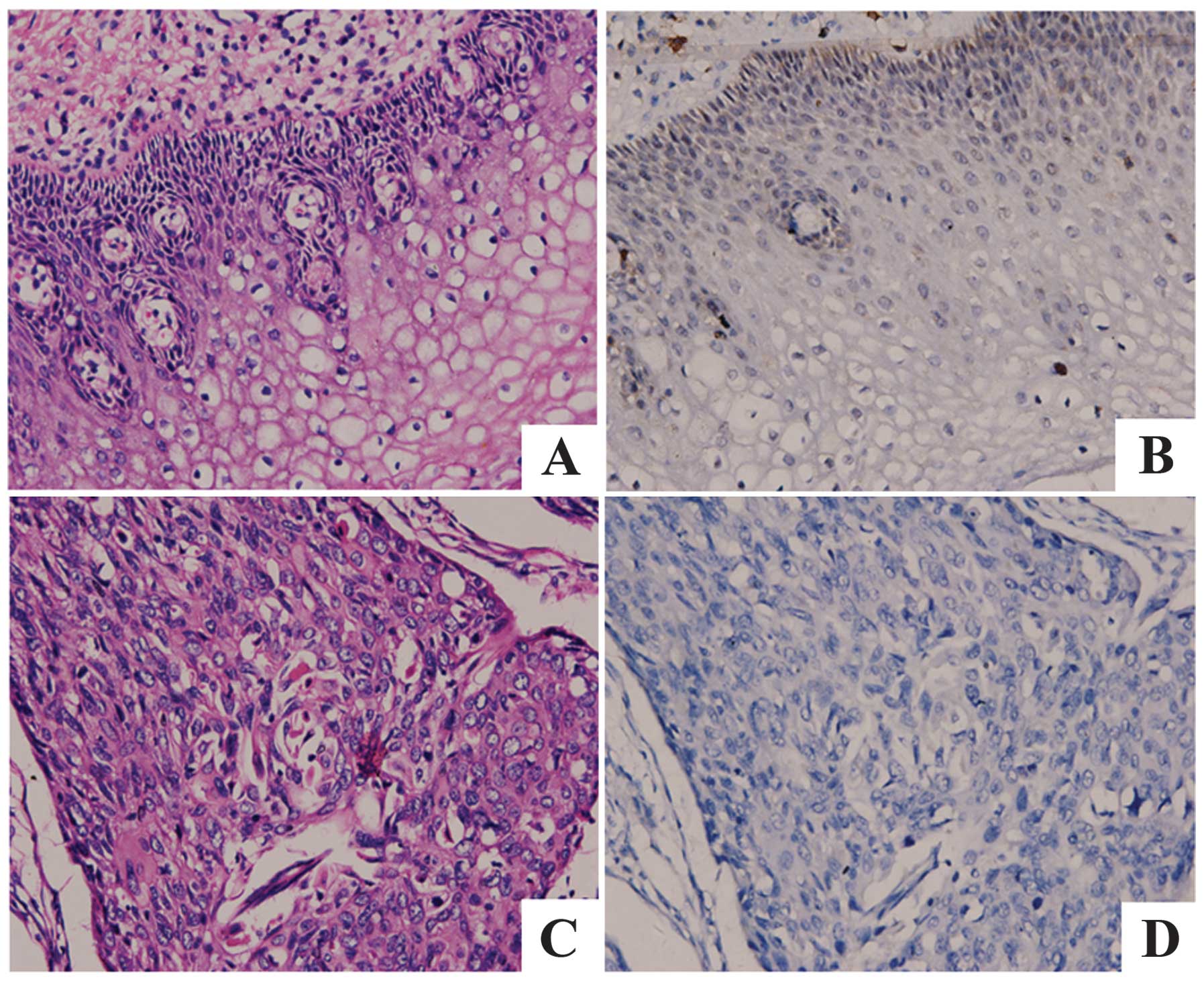
IHC analysis of protein expression levels
of Fra-1 in cervical cancer
IHC was carried out with antibodies against Fra-1
protein in cervical cancer and the adjacent non-cancerous tissues.
Fra-1 was identified as differentially expressed between cervical
cancer tissues versus the adjacent non-cancerous tissues. IHC
showed a similar pattern in protein expression with RT-qPCR
results. There was 10.0% (2/10) high score of Fra-1 in cervical
cancer tissues and 45% (9/20) in the adjacent non-cancerous
tissues. The distribution of low score was 65.0% (13/20) and 15.0%
(3/20) in cervical cancer and the adjacent non-cancerous tissues,
respectively (p=0.004 <0.05) (Fig.
1 and Table III).
 | Table IIIThe difference of Fra-1 expression
between cervical cancer and the adjacent non-cancerous tissues. |
Table III
The difference of Fra-1 expression
between cervical cancer and the adjacent non-cancerous tissues.
| | Score |
|---|
| |
|
|---|
| No. | Low (0–2) | Moderate (3–4) | High (5–6) | P |
|---|
| Cervical
cancer | 20 | 13 (65.0%) | 5 (25.0%) | 2 (10.0%) | 0.004 |
| Non-cancerous
tissues | 20 | 3 (15.0%) | 7 (35.0%) | 9 (45.0%) | 0.004 |
Analysis of protein expression levels of
Fra-1 in cervical cancer by western blotting
To determine whether the Fra-1 had lower expression
level in cervical cancer than the adjacent non-cancerous tissues,
we further examined the protein expression levels of Fra-1 in
cervical cancer and the adjacent non-cancerous tissues by western
blotting. In comparison with the control, the expression level was
low in cervical cancer tissues (Fig.
2). It corresponded to the results of RT-qPCR and IHC. It
confirmed that Fra-1 expression is low in cervical cancer.
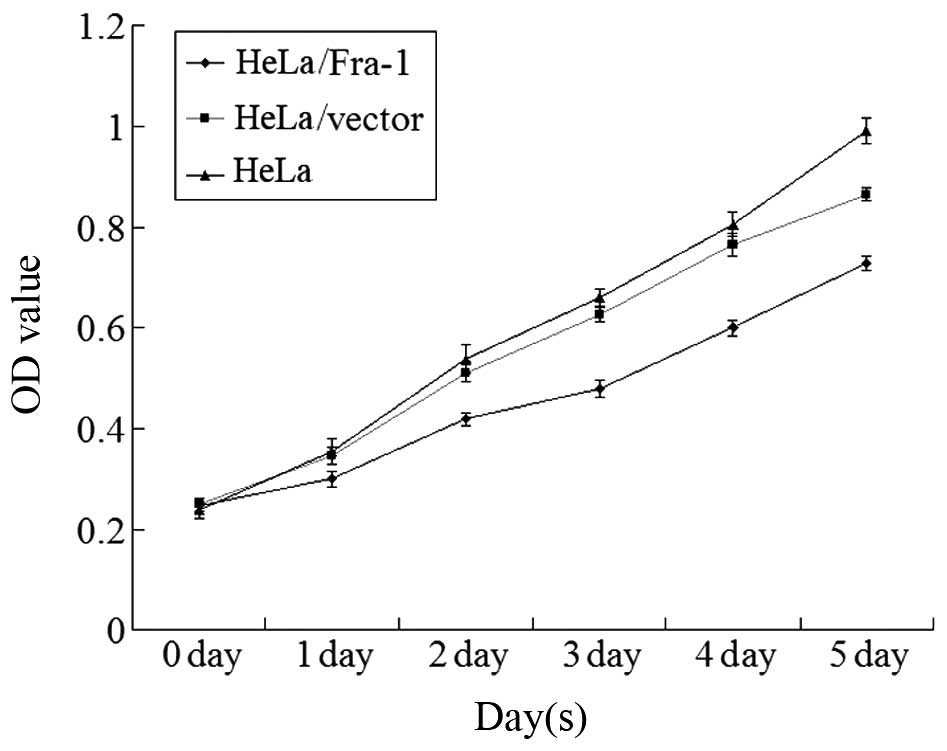
Fra-1 inhibits the growth of cervical
cancer cells in vitro
To elucidate the function of Fra-1 in the growth of
cervical cancer cells, the HeLa cells were transfected with the
plasmid pEGFP-N1/Fra-1 or control vector to generate Fra-1-stable
expressing HeLa/Fra-1, control HeLa/vector cell lines. After
demonstrating Fra-1 protein by western blotting, the spontaneous
proliferation of HeLa, HeLa/vector, and HeLa/Fra-1 cells was
determined by the MTT assays, respectively. Clearly, Fra-1
significantly inhibited the proliferation of HeLa cells (Fig. 3). Therefore, endogenous Fra-1
overexpression inhibited the proliferation of cervical cancer cells
in vitro.
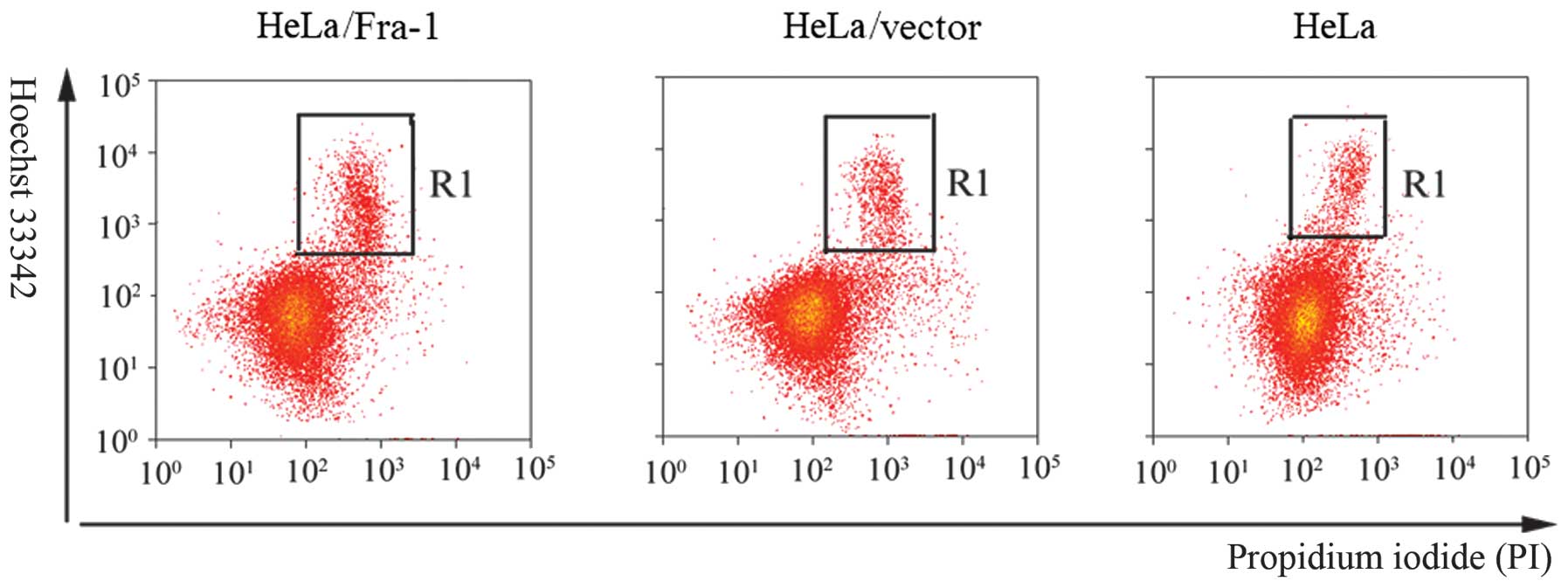
Fra-1 induces cervical cancer cell
apoptosis
Inhibition of cell proliferation usually is mediated
by inducing cell apoptosis. To determine whether apoptosis mediated
the growth in HeLa, HeLa/vector, and HeLa/Fra-1 cells, we performed
a Hoechst 33342/PI double staining experiment. A considerable
increase in apoptotic cells was observed for HeLa/Fra-1 cells
(15.36±0.48%), HeLa cells (8.97±0.91%), and HeLa/vector cells
(9.22±0.85%) (Fig. 4).
Fra-1 is correlated with dysregulation of
p53 signaling pathway in cervical cancer tissues in vitro
To uncover the possible mechanism of Fra-1 in
cervical cancer, we tested the expression levels of key molecules
in p53 signaling pathway by western blotting technology. p53 was
downregulated in cervical cancer compared with the adjacent
non-cancerous tissues, whereas, MDM2 proto-oncogene, E3 ubiquitin
protein ligase (MDM2) was upregulated in cervical cancer
(Fig. 5). Combined with the above
result showing low Fra-1 expression in cervical cancer, we inferred
that Fra-1 is correlated with dysregulation of p53 signaling
pathway in cervical cancer tissues in vitro.
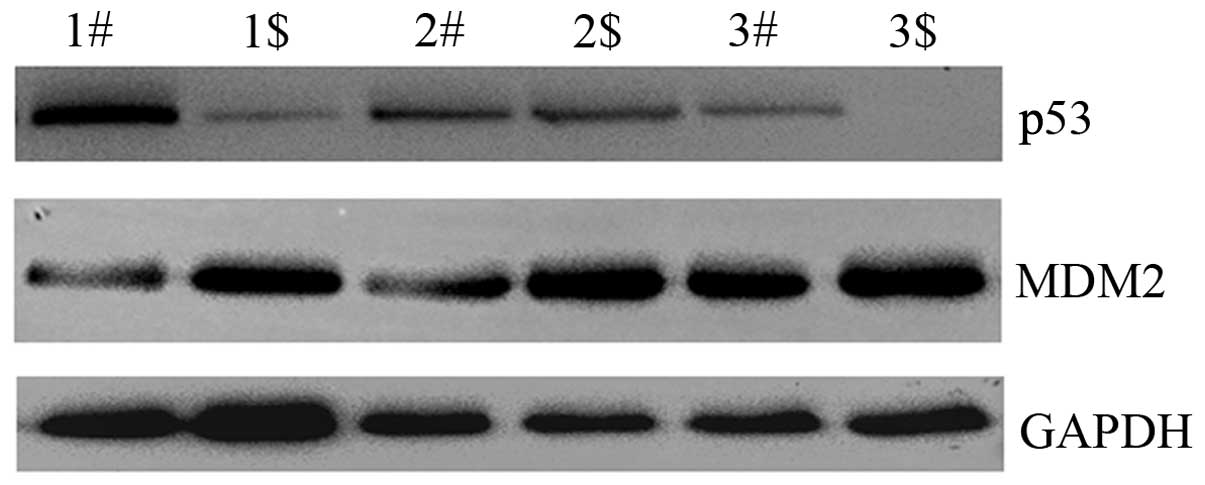
Fra-1 overexpression affects the
expression of p53 and MDM2 in vivo
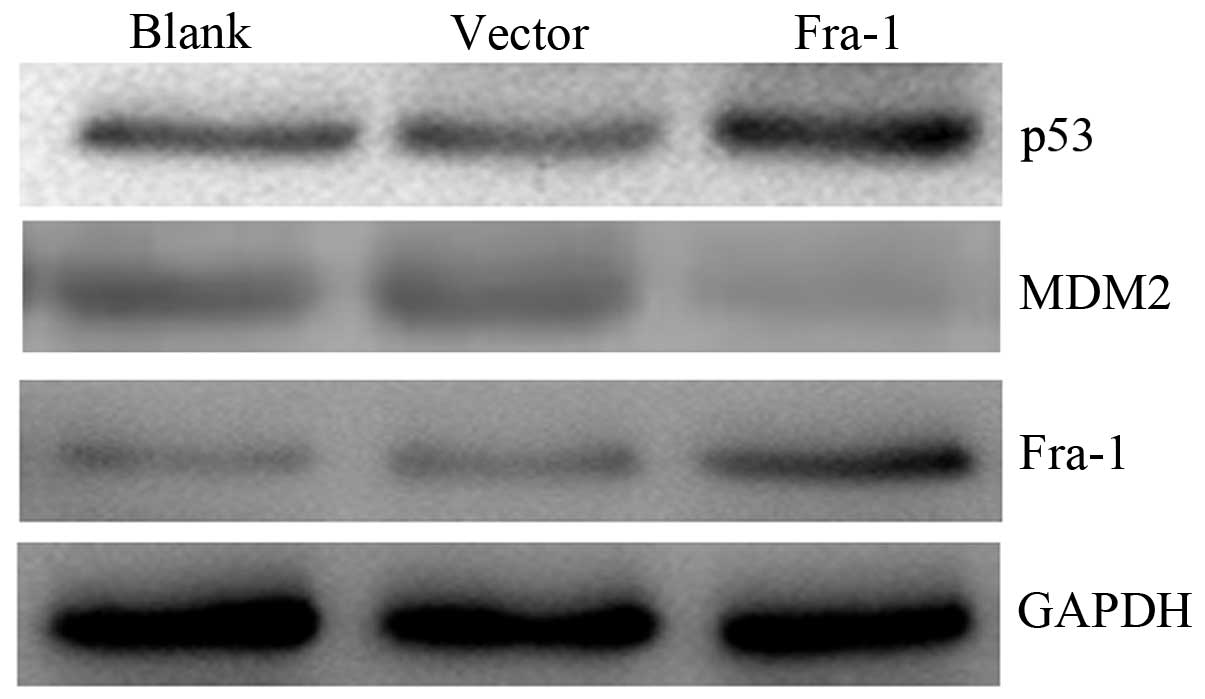
To confirm whether Fra-1 affects the expression of
p53 and MDM2 in vivo, the HeLa cells were transfected with
the plasmid pEGFP-N1/Fra-1 or control vector to generate
Fra-1-stable expressing HeLa/Fra-1, control HeLa/vector cell lines.
We harvested the cells and tested the expression levels of p53 and
MDM2 proteins in vivo. The p53 was upregulated in HeLa cells
with Fra-1 overexpression, but MDM2 was downregulated (Fig. 6). Our results suggested that Fra-1
overexpression affected the expression of p53 and MDM2 in
vivo.
Discussion
Cervical cancer that has been proven to be
associated with HPV is the second most common cancer in women
worldwide and is a leading cause of cancer deaths in women in
developing countries (45,46). Therefore, it is necessary and
urgent to study the etiology of cervical cancer.
In this study, we chose 20 cervical cancer tissues
and the adjacent non-cancerous tissues to perform real-time
quantitative RT-PCR of Fra-1 gene. The results showed that
the expression of Fra-1 gene was downregulated in cervical
cancer. The normalized Fra-1 gene expression in cervical
cancer was 0.32-fold compared with the control samples. Results of
IHC and western blotting showed a similar pattern in protein
expression with RT-qPCR results. Thus, we confirmed low Fra-1
expression in cervical cancer tissues. Kehrmann et al found
that Fra-1 was downregulated in the tumorigenic cell lines CGL3 and
HeLa compared to the non-tumorigenic 444 cells (28). The results of Soto et al
showed that Fra-1 has tumor-suppressing function upon micro-cell
transfer in HPV-16- and HPV-18-positive cervical-carcinoma cells
(29). Our data are consistent
with the above observations and suggest that Fra-1 may play an
important role in cervical cancer.
To elucidate the function of Fra-1 in the growth of
cervical cancer cells, our results showed that Fra-1 significantly
inhibited the proliferation of HeLa cells by MTT assay. Inhibition
of cell proliferation is usually mediated by inducing cell
apoptosis. Therefore, we tested apoptosis of Fra-1 overexpression
in HeLa cell lines and a considerable increase in apoptotic cells
was observed. Our data suggested that Fra-1 may affect the
proliferation of cervical cancer cells by mediated cell apoptosis.
Song et al found that Irisin promoted human umbilical vein
endothelial cell proliferation by partly suppressing cell apoptosis
(47). Yang et al confirmed
that downregulation of SIRT3 expression affected the proliferation
and apoptosis in esophageal squamous cell carcinoma EC9706 cells
(48). Above all, Fra-1 can affect
proliferation and apoptosis of HeLa cells.
To uncover the possible mechanism of Fra-1 in
cervical cancer, we detected the expression levels of p53 and MDM2
in cervical cancer tissues and in HeLa cells with Fra-1
overexpression by western blotting technology. We found that p53
was downregulated and MDM2 was upregulated in cervical cancer
compared with the adjacent non-cancerous tissues, whereas, the p53
was upregulated and MDM2 was downregulated in HeLa cells with Fra-1
overexpression. Degradation of p53 is regulated by its interaction
with specific E3 ubiquitin ligases, the best known one being
encoded by MDM2 (49). A greater
increase in p53 content and activation of p53 via additional
modification occur when the cell is exposed to various stress
factors, such as irradiation or DNA damage (50). Damage to p53-dependent mechanism is
often caused by overexpression of MDM2, which codes for a
p53-regulating protein (51).
Combined with the above result where Fra-1 expression was low in
cervical cancer, we inferred Fra-1 was correlated with
dysregulation of p53 signaling pathway in cervical cancer tissues
in vitro and Fra-1 overexpression affected the expression of
p53 and MDM2 in vivo.
In summary, our results showed that Fra-1 expression
was low in cervical carcinoma tissues and it plays an important
role in dysregulation of the p53 signaling pathway.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (81272975, 81402270); Key Project of
Hunan Provincial Natural Science Foundation (12JJ2044); the Key
Planned Science and Technology Project of Hunan Province
(2012FJ2014); the Planned Science and Technology Project of Hunan
Province (2011FJ3153); the Planned Project of Development and
Reform Commission of Hunan Province (2012-1493-1); the Planned
Project of Department of Health of Hunan Province (B2011-030,
B2012-029); the Planned Project of Key Subject Construction of the
Third Xiangya Hospital, Central South University; the Open-End Fund
for the Valuable and Precision Instruments of Central South
University.
Abbreviations:
|
Fra-1
|
FOS-like antigen-1
|
|
GAPDH
|
glyceraldehyde 3-phosphate
dehydrogenase
|
|
HPV
|
human papillomavirus
|
|
MDM2
|
MDM2 proto-oncogene, E3 ubiquitin
protein ligase
|
|
TP53
|
tumor protein p53
|
|
CI
|
confidence interval
|
|
IHC
|
immunohistochemistry
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boutas I, Sofoudis C, Kalampokas E,
Anastasopoulos C, Kalampokas T and Salakos N: Fertility
preservation in women with early stage cervical cancer. Review of
the literature. Eur J Gynaecol Oncol. 35:373–377. 2014.PubMed/NCBI
|
|
3
|
He L, Wu L, Su G, Wei W, Liang L, Han L,
Kebria M, Liu P, Chen C, Yu Y, Zhong M and Wang W: The efficacy of
neoadjuvant chemotherapy in different histological types of
cervical cancer. Gynecol Oncol. 134:419–425. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Georgieva S, Iordanov V and Sergieva S:
Nature of cervical cancer and other HPV-associated cancers. J BUON.
14:391–398. 2009.PubMed/NCBI
|
|
5
|
Lazcano-Ponce E and Allen-Leigh B:
Innovation in cervical cancer prevention and control in Mexico.
Arch Med Res. 40:486–492. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boshart M, Gissmann L, Ikenberg H,
Kleinheinz A, Scheurlen W and zur Hausen H: A new type of
papillomavirus DNA, its presence in genital cancer biopsies and in
cell lines derived from cervical cancer. EMBO J. 3:1151–1157.
1984.PubMed/NCBI
|
|
7
|
Dürst M, Gissmann L, Ikenberg H and zur
Hausen H: A papillomavirus DNA from a cervical carcinoma and its
prevalence in cancer biopsy samples from different geographic
regions. Proc Natl Acad Sci USA. 80:3812–3815. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheng JX, Yuan M, Li AL, Zhou P, Shen GQ
and Zhang Y: Quantitative analysis of P16 gene CpG methylation in
Uyghur patients with cervical squamous cell carcinoma and its
relationship with HPV16 infection. Genet Mol Res. 13:7428–7436.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Andersson S, Mints M, Gyllensten U,
Lindell M, Gustavsson I, Lambe M and Wilander E: Uneven
distribution of human papillomavirus 16 in cervical carcinoma in
situ and squamous cell carcinoma in older females: A retrospective
database study. Oncol Lett. 8:1528–1532. 2014.PubMed/NCBI
|
|
10
|
McLaughlin-Drubin ME and Munger K: Viruses
associated with human cancer. Biochim Biophys Acta. 1782:127–150.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Deivendran S, Marzook KH and Radhakrishna
Pillai M: The role of inflammation in cervical cancer. Adv Exp Med
Biol. 816:377–399. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ramdass B, Chowdhari A and Koka P:
Cancer-initiating cells as target for prevention of recurring
disease etiology: role of these malignant putative progenitor cells
in relapse or metastasis of human cervical carcinoma. J Stem Cells.
8:233–251. 2013.
|
|
13
|
Luo Y, Zhou H, Mizutani M, Mizutani N,
Reisfeld RA and Xiang R: Transcription factor Fos-related antigen 1
is an effective target for a breast cancer vaccine. Proc Natl Acad
Sci USA. 100:8850–8855. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo Y, Zhou H, Mizutani M, Mizutani N, Liu
C, Xiang R and Reisfeld RA: A DNA vaccine targeting Fos-related
antigen 1 enhanced by IL-18 induces long-lived T-cell memory
against tumor recurrence. Cancer Res. 65:3419–3427. 2005.PubMed/NCBI
|
|
15
|
Cohen DR and Curran T: fra-1: a
serum-inducible, cellular immediate-early gene that encodes a
fos-related antigen. Mol Cell Biol. 8:2063–2069. 1988.PubMed/NCBI
|
|
16
|
Schreiber M, Poirier C, Franchi A,
Kurzbauer R, Guenet JL, Carle GF and Wagner EF: Structure and
chromosomal assignment of the mouse fra-1 gene, and its exclusion
as a candidate gene for oc (osteosclerosis). Oncogene.
15:1171–1178. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Desmet CJ, Gallenne T, Prieur A, Reyal F,
Visser NL, Wittner BS, Smit MA, Geiger TR, Laoukili J, Iskit S,
Rodenko B, Zwart W, Evers B, Horlings H, Ajouaou A, Zevenhoven J,
van Vliet M, Ramaswamy S, Wessels LF and Peeper DS: Identification
of a pharmacologically tractable Fra-1/ADORA2B axis promoting
breast cancer metastasis. Proc Natl Acad Sci USA. 110:5139–5144.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu D, Chen S, Tan X, Li N, Liu C, Li Z,
Liu Z, Stupack DG, Reisfeld RA and Xiang R: Fra-1 promotes breast
cancer chemo-sensitivity by driving cancer stem cells from
dormancy. Cancer Res. 72:3451–3456. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Casalino L, De Cesare D and Verde P:
Accumulation of Fra-1 in ras-transformed cells depends on both
transcriptional autoregulation and MEK-dependent posttranslational
stabilization. Mol Cell Biol. 23:4401–4415. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zippo A, De Robertis A, Serafini R and
Oliviero S: PIM1-dependent phosphorylation of histone H3 at serine
10 is required for MYC-dependent transcriptional activation and
oncogenic transformation. Nat Cell Biol. 9:932–944. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Casalino L, Bakiri L, Talotta F, Weitzman
JB, Fusco A, Yaniv M and Verde P: Fra-1 promotes growth and
survival in RAS-transformed thyroid cells by controlling cyclin A
transcription. EMBO J. 26:1878–1890. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Adiseshaiah P, Papaiahgari SR, Vuong H,
Kalvakolanu DV and Reddy SP: Multiple cis-elements mediate the
transcriptional activation of human fra-1 by
12-O-tetradecanoylphorbol-13-acetate in bronchial epithelial cells.
J Biol Chem. 278:47423–47433. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adiseshaiah P, Peddakama S, Zhang Q,
Kalvakolanu DV and Reddy SP: Mitogen regulated induction of FRA-1
proto-oncogene is controlled by the transcription factors binding
to both serum and TPA response elements. Oncogene. 24:4193–4205.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gruda MC, Kovary K, Metz R and Bravo R:
Regulation of Fra-1 and Fra-2 phosphorylation differs during the
cell cycle of fibroblasts and phosphorylation in vitro by MAP
kinase affects DNA binding activity. Oncogene. 9:2537–2547.
1994.PubMed/NCBI
|
|
25
|
Hurd TW, Culbert AA, Webster KJ and Tavaré
JM: Dual role for mitogen-activated protein kinase (Erk) in
insulin-dependent regulation of Fra-1 (fos-related antigen-1)
transcription and phosphorylation. Biochem J. 368:573–580. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Milde-Langosch K: The Fos family of
transcription factors and their role in tumourigenesis. Eur J
Cancer. 41:2449–2461. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Belguise K, Milord S, Galtier F,
Moquet-Torcy G, Piechaczyk M and Chalbos D: The PKCθ pathway
participates in the aberrant accumulation of Fra-1 protein in
invasive ER-negative breast cancer cells. Oncogene. 31:4889–4897.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kehrmann A, Truong H, Repenning A, Boger
R, Klein-Hitpass L, Pascheberg U, Beckmann A, Opalka B and
Kleine-Lowinski K: Complementation of non-tumorigenicity of
HPV18-positive cervical carcinoma cells involves differential mRNA
expression of cellular genes including potential tumor suppressor
genes on chromosome 11q13. Cancer Genet. 206:279–292. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Soto U, Denk C, Finzer P, Hutter KJ, zur
Hausen H and Rösl F: Genetic complementation to non-tumorigenicity
in cervical-carcinoma cells correlates with alterations in AP-1
composition. Int J Cancer. 86:811–817. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Missero C and Antonini D: Crosstalk among
p53 family members in cutaneous carcinoma. Exp Dermatol.
23:143–146. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bertheau P, Lehmann-Che J, Varna M, Dumay
A, Poirot B, Porcher R, Turpin E, Plassa LF, de Roquancourt A,
Bourstyn E, de Cremoux P, Janin A, Giacchetti S, Espié M and de Thé
H: p53 in breast cancer subtypes and new insights into response to
chemotherapy. Breast. 22(Suppl 2): S27–S29. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tassone P, Old M, Teknos TN and Pan Q:
p53-based therapeutics for head and neck squamous cell carcinoma.
Oral Oncol. 49:733–737. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ku JH, Byun SS, Jeong H, Kwak C, Kim HH
and Lee SE: The role of p53 on survival of upper urinary tract
urothelial carcinoma: a systematic review and meta-analysis. Clin
Genitourin Cancer. 11:221–228. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tornesello ML, Buonaguro L and Buonaguro
FM: Mutations of the TP53 gene in adenocarcinoma and squamous cell
carcinoma of the cervix: a systematic review. Gynecol Oncol.
128:442–448. 2013. View Article : Google Scholar
|
|
35
|
Liu J, Ma Q, Zhang M, Wang X, Zhang D, Li
W, Wang F and Wu E: Alterations of TP53 are associated with a poor
outcome for patients with hepatocellular carcinoma: evidence from a
systematic review and meta-analysis. Eur J Cancer. 48:2328–2338.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mitchell S, Mayer E and Patel A:
Expression of p53 in upper urinary tract urothelial carcinoma. Nat
Rev Urol. 8:516–522. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nayak SK, Panesar PS and Kumar H:
Non-genotoxic p53-activators and their significance as antitumor
therapy of future. Curr Med Chem. 18:1038–1049. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xiao S, Zhou Y, Jiang J, Yuan L and Xue M:
CD44 affects the expression level of FOS-like antigen 1 in cervical
cancer tissues. Mol Med Rep. 9:1667–1674. 2014.PubMed/NCBI
|
|
39
|
Liao S, Xiao S, Zhu G, Zheng D, He J, Pei
Z, Li G and Zhou Y: CD38 is highly expressed and affects the
PI3K/Akt signaling pathway in cervical cancer. Oncol Rep.
32:2703–2709. 2014.PubMed/NCBI
|
|
40
|
Zhu W, Li J, Su J, Li J, Li J, Deng B, Shi
Q, Zhou Y and Chen X: FOS-like antigen 1 is highly expressed in
human psoriasis tissues and promotes the growth of HaCaT cells in
vitro. Mol Med Rep. 10:2489–2494. 2014.PubMed/NCBI
|
|
41
|
Xiao S, Liao S, Zhou Y, Jiang B, Li Y and
Xue M: High expression of octamer transcription factor 1 in
cervical cancer. Oncol Lett. 7:1889–1894. 2014.PubMed/NCBI
|
|
42
|
Zhou Y, Wang W, Zheng D, Peng S, Xiong W,
Ma J, Zeng Z, Wu M, Zhou M, Xiang J, Xiang B, Li X, Li X and Li G:
Risk of nasopharyngeal carcinoma associated with polymorphic
lacto-transferrin haplotypes. Med Oncol. 29:1456–1462. 2012.
View Article : Google Scholar
|
|
43
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
44
|
Hara A and Okayasu I: Cyclooxygenase-2 and
inducible nitric oxide synthase expression in human astrocytic
gliomas: correlation with angiogenesis and prognostic significance.
Acta Neuropathol. 108:43–48. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Salehi M, Taheri T, Mohit E, Zahedifard F,
Seyed N, Taslimi Y, Sattari M, Bolhassani A and Rafati S:
Recombinant Leishmania tarentolae encoding the HPV type 16 E7 gene
in tumor mice model. Immunotherapy. 4:1107–1120. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sahiner F, Gümral R, Sener K, Yiğit N,
Dede M, Yapar M and Kubar A: Investigation of HPV-DNA in cervical
smear samples by two different methods: MY09/11 consensus PCR and
type-specific real-time PCR. Mikrobiyol Bul. 46:624–636. 2012.(In
Turkish). PubMed/NCBI
|
|
47
|
Song H, Wu F, Zhang Y, Zhang Y, Wang F,
Jiang M, Wang Z, Zhang M, Li S, Yang L, Wang XL, Cui T and Tang D:
Irisin promotes human umbilical vein endothelial cell proliferation
through the ERK signaling pathway and partly suppresses high
glucose-induced apoptosis. PLoS One. 9:e1102732014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang M, Yang C and Pei Y: Effects of
downregulation of SIRT3 expression on proliferation and apoptosis
in esophageal squamous cell carcinoma EC9706 cells and its
molecular mechanisms. Biomed Mater Eng. 24:3883–3890.
2014.PubMed/NCBI
|
|
49
|
Chumakov PM: Function of the p53 gene:
choice between life and death. Biochemistry (Mosc). 65:28–40.
2000.
|
|
50
|
Harris SL and Levine AJ: The p53 pathway:
positive and negative feedback loops. Oncogene. 24:2899–2908. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chipuk JE and Green DR: Dissecting
p53-dependent apoptosis. Cell Death Differ. 13:994–1002. 2006.
View Article : Google Scholar : PubMed/NCBI
|