Introduction
Epithelial ovarian cancer (EOC) consists of multiple
histotypes differing in etiology and clinical course. The most
prevalent histotype is serous ovarian cancer (SOC), which is highly
lethal (1,2). Because of the absence of
characteristic symptoms and the lack of effective biomarkers, the
majority of SOC patients have widely dispersed intra-peritoneal
and/or lymph node metastatic disease at the time of diagnosis
(3,4). Despite advances in surgery and
chemotherapy over the last few decades, the 5-year survival rate
for SOC patients with advanced disease remains <30% (2,5).
Such an unfavourable prognosis has been largely correlated with
tumour metastasis. Therefore, strategies for elucidating the
mechanism underlying metastasis and identifying novel biomarkers
for SOC are urgently needed.
Long non-coding RNAs (lncRNAs, >200 nt in
length), previously disregarded as transcriptional noise, are
emerging as new regulators in the cancer paradigm (6,7). An
increasing number of discoveries of misregulated lncRNA expression
across numerous cancer types have suggested that aberrant lncRNA
expression may be a major contributor to tumourigenesis. Further,
lncRNAs may add another layer to cancer research because their
potential usefulness as biomarkers (8–15).
Therefore, to fully understand the complex mechanisms underlying
carcinogenesis and cancer metastasis, the role of lncRNAs must be
considered.
The antisense non-coding RNA in the INK4 locus
(ANRIL) is a recently discovered lncRNA encoded in the chromosome
9p21 region, which has been highlighted as a hotspot for cancer
research because this gene locus is often homozygously deleted or
transcriptionally silenced in ~40% of human cancers (16). The data gathered to date strongly
implicate ANRIL in the epigenetic regulation of INK4b/ARF/INK4a
tumour suppressors (17–19). In addition, ANRIL has been reported
to contribute to a number of cellular events in many cancers
facilitating cell proliferation and senescence, suggesting it has a
pro-cancerogenic role (20,21).
To date, upregulation of ANRIL has gradually become known as a
primary feature of many human solid carcinomas, including melanoma
(19), breast cancer (22), pancreatic carcinoma (23), nasopharyngeal carcinoma (24), basal cell carcinoma (25), glioma (26) and leukemia (27). However, its involvement in SOC
remains poorly investigated.
In the present study, we detected the expression of
ANRIL in SOC tissues and assessed its association with
clinicopathological factors as well as patient prognosis.
Additionally, using in vitro assays, we determined its role
in metastasis and invasion during SOC progression. We also employed
metastasis-related gene mRNA microarrays to investigate the
downstream molecular events involving ANRIL and SOC metastasis. Our
study highlights the significance of ANRIL in SOC metastasis and
prognosis.
Materials and methods
Patients and tissue samples
A total of 68 SOC tissue samples were obtained from
patients admitted to the Department of Gynaecology, Obstetrics and
Gynaecology Hospital of Fudan University between August 2005 and
December 2008. All diagnoses were confirmed by histology. Patients
with borderline ovarian tumours, patients with two or more
different malignancies and patients who had received preoperative
radiotherapy, chemotherapy, or hormonal therapy were excluded. The
control group consisted of 30 normal ovarian epithelial tissue
samples obtained from participants diagnosed with uterine fibroids
scheduled to undergo hysterectomy and oophorectomy. Patients with
ovarian cysts, patients who had experienced ovarian pathology and
patients who had undergone previous ovarian surgery were excluded.
All samples were frozen immediately in liquid nitrogen and stored
at −80°C until analysis.
Clinicopathological details of SOC patients were
evaluated by reviewing medical charts and the original pathology
reports. Staging and grading were determined in accordance with the
criteria of the International Federation of Gynaecologists and
Obstetricians (FIGO) and the World Health Organization (WHO).
Follow-up data were obtained by reviewing the outpatient charts or
via correspondence. Overall survival (OS) was defined as the time
interval between the date of surgery and the date of death or end
of follow-up (January 2013). This study was approved by the
Research Ethics Committee of Fudan University, China. Informed
consent was obtained from all of the patients.
Cell line and cell culture
Two paired human SOC cell lines: parental (SK-OV-3,
HO8910) and highly metastatic sublines (SK-OV-3.ip1, HO8910-PM)
(28–31) were gifts from the University of
Texas M.D. Anderson Cancer Center (Houston, TX, USA). All cells
were cultured in RPMI-1640 medium (Gibco BRL, Gaithersburg, MD,
USA) containing 10% fetal bovine serum (FBS; Gibco) with 100 U/ml
penicillin and 100 mg/ml streptomycin and maintained in a
humidified 5% CO2 incubator at 37°C.
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from cancerous/non-cancerous
specimens or cell lines using TRIzol reagent (Invitrogen, Carlsbad,
CA, USA). RNA was reverse transcribed into cDNAs using a
Prime-Script™ one step RT-PCR kit (Takara, Dalian, China). QRT-PCR
reactions were performed using an ABI7500 System (Applied
Biosystems, Foster City, CA, USA) and SYBR Green PCR Master Mix
(Takara). The primer sequences for ANRIL were
5′-TGTACTTAACCACTGGACTACCTGCC-3′ (forward) and
5′-CATTCTGATTCAACAGCAGAGATCAAAG-3′ (reverse). Glyceraldehyde
3-phosphate dehydrogenase (GAPDH) was applied as an internal
control, the primers for which were 5′-GTCAACGGATTTGGTCTGTATT-3′
(forward) and 5′-AGTCTTCTGGGTGGCAGTGAT-3′ (reverse). Each assay was
performed in triplicate, and the average was calculated. ANRIL
expression levels were normalised to GAPDH.
Small interfering RNAs (siRNAs) and
transfection
For the in vitro study, SK-OV-3.ip1 and
HO8910-PM cells were transfected with either 50 nM siRNAs targeting
ANRIL or scrambled negative controls (GenePharma, Shanghai, China)
using Lipofectamine 2000 transfection reagent (Invitrogen)
according to the instructions provided by the manufacturer. The two
siRNA sequences targeting 2 different sections of ANRIL were
5′-GCAAGAAACATTGCTGCTAGC-3′ and 5′-GCCCAATTATGCTGTGGTAAC-3′. After
48 h of transfection, knockdown of ANRIL was confirmed via
qRT-PCR.
Wound-healing assay
SK-OV-3.ip1 and HO8910-PM cells were transfected
with either 50 nM siRNAs targeting ANRIL or a scrambled negative
control (si-NC). When cell confluence reached ~80% at 24 h
posttransfection, wounds were created in confluent cells using a
200-μl pipette tip. Cells were then rinsed with medium to remove
any free-floating cells and debris. Medium was added, and the
culture plates were incubated at 37°C. Different stages of wound
healing were observed along the scrape line, and representative
scrape lines were photographed. Duplicate wells for each condition
were examined, and each experiment was repeated in triplicate.
Matrigel invasion assay
SK-OV-3.ip1 and HO8910-PM cells were transfected
with 50 nM siRNAs targeting ANRIL or a scrambled negative control
(si-NC). At 24 h post-infection, infected cells were harvested and
subjected to the following assays. Infected cells
(1×105) were plated in the top chamber of Transwell
assay inserts (Millipore, Billerica, MA, USA) with a Matrigel
coated membrane containing pores with a diameter of 8 μm in 200 ml
of serum-free RPMI-1640. The assays were conducted in triplicate.
Inserts were then placed into the bottom chamber wells of a 24-well
plate containing RPMI-1640 with 10% FBS as a chemo-attractant.
After 48 h of incubation, the remaining cells were removed from the
top layer of the insert by scrubbing with a sterile cotton swab.
Invading cells from the bottom surface were stained with 0.1%
crystal violet prior to being examined, counted, and photographed
using digital microscopy. Cell numbers were calculated in five
random fields for each chamber, and the average value was
calculated.
Western blotting
The cells were lysed with RIPA buffer [50 mM
Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 0.5%
Na-deoxycholate] containing protease inhibitors (Roche, Complete
Mini). Lysate aliquots (20–30 μg) were separated on 10% sodium
dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gels
and transferred to a polyvinyl difluoride (PVDF) membrane. The
membranes were incubated with the primary antibodies (Cell
Signalling) rabbit anti-MMP3, anti-MET and anti-GAPDH overnight at
4°C. Primary antibody incubation was followed with HRP-conjugated
secondary antibody incubation. The bound antibodies were detected
with ECL kit (Pierce, PI32209).
Tumour metastasis real-time PCR
array
Total RNA was extracted from
SK-OV-3.ip1-ANRIL-siRNA1 cells and SK-OV-3.ip1-si-NC cells with an
RNeasy mini kit (Qiagen) and further purified using an RNeasy
MinElute™ Cleanup kit (Qiagen). An RT2 First Strand kit
(Qiagen) was employed to produce a cDNA library for the total RNA
extracted. Following the manufacturer’s protocol, the cDNA was then
processed to perform a Human Tumour Metastasis RT2
Profiler™ PCR array (Qiagen, Mississauga, ON, Canada) containing 84
genes known to be related to tumour metastasis, five housekeeping
genes were used for a genomic DNA control, and three positive
controls to ensure high-quality data normalisation across samples.
The results were analysed using SA Biosciences software. For
relative quantification, 2−ΔΔCt was calculated and used
as an indication of the relative expression level.
Statistical analysis
All of the statistical analyses were performed using
SPSS for Windows v.16.0 (SPSS, Chicago, IL, USA). The continuous
data were analysed using an independent t-test between the two
groups, whereas categorical data were analysed by the χ2
test or Fisher’s exact test, as appropriate. OS curves were plotted
according to the Kaplan-Meier method, and the log-rank test was
applied for comparison. The variables were used in multivariate
analysis on the basis of the Cox proportional hazards model.
P-values <0.05 were considered statistically significant
(P<0.05).
Results
ANRIL is overexpressed and correlates
with poor prognosis in SOC
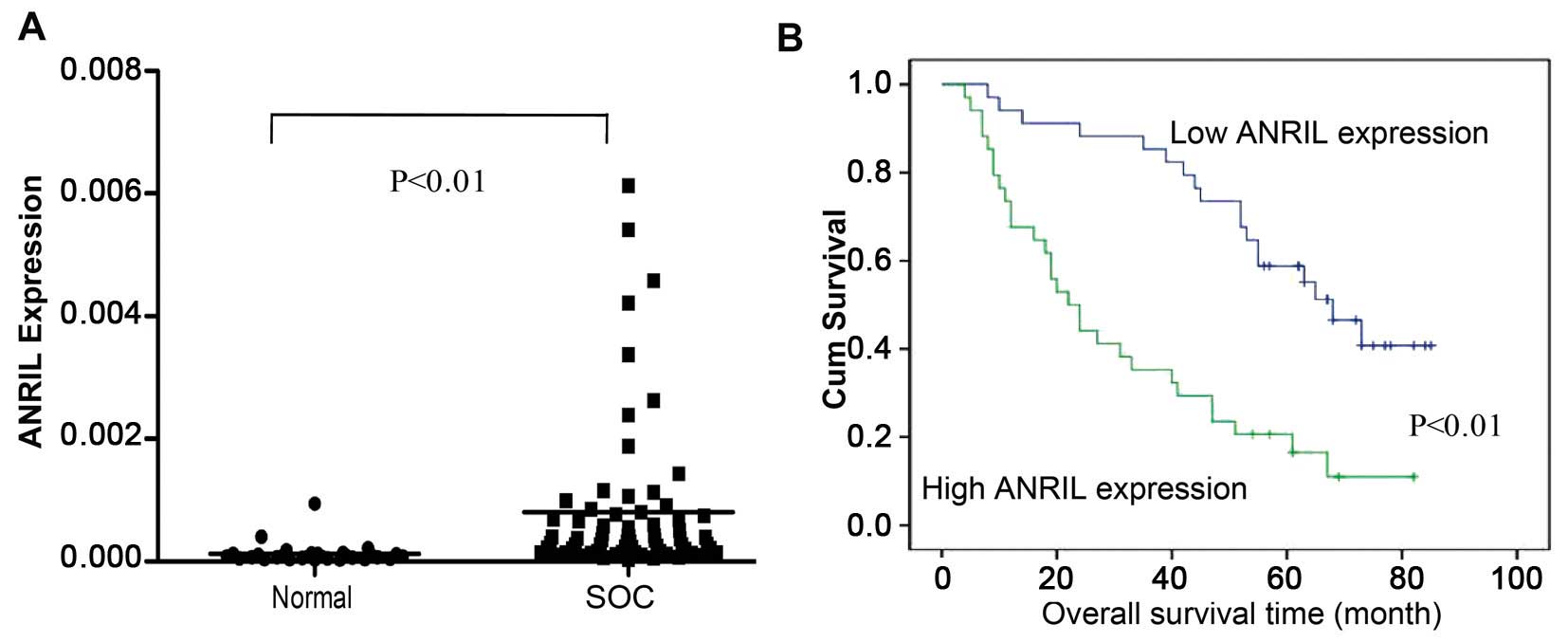
ANRIL expression levels in 68 SOC and 30
non-cancerous tissues were examined using qRT-PCR. The results
showed that ANRIL levels in SOC tissues were significantly higher
than those in non-cancerous tissues (P<0.01; Fig. 1A).
For the clinicopathological correlation analysis,
the 68 SOC patients were divided into two groups according to the
median relative ANRIL expression value that was used as the cut-off
(10): high ANRIL group (n=34):
ANRIL expression value ≥ the 50th percentile (with an average ΔCt
expression value of 10.046 compared to GAPDH); low ANRIL group
(n=34): ANRIL expression value less than the 50th percentile (with
an average ΔCt expression value of 12.584 compared to GAPDH). As
shown in Table I, elevated ANRIL
expression was correlated with advanced FIGO stage, high
histological grade and lymph node metastasis, but not with age,
residual tumour diameter, CA125 level or ascites.
 | Table IAssociation of ANRIL expression with
clinicopathological variables in 68 SOC patients. |
Table I
Association of ANRIL expression with
clinicopathological variables in 68 SOC patients.
| Low ANRIL expression
(n=34) | High ANRIL expression
(n=34) | |
|---|
|
|
| |
|---|
| Variables | n (%) | n (%) | P-value |
|---|
| Age (years) | | | |
| <50 | 11 (42.3) | 15 (57.7) | 0.318 |
| ≥50 | 23 (54.8) | 19 (45.2) | |
| FIGO stage | | | |
| I–II | 15 (78.9) | 4 (21.1) | 0.006 |
| III–IV | 19 (38.8) | 30 (61.2) | |
| Histological
grade | | | |
| G1–G2 | 16 (66.7) | 8 (33.3) | 0.042 |
| G3 | 18 (40.9) | 26 (59.1) | |
| Residual tumour
diameter (cm) | | | |
| <1 | 26 (56.5) | 20 (43.5) | 0.12 |
| ≥1 | 8 (36.4) | 14 (63.6) | |
| Lymph node
metastasis | | | |
| Absent | 21 (75.0) | 7 (25.0) | 0.001 |
| Present | 13 (32.5) | 27 (67.5) | |
| CA125 level
(U/ml) | | | |
| <600 | 15 (48.4) | 16 (51.6) | 0.808 |
| ≥600 | 19 (51.4) | 18 (48.6) | |
| Ascites | | | |
| <100 | 16 (61.5) | 10 (38.5) | 0.134 |
| ≥100 | 18 (42.9) | 24 (57.1) | |
The OS curves calculated using the Kaplan-Meier
method according to ANRIL expression are shown in Fig. 1B. According to the univariate
analysis, ANRIL expression was correlated with OS (P<0.001,
Table II). Using a multivariate
Cox regression analysis, ANRIL expression, in addition to FIGO
stage, histological grade and lymph node metastasis, was an
independent predictor of OS (P<0.01, Table II).
 | Table IIUnivariate and multivariate analysis
of overall survival in 68 SOC patients. |
Table II
Univariate and multivariate analysis
of overall survival in 68 SOC patients.
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| Overall survival
(months) | Overall
survival |
|---|
|
|
|
|---|
| Variables | Mean ± SE | P-value | β | SE | Wald | P-value | Exp (β) | 95% CI |
|---|
| Age (years) | | | | | | | | |
| <50 | 42.67±4.83 | 0.341 | - | - | - | - | - | - |
| ≥50 | 49.29±4.68 | | - | - | - | - | - | - |
| FIGO stage | | | | | | | | |
| I–II | 78.96±2.60 | <0.001 | - | - | - | - | - | - |
| III–IV | 35.00±3.44 | | 1.489 | 0.674 | 4.877 | 0.027 | 4.431 | 1.182–16.605 |
| Histological
grade | | | | | | | | |
| G1–G2 | 66.02±5.10 | <0.001 | - | - | - | - | - | - |
| G3 | 36.82±3.85 | | 0.918 | 0.38 | 5.844 | 0.016 | 2.504 | 1.190–5.271 |
| Residual tumour
diameter (cm) | | | | | | | | |
| <1 | 54.36±4.39 | 0.001 | - | - | - | - | - | - |
| ≥1 | 33.24±4.88 | | 0.307 | 0.313 | 0.962 | 0.327 | 1.359 | 0. 736–2.508 |
| CA125 level
(U/ml) | | | | | | | | |
| <600 | 53.61±4.92 | 0.142 | - | - | - | - | - | - |
| ≥600 | 42.37±4.89 | | - | - | - | - | - | - |
| Lymph node
metastasis | | | | | | | | |
| Absent | 71.97±3.53 | <0.001 | - | - | - | - | - | - |
| Present | 29.28±3.18 | | 1.523 | 0.47 | 10.479 | 0.001 | 4.584 | 1.823–11.523 |
| Ascites | | | | | | | | |
| <100 | 51.55±5.50 | 0.511 | - | - | - | - | - | - |
| ≥100 | 44.78±4.56 | | - | - | - | - | - | - |
| ANRIL
expression | | | | | | | | |
| Low | 62.44±4.19 | <0.001 | - | - | - | - | - | - |
| High | 32.05±4.31 | | 0.639 | 0.317 | 4.059 | 0.044 | 1.895 | 1.018–3.530 |
These results suggest that ANRIL can be used as a
powerful independent prognostic factor and that overexpression of
ANRIL might have an important role in SOC progression.
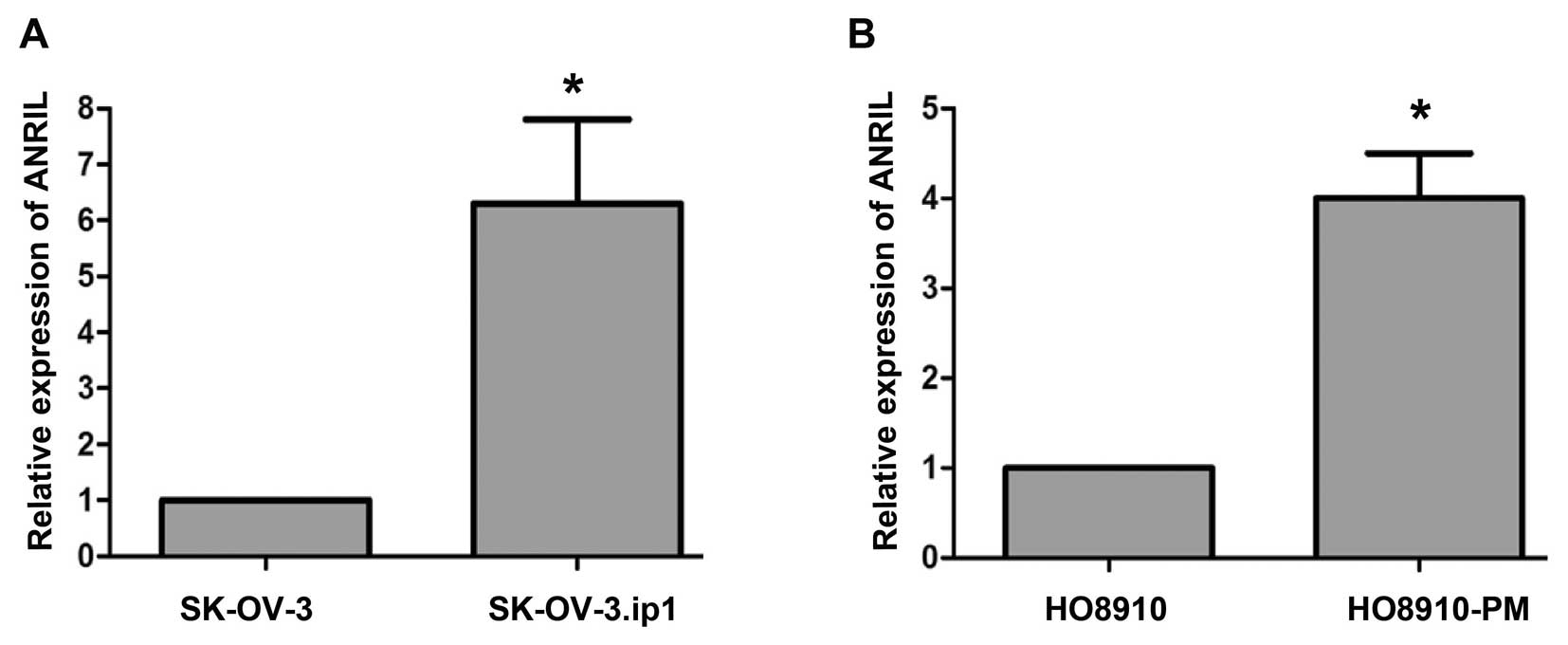
ANRIL expression in SOC cell lines
Because overexpression of ANRIL was associated with
lymph node metastasis and poor prognosis, we speculated that ANRIL
might play a role in mediating SOC metastasis. To explore this
possibility, we examined ANRIL levels in two paired SOC cell lines:
parental (SK-OV-3, HO8910) and highly metastatic sublines (SK-OV-3.
ip1, HO8910-PM). The results showed that SK-OV-3.ip1 and HO8910-PM
expressed higher levels of ANRIL than their parental cell lines
(Fig. 2), suggesting that ANRIL
was associated with SOC metastasis.
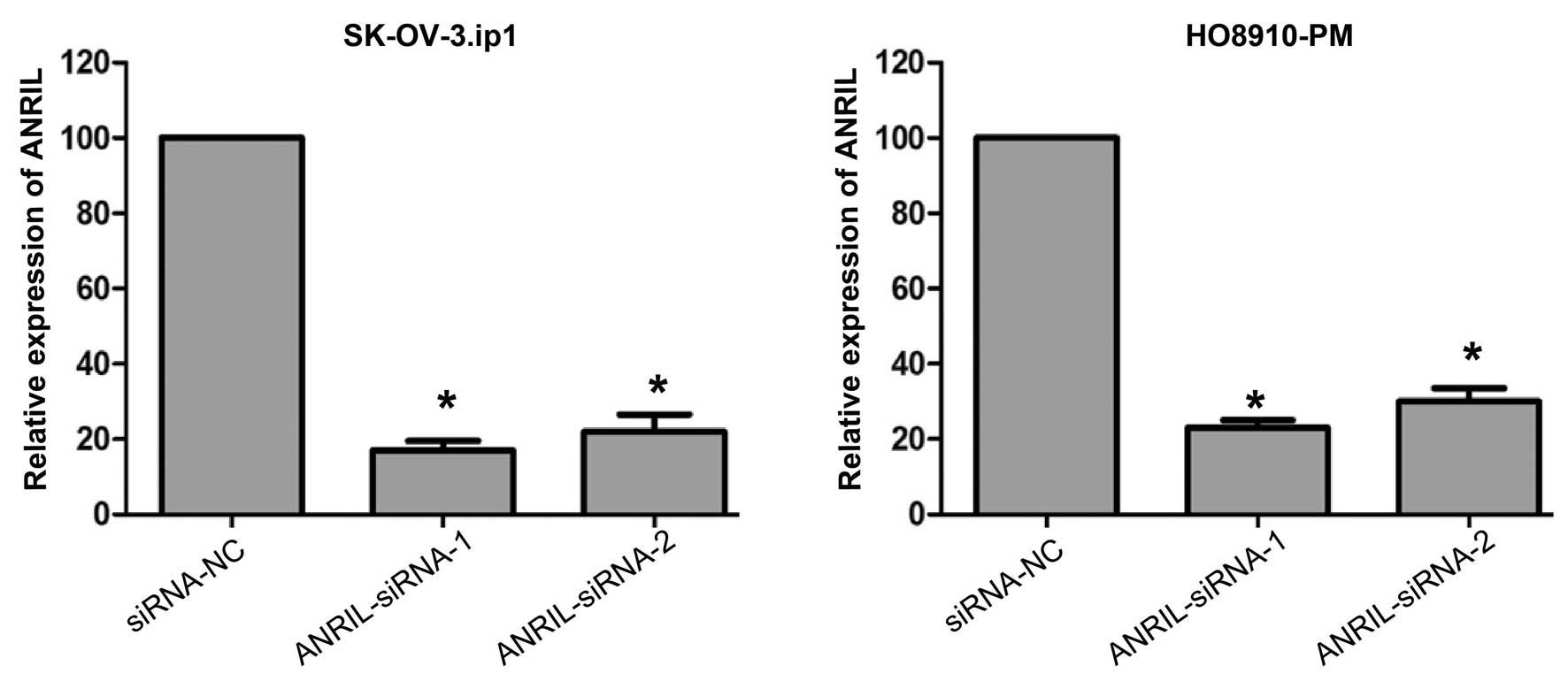
ANRIL silencing attenuates migration and
invasion of SOC cells
To further investigate the role of ANRIL in SOC
metastasis, the effects of siRNA-mediated knockdown of ANRIL
expression were first investigated in SK-OV-3.ip1 and HO8910-PM
cells. Forty-eight hours following treatment, both siRNAs
efficiently knocked down ANRIL expression in these cells (Fig. 3).
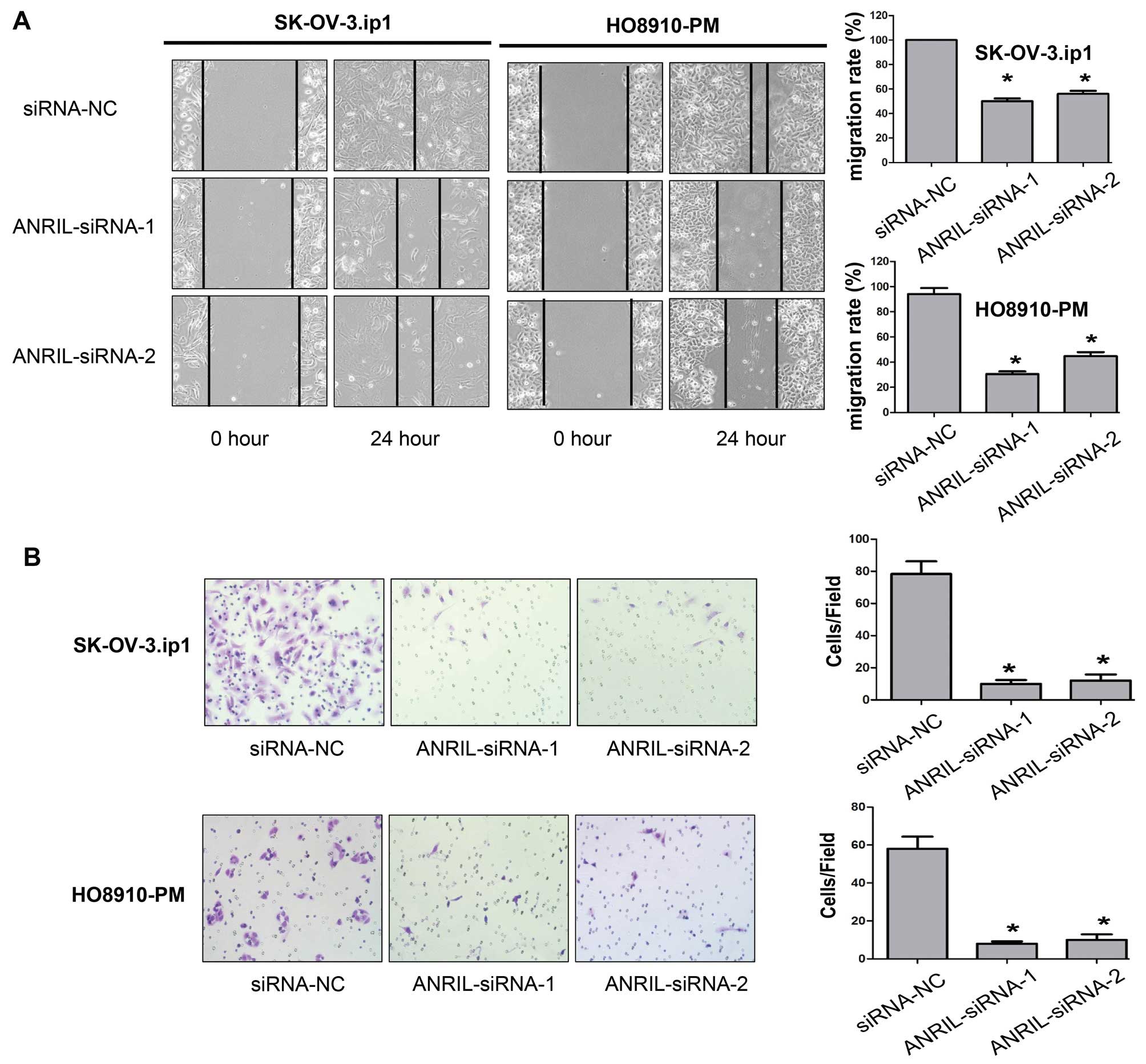
We then assessed the effects of ANRIL on the
migratory and invasive behaviour of SOC cells. Wound-healing assays
showed that knockdown of ANRIL caused an apparent suppression of
cell migration in both SK-OV-3.ip1 and HO8910-PM cells (P<0.01,
Fig. 4A). Matrigel invasion assays
also demonstrated that depletion of ANRIL markedly reduced the
invasive ability of both SK-OV-3.ip1 and HO8910-PM cells
(P<0.01, Fig. 4B). We therefore
concluded that ANRIL promotes SOC cell migration and invasion in
vitro.
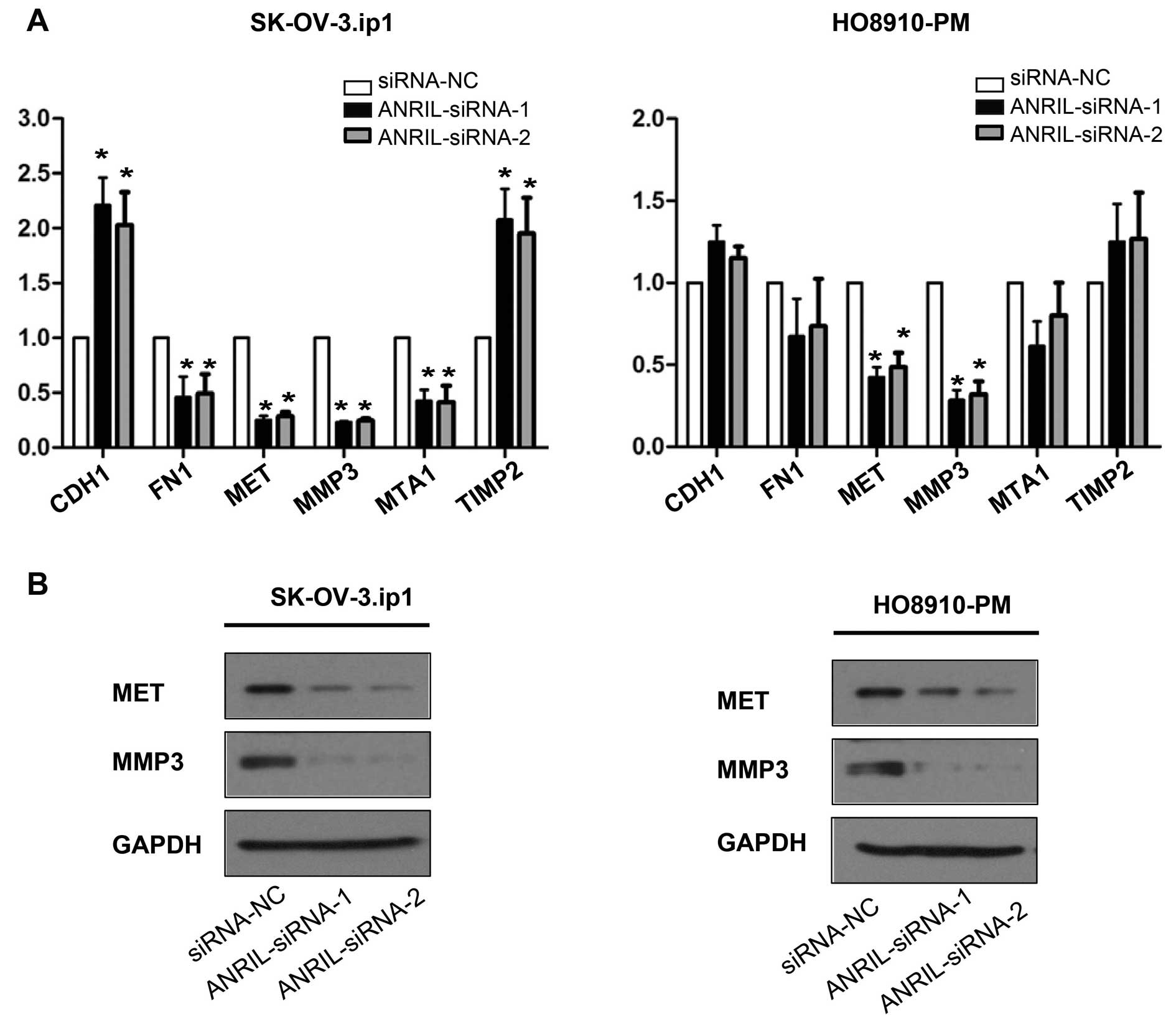
MET and MMP3 are the key downstream genes
of ANRIL involved in SOC cell metastasis
To further study possible mechanisms through which
ANRIL alters migration and invasion in SOC cells, tumour
metastasis-related gene expression profiles of
SK-OV-3.ip1-ANRIL-siRNA1 cells were first compared with those of
SK-OV-3.ip1-si-NC cells using real-time PCR array analysis. The
results showed that six genes were markedly dysregulated
(>2-fold) after ANRIL silencing in SK-OV-3.ip1 cells,
specifically four downregulated (MMP3, MTA1, FN1 and MET) and two
upregulated (CDH1 and TIMP2) genes (Table III).
 | Table IIIGenes dysregulated >2-fold after
ANRIL silencing in SK-OV-3.ip1 cells identified by array. |
Table III
Genes dysregulated >2-fold after
ANRIL silencing in SK-OV-3.ip1 cells identified by array.
| Gene name | GeneBank ID | Description | Function | Fold change |
|---|
| CDH1 | NM_004360 | Cadherin 1, type 1,
E-cadherin | Inhibits tumour
metastasis | 2.38 |
| FN1 | NM_002026 | Fibronectin 1 | Participates in
cell adhesion | −2.25 |
| MET | NM_000245 | Met
proto-oncogene | Protooncogene,
promotes cell proliferation | −3.21 |
| MMP3 | NM_002422 | Matrix
metallopeptidase 3 | Promotes
metastasis | −3.83 |
| MTA1 | NM_004689 | Metastasis
associated 1 | Promotes
metastasis | −2.36 |
| TIMP2 | NM_003255 | TIMP
metallopeptidase inhibitor 2 | Inhibits
metastasis | 2.19 |
These downstream genes were further validated by
qRT-PCR and western blotting assays in both SK-OV-3. ip1 and
HO8910-PM cells. Consistent with the array results, decreased MET
and MMP3 mRNA and protein levels were detected by qRT-PCR and
western blotting in both SK-OV-3. ip1 and HO8910-PM cells after
ANRIL silencing with both siRNAs (Fig.
5).
Taken together, these data indicate that ANRIL
regulates SOC cell migration and invasion, at least in part,
through the regulation of MET and MMP3.
Discussion
Mounting evidence indicates that eukaryotic
transcriptomes and genomes are not the simple substrates of
protein-coding gene transcription that they were once thought to
be, but rather exhibit extensive non-coding RNA (ncRNA) expression
(6). Recent studies have
highlighted the role of long ncRNAs (lncRNAs) in carcinogenesis and
have suggested that this class of genes might be used as biomarkers
in cancer. For example, HOTAIR has been demonstrated to be
upregulated in primary breast tumours and metastases, and elevated
HOTAIR expression is an indispensable predictor of eventual
metastasis and death (8). MALAT-1
has been found to promote cell motility and predict poorer clinical
outcome in lung cancer (9). HULC
has been implicated in the regulation of hepatoma cancer cell
proliferation, and higher HULC expression can be used as a
non-invasive promising novel biomarker for diagnosis and/or
prognosis in hepatocellular carcinoma (32,33).
These studies emphasise the roles and clinical significance of
lncRNAs in cancer biology.
ANRIL, transcribed as a 3.8-kb lncRNA in the
opposite direction from the INK4b/ARF/INK4a gene cluster, was first
identified following genetic analysis of familial melanoma patients
with neural tumours (19).
Recently, some genome-wide association studies have identified
ANRIL as a risk locus for several other cancers, including breast
cancer, pancreatic carcinoma, nasopharyngeal carcinoma, basal cell
carcinoma, glioma and leukemia (22–27).
Inspired by these lines of evidence, we investigated ANRIL
expression in SOC and analysed its clinical significance in the
present study. Our data revealed that ANRIL expression levels in
SOC tissues were clearly higher than those in non-cancerous
tissues, strongly suggesting the possibility that ANRIL can be used
as a potential biomarker to detect SOC. Furthermore, elevated ANRIL
expression was associated with advanced FIGO stage, high
histological grade and lymph node metastasis, indicating that
overexpression of ANRIL may facilitate a more malignant ovarian
cancer phenotype as well as metastasis. Importantly, the univariate
and multivariate survival analyses showed that overexpression of
ANRIL was an independent factor for predicting OS in SOC patients,
demonstrating that ANRIL may act as a crucial prognostic biomarker
for SOC patients. Thus, the examination of ANRIL expression could
be used as an additional tool in identifying those SOC patients at
increased risk for tumour metastasis and/or a poorer prognosis.
Previous studies have revealed that ANRIL
contributes to various cellular events in many cancers, such as
facilitated cell proliferation and senescence (20,21),
suggesting a pro-cancerogenic role of the transcript. Encouraged by
these earlier studies and our findings that overexpression of ANRIL
was associated with lymph node metastasis and poor prognosis, it is
therefore a logically hypothesis that ANRIL may be involved in SOC
metastasis. Consistent with this hypothesis, our data demonstrated
that ANRIL expression levels in highly metastatic SOC cell sublines
(SK-OV-3.ip1 and HO8910-PM) were significantly higher than those in
parental cells (SK-OV-3 and HO8910), indicating the metastatic
potential of ANRIL. Subsequent wound-healing and Matrigel invasion
assays showed that siRNA-mediated knockdown of ANRIL attenuated the
ability of either cell migration or invasion in both SK-OV-3.ip1
and HO8910-PM cells. These results collectively suggest that ANRIL
is an important factor for cell migration and invasion of SOC.
To date, the regulatory molecular events associated
with ANRIL are not clear. Several previous reports have established
that ANRIL has a regulatory effect on its neighbours CDKN2A/B
(20,21,34);
however, there is also evidence that ANRIL acts on certain genes
that do not appear to be downstream to CDKN2A/B (35,36),
reflecting a very complex regulatory panorama for this lncRNA. To
investigate the downstream molecular events involving ANRIL and SOC
invasiveness and/or metastasis in the current study, we compared
SK-OV-3. ip1-ANRIL-siRNA1 cells and SK-OV-3.ip1-si-NC cells using a
human tumour metastasis real-time PCR array, containing 84
well-known cell invasion/metastasis-related genes. Notably, the
mRNAs of six genes were differentially expressed (>2-fold; i.e.,
MMP3, MTA1, FN1 and MET were downregulared, and CDH1 and TIMP2 were
upregulated). Subsequently, downregulation of MET and MMP3 protein
was confirmed by western blotting in both SK-OV-3.ip1 and HO8910-PM
cells. Taken together, these results suggest that ANRIL might
regulate SOC cell migration/invasion by regulating MET and MMP3.
Future efforts will be devoted to exploring the underlying
molecular mechanism through which ANRIL regulates MET and MMP3.
In conclusion, ANRIL is overexpressed in SOC, and
its overexpression correlates with an aggressive/poor prognostic
phenotype of the tumour. Furthermore, functional studies suggest a
critical role of ANRIL in the control of SOC cell
migration/invasion at least in part through regulation of MET and
MMP3. These data highlight the significance of ANRIL in SOC
progression, suggesting that ANRIL may be a crucial predictor for
SOC metastasis/poor prognosis and a potential therapeutic
target.
Acknowledgements
This study was supported by funding from the
National Natural Science Foundation of China (81370689; to K.-Q.H.)
and from Shanghai Science and Technology Development Funds for the
Talents (15YF1401400; to J.-J.Q.).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bowtell DD: The genesis and evolution of
high-grade serous ovarian cancer. Nat Rev Cancer. 10:803–808. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Feigenberg T, Clarke B, Virtanen C,
Plotkin A, Letarte M, Rosen B, Bernardini MQ, Kollara A, Brown TJ
and Murphy KJ: Molecular profiling and clinical outcome of
high-grade serous ovarian cancer presenting with low- versus
high-volume ascites. Biomed Res Int. 2014:3671032014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lengyel E: Ovarian cancer development and
metastasis. Am J Pathol. 177:1053–1064. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rustin G, van der Burg M, Griffin C, Qian
W and Swart AM: Early versus delayed treatment of relapsed ovarian
cancer. Lancet. 377:380–381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar :
|
|
10
|
Yuan SX, Yang F, Yang Y, Tao QF, Zhang J,
Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al: Long noncoding RNA
associated with microvascular invasion in hepatocellular carcinoma
promotes angiogenesis and serves as a predictor for hepatocellular
carcinoma patients’ poor recurrence-free survival after
hepatectomy. Hepatology. 56:2231–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
de Kok JB, Verhaegh GW, Roelofs RW,
Hessels D, Kiemeney LA, Aalders TW, Swinkels DW and Schalken JA:
DD3(PCA3), a very sensitive and specific marker to detect prostate
tumors. Cancer Res. 62:2695–2698. 2002.PubMed/NCBI
|
|
12
|
Sun M, Xia R, Jin F, Xu T, Liu Z, De W and
Liu X: Downregulated long noncoding RNA MEG3 is associated with
poor prognosis and promotes cell proliferation in gastric cancer.
Tumour Biol. 35:1065–1073. 2014. View Article : Google Scholar
|
|
13
|
Quagliata L, Matter MS, Piscuoglio S,
Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z,
Boldanova T, et al: Long noncoding RNA HOTTIP/HOXA13 expression is
associated with disease progression and predicts outcome in
hepatocellular carcinoma patients. Hepatology. 59:911–923. 2014.
View Article : Google Scholar :
|
|
14
|
Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H
and Jia W: Overexpression of long noncoding RNA PCAT-1 is a novel
biomarker of poor prognosis in patients with colorectal cancer. Med
Oncol. 30:5882013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW,
Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, et al: Rapid identification
of UCA1 as a very sensitive and specific unique marker for human
bladder carcinoma. Clin Cancer Res. 12:4851–4858. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tano K and Akimitsu N: Long non-coding
RNAs in cancer progression. Front Genet. 3:2192012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu W, Gius D, Onyango P, Muldoon-Jacobs K,
Karp J, Feinberg AP and Cui H: Epigenetic silencing of tumour
suppressor gene p15 by its antisense RNA. Nature. 451:202–206.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
El Messaoudi-Aubert S, Nicholls J,
Maertens GN, Brookes S, Bernstein E and Peters G: Role for the
MOV10 RNA helicase in polycomb-mediated repression of the INK4a
tumor suppressor. Nat Struct Mol Biol. 17:862–868. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pasmant E, Laurendeau I, Héron D, Vidaud
M, Vidaud D and Bièche I: Characterization of a germ-line deletion,
including the entire INK4/ARF locus, in a melanoma-neural system
tumor family: Identification of ANRIL, an antisense noncoding RNA
whose expression coclusters with ARF. Cancer Res. 67:3963–3969.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kotake Y, Nakagawa T, Kitagawa K, Suzuki
S, Liu N, Kitagawa M and Xiong Y: Long non-coding RNA ANRIL is
required for the PRC2 recruitment to and silencing of p15(INK4B)
tumor suppressor gene. Oncogene. 30:1956–1962. 2011. View Article : Google Scholar :
|
|
21
|
Yap KL, Li S, Muñoz-Cabello AM, Raguz S,
Zeng L, Mujtaba S, Gil J, Walsh MJ and Zhou MM: Molecular interplay
of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by
polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell.
38:662–674. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Turnbull C, Ahmed S, Morrison J, Pernet D,
Renwick A, Maranian M, Seal S, Ghoussaini M, Hines S, Healey CS, et
al: Breast Cancer Susceptibility Collaboration (UK): Genome-wide
association study identifies five new breast cancer susceptibility
loci. Nat Genet. 42:504–507. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen J, Li D, Wei C, Sen S, Killary AM,
Amos CI, Evans DB, Abbruzzese JL and Frazier ML: Aurora-A and p16
polymorphisms contribute to an earlier age at diagnosis of
pancreatic cancer in Caucasians. Clin Cancer Res. 13:3100–3104.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bei JX, Li Y, Jia WH, Feng BJ, Zhou G,
Chen LZ, Feng QS, Low HQ, Zhang H, He F, et al: A genome-wide
association study of nasopharyngeal carcinoma identifies three new
susceptibility loci. Nat Genet. 42:599–603. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stacey SN, Sulem P, Masson G, Gudjonsson
SA, Thorleifsson G, Jakobsdottir M, Sigurdsson A, Gudbjartsson DF,
Sigurgeirsson B, Benediktsdottir KR, et al: New common variants
affecting susceptibility to basal cell carcinoma. Nat Genet.
41:909–914. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rajaraman P, Melin BS, Wang Z,
McKean-Cowdin R, Michaud DS, Wang SS, Bondy M, Houlston R, Jenkins
RB, Wrensch M, et al: Genome-wide association study of glioma and
meta-analysis. Hum Genet. 131:1877–1888. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sherborne AL, Hosking FJ, Prasad RB, Kumar
R, Koehler R, Vijayakrishnan J, Papaemmanuil E, Bartram CR,
Stanulla M, Schrappe M, et al: Variation in CDKN2A at 9p21.3
influences childhood acute lymphoblastic leukemia risk. Nat Genet.
42:492–494. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu D, Wolf JK, Scanlon M, Price JE and
Hung MC: Enhanced c-erbB-2/neu expression in human ovarian cancer
cells correlates with more severe malignancy that can be suppressed
by E1A. Cancer Res. 53:891–898. 1993.PubMed/NCBI
|
|
29
|
Bai F, Feng J, Cheng Y, Shi J, Yang R and
Cui H: Analysis of gene expression patterns of ovarian cancer cell
lines with different metastatic potentials. Int J Gynecol Cancer.
16:202–209. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Y, Dong L, Cui H, Shen DH, Wang Y,
Chang XH, Fu TY, Ye X and Yao YY: Up-regulation of mitochondrial
antioxidation signals in ovarian cancer cells with aggressive
biologic behavior. J Zhejiang Univ Sci B. 12:346–356. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shenhua X, Lijuan Q, Hanzhou N, Xinghao N,
Chihong Z, Gu Z, Weifang D and Yongliang G: Establishment of a
highly metastatic human ovarian cancer cell line (HO-8910PM) and
its characterization. J Exp Clin Cancer Res. 18:233–239.
1999.PubMed/NCBI
|
|
32
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M, et al: Characterization of HULC, a novel gene with
striking up-regulation in hepatocellular carcinoma, as noncoding
RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Du Y, Kong G, You X, Zhang S, Zhang T, Gao
Y, Ye L and Zhang X: Elevation of highly up-regulated in liver
cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell
proliferation via down-regulating p18. J Biol Chem.
287:26302–26311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Congrains A, Kamide K, Oguro R, Yasuda O,
Miyata K, Yamamoto E, Kawai T, Kusunoki H, Yamamoto H, Takeya Y, et
al: Genetic variants at the 9p21 locus contribute to
atherosclerosis through modulation of ANRIL and CDKN2A/B.
Atherosclerosis. 220:449–455. 2012. View Article : Google Scholar
|
|
35
|
Sato K, Nakagawa H, Tajima A, Yoshida K
and Inoue I: ANRIL is implicated in the regulation of nucleus and
potential transcriptional target of E2F1. Oncol Rep. 24:701–707.
2010.PubMed/NCBI
|
|
36
|
Congrains A, Kamide K, Katsuya T, Yasuda
O, Oguro R, Yamamoto K, Ohishi M and Rakugi H: CVD-associated
non-coding RNA, ANRIL, modulates expression of atherogenic pathways
in VSMC. Biochem Biophys Res Commun. 419:612–616. 2012. View Article : Google Scholar : PubMed/NCBI
|