Introduction
The growth and metastatic spread of malignant tumors
cannot proceed without the development of a vascular supply.
Vascular endothelial growth factor-A (VEGF-A) plays a key role in
tumor angiogenesis (1–4). The significant amount of VEGFR
expression in the tumor vasculature presents a unique opportunity
for therapeutic intervention. VEGF and its receptor VEGFR-1/VEGFR-2
provide an alternative approach for destroying tumor endothelium
through targeting in combination with agents that kill cells,
making them targets for the delivery of potent toxins to tumor
endothelial cells (5,6). VEGF mRNA is alternatively spliced,
leading to proteins that are 208, 189, 165, or 121 amino acids in
length (7). VEGF165 and
VEGF121 are secreted as soluble factors; however,
VEGF208 and VEGF189 are secreted while
binding to the extracellular matrix (8). Compared with VEGF121,
VEGF165 retains a heparin-binding domain, which induces
binding to the cell surface receptor. Furthermore,
VEGF165 is the most abundantly expressed splice variant
(9). In the present study, we
chose melittin for fusion with VEGF. This fusion protein, denoted
as VEGFR165-melittin, was shown to potently inhibit
hepatocellular carcinoma and pancreatic cancer in vivo and
in vitro.
Melittin is the principal toxic component in venom
from the European honey bee Apis mellifera. This protein is
a cationic, hemolytic and small linear peptide composed of 26 amino
acid residues. Notably, the N-terminus is predominantly hydrophobic
and the C-terminus is hydrophilic. Melittin has various effects,
including antibacterial, antiviral, and anti-inflammatory effects,
in various cell types (10). It
has been reported that melittin can induce apoptosis, cell cycle
arrest and growth-inhibition in different tumor cells (11–13).
However, the significant toxicity of melittin is achieved through a
highly non-specific cytolytic attack of lipid membranes (14). The principle of the melittin
toxicity is its physical and chemical destruction of cellular
membranes, leading to a profound increase in the cell permeability
barrier and leakage of cell contents (15,16),
thereby precluding any meaningful therapeutic benefit. An
alternative approach for achieving practical therapeutic
applications would be designing a new paradigm for the targeted
delivery of potent toxins to tumor cells. Moreover, it has been
reported that melittin suppresses tumor growth by targeting VEGF
(17,18). Therefore, melittin as a fusion
partner should work well with VEGF.
In the present study, we prepared a novel fusion
protein, VEGF165-melittin, in Pichia pastoris. We
generated an effective method for producing the recombinant protein
in large quantities with high purity. Our results demonstrate that
VEGF165-melittin retains functional activities including
cytotoxicity and growth inhibition in HepG-2 and MHCC97-H human
hepatocellular carcinoma cells in vitro. Furthermore, the
fusion toxin was able to inhibit tumor growth in vivo. This
fusion protein has the potential to be used as a new paradigm for
the targeted delivery of cell-penetrating toxins to kill cancer
cells in vitro and in vivo.
Materials and methods
Reagents and materials
Pichia pastoris X-33, the pPICZαC vector, and
Zeocin antibiotic were obtained from Invitrogen (Carlsbad, CA,
USA). Restriction enzymes, T4 DNA ligase, DNA marker, and the
pMD-18T vector were purchased from Takara (Dalian, China). The
protein marker was purchased from Thermo Fermentas and New England
Biolabs (Guangzhou, China). All primers were synthesized by
Shanghai Sangon Biotechnology Corp. (Shanghai, China).
Anti-VEGF165, anti-VEGFR-1, anti-VEGFR-2, anti-melittin,
HRP-goat anti-rabbit conjugate and HRP-goat anti-mouse conjugate
were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Melittin was purchased from Nanning Innovation and Technology
Pharmaceutical Co., Ltd. (Guangxi, China). Anti-VEGF blocking
antibody was purchased from R&D Systems (Minneapolis, MN, USA).
VEGFR-2/KDR gene was purchased from Sino Biological Inc. (Beijing,
China).
Human hepatocellular carcinoma cell lines (HepG-2
and MHCC97-H), a human pancreatic adenocarcinoma cell line
(AsPC-1), and 293 human primary embryonic kidney cells were
obtained from the American Type Culture Collection. All the cells
were passaged according to their protocol from ATCC, and no more
than 6 months elapsed after the resuscitation and culturing of the
cells. Serum and culture medium were purchased from Invitrogen.
BALB/c mice and BALB/c nude mice (4–5 weeks) were obtained from the
Experimental Animal Research Centre of Zhongshan University and
raised in its laboratory. All animal protocols followed the
National Guidelines for the Care and Use of Animals.
Yeast culture media
Pichia pastoris was cultured in YPD medium
containing 10 g/l yeast extract, 20 g/l peptone and 20 g/l
D-glucose. To prepare YPD plates, 2% agar (w/v) was added into YPD
medium. YPD-Zeocin plates containing 0.1 mg/ml Zeocin were used for
the selection of transformants. The Pichia pastoris cells
were grown in BMGY medium (1% yeast extract, 2% peptone, 1%
glycerol, 1.34% yeast nitrogen base and 0.1 M potassium phosphate,
pH 6.0) and BMMY medium (1% yeast extract, 2% peptone, 0.5%
methanol, 1.34% yeast nitrogen base and 0.1 M potassium phosphate,
pH 6.0) for induction.
Construction of expression vector
containing pPICZαC/VEGF165-melittin
A DNA insert encoding melittin was prepared via
artificial synthesis. A linker containing (GGGGS)4,
EcoRI, ApaI, AccI and XbaI sequences
were appended when the synthetic fragment was designed. Then, the
melittin DNA fragment was digested with EcoRI and
XbaI and ligated into a linearized pPICZαC vector to
generate the plasmid pPICZαC/melittin.
To clone the VEGF165 gene,
reverse-transcription poly-merase chain reaction (RT-PCR) was
performed with the primers 5′-ATT CTC GAG AAG AGA GCA CCC ATG GCA
GAA GGA G-3′ (forward) and 5′-GTA GAA TTC CCG CCT CGG CTT GTC ACA
TTT TTA-3′ (reverse), and total RNA extracted from human hepatoma
(HepG2) cells served as the template. Following digestion with
XhoI and EcoRI, the PCR fragment was cloned into
pPICZαC/melittin and treated with the same endonucleases to
generate the recombinant eukaryotic expression plasmid
pPICZαC/VEGF165-melittin. The recombinant plasmid was
confirmed by restriction analysis and sequencing.
Transformation and screening of
recombinant strains
Recombinant plasmid DNA was linearized with
SacI and then transformed into Pichia pastoris X-33
by electroporation using a MicroPulser (Bio-Rad Laboratories,
Hercules, CA, USA) following the pPICZαC vector manual. The yeast
strains transformed with empty vector pPICZαC plasmid served as a
negative control. The cells were spread on YPD plates containing
Zeocin at 100, 250, 500 and 1,000 mg/ml and incubated at 28°C.
Colonies appeared after 2–3 days of incubation at 28°C. The
inserted foreign gene in the genomic DNA of transformants were
detected by PCR assay using the primers mentioned above. Thirty
cycles of PCR were performed with incubations for 30 sec at 94°C,
30 sec at 55°C and 1.5 min at 72°C.
Optimized expression of the fusion
protein in P. pastoris
To confirm the optimal expression conditions for the
fusion protein, various culture parameters, including induction
time-points and pH values (pH 3.0–7.0 with 0.5 pH intervals), were
evaluated. The processes were the same as above. At specific
intervals, 0.5 ml cell suspensions were removed and then
substituted with the same volume of fresh medium. The cell culture
supernatant was tested by ELISA assays.
Purification of
VEGF165-melittin
VEGF165-melittin production was scaled up
in 2 l BMGY medium based on the process introduced in the
Invitrogen manual (19).
Transformants were cultured at 28°C (pH 6.0) until the culture
reached OD600=2.0–6.0, the cells were harvested by
centrifugation, redissolved in 2 l BMMY medium, and cultured at
28°C with oscillation for 72 h. The fermentation broth was
supplemented every 24 h with 10 ml methanol to maintain the induced
control.
Fermentation supernatant was collected by filtration
(0.45 μm) after harvesting by centrifugation at 12,000 r/min for 15
min. A Ni2+ NTA column (GE Healthcare, Piscataway, NJ,
USA) was equilibrated in binding buffer (20 mM
Na3PO4·12 H2O, pH 7.4, 0.5 M NaCl
and 30 mM imidazole). The supernatant was diluted 3-fold with
binding buffer and loaded onto a Ni2+ NTA column at a
speed of 0.5 ml/min. Then, the column was washed with the same
buffer at a rate of 1.0 ml/min to eliminate unbound proteins. Bound
protein was then eluted from the column with 20 mM
Na3PO4·12 H2O, pH 7.4, 0.5 M NaCl
and 0.2 M imidazole at a rate of 0.8 ml/min. Eluted protein was
then transferred to storage buffer (1X PBS) by chromatography using
a Thermo Scientific Zeba desalting column (Thermo Fisher
Scientific, Waltham, MA USA).
Protein assay
The protein concentrations of the samples were
measured using the Bradford assay with bovine serum albumin as a
standard.
Enzyme-linked immunosorbent assay
Individual wells of ELISA plates (Costar) were
coated with fusion toxin sample supernatants and coating buffer
(Na2CO3-NaHCO3, pH 9.6, dilution:
2 μg/100 μl) overnight at 4°C. The plates were blocked with 2% BSA
in TPBS (PBS1, 0.1% Tween-20, pH 7.2) and incubated for
2 h at room temperature. The primary antibody against rabbit was
used at 1:1,000 and precoated for 2 h at 37°C. After several washes
with TPBS, the plates were incubated with goat anti-rabbit IgG
conjugated to HRP (1:2,000 dilutions at blocking buffer) for 2 h.
The color reaction was implemented with OPD zymolyte containing
0.02% H2O2, and the plates were incubated for
15 min at room temperature in the dark. Then, 50 μl of
H2SO4 solution (2 M) was used to stop the
reaction. Absorbance values at 490 nm were read using an ELX800
microplate reader (Bio-Tek Instruments Inc., Winooski, VT, USA).
After adding stop solution, plate reads were completed within 2
h.
SDS-PAGE and western blot assays
Cell lysates were separated by SDS-PAGE in 10% gels
and transferred to a polyvinylidene difluoride (PVDF) membrane
(Millipore) using a semi-dry electroblotting apparatus (Bio-Rad
Laboratories) at 200 mA for 1 h in Towbin transfer buffer (25 mM
Tris and 192 mm glycine). The membrane was blocked with 2% BSA for
1.5 h at room temperature. Then, the membrane was incubated with
primary antibodies against rabbit for 12 h. After washing, the
membrane was incubated with a goat anti-rabbit IgG antibody
conjugated to HRP (Weijia, Shaanxi, China) that was diluted 1:250.
The bound antibody was developed with 3,30-diaminobenzidine
(DAB).
N-terminal amino acid sequence and mass
spectrometric analyses
The N-terminal amino acid sequence of the
VEGF165-melittin fusion protein was determined by
automated Edman degradation, which was performed with a model
protein sequencer-491 (Applied Biosystems, Foster City, CA, USA).
The purified protein was adsorbed onto a PVDF membrane (ProSorb)
and sequenced using established protocols. Mass spectrometric
analysis of VEGF165-melittin was performed with an
autoflex speed MALDI-TOF/TOF MS (Brucker Daltonics, Billerica, MA,
USA).
Reverse transcription-polymerase chain
reaction
Total cellular RNA was extracted from cell cultures
using the RNAiso reagent (Takara, Tokyo, Japan) according to the
manufacturer's protocol. RNA concentration was detected using a
BioPhotometer (Eppendorf Scientific, Hamburg, Germany). Reverse
transcription of total RNA primed with an oligo(dT) oligonucleotide
was done with M-MLV reverse transcriptase (Promega, Mannheim,
Germany) according to the instructions of the manufacturer.
First-strand complementary DNA was amplified using Takara Ex Taq
(Takara).
The primers for the respective genes were designed
as follows: VEGF, 5′-GCA CCC ATG GCA GAA GGA-3′ (forward) and
5′-TTC TGT ATC AGT CTT TCC-3′ (reverse); VEGFR-1, 5′-GAA GGC ATG
AGG ATG AGA-3′ (forward) and 5′-CAG GCT CAT GAA CTT GAA-3′
(reverse); KDR/VEGFR-2, 5′-CAT GTA CGG TCT ATG CCA-3′ (forward) and
5′-CGT TGG CGC ACT CTT CCT-3′ (reverse); and β-actin, 5′-TTC CTG
GGC ATG GAG TCC-3′ (forward) and 5′-CGC CTA GAA GCA TTT GCG-3′
(reverse). RT-PCR products were analyzed by electrophoresis on a 1%
agarose gel.
Cytotoxicity assay
Cells were seeded in 96-well plates at
5–10×104 cells/well. Cells were then starved with phenol
red-free Dulbecco's modified Eagle's medium plus 1% dialyzed fetal
calf serum (A15-107; PAA Laboratories, Dartmouth, MA, USA) for 24
h. The experiment included six VEGF165-melittin fusion
protein groups (0, 0.8, 1.6, 3.2, 6.4 and 12.8 μg/ml). Human
primary embryonic kidney cells (n=293) were used for control. Cell
growth was induced by the fusion toxin for 48 h and then measured
with the MTT assay. Absorbance at 570 nm was detected with a
reference at 630 nm serving as a blank. The influence of the fusion
toxin on cell activity was evaluated and compared with control. The
control cells were set to 100% activity. The mean value of 5 wells
was counted, and triplicates were used in each experiment.
To test VEGFR-mediated effects of
VEGF165-melittin fusion protein on the proliferation and
viability of human cancer cells, HepG-2 cells was used for
subsequent studies. HepG-2 cells were cultured as described above.
Five experimental groups were designed and HepG-2 untreated was the
control group.
Inhibitory effects of
VEGF165-melittin on hepatocellular carcinoma and
pancreatic cancer xenografts in nude mice
After hypodermic injection of 2.5×107
HepG-2 or 5×106 AsPC-1 cells in BALB/c athymic nude
mice, initial tumors were observed on day 21. Afterward, all mice
in the experimental groups were intravenously injected with 0.2 mg
VEGF165-melittin daily for 28 days, and PBS was used as
a control. The subcutaneous tumor parameters were measured every
day, including the length, width and height. The tumor volume
(mm3) was estimated according to the equation
a2b/2, where a is the short diameter (mm) and b is the
long diameter (mm). The tumor weights were measured after the mice
were sacrificed. The tumor samples were maintained in formalin, and
an assessment of mortality was performed.
Results
VEGF165-melittin expression
and optimization
A plasmid was created to express the
VEGF165 fragment fused to melittin to generate a 25 kD
VEGF165-melittin fusion toxin. The structure of the
details of VEGF165-melittin is shown in Fig. 1A. pPICZαC/melittin is based on the
Pichia pastoris expression vector pPICZαC. This vector was
used to express the VEGF165-melittin fusion protein,
which is composed of the melittin fragment cut from the ApaI
and XbaI sites following the VEGF165 fragment. A
linker, (GGGGS)4, was synthesized for spatial
configuration of the fusion toxin. The VEGF165 sequence
was amplified and inserted into the pPICZαC/melittin expression
vector to create pPICZαC/VEGF165-melittin. Sequence
analysis of the plasmid DNA was used to confirm integration in
positive colonies.
After electroporation with SacI-linearized
pPICZαC/VEGF165-melittin, 90% of transformants were
Mut+. PCR analysis of genomic DNA demonstrated that the
gene of interest was integrated into the stable transformants, and
no similar bands were observed for negative control samples.
The positive transformants were germinated in BMGY
medium and induced in BMMY medium at 28°C for 7 days. The volume of
the culture medium was 10 ml. After 3 days, the culture
supernatants were analyzed by SDS-PAGE. The results indicated that
the molecular weight of VEGF165-melittin was consistent
with the predicted size of 25 kDa (Fig. 1B).
Transformants expressing a high level of fusion
protein were selected, and one was chosen for the scaling up. Based
on analysis of optimized expression conditions, the parameters used
were as follows: pH: 6.0, induction time-point: 72 h, and final
methanol concentration: 0.5% (v/v) (Fig. 1C and D).
VEGF165-melittin fermentation
and purification
VEGF165-melittin supernatant was purified
by Ni2+ affinity chromatography and Thermo Scientific
Zeba desalting column chromatography. Following these processes,
~160 mg pure recombinant protein was obtained from 2 l fermentation
liquor. SDS-PAGE analysis demonstrated that the purity of
VEGF165-melittin was ~95% (Fig. 2A). At every step of purification,
the recovery, purity and yield of the fusion toxin were estimated
as shown in Table I.
 | Table ISummary of purification process of
VEGF165-melittin from 2 liters of culture supernatant
purification. |
Table I
Summary of purification process of
VEGF165-melittin from 2 liters of culture supernatant
purification.
| Purification
steps | Total protein
(mg/l) |
VEGF165-melittin (mg/l) | Purity (%) | Recovery (%) |
|---|
| Supernatants | 256.5 | 154.2 | 60.1 | |
|
Ni2+-NTA | 121.1 | 115.3 | 95.2 | 74.8 |
| Desalting
column | 84.3 | 80.3 | 95.3 | 52.1 |
Western blot assays were used to preliminarily
evaluate the purified recombinant protein. The identity of
VEGF165-melittin was confirmed by immunoreactivity with
a rabbit anti-human VEGF165 polyclonal antibody
(Fig. 2B). The results were
consistent with our expectations. No band was observed in lane 1,
which contains the supernatant of the X33 pPICZαC transformant.
Molecular weight and N-terminal
sequencing analyses
To verify the molecular weight and integrity of the
recombinant protein, mass spectrometry was performed using purified
VEGF165-melittin. The expected molecular mass
VEGF165-melittin is 221 amino acids, and it primarily
exists in solution as a homodimer due to a disulfide linkage in the
linker. The results of the molecular weight analysis of the fusion
toxin are shown in Fig. 3, and
they are in accordance with our previous results, indicating that
the purified recombinant toxin is the expected
VEGF165-melittin protein.
According to N-terminal sequencing analysis, the
first 15 amino acids of the purified peptide were A P M A E G G G Q
N H H E V V. These were consistent with the N-terminal sequence of
VEGF165-melittin, thus indicating successful expression
and purification of this protein.
Cytotoxicity assay
The effects of VEGF165-melittin on the
proliferation and viability of human hepatocellular carcinoma cell
lines (HepG-2 and MHCC97-H), human pancreatic adenocarcinoma cell
lines AsPC-1 and human primary embryonic kidney cells 293 were
studied for a 72-h period. The fusion protein was applied to the
cells at seven different final concentrations. Fig. 4 shows the proliferation and
viability changes that occurred during treatment. Cell counts and
an MTT-assay indicated that the fusion toxin influenced the
proliferation of HepG-2 and MHCC97-H cells more significantly than
that of AsPC-1 cells. The proliferation of the HepG-2 cells
significantly decreased by 55% in an MTT assay (P<0.01).
However, an effect of the fusion toxin on the viability of 293
cells was not observed, even at the highest
VEGF165-melittin dose.
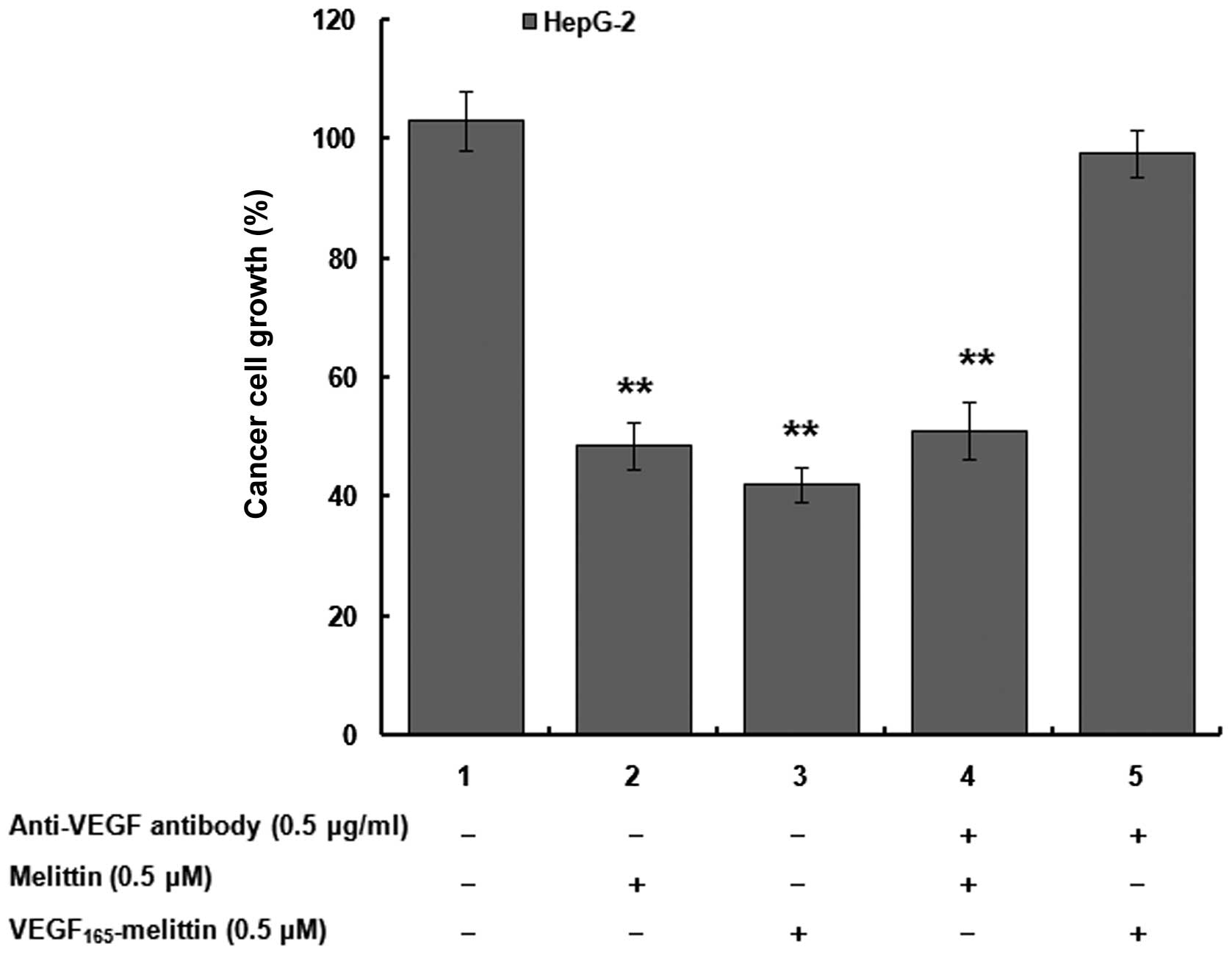
To further assess the mediation effects between
VEGF165-melittin and VEGFR inhibition, human
hepatocellular carcinoma cells HepG-2 were incubated with
VEGF165-melittin or melittin for 48 h in the presence or
absence of 0.5 μg/ml anti-VEGF antibody. Fig. 5 shows the proliferation of the
HepG-2 cells significantly decreased when melittin or
VEGF165-melittin was added (P<0.01). However, in
VEGF165-melittin groups, the inhibitory activity was not
observed after incubated with the anti-VEGF antibody. This
sensitivity of HepG-2 might be mediated by VEGFR present on HepG-2
cells, since 293 cells without known VEGF receptors were not
affected by VEGF165-melittin at high concentrations
(Fig. 4). Presence of VEGFR
appears to be necessary for induction of HepG-2 cell death by
VEGF165-melittin.
VEGF165-melittin-mediated
tumor growth inhibition in vivo
In the HepG-2 xenograft nude mouse model, the
average tumor volume in VEGF165-melittin mice was 843
mm3, and it was 1,769 mm3 in control mice
(Fig. 6A). Therefore, the
inhibitory rate of the average tumor volume was 52.3%. Twenty-eight
days after treatment with the fusion toxin, the survival was 100%
for VEGF165-melittin mice and 60% for control mice
(Fig. 6B). In the AsPC-1 xenograft
nude mouse model, inhibition of the average tumor volume in the
experimental group was 34.4% as compared with the control group.
Significant VEGF165-melittin-mediated inhibition of
tumor growth was demonstrated. Based on these results, it was
obvious that there were stronger effects in HepG-2 compared with
AsPC-1 cells, which suggests that the high expression level of
VEGFR-2 in HepG-2 might mediate this influence.
Specific toxicity of
VEGF165-melittin targeting VEGFR-2
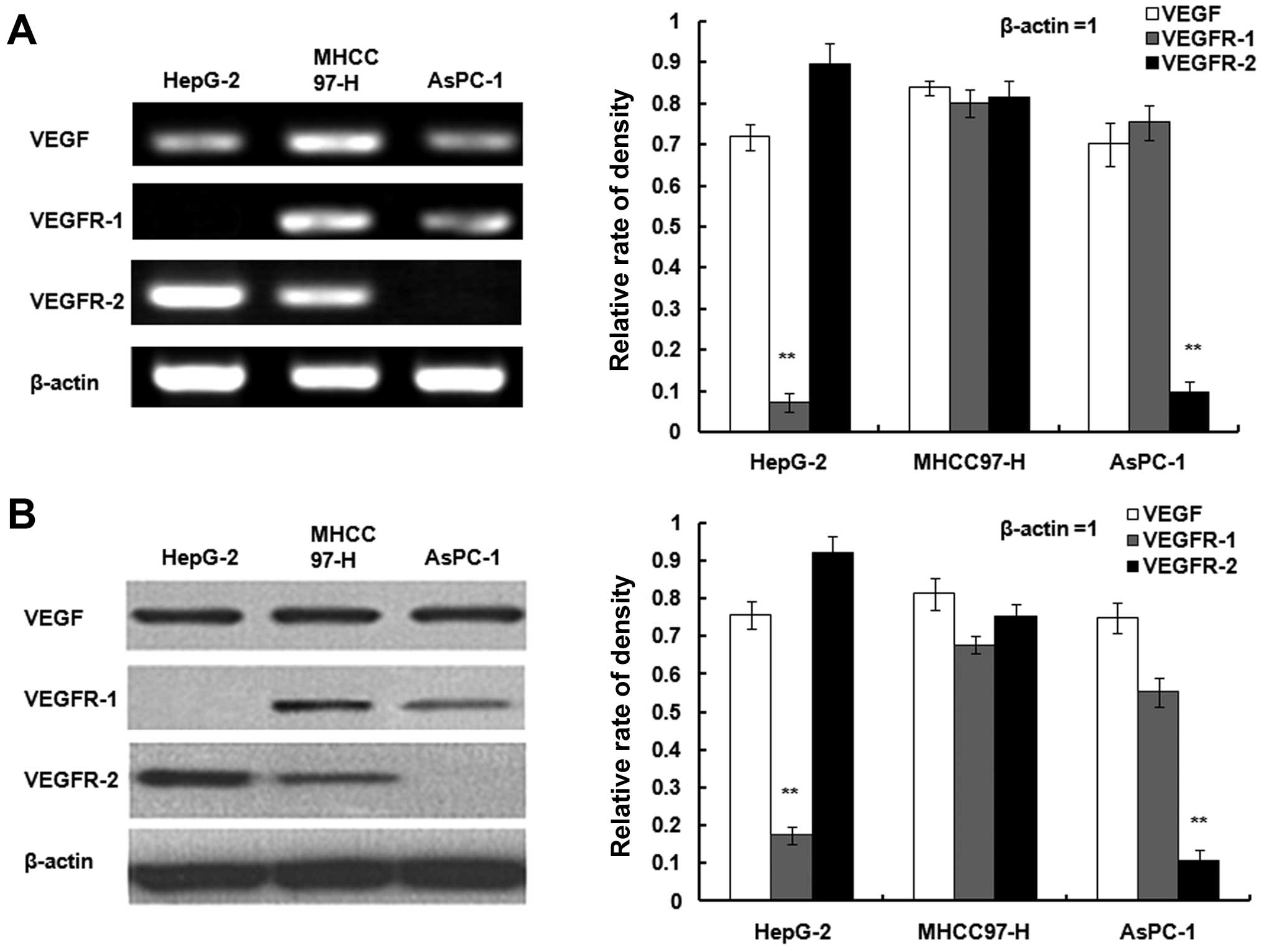
Expression of VEGF, VEGFR-1 and KDR/VEGFR-2 in
HepG-2 and MHCC97-H cells as well as AsPC-1 cells were determined
with RT-PCR and western blot assays (Fig. 7A and B). All three cell lines
exhibited VEGF. MHCC-97H cells were positive for VEGFR-1 and
KDR/VEGFR-2. KDR/VEGFR-2 was expressed in HepG-2 and VEGFR1 was
expressed in AsPC-1.
Furthermore, KDR/VEGFR-2 was overexpressed in 293
cells to evaluate the specific targeting of fusion protein
(Fig. 8A). The effect of
VEGF165-melittin on the proliferation and viability of
293 cells, 293 transfected with pCMVp-NEO-BAN/KDR plasmid (293/KDR)
and HepG-2 cells was studied. The 293 cells transfected with
pCMVp-NEO-BAN empty plasmid (293/pCMV) were used as control. In
targeted cells, the cytotoxicity of VEGF165-melittin was
strongly dependent on the VEGFR-2 density. Fig. 8B shows proliferation and viability
changes using the MTT assay.
Statistical analysis
Statistical analysis was performed using Statistical
Package for Social Sciences (SPSS) 13.0 software. Data are
presented as the means ± SD. Statistical significance was
determined by one-way analysis of variance or the t-test. P-values
<0.05 were considered to be statistically significant.
Discussion
It is well known that tumor cell-derived VEGF is a
key factor that acts on endothelial cells to promote angiogenesis,
tumor growth and metastasis. Targeting proangiogenic mediators such
as VEGF/VEGFR has emerged as a promising anti-cancer treatment
strategy. In particular, VEGF fusion proteins have become an
important aspect in novel cancer treatment strategies (20). In the present study, we constructed
a protein containing VEGF165 fused to melittin
(VEGF165-melittin). Successful expression of active
VEGF165-melittin was achieved in Pichia pastoris
with yields >80 mg/l. N-terminal sequencing and mass
spectrometric analysis verified that the fusion toxin was expressed
and purified as expected. MTT and xenografts assays demonstrated
that VEGF165-melittin inhibited tumor growth in
vivo and in vitro.
Among the identified proangiogenic regulators, VEGF,
particularly VEGF-A and its two tyrosine kinase receptors, fms-like
tyrosine kinase receptor (Flt1 and VEGFR-1) and kinase insert
domain-containing receptor (KDR/FLK1 and VEGFR-2), have been
identified as key mediators of the regulation of pathologic blood
vessel growth and maintenance (21). In our results,
VEGF165-melittin was more effective in HepG-2 than
MHCC97-H cells. This diversity may be caused by differences in the
VEGFR-1 and VEGFR-2 proteins expressed in HepG-2 and MHCC97-H
cells. In subsequent studies, the expression of VEGF and the VEGF
receptors (VEGFR-1 and VEGFR-2) was evaluated in HepG-2 and
MHCC97-H cells by RT-PCR and western blot assays. Compared with the
results we reported here, the VEGF165-melittin fusion
toxin should be selective in targeting tumor cells that overexpress
VEGFR-2. We hypothesize that the enhanced efficacy of VEGF fusion
toxin may be due to the overexpression of VEGFR-2 in growing cells.
Subsequently, 293 human primary embryonic kidney cells (293/KDR)
overexpressing VEGFR-2 was constructed in our laboratory.
VEGF165-melittin inhibited growth of 293/KDR cells at a
dose of 6.4 μg/ml. These effects were mediated by VEGFR-2, since
the parental 293 cells lacking VEGFR-2 were not inhibited by fusion
protein.
Melittin is a main component of bee venom. It is a
small peptide with a linear structure composed of 26 amino acids
(22). Bee venom has a wide range
of effects including antibacterial, antiviral and anti-inflammatory
effects; thus, it has been extensively used in the field of
traditional medicine including treatments for back pain, rheumatism
and skin diseases (23,24). Furthermore, it has been shown that
bee venom and/or melittin have inhibitory effects on the tumor
growth of cervical, prostate, renal, breast, ovarian and liver
tumor cells (10,25,26).
The results of our research are in accordance with previous studies
demonstrating the suppression of melittin on the growth of human
hepatic carcinoma cell lines (11,27).
Additionally, the fusion toxin VEGF165-melittin
inhibited the proliferation of human hepatocellular carcinoma cell
lines (HepG-2 and MHCC97-H) in a concentration-dependent manner.
The most effective inhibitory concentration of
VEGF165-melittin was 6.4 μg/ml, resulting in an
inhibition ratio of 52.3%. The remarkable suppressive effects on
cell proliferation were observed after 48 h in the experimental
group. The present study indicated that the fusion toxin directly
inhibits the growth of hepG-2 human hepatocellular carcinoma cells
in vitro and in vivo. In follow-up experiments, more
studies will be designed to detect the antitumor activity and
mechanism of this fusion protein.
As a lower eukaryote, Pichia pastoris was
identified as a suitable expression system for various recombinant
proteins that retains biological activity with high quantity
yields, and it also offers the benefits of E. coli
(cost-effective and easy scale-up). In addition, the advantages of
expression in a eukaryotic system include proper protein
processing, folding and post-translational modifications (28,29).
In addition, Pichia pastoris does not secrete large amounts
of intrinsic proteins, resulting in the easy isolation of foreign
proteins. In the present study, VEGF165-melittin
production was performed in a 2-liter fermentor, with yields >80
mg/l. The successful expression and purification of the recombinant
fusion toxin VEGF165-melittin and its activity in human
hepatocellular carcinoma cells demonstrates that the fusion protein
has the potential to be used as a novel cancer treatment strategy.
This is the first report to describe the secretory expression of a
human vascular endothelial growth factor fused to melittin in
Pichia pastoris.
Acknowledgements
The present study was supported by grants from the
Foundation for Distinguished Young Talents in Higher Education in
Guangdong, China (LYM11080), the National Nature Science Foundation
of China (No. 81101542) and the Guangdong Provincial Key Laboratory
of Biotechnology Candidate Drug Research.
Abbreviations:
|
PBS
|
phosphate-buffered saline
|
|
OPD
|
ortho-phenyl-enediamine
|
|
DAB
|
3,3′-diaminobenzidine
|
|
BSA
|
bovine serum albumin
|
References
|
1
|
Carmeliet P and Jain RK: Angiogenesis in
cancer and other diseases. Nature. 407:249–257. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhu W, Kato Y and Artemov D: Heterogeneity
of tumor vasculature and antiangiogenic intervention: Insights from
MR angiography and DCE-MRI. PLoS One. 9:e865832014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferrara N and Ferrara N: Vascular
endothelial growth factor: Molecular and biological aspects. Curr
Top Microbiol Immunol. 237:1–30. 1999.PubMed/NCBI
|
|
4
|
Hicklin DJ and Ellis LM: Role of the
vascular endothelial growth factor pathway in tumor growth and
angiogenesis. J Clin Oncol. 23:1011–1027. 2005. View Article : Google Scholar
|
|
5
|
Zhang D, Li B, Shi J, Zhao L, Zhang X,
Wang C, Hou S, Qian W, Kou G, Wang H, et al: Suppression of tumor
growth and metastasis by simultaneously blocking vascular
endothelial growth factor (VEGF)-A and VEGF-C with a
receptor-immunoglobulin fusion protein. Cancer Res. 70:2495–2503.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen AI and Advani RH: Beyond the
guidelines in the treatment of peripheral T-cell lymphoma: New drug
development. J Natl Compr Canc Netw. 6:428–435. 2008.PubMed/NCBI
|
|
7
|
Tischer E, Mitchell R, Hartman T, Silva M,
Gospodarowicz D, Fiddes JC and Abraham JA: The human gene for
vascular endothelial growth factor. Multiple protein forms are
encoded through alternative exon splicing. J Biol Chem.
266:11947–11954. 1991.PubMed/NCBI
|
|
8
|
Houck KA, Leung DW, Rowland AM, Winer J
and Ferrara N: Dual regulation of vascular endothelial growth
factor bioavailability by genetic and proteolytic mechanisms. J
Biol Chem. 267:26031–26037. 1992.PubMed/NCBI
|
|
9
|
Koutsioumpa M, Poimenidi E, Pantazaka E,
Theodoropoulou C, Skoura A, Megalooikonomou V, Kieffer N, Courty J,
Mizumoto S, Sugahara K, et al: Receptor protein tyrosine
phosphatase beta/zeta is a functional binding partner for vascular
endothelial growth factor. Mol Cancer. 14:19–34. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oršolić N: Bee venom in cancer therapy.
Cancer Metastasis Rev. 31:173–194. 2012. View Article : Google Scholar
|
|
11
|
Wang C, Chen T, Zhang N, Yang M, Li B, Lü
X, Cao X and Ling C: Melittin, a major component of bee venom,
sensitizes human hepatocellular carcinoma cells to tumor necrosis
factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis
by activating CaMKII-TAK1-JNK/p38 and inhibiting IκBα kinase-NF-κB.
J Biol Chem. 284:3804–3813. 2009. View Article : Google Scholar
|
|
12
|
Gajski G and Garaj-Vrhovac V: Melittin: A
lytic peptide with anti-cancer properties. Environ Toxicol
Pharmacol. 36:697–705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu S, Yu M, He Y, Xiao L, Wang F, Song C,
Sun S, Ling C and Xu Z: Melittin prevents liver cancer cell
metastasis through inhibition of the Rac1-dependent pathway.
Hepatology. 47:1964–1973. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hoskin DW and Ramamoorthy A: Studies on
anticancer activities of antimicrobial peptides. Biochim Biophys
Acta. 1778:357–375. 2008. View Article : Google Scholar
|
|
15
|
Soman NR, Baldwin SL, Hu G, Marsh JN,
Lanza GM, Heuser JE, Arbeit JM, Wickline SA and Schlesinger PH:
Molecularly targeted nanocarriers deliver the cytolytic peptide
melittin specifically to tumor cells in mice, reducing tumor
growth. J Clin Invest. 119:2830–2842. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee MT, Chen FY and Huang HW: Molecular
mechanism of Peptide-induced pores in membranes. Biochemistry.
43:3590–3599. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huh JE, Kang JW, Nam D, Baek YH, Choi DY,
Park DS and Lee JD: Melittin suppresses VEGF-A-induced tumor growth
by blocking VEGFR-2 and the COX-2-mediated MAPK signaling pathway.
J Nat Prod. 75:1922–1929. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shin JM, Jeong YJ, Cho HJ, Park KK, Chung
IK, Lee IK, Kwak JY, Chang HW, Kim CH, Moon SK, et al: Melittin
suppresses HIF-1α/VEGF expression through inhibition of ERK and
mTOR/p70S6K pathway in human cervical carcinoma cells. PLoS One.
8:e693802013. View Article : Google Scholar
|
|
19
|
Pichia Expression Kit Version F. A Manual
of Methods for Expression of Recombinant Proteins in Pichia
pastoris. Invitrogen. 2002
|
|
20
|
Ciomber A, Smagur A, Mitrus I, Cichoń T,
Smolarczyk R, Sochanik A, Szala S and Jarosz M: Antitumor effects
of recombinant antivascular protein ABRaA-VEGF121 combined with
IL-12 gene therapy. Arch Immunol Ther Exp (Warsz). 62:161–168.
2014. View Article : Google Scholar
|
|
21
|
Xu WW, Li B, Lam AK, Tsao SW, Law SY, Chan
KW, Yuan QJ and Cheung AL: Targeting VEGFR1- and VEGFR2-expressing
non-tumor cells is essential for esophageal cancer therapy.
Oncotarget. 6:1790–1805. 2015.PubMed/NCBI
|
|
22
|
Liu M, Zong J, Liu Z, Li L, Zheng X, Wang
B and Sun G: A novel melittin-MhIL-2 fusion protein inhibits the
growth of human ovarian cancer SKOV3 cells in vitro and in vivo
tumor growth. Cancer Immunol Immunother. 62:889–895. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee MT, Sun TL, Hung WC and Huang HW:
Process of inducing pores in membranes by melittin. Proc Natl Acad
Sci USA. 110:14243–14248. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sommer A, Fries A, Cornelsen I, Speck N,
Koch-Nolte F, Gimpl G, Andrä J, Bhakdi S and Reiss K: Melittin
modulates keratinocyte function through P2 receptor-dependent ADAM
activation. J Biol Chem. 287:23678–23689. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qu L, Jiang M, Li Z, Pu F, Gong L, Sun L,
Gong R, Ji G and Si J: Inhibitory effect of biosynthetic nanoscale
peptide Melittin on hepatocellular carcinoma, driven by survivin
promoter. J Biomed Nanotechnol. 10:695–706. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jo M, Park MH, Kollipara PS, An BJ, Song
HS, Han SB, Kim JH, Song MJ and Hong JT: Anti-cancer effect of bee
venom toxin and melittin in ovarian cancer cells through induction
of death receptors and inhibition of JAK2/STAT3 pathway. Toxicol
Appl Pharmacol. 258:72–81. 2012. View Article : Google Scholar
|
|
27
|
Liu H, Han Y, Fu H, Liu M, Wu J, Chen X,
Zhang S and Chen Y: Construction and expression of sTRAIL-melittin
combining enhanced anticancer activity with antibacterial activity
in Escherichia coli. Appl Microbiol Biotechnol. 97:2877–2884. 2013.
View Article : Google Scholar
|
|
28
|
Su M, Chang W, Cui M, Lin Y, Wu S and Xu
T: Expression and anticancer activity analysis of recombinant human
uPA1-43-melittin. Int J Oncol. 46:619–626. 2015.
|
|
29
|
Wang DD, Su MM, Sun Y, Huang SL, Wang J
and Yan WQ: Expression, purification and characterization of a
human single-chain Fv antibody fragment fused with the Fc of an
IgG1 targeting a rabies antigen in Pichia pastoris. Protein Expr
Purif. 86:75–81. 2012. View Article : Google Scholar : PubMed/NCBI
|