Introduction
Breast cancer is a heterogeneous group of malignant
tumors (1). Clinicopathological
surrogate definitions of subtypes have been used for a long time.
However, these subtypes even have subtypes considering their
distinct responses to available therapy and clinical outcomes
(1,2). Although accumulating evidence
supports the use of multi-gene signatures to make distinctions
among breast cancer patients, the cost of these assays remains
prohibitive (3). The heterogeneity
in tumor cell phenotypes make breast tumor categorization a
challenging task (1).
The phosphoinositide 3-kinase (PI3K) pathway
provides proliferative and migratory signals and is frequently
activated in human breast cancer (4–7). The
PI3K family of enzymes encompasses class I, II and III, with only
class I being involved in human cancer (8–11).
Class IA PI3K consists of a catalytic subunit (p110α as a key
subunit) and a regulatory subunit (p85α as a key subunit decoded by
PIK3R1) (11–13). When lacking upstream signals, p85
stabilizes p110 and suppress its catalytic activities (14). Uchino et al (7) reported that PIK3R1 was
significantly downregulated in MDA-MB-231 cells and MCF-7 invasive
clone compared with MCF-7 cells, thereby possibly contributing to
metastasis development. Another study demonstrated that p85α
downregulation was an independent prognostic marker in breast
cancer (15). Although the
importance of the PI3K/AKT pathway in breast cancer is well known,
the function of p85α in breast cancer has not been widely
studied.
miR-21-5p (previously named miR-21) is one of the
most overexpressed miRNAs in numerous malignancies (16–19).
miR-21 targets many important tumor suppressors to promote breast
cancer growth, proliferation, migration and metastasis (20–22).
We have previously shown that miR-21 was overexpressed in breast
cancer and associated with inferior survival (23). We have reported on human genome
microarray to screen potential targets of miR-21 (24).
In the present study, to elucidate the mechanisms by
which miR-21 regulate breast tumor migration and invasion, we
applied pathway enrichment analysis and target-predicting
algorithms for the screening target of miR-21. PIK3R1 was
predicted to be a functional target of miR-21. We further
investigated the regulation of PIK3R1 coding protein p85α by
miR-21, the impact of changes in antimiR-21 mediated p85α
expression and the clinicopathological and prognostic significance
of p85α in breast cancer patients.
Materials and methods
Cell lines
Human breast cancer cell lines (MCF-10A, MDA-MB-231
and BT-474) were purchased from the American Type Culture
Collection and cultured according to specifications. Human breast
cancer cell lines (MCF-7, BT-549, T47D and SK-BR-3) were purchased
from the Cell Bank of Chinese Academy of Sciences. All cells were
used within 2 months after resuscitation of frozen aliquots.
Quantification of miRNA and mRNA
Total RNA was isolated from cells and tissues using
the Total RNA Purification kit (Norgen Biotek Corp., Thorold, ON,
Canada). miR-21 expression was assessed by quantitative reverse
transcription-polymerase chain reaction (RT-qPCR) analysis using
microRNA PCR system (Exiqon A/S) according to the manufacturer's
instructions. RT-qPCR was utilized to analyze expression changes of
potential miR-21 targets as previously described (23). Primers for PCR amplifications
(Table I) were designed using
Primer5.0 Input (version 0.4.0). Relative mRNA levels were
calculated using the 2−Δ Δ CT method (25).
 | Table ISequences of RNA and DNA
oligonucleotides. |
Table I
Sequences of RNA and DNA
oligonucleotides.
| Name | Sense strand/sense
primer (5′-3′) | Antisense
strand/antisense primer (5′-3′) |
|---|
| Primers for
RT-PCR |
| PIK3R1 |
TTTGCCGAGCCCTATAACT |
TGCATATACTGGGTAGGCTAGT |
| 18s rRNA |
CCTGGATACCGCAGCTAGGA |
GCGGCGCAATACGAATGCCCC |
| Primers for the
3′-UTR of PIK3R1 cloning |
| PIK3R1
XhoIF |
ccgctcgagAGCGCTTACTCTTTGATCCTTCTCC | |
| PIK3R1
NotIR |
CTATTAGGGTAGTGACCATATTATGGTTG | |
| mutPIK3R1-1 |
GTTTTAAATGTACCTTCAGATATTCGATCCCCACCCCAGTTTTTGTT |
AACAAAAACTGGGGTGGGGATCGAATATCTGAAGGTACATTTAAAAC |
| mutPIK3R1-2 |
GTTTTGTTGGGCAGTGCCTGTATTCGATCAAAGCTGCTTTATTCAAT |
ATTGAATAAAGCAGCTTTGATCGAATACAGGCACTGCCCAACAAAAC |
| siRNA duplexes |
| PIK3R1-siRNA1 |
CAAAGGAUUAUGCAUAAUUdTdT |
AAUUAUGCAUAAUCCUUUGdTdT |
| PIK3R1-siRNA2 |
CCAAUAUUCACUGGUGGAAdTdT |
UUCCACCAGUGAAUAUUGGdTdT |
| PIK3R1-siRNA3 |
CUAUUGAAGCAUUUAAUCAdTdT |
UCAUUAAAUGCUUCAAUAGdTdT |
| Control-siRNA |
UUCUCCGAACGUGUCACGUTT |
ACGUGACACGUUCGGAGAATT |
Luciferase reporter assay
The 3′-untranslated region (UTR) of PIK3R1
containing the putative miR-21 target sites was amplified by PCR
from genome DNA derived from HEK293T cells. The synthetic mutant
3′-UTR of PIK3R1 was produced by PCR, and then the PCR
products were cloned into psiCHECK-2 vector. After digestion by
XhoI and NotI, the fragment containing 3′-UTR of
PIK3R1 was cloned into psiCHECK-2 vector (Promega, Madison,
WI, USA). All inserts were sequenced to verify polymerase fidelity.
The PCR primers are listed in Table
I. HEK293T cells were cultured in 24-well plates and
cotransfected with 200 ng of psiCHECK-2 vector containing 3′-UTR of
PIK3R1 and 50 nM of miRNA mimic (Exiqon A/S) per well.
Transfections were performed using Lipofectamine® 2000
(Invitrogen, Carlsbad, CA, USA). The luciferase analysis was
performed 48 h later using the Dual-luciferase reporter assay
system (cat. no. E1910; Promega) according to the manufacturer's
protocol. Firefly luciferase activity was normalized to
Renilla luciferase activity. miRNA mimic negative control
was used as the control miRNA. Experiments were carried out in
triplicate.
Cell transfection and transduction
For transient miR-21 knockdown, the LNA-antimiR-21
or LNA-control (Exiqon A/S, Vedbaek, Denmark) were delivered at a
final concentration of 50 nM using Lipofectamine 2000 (Invitrogen).
For PIK3R1 knockdown, three siRNAs (Sigma-Aldrich, St.
Louis, MO, USA) designed against PIK3R1 (GenBank accession
no. NM_181523) were included (Table
I). One control siRNA (Sigma-Aldrich) exhibiting no significant
sequence similarity to human, mouse or rat gene sequence served as
a negative control. Transfection was performed with Lipofectamine
2000 (Invitrogen) according to the manufacturer's instructions. For
PIK3R1 overexpression, lentivirus was produced by transfecting HEK
293T packaging cells in DMEM (HyClone, Logan, UT, USA; cat. no.
SH30022.01B) with a 3-plasmid system. DNA for transfection was
prepared by mixing pHelper 1.0, pHelper 2.0 and
pLVX-IRES-Neo-PIK3R1. The empty vector pLVX-IRES-Neo was purchased
from Clontech Laboratories (Mountain View, CA, USA; cat. no.
632184), and the plasmid pLVX-IRES-Neo-PIK3R1 was generated by
insertion of PIK3R1 sequence. MDA-MB-231 cells were
transduced with lentivirus in the presence of 6 μg/ml polybrene
(Sigma-Aldrich) for 24 h. Cells were then selected for 7 days in
2.5 mg/ml neomycin. Overexpression of PIK3R1 was confirmed by
western blot analysis.
Cell viability and clonogenic assays
Cell growth and viability were measured by
MTS-formazan reduction using CellTiter 96 Aqueous One Solution Cell
Proliferation Assay (Promega) at 24, 48, 72 and 96 h
post-transfection with a vector (empty pcDNA3.1) or PIK3R1.
Absorbance was measured at 490 nm using a Multiskan plate reader
(Thermo Labsystems, Beverly, MA, USA). Raw values were averaged,
and background absorbance (medium without cells) subtracted. For
this assay cells were plated at 10,000 cells/well in triplicate for
each transfection condition and time-point. Raw values were
averaged, and background absorbance (medium without cells)
subtracted. The cellular effects of these manipulations were
further investigated in MDA-MB-231 and BT-474 cells using
clonogenic assays. Briefly, cells were plated on 6-well plates at
100 and 200 cells/well in triplicate and incubated at 37°C under 5%
CO2 for 2 days post-transfection. After 2 weeks, plates
were washed, fixed in 50% methanol and stained with 0.1% crystal
violet and then the number of colonies was counted.
In vivo tumorigenicity assays
Five-week-old female BALB/c-nude mice, provided by
Shanghai Laboratory Animal Center, Chinese Academy Sciences
(Shanghai, China) were used. Equivalent amounts of MDA-MB-231 cells
transfected with PIK3R1 or vector were injected
subcutaneously (107 cells/tumor) into the left axilla of
nude mice. Mice were weighed, and the longest and the shortest
diameters of the tumor were measured every day. The tumor volume
(V) was calculated according to the following equation: V =
a×b2/2, where a is the longest diameter and b is the
shortest diameter of the tumor (26). Thirty-six days after the initial
injection, the animals were sacrificed and tumors were extracted
and weighed. The ethics guidelines for investigations in conscious
animals were followed in all experiments.
Wound healing/migration assay
To assay the migratory response of breast cancer
cells to miR-21 inhibitor or PIK3R1 expression, the cellular
effects of these manipulations were further investigated using a
wound healing assay as previously described (24). Cells were allowed to reach
confluence before dragging a 1-ml sterile pipette tip (Axygen
Scientific, Inc., Union City, CA, USA) through the monolayer. Cells
were washed with PBS to remove cellular debris and allowed to
migrate for 48 h. Images were acquired at 0, 6, 24 and 48 h
post-wounding with a digital camera system (Leica DFC480; Leica
Microsystems, Bannockburn, IL, USA). Cell-free areas were measured
with ImageJ software (National Institutes of Health, Bethesda, MD,
USA) and were expressed as the percentage of migration compared to
control, arbitrarily set at 100% (27). All experiments were carried out in
triplicate.
In vitro invasion assay
Invasion of cells in vitro was assayed using
the BD BioCoat Matrigel Invasion Chambers and Control Inserts (BD
Biosciences, Bedford, MA, USA) respectively. Each well of a 24-well
plate contained an insert with an 8-μm pore size polyethylene
terephthalate membrane. Cells (1×105 per Transwell) were
suspended in serum-free DMEM and seeded into the upper chamber.
DMEM containing 2% fetal bovine serum was then added to the bottom
chamber of 24-well plates to serve as a chemoattractant. After 48 h
of incubation, cells on the upper surface of the filter were
removed, and cells that migrated to the lower surface were fixed
and stained with 1% toluidine blue. For quantification of cell
invasion, 10 fields per experimental condition were randomly
selected as previously described (28) and micrographed with IX71 microscope
(Olympus, Tokyo, Japan). Images are representative of at least
three independent experiments.
Western blots
Cells were harvested and lysed in
radio-immunoprecipitation buffer (Upstate Biotechnology, Inc., Lake
Placid, NY, USA). Antibodies used for immunoblot analysis were p85α
1:1,000 (Cell Signaling Technology, 13666), p110α 1:1,000 (Cell
Signaling Technology, 42336), p-AKT (Ser473) 1:2,000 (Cell
Signaling Technology, 4060), AKT 1:1,000 (Cell Signaling
Technology, 9272), E-cadherin 1:1,000 (Cell Signaling Technology,
3195), N-cadherin 1:1,000 (Cell Signaling Technology, 13116),
vimentin 1:1,000 (Abcam, 92547), FSP1 1:1,000 (Cell Signaling
Technology, 13018), snail 1:1,000 (Cell Signaling Technology, 3879)
and slug 1:1,000 (Cell Signaling Technology, 9585). GAPDH 1:3,000
(Santa Cruz Biotechnology, sc-32233) or β-actin 1:1,000 (Cell
Signaling Technology, 8457) were used as loading controls. All
bands were detected using a SuperSignal West Pico Chemiluminescent
Substrate (Pierce, Rockford, IL, USA).
Tissue specimens
Eligible patients were women with invasive breast
cancer, no special type; operable; no previous chemotherapy;
adequate formalin-fixed paraffin-embedded (FFPE) tumor specimens
from the pre-treatment biopsy or surgery sample for representation
in tissue microarrays (TMAs); outcome data available. Patients with
distant metastases or a history of a previous or concomitant
malignancy were excluded. The archived FFPE tissues were obtained
from the Department of Pathology, Guangdong General Hospital
between 2009 and 2012. A consensus diagnosis of invasive breast
cancer was confirmed by two expert pathologists according to the
fourth edition of the World Health Organization (WHO)
classification of tumors of the breast, published in 2012 (29). The surrogate definition of
intrinsic subtypes of breast cancer was according to the St Gallen
International Expert Consensus 2013 (3). The clinicopathological
characteristics of the patients are summarized in Table II. Median follow-up time was 36
months (range, 5–68 months). The Research Ethics Committee of
Guangdong General Hospital and Guangdong Academy of Medical Science
reviewed and approved the study (no. GDREC2012022H) according to
the principles expressed in the Declaration of Helsinki. The
Research Ethics Committee specifically waived the need for informed
consent for this retrospective study.
 | Table IIPatient clinicopathological
characteristics. |
Table II
Patient clinicopathological
characteristics.
| Patients
(N=320) |
|---|
|
|
|---|
|
Characteristics | No. of
patients | % |
|---|
| Median age (range),
years | 50 (25–91) | |
| Clinical stage at
diagnosis |
| I | 109 | 34.1 |
| II | 144 | 45.0 |
| III | 67 | 20.9 |
| Tumor stage (size
cm) |
| T1 (≤2.0) | 157 | 49.1 |
| T2 (>2.0 to
≤5.0) | 131 | 40.9 |
| T3 (>5.0) | 26 | 8.1 |
| T4a | 6 | 1.9 |
| Nodal stage |
| N0 (node
negative) | 178 | 55.6 |
| N1 (1–3 positive
nodes) | 83 | 25.9 |
| N2 (4–9 positive
nodes) | 36 | 11.3 |
| N3 (≥10 positive
nodes) | 23 | 7.2 |
| Histological
grade |
| Grade 1 | 16 | 5.0 |
| Grade 2 | 176 | 55.0 |
| Grade 3 | 128 | 40.0 |
| Subtypes of breast
cancer |
| Luminal
A-like | 71 | 22.2 |
| Luminal
B-like | 186 | 58.1 |
| HER2 positive
(non-luminal) | 27 | 8.4 |
| Triple negative
(ductal) | 31 | 9.7 |
| Not known | 5 | 1.6 |
| ER status |
| Negative | 66 | 20.6 |
| Positive | 254 | 79.4 |
| PR status |
| Negative | 78 | 24.4 |
| Positive | 242 | 75.6 |
| HER2 status |
| Negative | 238 | 74.4 |
| Positive | 69 | 21.6 |
| Not known | 13 | 4.1 |
| Surgery |
| Mastectomy | 281 | 87.8 |
| Breast
conservation | 39 | 12.2 |
| Chemotherapy |
| Neoadjuvant
chemotherapy | 70 | 21.9 |
| Adjuvant
chemotherapy | 165 | 51.6 |
| Not given | 85 | 26.6 |
| Targeted
therapy |
| Herceptin | 12 | 3.8 |
| No herceptin | 308 | 96.3 |
TMA construction and immunohistochemistry
(IHC)
MAs that contained three representative 2.0-mm cores
from each tumor of the cases were prepared with a tissue
microarrayer (Beecher Instruments, Silver Spring, MD, USA).
Immunohistochemical staining was performed using Real EnVision kit
(K5007; Dako, Carpinteria, CA, USA) on an automated immunostaining
instrument (Leica Bond-Max; Leica Microsystems GmbH, Wetzlar,
Germany) according to the manufacturer's instructions. Internal
control cores were present in each TMA. Sections were subjected to
staining protocols with the anti-PI3 kinase p85α antibody (EP380Y)
(Abcam; cat. no. ab40755). A negative control was performed in all
cases by omitting the primary antibody, which in all instances
resulted in negative immunoreactivity. Positive immunohistochemical
staining was defined as a brown cytoplasmic staining for p85α. A
semi-quantitative intensity scale ranging from 0 (no staining) to
3+ (the most intense staining) was used by comparing neoplastic
cells to adjacent breast cells belonging to normal terminal duct
lobular units as previously described (15). p85α downregulation was defined by
an IHC score 0, and p85α overexpression by an IHC score 1+ to 3+
(15). The localization and
intensity of staining were assessed by two independent
pathologists. Hormonal receptors were evaluated with the 1D5
antibody for the estrogen receptor (ER; Dako) and antibody PGR-1A6
for the progesterone receptor (PR; Dako). The human epidermal
growth factor receptor 2 (HER2/neu) was detected with CB11 (Dako).
Hormonal receptors and gene copy number of HER2 were assessed by
IHC staining on 4-μm thick tumor sections from FFPE blocks.
Fluorescein in situ hybridization
(FISH)
HER2 amplification status was detected by PathVysion
kit (Abbott) according to the manufacturer's instructions. HER2 was
defined as amplified when the FISH ratio was 2 or greater.
Statistical analysis
Statistical analysis was prepared using the
Statistical Package of MedCalc statistical software (version
12.7.4; MedCalc Software, Mariakerke, Belgium) and Social Sciences
(version 20.0; SPSS, Inc., Chicago, IL, USA). The receiver
operating characteristic curves were constructed to estimate the
optimal cut-off points for of p85α protein and miR-21 as the
predictors for disease-free survival (DFS) and overall survival
(OS). Pearson's Chi-square test and Spearman rank correlation
analysis were used to determine association and correlation between
variables. Survival analyses were plotted using Kaplan-Meier curves
and compared using the log-rank test. Univariate and multivariate
survival analyses were analyzed by Cox proportional hazards
regression models. The results were considered statistically
significant when two-sided P<0.05.
Results
PIK3R1 suppresses growth, invasiveness
and metastatic properties of breast cancer cells
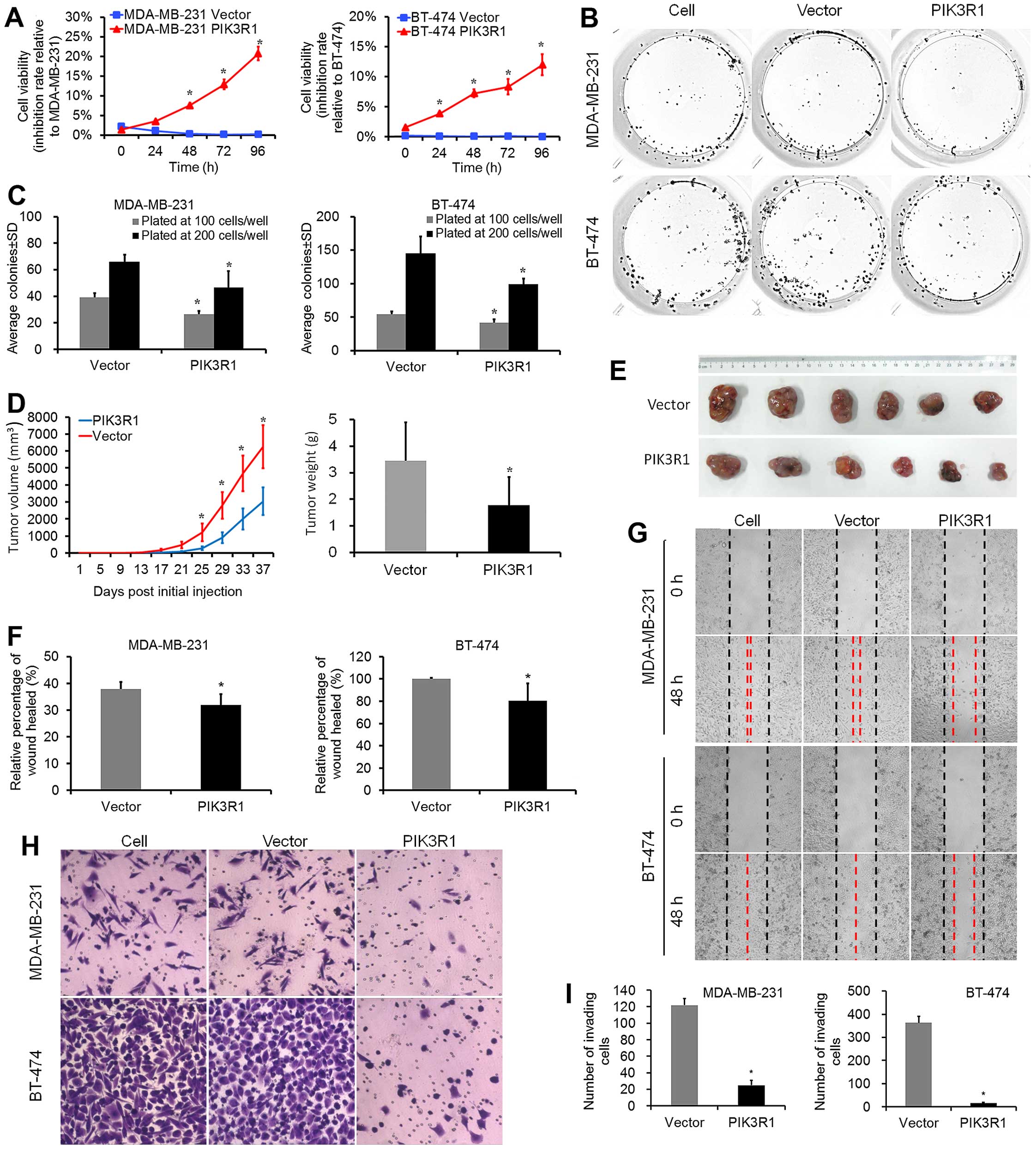
PIK3R1 overexpression significantly reduced
proliferation and colony formation capabilities in MDA-MB-231 and
BT-474 cell lines as compared to control cells (Fig. 1A–C). In vivo study showed
that at the 36th day, the average tumor volume in the PIK3R1
group (3040±812 mm3, mean ± SD) was significantly
smaller than that in the control group (Fig. 1D and E; 6258±1263 mm3,
P=0.008). Moreover, the average tumor weight in the PIK3R1
group (1.78±1.05 g) was lower than that in the control group
(Fig. 1D; 3.46±1.43 g, P=0.046).
PIK3R1 overexpression reduced the average percentage of
wound healed in both MDA-MB-231 and BT-474 cell lines as measured
at 48 h (Fig. 1F and G; P<0.001
for both lines as compared to control lines). We used the BD
Biocoat Matrigel Invasion Assay to test the invasive capabilities
of MDA-MB-231 and BT-474 cells expressing PIK3R1. For the 2
lines, PIK3R1 strongly reduced the number of invaded cells
vs. controls, with the lowest percent invasion in PIK3R1-BT-474
lines (Fig. 1H and I; 4.4%). These
data suggest that PIK3R1 plays an important role in the
suppression of cell proliferation, migration and invasion of breast
cancer cells.
PIK3R1 is a direct target of miR-21
We have previously identified miR-21 as an oncomiR
in breast cancer and have used human genome microarray to identify
potential targets of miR-21 (24).
In the present study, to biologically and metabolically interpret
of the array data, we applied pathway enrichment analysis with KEGG
(http://www.genome.jp/kegg/), GenMAPP
(http://www.genmapp.org/), and BioCarta
(http://www.biocarta.com/), and identified a set
of interesting genes, including PIK3R1, NFKB2,
STAT3 and AK3 (Table
III). To narrow down candidate target genes, we applied mRNA
target-predicting algorithms (TargetScan, picTar, miRDB, PITA and
microRNA.org) based on the presence of binding sites in the 3′-UTR.
All the five algorithms identified PIK3R1 as the potential
target of miR-21.
 | Table IIITop three signaling pathways for
PIK3R1 in breast cancer cells. |
Table III
Top three signaling pathways for
PIK3R1 in breast cancer cells.
| Pathway
analysis | Pathway name | Total | P-value | Q-value | Gene |
|---|
| KEGG | Regulation of actin
cytoskeleton | 7 | 0.012 | 0.002 | LIMK1,
SLC9A1, GNG12, WASF2, PIK3R1,
ARPC4, ACTN4 |
| Insulin signaling
pathway | 5 | 0.024 | 0.003 | FLOT2,
PRKCI, PPP1R3C, PIK3R1, PHKA2 |
| Apoptosis | 4 | 0.025 | 0.003 | APAF1,
TRAF2, NFKB2, PIK3R1 |
| GenMAPP | Lipid binding | 9 | 0.001 | <0.001 | PRKCI,
ANXA6, SCP2, STARD3, WDFY1,
ANXA2, PREX1, PIK3R1, BPI |
| Kinase
activity | 5 | 0.044 | 0.005 | ADCK4,
GALK1, AK3, CARKL, PIK3R1,
APAF1, IRF1, TRAF2, PIK3R1 |
| Apoptosis | 4 | 0.023 | 0.003 |
| BioCarta | Role of PI3K
subunit p85 in regulation of actin organization and cell
migration | 3 | 0.001 | <0.001 | ACTR2,
PIK3R1, ARPC4 |
| EGF signaling
pathway | 3 | 0.004 | 0.001 | STAT3,
PIK3R1, MEF2D |
| PDGF signaling
pathway | 3 | 0.004 | 0.001 | STAT3,
PIK3R1, MEF2D |
| Signaling of
hepatocyte growth factor receptor | 3 | 0.009 | 0.002 | STAT3,
PIK3R1, MEF2D |
| Mechanism of gene
regulation by peroxisome proliferators via PPARα | 3 | 0.020 | 0.003 | PPARBP,
EHHADH, PIK3R1 |
Interestingly, p85α has previously been shown to
exert tumor suppressor properties through negative regulation of
growth factor signaling (30).
PIK3R1 expression was significantly decreased by 18% in breast
cancer tissues (31) and cell
lines (7), and was associated with
decreased survival in breast cancer patients (15). Therefore, we conducted analyses to
determine whether miR-21 might target PIK3R1. First, we
examined miR-21 and PIK3R1 mRNA in a range of metastatic
(BT-474, MDA-MB-231 and BT-549), and non-metastatic (MCF-7, SK-BR-3
and T-47D) human breast cancer cell lines and breast epithelial
cell line MCF-10A. All breast cancer lines tested, except SK-BR-3
and T-47D, exhibited elevated levels of miR-21 compared to MCF-10A
cells, with corresponding reductions in PIK3R1 levels
(Fig. 1A). Next, to establish a
direct relationship between miR-21 and the predicted target gene, a
luciferase construct containing the 3′-UTR of PIK3R1 was
transfected with a miR-21 mimic, or a miRNA-negative control mimic
(Fig. 1B); a 44% reduction in
luciferase activity was observed only with the miR-21 mimic
(Fig. 1C). To further show that
miR-21 interacts directly with two seed-binding regions within the
3′-UTR of PIK3R1, two point mutations were generated in each
seed-binding region and were denoted as Mut845 and Mut1091
(Fig. 1B). Although a significant
reduction in luciferase activity was observed for the WT construct,
high luciferase activity was maintained in all of the mutants
(Fig. 1C), thereby supporting the
direct interaction between miR-21 and these two targeted regions
within the PIK3R1 3′-UTR.
AntimiR-21 suppresses tumor growth,
invasiveness and metastasis by targeting PIK3R1 via PI3K/AKT
signaling
We previously found that LNA-antimiR-21 suppressed
breast cancer cell growth and migration in vitro (24). In order to determine whether
antimiR-21-induced suppression of growth, invasiveness and
metastases of breast cancer cells are indeed executed via
PIK3R1, we utilized MDA-MB-231 and BT-474 cells, which
express high levels of endogenous miR-21, and transfected them with
LNA-antimiR-21. Indeed, inhibition of miR-21 in breast cancer cells
resulted in a 7- to 9-fold increase in PIK3R1 mRNA levels
(Fig. 3A) and an approximate
3-fold increase in protein (p85α) levels (Fig. 3B). Furthermore, over-expression of
miR-21 resulted in a 30–50% reduction in PIK3R1 mRNA levels
(Fig. 3A) and an approximate 30%
reduction in protein levels (Fig.
3B) in both MDA-MB-231 and BT-474 cells. Concomitant with the
increase in p85α, a decrease in PI3K pathway activation was
observed, as evidenced by decreased p-AKT expression (Fig. 3C). These results suggest that
miR-21-dependent proto-oncogene PI3K/AKT pathway is active in
breast cancer cell lines.
Moreover, PIK3R1 overexpression phenocopied
the suppression effects of LNA-antimiR-21 on cell proliferation and
colony formation capabilities. Notably, PIK3R1 knockdown abrogated
LNA-antimiR-21-induced suppression of cell proliferation and colony
formation capabilities (Fig.
4A–C). LNA-antimiR-21 reduced the average percentage of wound
healed in both cell lines as measured at 48 h (P<0.001). In
BT-474 cells, PIK3R1 knockdown significantly abrogated
LNA-antimiR-21-mediated cell migration (P=0.007). Although not in a
statistically significant manner, PIK3R1 knockdown also
abrogated LNA-antimiR-21-mediated cell migration in MDA-MB-231
cells (Fig. 4D and E). We used the
BD Biocoat Matrigel Invasion Assay to test the invasive capability
of MDA-MB-231 and BT-474 cells lacking miR-21. For these lines,
LNA-antimiR-21 strongly reduced the number of invaded cells vs.
controls, with the lowest percent invasion in the PIK3R1-BT-474
line (Fig. 4F and G; 4.4%).
Furthermore, PIK3R1 knockdown significantly abrogated
LNA-antimiR-21-mediated cell invasion in MDA-MB-231 (P=0.004) and
BT-474 lines (P<0.001). Together, these data support the
hypothesis that miR-21 by targeting PIK3R1 promotes breast
cancer cell growth, invasion and migration.
 | Figure 4AntimiR-21-induced suppression of
proliferation, clonogenicity, invasiveness, and metastatic
properties of breast cancer cells is mediated by direct repression
of PIK3R1. (A) MTS assays were conducted on breast cancer
cells after transfection with antimiR-21 (50 nmol/l), antimiR-21 +
PIK3R1-shRNA or control. At 48, 72 and 96 h, the antimiR-21 lines
showed significantly reduced levels of proliferation as compared to
control lines. PIK3R1-shRNA reversed the effect of antimiR-21 on
cells. (B) Representative images depicting clonogenic assays
performed with cells plated at 200 cells/well. (C) In MDA-MB-231
and BT-474 lines, antimiR-21 resulted in a decrease in colony
number as compared to control lines. PIK3R1-shRNA reversed the
effect of antimiR-21 on cells. (D) Representative images depicting
cell migration assays. (E) Cell migration was quantitated as
percentage of wound-healed area from corresponding control and
transfected cells. (F) Invasion assays in these control and
transfected cells. (G) For each cell line, antimiR-21 resulted in
reduced invasion as compared to controls. PIK3R1 knockdown
reversed the effect of antimiR-21 on cell migration in both cell
lines. (H) MDA-MB-231 and BT-474 lines were transfected with
PIK3R1, antimiR-21, antimiR-21 + PIK3R1-shRNA or control, followed
by western blot analysis of the indicated EMT-related proteins.
Relative E-cadherin, N-cadherin, vimentin, FSP1, snail and slug
levels were normalized to the β-actin level. (I) Breast cancer
lines were transfected with PIK3R1, antimiR-21, antimiR-21 +
PIK3R1-shRNA or control, followed by RT-qPCR analysis of the
indicated EMT-related mRNAs. Data represent mean ± SD.
*P<0.05; **P<0.01. |
AntimiR-21 reverses the
epithelial-mesenchymal transition (EMT) target PIK3R1 suppression
of invasiveness in breast cancer
To determine whether
antimiR-21/PIK3R1-induced suppression of invasiveness in
breast cancer cells is mediated by reversing EMT, we transfected
the MDA-MB-231 and BT-474 cell lines, which exhibit a mesenchymal
phenotype, with antimiR-21 or PIK3R1. Transfection of breast
cancer cells with antimiR-21 or PIK3R1 resulted in reversal
of EMT, as evidenced by repression of the mesenchymal markers
N-cadherin, vimentin, FSP1, snail and slug and induction of the
epithelial marker E-cadherin. Furthermore, PIK3R1 shRNA
reversed the effect of antimiR-21 or PIK3R1 on EMT (Fig. 4H and I).
p85α downregulation in patient tumor
specimens
To establish the relevance of our findings in the
patient tumors, we analyzed the expression of miR-21 by RT-qPCR and
p85α by IHC in 320 primary human invasive breast cancers, and the
adjacent non-tumor-affected epidermis. Alteration of p85α was also
verified at the protein level by IHC staining on TMAs. Positive
staining of p85α was found in the cytoplasm (Fig. 5A). Tumor cells showed p85α moderate
expression, while residual normal mammary epithelial cells
presented strong IHC staining intensity (Fig. 5B).
Staining scores and log2 of CT values
were analyzed using MedCalc statistical software to determine the
optimal survival cut-off points for dichotomizing expression of
p85α protein and miR-21. The cut points correspond to the maximum
Chi-square value of the Kaplan-Meier test for OS between groups
above and below the cut-point threshold. p85α down-regulation was
found in 25 (7.8%) of the 320 breast cancer patients. miR-21 high
expression was found in 119 (37.2%) of 320 patients. Next, we
investigated the negative regulation of endogenous p85α protein by
endogenous miR-21. Correlation analysis demonstrated that
endogenous p85α protein levels were not statistically correlated
with miR-21 in the patient tumor specimens (Fig. 5C; rs=−0.109, P=0.052, Spearman's
correlation analysis).
Correlation of p85α expression with
breast cancer clinicopathological characteristics and
prognosis
p85α downregulation was associated with PR positive
status (Table IV; P=0.047). No
significant correlation was observed between p85α and clinical
stage, tumor size, node status, histological grade, ER or HER2
status. miR-21 overexpression was associated with high clinical
stage (Table IV; P=0.016). No
correlation was observed between miR-21 and other
characteristic.
 | Table IVCorrelation between p85α protein
expression and clinicopathological parameters of breast cancer
patients. |
Table IV
Correlation between p85α protein
expression and clinicopathological parameters of breast cancer
patients.
| p85α | miR-21 |
|---|
|
|
|
|---|
|
Characteristics | Overexpression
(n=295)
N (%) | Downregulation
(n=25)
N (%) | P-value | Low
(n=201)
N (%) | High
(n=119)
N (%) | P-value |
|---|
| Clinical stage |
| I | 100 (34) | 9 (36) | 0.860 | 73 (36) | 36 (30) | 0.016 |
| II | 134 (45) | 10 (40) | | 96 (48) | 48 (40) | |
| III | 61 (21) | 6 (24) | | 32 (16) | 35 (29) | |
| Tumor size
(cm) |
| ≤2 | 144 (49) | 13 (52) | 0.760 | 97 (48) | 66 (55) | 0.213 |
| >2 | 151 (51) | 12 (48) | | 104 (52) | 53 (45) | |
| Node |
| Negative | 164 (56) | 14 (56) | 0.969 | 118 (59) | 60 (50) | 0.149 |
| Positive | 131 (44) | 11 (44) | | 83 (41) | 59 (50) | |
| Histological
grade |
| 1 | 13 (4) | 3 (12) | 0.245 | 13 (6) | 3 (3) | 0.110 |
| 2 | 163 (55) | 13 (52) | | 103 (51) | 73 (61) | |
| 3 | 119 (40) | 9 (36) | | 85 (42) | 43 (36) | |
| Subtypes of breast
cancer |
| Luminal
A-like | 63 (21) | 8 (32) | 0.095a | 43 (21) | 28 (23) | 0.095a |
| Luminal
B-like | 170 (58) | 16 (64) | | 113 (56) | 73 (61) | |
| HER2 positive | 27 (9) | 0 (0) | | 18 (9) | 9 (8) | |
| Triple
negative | 30 (10) | 1 (4) | | 24 (12) | 7 (6) | |
| Not known | 5 (2) | 0 (0) | | 3 (2) | 2 (2) | |
| ER |
| Negative | 64 (22) | 2 (8) | 0.104 | 47 (23) | 19 (16) | 0.113 |
| Positive | 231 (78) | 23 (92) | | 154 (77) | 100 (84) | |
| PR |
| Negative | 76 (26) | 2 (8) | 0.047 | 55 (27) | 23 (19) | 0.106 |
| Positive | 219 (74) | 23 (92) | | 146 (73) | 96 (81) | |
| HER2 |
| Negative | 218 (74) | 20 (80) | 0.467b | 154 (77) | 84 (71) | 0.591b |
| Positive | 65 (22) | 4 (16) | | 42 (21) | 27 (23) | |
| Not known | 12 (4) | 1 (4) | | 5 (2) | 8 (7) | |
Next, we investigated the prognostic impact of p85α
and miR-21 expression on breast cancer patients. The survival
curves showed that p85α downregulation was significantly associated
with inferior 5-year DFS and OS of breast cancer patients (Fig. 6A and B; DFS: P=0.005, OS: P=0.021;
log-rank tests). Within early stage stratum, patients with p85α
downregulation had inferior 5-year DFS and OS compared to those
with p85α overexpression (Fig. 6C and
D; P<0.001 for DFS, P=0.004 for OS, log-rank test). However,
within the late stage stratum, p85α expression was not related with
the patient survival (Fig. 6E and
F). Consistent with our previous study in another cohort, high
miR-21 expression was significantly associated with inferior 5-year
DFS and 5-year OS in this cohort (DFS: P=0.035; OS, P=0.028).
In univariate analysis, p85α downregulation, high
miR-21, high clinical stage, tumor size >2 cm, node positive,
high histological grade and breast conservation were associated
with inferior 5-year DFS and 5-year OS of the breast cancer
patients (Table V). While,
subtypes of breast cancer, hormone receptor status, HER2 status,
and chemotherapy were not associated with inferior 5-year DFS or
5-year OS. Multivariate Cox regression model that incorporated
significant factors in the univariate analyses showed that only
p85α downregulation and high clinical stage maintained independent
prognostic factors for both inferior 5-year OS and DFS (Table VI).
 | Table VUnivariate Cox models for patients
with invasive breast cancer. |
Table V
Univariate Cox models for patients
with invasive breast cancer.
| OS | DFS |
|---|
|
|
|
|---|
|
Characteristics | HR | 95% CI | P | HR | 95% CI | P-value |
|---|
| p85α overexpression
vs. downregulation | 3.06 | 1.13–8.31 | 0.028 | 2.68 | 1.30–5.54 | 0.008 |
| miR-21 low vs.
high | 2.47 | 1.08–5.65 | 0.033 | 1.80 | 1.03–3.12 | 0.038 |
| Clinical stage I
vs. II vs. III | 4.20 | 2.15–8.20 |
<0.001 | 3.59 | 2.35–5.50 |
<0.001 |
| Tumor size (cm) ≤2
vs. >2 | 3.44 | 1.28–9.27 | 0.015 | 3.82 | 1.95–7.46 |
<0.001 |
| Node negative vs.
positive | 5.38 | 2.00–14.45 | 0.001 | 3.76 | 2.02–6.98 |
<0.001 |
| Histological grade
1 vs. 2 vs. 3 | 2.57 | 1.19–5.55 | 0.017 | 2.25 | 1.47–3.45 |
<0.001 |
| Subtypes of breast
cancer | 1.25 | 0.77–2.04 | 0.371 | 1.01 | 0.72–1.43 | 0.944 |
| ER negative vs.
positive | 0.63 | 0.25–1.59 | 0.327 | 0.95 | 0.46–1.97 | 0.891 |
| PR negative vs.
positive | 0.96 | 0.36–2.58 | 0.933 | 1.19 | 0.57–2.46 | 0.646 |
| HER2 negative vs.
positive | 0.70 | 0.20–2.39 | 0.577 | 0.99 | 0.48–2.07 | 0.991 |
| Mastectomy vs.
breast conservation | 0.50 | 0.26–0.95 | 0.033 | 0.63 | 0.42–0.95 | 0.026 |
| Neoadjuvant
chemotherapy vs. adjuvant chemotherapy vs. not given | 0.33 | 0.05–2.48 | 0.282 | 0.55 | 0.20–1.52 | 0.246 |
 | Table VIMultivariate Cox model for patients
with invasive breast cancer. |
Table VI
Multivariate Cox model for patients
with invasive breast cancer.
| 5-year OS | 5-year DFS |
|---|
|
|
|
|---|
|
Characteristics | HR | 95% CI | P-valuea | HR | 95% CI | P-valuea |
|---|
| p85α overexpression
vs. downregulation | 3.42 | 1.24–9.41 | 0.017 | 2.90 | 1.39–6.04 | 0.004 |
| Clinical stage I
vs. II vs. III | 4.59 | 2.27–9.31 |
<0.001 | 3.34 | 2.19–5.10 |
<0.001 |
Discussion
In the present study, we present evidence that
PIK3R1 is a direct miR-21 target. PIK3R1 phenocopies
the effect of miR-21 knockdown. Furthermore, we expanded our
previous findings that miR-21 knockdown suppresses cell growth,
migration and invasion by inhibiting PI3K/AKT activation via
targeting PIK3R1. AntimiR-21/PIK3R1-induced
suppression of invasiveness in breast cancer cells is mediated by
reversing EMT. Additionally, we show an inverse correlation between
p85α expression levels and PR expression in patient tumors.
Finally, we demonstrate that p85α is downregulated in patients with
invasive breast cancer, indicating an inferior prognosis. Taken
together, our data provide novel insight into the regulation of
p85α expression in breast cancer and its potential role on
prognosis predication.
miR-21 is an oncomiR in breast cancer and targets
several tumor suppressor genes important for various cellular
processes (22). Here, we show
that p85α is downregulated in 7.8% of breast cancer tumors, and is
a direct target of miR-21. This finding is consistent with a recent
study by Toste et al (32).
They demonstrated a direct regulation of p85α by miR-21 and an
inverse correlation between miR-21 and p85α expression levels in
human pancreatic tumors. However, we did not find a statistically
significant correlation between miR-21 and p85α expression levels
in patient tumors (P=0.052). We speculate that patient tumor
sections for quantitative detection of miR-21, which inevitably
contain both normal and malignant cells, are the most possible
reason for this inconsistent result.
The protein p85α is necessary for stabilization and
membrane recruitment of the p110α subunit of PI3K (6). Loss of the p85α protein leads to
downstream PI3K pathway activation (30,32–35).
Therefore, the impact of p85α down-regulation on pathway signaling
could be caused by the loss of the inhibitory effect of p85α on
p110α and PI3K pathway activity (33,36).
p85α protein has also been reported to be a positive regulator of
PTEN via stabilization of this protein (37,38).
Besides, several studies evidenced that PTEN is one of miR-21
targets (21,38,39).
These studies support the notion that miR-21 actives PI3K pathway
via multiple targets. Our finding that p-AKT levels are decreased
after p85α overexpression in breast cancer cells is consistent with
these previous observations. In addition, PIK3R1 overexpression
phenocopies the effect of miR-21 knockdown on breast cancer cells
and PIK3R1 knockdown inversely abrogates
LNA-antimiR-21-mediated cell growth and invasion suppression. These
findings suggest that PIK3R1 exerts tumor suppressor
properties in breast cancer. Furthermore, the concept that p85α
downregulation can be protumorigenic (30) is supported by our finding that p85α
downregulation is seen in breast cancer tissues when compared with
normal tissues. In the present study, this newly identified p85α
downregulation by miR-21 has significant importance for
interpretation of miR-21 promoting breast cancer cells growth,
migration and invasion through the PI3K/AKT pathway.
Prognosis of invasive breast cancer, no special
type, is influenced by the classical variables of histological
grade, tumor size, lymph node status and clinical stage (14,29,40,41).
However, heterogeneity in tumor cell phenotypes make breast tumor
categorization a challenging task, especially as it is relevant to
therapeutic responses and patient prognosis (1). Our previous study and other research
demonstrated that elevated miR-21 could predict unfavorable
prognosis in breast cancer patients (23,42–44).
In this study, we performed an evaluation of the prognostic
significance of p85α, as well as miR-21, in a 320 patient cohort,
and confirmed that miR-21 was a prognostic marker for inferior
5-year DFS and 5-year OS in breast cancer patients. Noticeably,
p85α downregulation was a prognostic marker for inferior clinical
stage. This finding is consistent with the association between p85α
downregulation and an inferior prognosis not only in breast cancer
(15) but also pancreatic cancer
(32,45), hepatocellular cancers (30), neuroblastoma (46) and lung cancers (47). All these results support the notion
that p85α plays as a tumor suppressor gene in invasive breast
cancer tumors. Additional in vivo studies will be necessary
to confirm the relationship between miR-21 and p85α, and the role
of p85α in breast cancer.
In conclusion, we provided evidence that
PIK3R1 is a direct target of miR-21. miR-21 knockdown
induced increased p85α level, accompanied by decreased p-AKT level.
miR-21 may play a role in breast cancer development by promoting
breast cancer cell growth, migration and invasion partly by
inhibiting PI3K/AKT activation via targeting PIK3R1 and
reversing EMT. Furthermore, alterations in miR-21 and p85α had a
complementary impact on breast cancer patient survival. Finally,
p85α downregulation defined a specific subgroup of breast cancer
with shorter 5-year DFS and OS, which may require more aggressive
treatment.
Acknowledgements
We thank Xin-Chuang Cai and Jia Fu for support with
TMAs construction and IHC staining. The present study was funded by
the National Natural Science Foundation of China (NSFC, http://www.nsfc.gov.cn/) (grant nos. 81202111 and
81302143), the Guangdong Natural Science Foundation (grant no.
S2012040006409), and a National Clinical Key Subject Construction
Project Foundation of China. All procedures performed in studies
involving human participants were in accordance with the ethical
standards of the institutional and national research committee and
with the 1964 Helsinki declaration and its later amendments or
comparable ethical standards. This research involved formalin-fixed
paraffin-embedded tissue specimens from breast cancer patients. For
retrospective studies formal consent was not required.
References
|
1
|
Polyak K: Heterogeneity in breast cancer.
J Clin Invest. 121:3786–3788. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Banerji S, Cibulskis K, Rangel-Escareno C,
Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY,
Sougnez C, Zou L, et al: Sequence analysis of mutations and
translocations across breast cancer subtypes. Nature. 486:405–409.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Goldhirsch A, Winer EP, Coates AS, Gelber
RD, Piccart-Gebhart M, Thürlimann B, Senn HJ, Albain KS, Andre F,
Bergh J, et al: Panel members: Personalizing the treatment of women
with early breast cancer: Highlights of the St Gallen International
Expert Consensus on the Primary Therapy of Early Breast Cancer
2013. Ann Oncol. 24:2206–2223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Elkabets M, Vora S, Juric D, Morse N,
Mino-Kenudson M, Muranen T, Tao J, Campos AB, Rodon J, Ibrahim YH,
et al: mTORC1 inhibition is required for sensitivity to PI3K p110α
inhibitors in PIK3CA-mutant breast cancer. Sci Transl Med.
5:196ra992013. View Article : Google Scholar
|
|
5
|
Myhre S, Lingjærde OC, Hennessy BT, Aure
MR, Carey MS, Alsner J, Tramm T, Overgaard J, Mills GB,
Børresen-Dale AL, et al: Influence of DNA copy number and mRNA
levels on the expression of breast cancer related proteins. Mol
Oncol. 7:704–718. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yuan TL and Cantley LC: PI3K pathway
alterations in cancer: Variations on a theme. Oncogene.
27:5497–5510. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Uchino M, Kojima H, Wada K, Imada M, Onoda
F, Satofuka H, Utsugi T and Murakami Y: Nuclear beta-catenin and
CD44 upregulation characterize invasive cell populations in
non-aggressive MCF-7 breast cancer cells. BMC Cancer. 10:4142010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Engelman JA: Targeting PI3K signalling in
cancer: Opportunities, challenges and limitations. Nat Rev Cancer.
9:550–562. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hennessy BT, Smith DL, Ram PT, Lu Y and
Mills GB: Exploiting the PI3K/AKT pathway for cancer drug
discovery. Nat Rev Drug Discov. 4:988–1004. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao L and Vogt PK: Class I PI3K in
oncogenic cellular transformation. Oncogene. 27:5486–5496. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Juric D, Castel P, Griffith M, Griffith
OL, Won HH, Ellis H, Ebbesen SH, Ainscough BJ, Ramu A, Iyer G, et
al: Convergent loss of PTEN leads to clinical resistance to a
PI(3)Kα inhibitor. Nature. 518:240–244. 2015. View Article : Google Scholar :
|
|
12
|
Brachmann SM, Ueki K, Engelman JA, Kahn RC
and Cantley LC: Phosphoinositide 3-kinase catalytic subunit
deletion and regulatory subunit deletion have opposite effects on
insulin sensitivity in mice. Mol Cell Biol. 25:1596–1607. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao L and Vogt PK: Helical domain and
kinase domain mutations in p110alpha of phosphatidylinositol
3-kinase induce gain of function by different mechanisms. Proc Natl
Acad Sci USA. 105:2652–2657. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Araújo TG, Paiva CE, Rocha RM, Maia YC,
Sena AA, Ueira-Vieira C, Carneiro AP, Almeida JF, de Faria PR,
Santos DW, et al: A novel highly reactive Fab antibody for breast
cancer tissue diagnostics and staging also discriminates a subset
of good prognostic triple-negative breast cancers. Cancer Lett.
343:275–285. 2014. View Article : Google Scholar
|
|
15
|
Cizkova M, Vacher S, Meseure D, Trassard
M, Susini A, Mlcuchova D, Callens C, Rouleau E, Spyratos F,
Lidereau R, et al: PIK3R1 underexpression is an independent
prognostic marker in breast cancer. BMC Cancer. 13:5452013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Visani M, de Biase D, Marucci G, Cerasoli
S, Nigrisoli E, Bacchi Reggiani ML, Albani F, Baruzzi A and Pession
A: PERNO study group: Expression of 19 microRNAs in glioblastoma
and comparison with other brain neoplasia of grades I–III. Mol
Oncol. 8:417–430. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jamali Z, Asl Aminabadi N, Attaran R,
Pournagiazar F, Ghertasi Oskouei S and Ahmadpour F: MicroRNAs as
prognostic molecular signatures in human head and neck squamous
cell carcinoma: A systematic review and meta-analysis. Oral Oncol.
51:321–331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang F, Yang Z, Cao M, Xu Y, Li J, Chen
X, Gao Z, Xin J, Zhou S, Zhou Z, et al: MiR-203 suppresses tumor
growth and invasion and down-regulates MiR-21 expression through
repressing Ran in esophageal cancer. Cancer Lett. 342:121–129.
2014. View Article : Google Scholar
|
|
19
|
Abue M, Yokoyama M, Shibuya R, Tamai K,
Yamaguchi K, Sato I, Tanaka N, Hamada S, Shimosegawa T, Sugamura K,
et al: Circulating miR-483-3p and miR-21 is highly expressed in
plasma of pancreatic cancer. Int J Oncol. 46:539–547. 2015.
|
|
20
|
Shi Z, Zhang J, Qian X, Han L, Zhang K,
Chen L, Liu J, Ren Y, Yang M, Zhang A, et al: AC1MMYR2, an
inhibitor of dicer-mediated biogenesis of Oncomir miR-21, reverses
epithelial-mesenchymal transition and suppresses tumor growth and
progression. Cancer Res. 73:5519–5531. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li LQ, Li XL, Wang L, Du WJ, Guo R, Liang
HH, Liu X, Liang DS, Lu YJ, Shan HL, et al: Matrine inhibits breast
cancer growth via miR-21/PTEN/Akt pathway in MCF-7 cells. Cell
Physiol Biochem. 30:631–641. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baffa R, Fassan M, Volinia S, O'Hara B,
Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM, et
al: MicroRNA expression profiling of human metastatic cancers
identifies cancer gene targets. J Pathol. 219:214–221. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan LX, Huang XF, Shao Q, Huang MY, Deng
L, Wu QL, Zeng YX and Shao JY: MicroRNA miR-21 overexpression in
human breast cancer is associated with advanced clinical stage,
lymph node metastasis and patient poor prognosis. RNA.
14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ,
Hou JH, Fu J, Zeng MS, Yun JP, Wu QL, et al: Knockdown of miR-21 in
human breast cancer cell lines inhibits proliferation, in vitro
migration and in vivo tumor growth. Breast Cancer Res. 13:R22011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gramantieri L, Ferracin M, Fornari F,
Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E,
Grazi GL, et al: Cyclin G1 is a target of miR-122a, a microRNA
frequently down-regulated in human hepatocellular carcinoma. Cancer
Res. 67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gao L, Song JR, Zhang JW, Zhao X, Zhao QD,
Sun K, Deng WJ, Li R, Lv G, Cheng HY, et al: Chloroquine promotes
the anti-cancer effect of TACE in a rabbit VX2 liver tumor model.
Int J Biol Sci. 9:322–330. 2013. View Article : Google Scholar
|
|
27
|
Sun L, Li H, Chen J, Dehennaut V, Zhao Y,
Yang Y, Iwasaki Y, Kahn-Perles B, Leprince D, Chen Q, et al: A
SUMOylation-dependent pathway regulates SIRT1 transcription and
lung cancer metastasis. J Natl Cancer Inst. 105:887–898. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
De Bacco F, Luraghi P, Medico E, Reato G,
Girolami F, Perera T, Gabriele P, Comoglio PM and Boccaccio C:
Induction of MET by ionizing radiation and its role in
radioresistance and invasive growth of cancer. J Natl Cancer Inst.
103:645–661. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sunil RL, Lan OE, Stuart JS, Puay HT and
Marc JV: WHO Classification of Tumours. Fred TB, Elain SJ, Sunil RL
and Hiroko O: IARC Press; Lyon: pp. 2402012
|
|
30
|
Taniguchi CM, Winnay J, Kondo T, Bronson
RT, Guimaraes AR, Alemán JO, Luo J, Stephanopoulos G, Weissleder R,
Cantley LC, et al: The phosphoinositide 3-kinase regulatory subunit
p85alpha can exert tumor suppressor properties through negative
regulation of growth factor signaling. Cancer Res. 70:5305–5315.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Richardson AL, Wang ZC, De Nicolo A, Lu X,
Brown M, Miron A, Liao X, Iglehart JD, Livingston DM and Ganesan S:
X chromosomal abnormalities in basal-like human breast cancer.
Cancer Cell. 9:121–132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Toste PA, Li L, Kadera BE, Nguyen AH, Tran
LM, Wu N, Madnick DL, Patel SG, Dawson DW and Donahue TR: p85α is a
microRNA target and affects chemosensitivity in pancreatic cancer.
J Surg Res. 196:285–293. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shekar SC, Wu H, Fu Z, Yip SC, Nagajyothi,
Cahill SM, Girvin ME and Backer JM: Mechanism of constitutive
phos-phoinositide 3-kinase activation by oncogenic mutants of the
p85 regulatory subunit. J Biol Chem. 280:27850–27855. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Luo J and Cantley LC: The negative
regulation of phosphoinositide 3-kinase signaling by p85 and it's
implication in cancer. Cell Cycle. 4:1309–1312. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rabinovsky R, Pochanard P, McNear C,
Brachmann SM, Duke-Cohan JS, Garraway LA and Sellers WR: p85
associates with unphosphorylated PTEN and the PTEN-associated
complex. Mol Cell Biol. 29:5377–5388. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jaiswal BS, Janakiraman V, Kljavin NM,
Chaudhuri S, Stern HM, Wang W, Kan Z, Dbouk HA, Peters BA, Waring
P, et al: Somatic mutations in p85alpha promote tumorigenesis
through class IA PI3K activation. Cancer Cell. 16:463–474. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cheung LW, Hennessy BT, Li J, Yu S, Myers
AP, Djordjevic B, Lu Y, Stemke-Hale K, Dyer MD, Zhang F, et al:
High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer
elucidates a novel mechanism for regulation of PTEN protein
stability. Cancer Discov. 1:170–185. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Darido C, Georgy SR, Wilanowski T, Dworkin
S, Auden A, Zhao Q, Rank G, Srivastava S, Finlay MJ, Papenfuss AT,
et al: Targeting of the tumor suppressor GRHL3 by a
miR-21-dependent proto-oncogenic network results in PTEN loss and
tumorigenesis. Cancer Cell. 20:635–648. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xiong B, Cheng Y, Ma L and Zhang C: MiR-21
regulates biological behavior through the PTEN/PI-3 K/Akt signaling
pathway in human colorectal cancer cells. Int J Oncol. 42:219–228.
2013.
|
|
40
|
Badwe R, Hawaldar R, Parmar V, Nadkarni M,
Shet T, Desai S, Gupta S, Jalali R, Vanmali V, Dikshit R, et al:
Single-injection depot progesterone before surgery and survival in
women with operable breast cancer: A randomized controlled trial. J
Clin Oncol. 29:2845–2851. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Uji K, Naoi Y, Kagara N, Shimoda M,
Shimomura A, Maruyama N, Shimazu K, Kim SJ and Noguchi S:
Significance of TP53 mutations determined by next-generation ‘deep’
sequencing in prognosis of estrogen receptor-positive breast
cancer. Cancer Lett. 342:19–26. 2014. View Article : Google Scholar
|
|
42
|
Müller V, Gade S, Steinbach B, Loibl S,
von Minckwitz G, Untch M, Schwedler K, Lübbe K, Schem C, Fasching
PA, et al: Changes in serum levels of miR-21, miR-210, and miR-373
in HER2-positive breast cancer patients undergoing neoadjuvant
therapy: A translational research project within the Geparquinto
trial. Breast Cancer Res Treat. 147:61–68. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Markou A, Yousef GM, Stathopoulos E,
Georgoulias V and Lianidou E: Prognostic significance of
metastasis-related microRNAs in early breast cancer patients with a
long follow-up. Clin Chem. 60:197–205. 2014. View Article : Google Scholar
|
|
44
|
Lee JA, Lee HY, Lee ES, Kim I and Bae JW:
Prognostic implications of MicroRNA-21 overexpression in invasive
ductal carcinomas of the breast. J Breast Cancer. 14:269–275. 2011.
View Article : Google Scholar
|
|
45
|
Donahue TR, Tran LM, Hill R, Li Y,
Kovochich A, Calvopina JH, Patel SG, Wu N, Hindoyan A, Farrell JJ,
et al: Integrative survival-based molecular profiling of human
pancreatic cancer. Clin Cancer Res. 18:1352–1363. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fransson S, Abel F, Kogner P, Martinsson T
and Ejeskär K: Stage-dependent expression of PI3K/Akt-pathway genes
in neuroblastoma. Int J Oncol. 42:609–616. 2013.
|
|
47
|
Lu Y, Lemon W, Liu PY, Yi Y, Morrison C,
Yang P, Sun Z, Szoke J, Gerald WL, Watson M, et al: A gene
expression signature predicts survival of patients with stage I
non-small cell lung cancer. PLoS Med. 3:e4672006. View Article : Google Scholar : PubMed/NCBI
|