Introduction
Epigenetics is defined as the inheritance of
information based on gene expression levels as opposed to genetics,
which refers to information inherited on the basis of gene
sequences. Recent studies have shown that altered epigenetic
control of gene expression plays a substantial role in many
different diseases, including tumor development and progression
(1–3). Unlike genetic mutations, changes in
the epigenome associated with cancer are potentially reversible.
Novel therapies targeting epigenetic pathways are already used in
clinical cancer treatment or are currently in preclinical and
clinical trials (3). Histone
acetylation is a key epigenetic mechanism. Histone proteins
assemble with DNA in nucleosomes that function as transcriptional
regulators. The N-terminal tails of histones undergo acetylation on
specific residues. Acetylation of lysines is regulated by the
opposite action of histone acetyltransferase (HAT) and histone
deacetylases (HDAC) enzymes.
CREB binding protein (CBP) and p300 belong to the
HAT family. CBP/p300 are transcriptional coactivators for many
important transcription factors. Through their HAT activity, they
relax chromatin structure and make chromosomal DNA accessible
(4). The HAT activity of CBP/p300
also acetylates a number of non-histone transcription factors,
which can positively or negatively modulate their activity through
diverse mechanisms. CBP/p300 have at least 400 interacting partners
and act as hubs in gene networks (5). Through their interaction with diverse
transcriptional factors, CBP/p300 are involved in many cellular
processes such as DNA repair, cell growth, differentiation, and
apoptosis (6).
Somatic mutations of CBP/p300 occur in some
malignancies. For example, chromosomal translocations directly
involving CBP or p300 genes are associated with leukemia (7,8).
Bi-allelic somatic mutations in the p300 gene have been
occasionally observed in gastric, colon and breast cancers, and
truncating mutations in p300 were detected in primary tumors and
tumor cell lines (9,10). Mice with mono-allelic inactivation
of the CBP gene, but not of the p300 gene, develop an increased
incidence of hematological malignancies (11). In a study of chimeric mice,
hematological malignancies emerged from both CBP−/− and
p300−/− cell populations. This finding suggests that
both CBP and p300 appear to play a role in suppressing
hematological tumor development (12). Accumulating scientific evidence
indicates that CBP/p300 play a role in cancer phenotype. CBP/p300
promoted cancer progression in colon cancer cell lines with
microsatellite instability (13).
Furthermore, p300 regulated p53-dependent apoptosis after DNA
damage in colon cancer cells (14)
and p300 disruption promoted epithelial to mesenchymal transition
and an aggressive cancer phenotype (15).
Hepatocellular carcinoma (HCC) is a global health
concern due to its prevalence and dismal prognosis. The role of
epigenetic deregulation in HCC is being increasingly recognized
(16–18). However, as far as we know, there
have been few studies on the functional significance of CBP/p300
HAT activity in HCC. The selective small molecule inhibitor C646 of
CBP/p300, was identified in a virtual ligand screen (19). C646 has been used to study p300 HAT
function in prostate cancer (20),
melanoma (21), non-small cell
lung cancer (22), and acute
myeloid leukemia (23). These
studies indicated that CBP/p300 play an important role in cancer
cell progression.
Therefore, in this study, we investigated the impact
of CBP/p300 HAT activity in HCC and the possibility that CBP/p300
may be a promising therapeutic target in HCC.
Materials and methods
Cell lines and reagents
The human HCC cell line SK-HEP1 was purchased from
the American Type Culture Collection (Rockville, MD, USA). The
human HCC cell lines HepG2, HLE and Huh7 were purchased from the
Health Science Research Resource Bank (Osaka, Japan). All cell
lines were cultured in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 1% penicillin/streptomycin and 10% fetal calf
serum (FCS) (all from Life Technologies, Tokyo, Japan).
The EP300 monoclonal antibody was purchased from
Abnova (Taipei, Taiwan). The CBP rabbit monoclonal antibody was
purchased from Cell Signaling Technology (Denver, MA, USA). C646
was purchased from Sigma-Aldrich (St. Louis, MO, USA), and was
dissolved in dimethyl sulfoxide (DMSO). Recombinant human TRAIL was
purchased from R&D Systems (Minneapolis, MN, USA).
Immunohistochemistry
Immunohistochemical staining for CBP and p300 was
performed on tissue arrays (Super Bio Chips, Seoul, Korea) using a
TSA Biotin system (Perkin Elmer, Waltham, MA, USA). Deparaffinized
sections were heated for 15 min in citrate buffer at 120°C in an
autoclave to reactivate the antigen and were then treated with 0.3%
H2O2 in methanol for 20 min to deactivate
endogenous peroxidases. Sections were blocked with TNB blocking
buffer (0.1 M Tris-HCl pH 7.5, 0.15 M NaCl, 0.5% blocking reagent)
for 30 min, covered with primary antibody at 4°C overnight, covered
with second-step biotinylated antibody for 60 min, and then
incubated with streptavidin-horseradish peroxidase (HRP) for 30
min. After washing, sections were incubated with the biotinyl
tyramide amplification reagent for 10 min. After washing, sections
were incubated with streptavidin-HRP for 30 min and were then
incubated with diaminobenzidine (DAB) (Dojindo, Rockville, MD,
USA)/0.15% H2O2 and counterstained with
hematoxylin (Muto Pure Chemicals, Tokyo, Japan). Staining was
observed under a microscope (BZ-X710; Keyence, Osaka, Japan).
Detection of histone acetylation by
immunoblotting
HCC cells were incubated at 37°C in a 5%
CO2 atmosphere. Cells were treated with C646 (0, 20 and
50 μM) for 6 h following which histones were isolated using the
EpiQuik Total Histone Extraction kit (Epigentek, Brooklyn, NY, USA)
according to the manufacturer's instructions. The protein content
of each sample was measured using the Bio-Rad protein assay kit
(Bio-Rad Laboratories, Hercules, CA, USA). Aliquots from each
sample containing equal amounts of protein were separated by sodium
dodecyl sulfate (SDS)-polyacrylamide gel (PAGE) electrophoresis and
transferred onto polyvinylidene difluoride (PVDF) membranes
(Millipore, Billerica, MA, USA) for immunoblotting as described
previously (24). The following
primary antibodies were used: anti-acetyl-histone H3 (1:10,000) and
anti-histone H3 (1:20,000) (Millipore). Signals were detected with
the appropriate second antibodies and an ECL kit (Amersham
Pharmacia Biotech, Bucks, UK).
Cell proliferation and viability
assay
MTT assay. HCC cells were plated at a density of
5×103 cells/well in 96-well microtiter plates (Corning
Glass Works, Corning, NY, USA), and each plate was incubated at
37°C in a 5% CO2 atmosphere. After incubation for 24 h,
50 μl of drug solution was added, and the plates were incubated for
an additional 24 or 48 h according to the experiments. The
live-cell count was determined using a Cell Titer 96 assay kit
(Promega, Madison, WI, USA) according to the manufacturer's
instructions. The absorbance of the contents of each well was
measured at 570 nm with a microtiter plate reader (Bio-Rad
Laboratories).
Detection of apoptosis-related proteins
by immunoblotting
Expression of survivin, XIAP and Bcl-xL in HCC cell
lines was analyzed using immunoblotting. Cells were harvested after
incubation with C646 (0, 5, 20 and 50 μM) for 24 h and were lysed
on ice with RIPA buffer (Thermo Fisher Scientific, Waltham, MA,
USA). Aliquots from each sample containing equal amounts of protein
were separated by SDS-PAGE and transferred onto PVDF membranes
(Millipore) for immunoblotting. The following primary antibodies
were used: anti-β-actin antibody (1:4,000) (ab8227; Abcam,
Cambridge, MA, USA), anti-XIAP antibody (1:2,000), anti-Bcl-xL
antibody (1:1,000) (both from BD Transduction Laboratories,
Lexington, KY, USA) and anti-survivin antibody (1:50) (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA). Signals were detected
with the appropriate second antibodies and an ECL kit (Amersham
Pharmacia Biotech).
Invasion assay
Changes in the invasion of HCC cells were analyzed
using a BioCoat Matrigel Invasion chamber (8 μm pore-size; Corning,
Bedford, MA, USA) according to the manufacturer's instructions. HCC
cells (5×104/well) suspended in serum free media were
seeded in the upper chamber with or without the indicated doses of
C646. Ten percent FCS, acting as a chemoattractant, was placed in
the lower chambers. After incubation for 22 h at 37°C in a 5%
CO2 atmosphere, the non-invaded cells on the upper
surface were removed with a cotton swab. The invaded cells on the
filter membrane were stained with Diff-Quick solution (Sysmex,
Kobe, Japan). The invaded cells were counted under a microscope
(Keyence) and five randomly chosen fields were counted for each
assay.
Real-time-polymerase chain reaction (PCR)
array
Changes in the expression of genes related to the
migration and invasion of Huh7 cells following exposure to C646 (0
or 20 μM) for 24 h were analyzed using an RT2 PCR array
(Human Extracellular Matrix and Adhesion Molecules) (SABiosciences
Corp., Frederick, MD, USA) according to the manufacturer's
instructions. Observed changes in mRNA expression were confirmed
using quantitative real-time PCR.
Quantitative reverse transcription
(RT)-PCR
Total RNA was extracted from ~107 Huh7
cells using an RNeasy mini kit (Qiagen, Tokyo, Japan), and cDNA was
synthesized by extension of oligo(dT) primers using PrimeScript
reverse transcriptase (Takara, Ohtsu, Japan). Quantitative
(qRT-PCR) was performed using an ABI PRISM 7300 Real-time PCR
system (Applied Biosystems, Foster City, CA, USA) with EagleTaq
Master Mix kits (Roche Molecular Systems, Branchburg, NJ, USA). The
expression levels of target genes were determined from triplicate
reactions by normalization of expression data to that of GAPDH. The
primer sets for human matrix metallopeptidase 15 (MMP15), human
laminin alpha 3 (LAMA3), human secreted phosphoprotein 1 (SPP1) and
human GAPDH were as follows: human MMP15 forward,
5′-GCTGCTCCTGGTGCTTCT-3′ and reverse, 5′-CTGAGGCAGGTAGCCATAAAG-3′;
human LAMA3 forward, 5′-AGCCCGGGAAGCACTTAT and reverse,
5′-TGTCCATAGAGGCCGTGAC-3′; human SPP1 forward,
5′-GGGCTTGGTTGTCAGCAG-3′ and reverse,
5′-CACTGCAATTCTCATGGTAGTGA-3′; human GAPDH forward,
5′-TATAAATTGAGCCCGCAGCC-3′ and reverse,
5′-TTCCCGTTCTCAGCCTTGAC-3′.
Statistical analysis
Cell number and gene expression data were compared
using the two-tailed Student's t-test. For analysis of differences
between rates, the Chi-square test for independence was used. Cell
invasion was compared using one-way ANOVA. A P-value <0.05 was
considered statistically significant.
Results
CBP and p300 are expressed in HCC
We first performed immunohistochemical staining for
CBP and p300 in tissue array samples. Results of the
immunohistochemical analyses of CBP are summarized in Table I. The positive rates for CBP
staining were extrahepatic metastatic HCC lesions > HCC primary
lesions > non-malignant liver tissues (P<0.01). However,
there were no correlations between the degrees of cancer cell
differentiation and positive staining for CBP. Fig. 1A–C show representative staining of
CBP in non-malignant liver tissue, HCC primary lesion and
extrahepatic metastatic HCC lesion, respectively. Results of the
immunohistochemical analyses of p300 are summarized in Table II. The positive rates for p300
staining were extrahepatic metastatic HCC lesions > HCC primary
lesions > non-malignant liver tissues (P<0.01). Additionally,
there were statistical correlations between the degree of cancer
cell differentiation and positive staining for p300 (P<0.05).
Poorly differentiated HCC showed stronger p300 staining. Fig. 1D–F show representative staining of
p300 in non-malignant liver tissue, HCC primary lesion and
extrahepatic metastatic HCC lesion, respectively. CBP and p300 were
also expressed in HepG2, Huh7, HLE and SK-HEP1 cells (data not
shown). These results suggest that positive staining of CBP and
p300 is a good marker of HCC and positive staining of p300 is a
better marker of the malignant character of HCC than positive
staining of CBP.
 | Table IImmunohistochemical analysis of CBP
expression. |
Table I
Immunohistochemical analysis of CBP
expression.
| Extent of positive
staining |
|---|
|
|
|---|
| 0–33% | 34–66% | 67–100% |
|---|
| HCC (primary
lesion) | 4 (7%) | 21 (34%) | 36 (59%) |
| Well
differentiated | 0 (0%) | 6 (10%) | 6 (10%) |
| Moderately
differentiated | 3 (5%) | 12 (20%) | 25 (41%) |
| Poorly
differentiated | 1 (2%) | 3 (5%) | 5 (8%) |
| Extrahepatic
metastatic | 0 (0%) | 2 (10%) | 19 (90%) |
| HCC lesion | | | |
| Non-malignant
liver | 2 (22%) | 5 (56%) | 2 (22%) |
 | Table IIImmunohistochemical analysis of p300
expression. |
Table II
Immunohistochemical analysis of p300
expression.
| Extent of positive
staining |
|---|
|
|
|---|
| 0–33% | 34–66% | 67–100% |
|---|
| HCC (primary
lesion) | 11 (18%) | 17 (28%) | 33 (54%) |
| Well
differentiated | 5 (8%) | 5 (8%) | 2 (3%) |
| Moderately
differentiated | 5 (8%) | 11 (18%) | 24 (39%) |
| Poorly
differentiated | 1 (2%) | 1 (2%) | 7 (11%) |
| Extrahepatic
metastatic | 0 (0%) | 1 (1%) | 19 (99%) |
| HCC lesion | | | |
| Non-malignant
liver | 8 (89%) | 1 (11%) | 0 (0%) |
C646 reduces histone H3 acetylation in
HCC cells
C646 has selective acetyltrasferase activity against
CBP/p300 HAT. To assess the effect of C646 on global histone H3
acetylation in HCC cells, we used immunoblotting. Exposure of
HepG2, Huh7, HLE and SK-HEP1 cells to C646 for 6 h resulted in
dose-dependent reduction in global histone H3 acetylation (Fig. 2).
C646 reduces the proliferation of HCC
cells
Next, to assess if C646 could affect HCC cell
proliferation, we examined the effect of C646 on cancer cell number
using an MTT assay (Fig. 3).
Exposure of HepG2, Huh7, HLE and SK-HEP1 cells to C646 for 24 h
resulted in decreased cell number in a dose-dependent manner.
Moreover, nuclear fragmentation stained with
4,6-diamino-2-phenylindole (DAPI) was scarcely observed in SK-HEP1
cells treated with C646 (data not shown), suggesting C646 mainly
reduced the proliferation of HCC cells rather than induced
apoptosis.
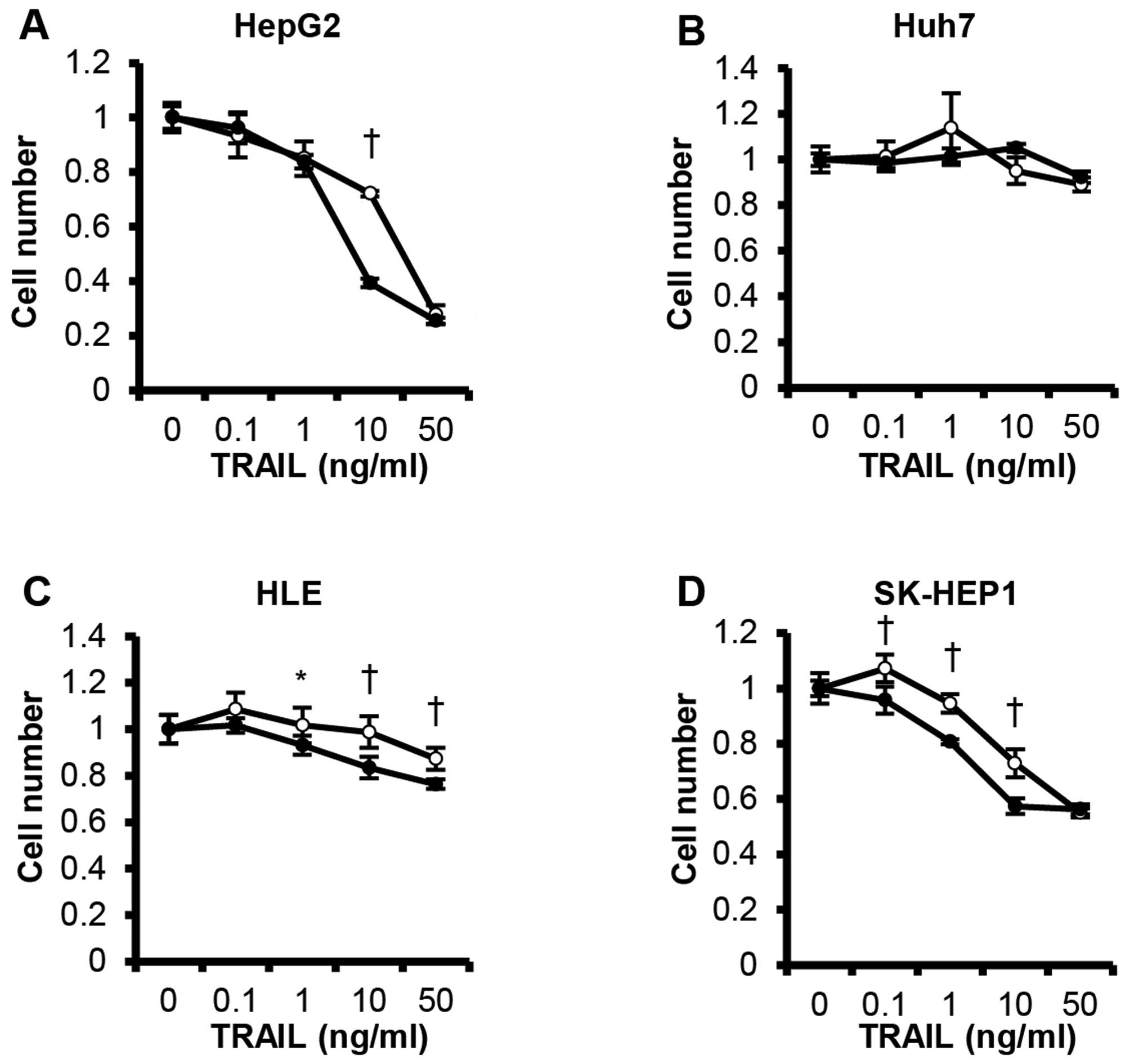
C646 augments TRAIL-induced apoptotic
sensitivity
Since most HCC cells are resistant to TRAIL, we
investigated if CBP/p300 may be involved in this resistance by
analysis of the effects of C646 on TRAIL-induced apoptosis. We
incubated HCC cells with different concentrations of TRAIL, with or
without C646 for 48 h. Although 20 μM C646 did not affect
TRAIL-induced apoptotic sensitivity of Huh7 cells (Fig. 4B), C646 augmented TRAIL-induced
apoptotic sensitivity in HepG2, HLE and SK-HEP1 cells (Fig. 4A, C and D).
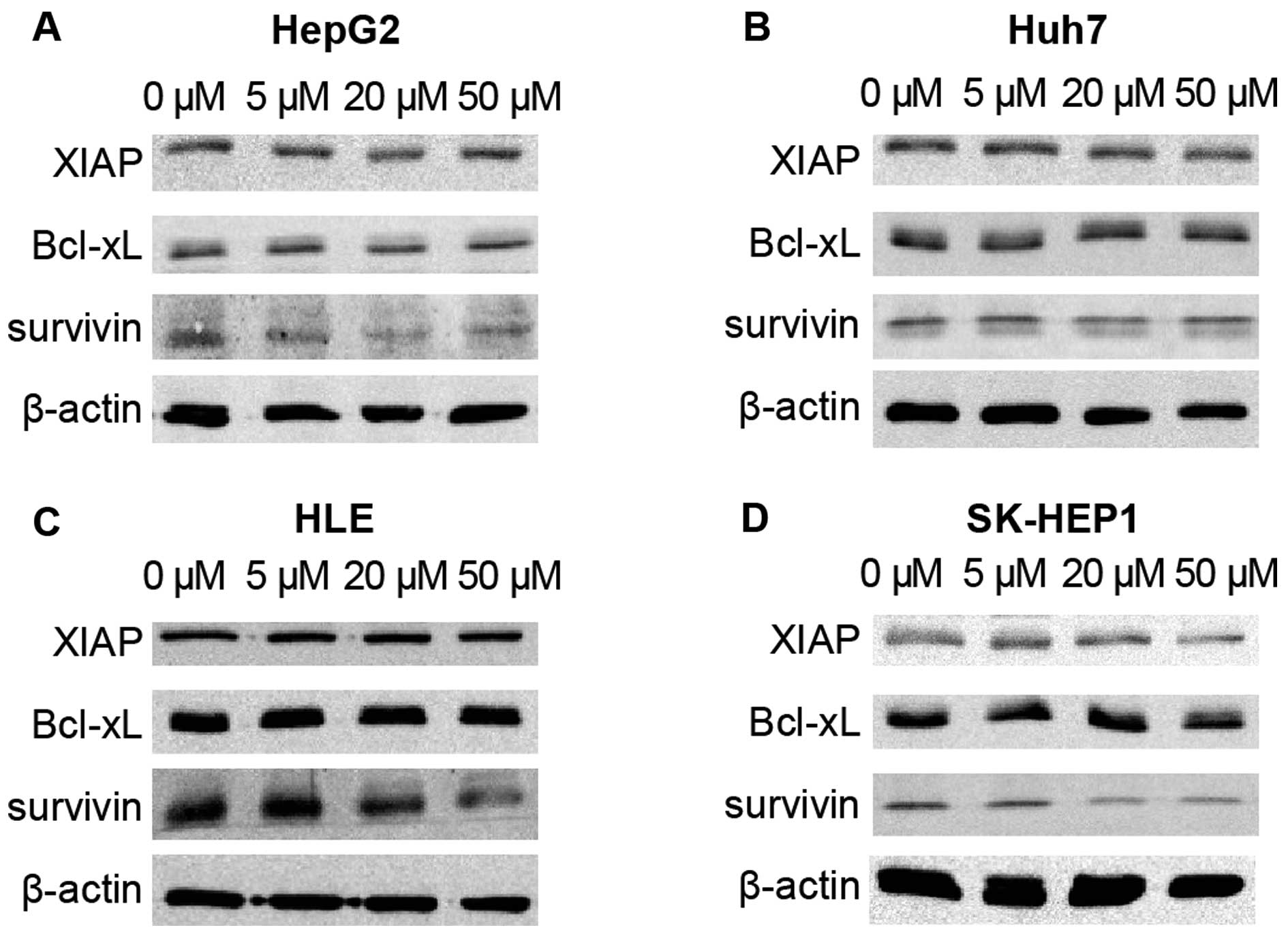
C646 suppresses expression of
survivin
In order to elucidate the mechanism of the effect of
C646 on TRAIL-induced apoptotic sensitivity, we next used
immunoblotting to investigate the effects of C646 on the
intracellular expression level of survivin, XIAP and Bcl-xL, as
these proteins play an important role in controlling apoptotic
pathways. In Huh7 cells, the levels of Bcl-xL, XIAP and survivin
did not change following C646 treatment (Fig. 5B). In HepG2, HLE, and SK-HEP1
cells, although the levels of Bcl-xL and XIAP did not change in
response to C646 treatment, the level of survivin was reduced in
response to C646 treatment in a dose-dependent manner (Fig. 5A, C and D). The levels of survivin
were reduced by C646 treatment in all cell lines except for Huh7,
providing a possible explanation for the low sensitivity of Huh7 to
C646 augmentation of TRAIL-induced apoptosis.
C646 inhibits the invasion of HCC
cells
Finally, we examined the effect of C646 on HCC cell
invasion using a Matrigel-coated invasion chamber. Interestingly,
culture with C646 (0, 10 or 20 μM) significantly inhibited the
invasion of Huh7 cells after 22 h in a dose-dependent manner
(Fig. 6B). Similar inhibition of
invasion following exposure to C646 was observed for HLE cells
(Fig. 6C) and SK-HEP1 cells
(Fig. 6D). However, culture with
C646 did not significantly inhibit the invasion of HepG2 cells
(Fig. 6A).
C646 changes invasion-related mRNA
expression in HCC cells
To elucidate the mechanism by which C646 suppresses
the invasion of HCC cells, we compared the mRNA expression of Huh7
cells cultured with 20 μM C646 with that of control cells using a
real-time PCR array. This experiment showed >1.5 fold-changes in
the mRNA expression levels of MMP15, LAMA3 and SPP1 between the
C646-treated and control cells and amplification of these mRNAs was
detected at fewer than 30 PCR cycles (data not shown). To confirm
these results, we examined C646-induced changes in the level of
MMP15, LAMA3 and SPP1 mRNA expression in Huh7 cells using qRT-PCR.
The level of MMP15 (P<0.01) mRNA expression was significantly
reduced, whereas the level of LAMA3 (P<0.05) and SPP1
(P<0.01) mRNA expression was significantly increased, in Huh7
cells following exposure to C646 (Fig.
7).
Discussion
In the current study, we observed that high
expression of CBP/p300 was more frequently observed in HCC tissues
than in non-malignant liver tissues and showed the highest
frequency in extrahepatic metastatic HCC lesions. Moreover,
expression of p300, but not that of CBP was strongly correlated
with the degree of cancer cell differentiation in HCC. These
results suggested that CBP/p300 have a critical role in
determination of the biological properties of HCC.
Previous studies indicate that increased expression
of CBP/p300 was observed in a variety of human cancers. In prostate
cancer, p300 expression correlates with nuclear alterations of
tumor cells, contributes to tumor growth and is a predictor of
aggressive features (25). In
colon cancer, both CBP and p300 are overexpressed in adenocarcinoma
tissues compared with normal mucosae (26). In nasopharyngeal carcinoma (NPC),
high expression of p300 is more frequently observed in NPC tissues
when compared to the non-neoplastic nasopharyngeal mucosal tissues
(27). Similarly, in HCC, high
expression of p300 is more frequently observed in HCC tissues when
compared to the adjacent liver tissues. Moreover, high expression
of p300 is associated with an aggressive feature of HCC and is a
strong and independent predictor of cancer-specific survival
(28). The above evidence strongly
supports the findings we obtained in this study.
We investigated the biological function of CBP/p300
in HCC cells using the small molecule competitive CBP/p300
inhibitor C646, which has been shown to selectively inhibit
CBP/p300 HAT activity (19). We
found that proliferation and invasion of HCC cells were
significantly inhibited by exposure to C646. Previous studies have
shown that C646 induces apoptosis in prostate cancer cells
(20) and leukemia cells (23). On the contrary, our results
indicated that C646 did not induce apoptosis directly, but
augmented TRAIL-induced apoptotic sensitivity. TRAIL, a member of
the tumor necrosis factor (TNF) family, selectively induces
apoptosis in various transformed cell lines. Furthermore, various
types of tumor cells including HCC show resistance to TRAIL-induced
signaling. In order to elucidate the molecular basis by which C646
augments TRAIL sensitivity, we examined the expression of the
inhibitor of apoptosis (IAPs) proteins that are overexpressed in
HCC cells and confer tumor cell survival and proliferation mainly
by inhibiting the caspase cascade (29–31).
We found that decreased cell proliferation by C646 correlated with
decreased levels of survivin. A previous study showed that survivin
promotes the proliferation of HCC cells (29). These results indicated that
downregulation of survivin protein levels by C646 contribute to its
anti-proliferation effects and to its augmentation of TRAIL-induced
apoptotic sensitivity.
In addition to antiproliferative and apoptotic
effects of C646, we found that C646 substantially inhibited the
invasion of HCC cell lines. A previous study showed that inhibition
of p300 in prostate cancer cells leads to a decrease in cell
migration and invasion (20),
which supports our results. To clarify the mechanism by which C646
inhibited the invasion of HCC cell lines, we examined the effect of
C646 on the mRNA expression of various invasion-related genes and
found that the mRNA expression of MMP15, LAMA3 and SPP1 mRNA was
significantly changed compared to control cells.
MMP15 is classified into the membrane type MMPs that
are important for pericellular proteolysis. In the present study,
the expression level of MMP15 was significantly decreased by C646
treatment. MMP-15 suppression was previously shown to induce
inhibitory effects on the 3D proliferation and in vivo tumor
growth of human fibrosarcoma and gastric cancer cells (32). Furthermore, MMP15 expression is
associated with tumor progression and angiogenesis in human lung
cancer (33). Our result is
consistent with these data. Laminin-5 is a heterotrimer of α3, β3
and γ2 subunits, which are encoded by LAMA3, LAMB3 and LAMC2 genes,
respectively. Our results showed that decreased invasion of HCC
cells correlated with an increased expression level of LAMA3.
Laminin β3 and γ2 chains accumulate intracellularly and play a role
in cancer progression, while epigenetic silencing of the laminin α3
chain may lead to an inability to synthesize the basement membrane
and may affect cancer cell invasion in gastric cancer (34). Furthermore, the overexpression of
p300 in breast epithelial cells leads to decreased levels of
expression of LAMA3 and LAMC2, resulting in decreased adhesion
(35). SPP1 (also known as
osteopontin) is a multifunctional cytokine that impacts cell
proliferation, survival, drug resistance, invasion, and stem-like
behavior. It is implicated in promoting the invasive and metastatic
progression of many cancers (36).
Increased expression of SPP1 was inconsistent with inhibitory
effects on HCC cell invasion. However, it has been recognized that
metastasis regulating genes, such as MMPs, play complex, and often
opposing roles in cancer progression.
We acknowledge that our experimental data were
obtained from cultured cell lines, and we did not clarify the
effect of C646 on normal hepatocytes and stromal cells. The
relationships between HCC cells and stromal cells remain to be
elucidated. Therefore further investigation is required. Also, the
precise mechanism through which C646 changes invasion-related mRNA
expression remains to be elucidated.
In conclusion, C646 inhibited cellular proliferation
and invasion, and augmented TRAIL-induced apoptotic sensitivity in
HCC cells. Our results suggest that CBP/p300 HAT activity has an
important role in malignant transformation, proliferation,
apoptotic sensitivity and invasion in HCC. CBP/p300 could be a
promising therapeutic target in HCC.
References
|
1
|
Waldmann T and Schneider R: Targeting
histone modifications - epigenetics in cancer. Curr Opin Cell Biol.
25:184–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dawson MA, Kouzarides T and Huntly BJ:
Targeting epigenetic readers in cancer. N Engl J Med. 367:647–657.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang F, Marshall CB and Ikura M:
Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis:
Structural and functional versatility in target recognition. Cell
Mol Life Sci. 70:3989–4008. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bedford DC, Kasper LH, Fukuyama T and
Brindle PK: Target gene context influences the transcriptional
requirement for the KAT3 family of CBP and p300 histone
acetyltransferases. Epigenetics. 5:9–15. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kalkhoven E: CBP and p300: HATs for
different occasions. Biochem Pharmacol. 68:1145–1155. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Borrow J, Stanton VP Jr, Andresen JM,
Becher R, Behm FG, Chaganti RS, Civin CI, Disteche C, Dubé I,
Frischauf AM, et al: The translocation t(8;16)(p11;p13) of acute
myeloid leukaemia fuses a putative acetyltransferase to the
CREB-binding protein. Nat Genet. 14:33–41. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chaffanet M, Gressin L, Preudhomme C,
Soenen-Cornu V, Birnbaum D and Pébusque MJ: MOZ is fused to p300 in
an acute monocytic leukemia with t(8;22). Genes Chromosomes Cancer.
28:138–144. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Muraoka M, Konishi M, Kikuchi-Yanoshita R,
Tanaka K, Shitara N, Chong JM, Iwama T and Miyaki M: p300 gene
alterations in colorectal and gastric carcinomas. Oncogene.
12:1565–1569. 1996.PubMed/NCBI
|
|
10
|
Gayther SA, Batley SJ, Linger L, Bannister
A, Thorpe K, Chin SF, Daigo Y, Russell P, Wilson A, Sowter HM, et
al: Mutations truncating the EP300 acetylase in human cancers. Nat
Genet. 24:300–303. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kung AL, Rebel VI, Bronson RT, Ch'ng LE,
Sieff CA, Livingston DM and Yao TP: Gene dose-dependent control of
hematopoiesis and hematologic tumor suppression by CBP. Genes Dev.
14:272–277. 2000.PubMed/NCBI
|
|
12
|
Rebel VI, Kung AL, Tanner EA, Yang H,
Bronson RT and Livingston DM: Distinct roles for CREB-binding
protein and p300 in hematopoietic stem cell self-renewal. Proc Natl
Acad Sci USA. 99:14789–14794. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ionov Y, Matsui S and Cowell JK: A role
for p300/CREB binding protein genes in promoting cancer progression
in colon cancer cell lines with microsatellite instability. Proc
Natl Acad Sci USA. 101:1273–1278. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Iyer NG, Chin SF, Ozdag H, Daigo Y, Hu DE,
Cariati M, Brindle K, Aparicio S and Caldas C: p300 regulates
p53-dependent apoptosis after DNA damage in colorectal cancer cells
by modulation of PUMA/p21 levels. Proc Natl Acad Sci USA.
101:7386–7391. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Krubasik D, Iyer NG, English WR, Ahmed AA,
Vias M, Roskelley C, Brenton JD, Caldas C and Murphy G: Absence of
p300 induces cellular phenotypic changes characteristic of
epithelial to mesenchyme transition. Br J Cancer. 94:1326–1332.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ozen C, Yildiz G, Dagcan AT, Cevik D, Ors
A, Keles U, Topel H and Ozturk M: Genetics and epigenetics of liver
cancer. N Biotechnol. 30:381–384. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pogribny IP and Rusyn I: Role of
epigenetic aberrations in the development and progression of human
hepatocellular carcinoma. Cancer Lett. 342:223–230. 2014.
View Article : Google Scholar :
|
|
18
|
Ma L, Chua MS, Andrisani O and So S:
Epigenetics in hepatocellular carcinoma: An update and future
therapy perspectives. World J Gastroenterol. 20:333–345. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bowers EM, Yan G, Mukherjee C, Orry A,
Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, et
al: Virtual ligand screening of the p300/CBP histone
acetyltransferase: Identification of a selective small molecule
inhibitor. Chem Biol. 17:471–482. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Santer FR, Höschele PP, Oh SJ, Erb HH,
Bouchal J, Cavarretta IT, Parson W, Meyers DJ, Cole PA and Culig Z:
Inhibition of the acetyltransferases p300 and CBP reveals a
targetable function for p300 in the survival and invasion pathways
of prostate cancer cell lines. Mol Cancer Ther. 10:1644–1655. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan G, Eller MS, Elm C, Larocca CA, Ryu B,
Panova IP, Dancy BM, Bowers EM, Meyers D, Lareau L, et al:
Selective inhibition of p300 HAT blocks cell cycle progression,
induces cellular senescence, and inhibits the DNA damage response
in melanoma cells. J Invest Dermatol. 133:2444–2452. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Oike T, Komachi M, Ogiwara H, Amornwichet
N, Saitoh Y, Torikai K, Kubo N, Nakano T and Kohno T: C646, a
selective small molecule inhibitor of histone acetyltransferase
p300, radio-sensitizes lung cancer cells by enhancing mitotic
catastrophe. Radiother Oncol. 111:222–227. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao XN, Lin J, Ning QY, Gao L, Yao YS,
Zhou JH, Li YH, Wang LL and Yu L: A histone acetyltransferase p300
inhibitor C646 induces cell cycle arrest and apoptosis selectively
in AML1-ETO-positive AML cells. PLoS One. 8:e554812013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fuke H, Shiraki K, Sugimoto K, Tanaka J,
Beppu T, Yoneda K, Yamamoto N, Ito K, Masuya M and Takei Y: Jak
inhibitor induces S phase cell-cycle arrest and augments
TRAIL-induced apoptosis in human hepatocellular carcinoma cells.
Biochem Biophys Res Commun. 363:738–744. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Isharwal S, Miller MC, Marlow C, Makarov
DV, Partin AW and Veltri RW: p300 (histone acetyltransferase)
biomarker predicts prostate cancer biochemical recurrence and
correlates with changes in epithelia nuclear size and shape.
Prostate. 68:1097–1104. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ishihama K, Yamakawa M, Semba S, Takeda H,
Kawata S, Kimura S and Kimura W: Expression of HDAC1 and CBP/p300
in human colorectal carcinomas. J Clin Pathol. 60:1205–1210. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liao ZW, Zhou TC, Tan XJ, Song XL, Liu Y,
Shi XY, Huang WJ, Du LL, Tu BJ and Lin XD: High expression of p300
is linked to aggressive features and poor prognosis of
nasopharyngeal carcinoma. J Transl Med. 10:1102012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li M, Luo RZ, Chen JW, Cao Y, Lu JB, He
JH, Wu QL and Cai MY: High expression of transcriptional
coactivator p300 correlates with aggressive features and poor
prognosis of hepatocellular carcinoma. J Transl Med. 9:52011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ito T, Shiraki K, Sugimoto K, Yamanaka T,
Fujikawa K, Ito M, Takase K, Moriyama M, Kawano H, Hayashida M, et
al: Survivin promotes cell proliferation in human hepatocellular
carcinoma. Hepatology. 31:1080–1085. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Takehara T, Liu X, Fujimoto J, Friedman SL
and Takahashi H: Expression and role of Bcl-xL in human
hepatocellular carcinomas. Hepatology. 34:55–61. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shiraki K, Sugimoto K, Yamanaka Y,
Yamaguchi Y, Saitou Y, Ito K, Yamamoto N, Yamanaka T, Fujikawa K,
Murata K, et al: Overexpression of X-linked inhibitor of apoptosis
in human hepatocellular carcinoma. Int J Mol Med. 12:705–708.
2003.PubMed/NCBI
|
|
32
|
Ito E, Yana I, Fujita C, Irifune A, Takeda
M, Madachi A, Mori S, Hamada Y, Kawaguchi N and Matsuura N: The
role of MT2-MMP in cancer progression. Biochem Biophys Res Commun.
393:222–227. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen L, Zhou Q, Xu B, Liu J, Shi L, Zhu D,
Wu C and Jiang J: MT2-MMP expression associates with tumor
progression and angiogenesis in human lung cancer. Int J Clin Exp
Pathol. 7:3469–3477. 2014.PubMed/NCBI
|
|
34
|
Ii M, Yamamoto H, Taniguchi H, Adachi Y,
Nakazawa M, Ohashi H, Tanuma T, Sukawa Y, Suzuki H, Sasaki S, et
al: Co-expression of laminin β3 and γ2 chains and epigenetic
inactivation of laminin α3 chain in gastric cancer. Int J Oncol.
39:593–599. 2011.PubMed/NCBI
|
|
35
|
Miller KA, Chung J, Lo D, Jones JC,
Thimmapaya B and Weitzman SA: Inhibition of laminin-5 production in
breast epithelial cells by overexpression of p300. J Biol Chem.
275:8176–8182. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shevde LA and Samant RS: Role of
osteopontin in the pathophysiology of cancer. Matrix Biol.
37:131–141. 2014. View Article : Google Scholar : PubMed/NCBI
|