Introduction
Pancreatic ductal adenocarcinoma (PDAC), a common
digestive system cancer, is highly malignant and has a poor disease
outcome. Despite the progress in the understanding of the molecular
and genetic basis of this disease, the 5-year survival rate has
remained low and usually does not exceed 5%. Only 20–25% of the
patients present with potentially resectable disease, and surgery
represents the only chance for a cure (1,2).
PDAC is considered as a systemic disease because of the high rate
of relapse after curative surgery in patients with resectable
disease at diagnosis. Enormous efforts have been made to identify
the special molecular markers for PDAC, which show vast application
prospect as targets for the disease treatment. The research fields
of molecular markers for PDAC include proteins, such as K-Ras, p16,
and SMAD4 (3–5); miRNAs, such as miR-210 and miR-221
(6–8); and the recently research hotspot the
lncRNAs.
Long non-coding RNAs (lncRNAs) refer to a group of
RNAs that are usually more than 200 nucleotides and are not
involved in protein generation (9). Recent studies have begun to associate
subsets of lncRNAs to specific regulatory mechanisms of important
biological processes, including cell proliferation, survival,
differentiation, and chromatin remodeling both in cis and in
trans (10–19). Many functional lncRNAs have been
shown to play key roles in organ development and cancer. Some
lncRNAs act as tumor suppressor; others participate in cellular
replicative immortality, or even regulate angiogenesis and
metastasis (20,21). Previous studies have reported
several lncRNA, such as HOTAIR, MALAT1 and PVT1 (22–24),
which revealed the significance of lncRNAs in the regulation of
multiple biological processes at different levels that may served
as molecular markers for several cancer. However, the roles of
lncRNAs in the progression of PDAC remain not well identified.
To evaluate the expression profile and identify the
special lncRNAs in PDAC, we interrogated the differentially
expression profiles of lncRNAs and mRNAs between 3 PDAC samples and
their matched adjacent non-tumor samples via microarray. Gene
ontology (GO) analysis, pathway analysis and network analysis was
done for further investigation. Quantitative reverse transcription
polymerase chain reaction (qRT-PCR) was used to validate several
random upregulated and downregulated lncRNAs in the 3 PDAC tissues.
Further, HOTAIRM1, one of thousands of deregulated lncRNAs we
identified, was further evaluated in 12 pairs of matched
tumor/non-tumor (T/N) tissues via qRT-PCR. This study uncovers the
aberrant expression of lncRNAs in PDAC tissues, and may contribute
to understanding of the mechanism of PDAC progression and provide
new potential molecular markers for diagnosis and treatment of
PDAC.
Materials and methods
Patients and tissue samples
A total of twelve PDAC tissue samples and their
matched adjacent non-tumor samples were obtained with informed
consent from PDAC patients at Department of Surgery, Sichuan
Provincial People's Hospital. The diagnosis of all patients was
confirmed based on the WHO classification and staged according to
the tumor node metastasis classification and were reviewed by two
pathologists. Clinical parameters were recorded for each sample,
included age, gender, location of tumor, vascular permeation, TNM
stage and differentiation. Samples were taken during surgery,
immediately frozen in liquid nitrogen, and stored at −80°C for
further analysis. Paired tumor and non-tumor tissues from three
PDAC patients were used for the micro-array assay. Twelve paired
PDAC tissues (not including the 3 paired tissues used for
microarray) were used for the qRT-PCR validation assay. The
analysis of human tissues were approved by the Human Research
Ethics Committee of Sichuan Provincial People's Hospital, and all
PDAC patients gave written informed consent for the use of clinical
samples for medical research.
RNA isolation
Total RNA was isolated from the 15 PDAC tissues and
paired non-tumor tissues using TRIzol reagent (Invitrogen, CA,
USA), and quantified using a NanoDrop ND-1000 spectrophotometer
(NanoDrop, DE, USA). The integrity of RNA was assessed by standard
denaturing agarose gel electrophoresis, and the purity was
estimated by the ratio of absorbance at 260–280 nm.
Microarray
Arraystar Human LncRNA Microarray V3.0 is designed
for the global profiling of human lncRNAs and protein-coding
transcripts, which is updated from the previous Microarray V2.0.
Approximately 30,586 lncRNAs and 26,109 coding transcripts can be
detected by our third-generation lncRNA microarray. The lncRNAs are
carefully constructed using the most highly respected public
transcriptome databases (including Refseq, UCSC known genes, and
Gencode), as well as landmark publications. Each transcript is
represented by a specific exon or splice junction probe, which can
identify individual transcript accurately. Positive probes for
housekeeping genes and negative probes are also printed onto the
array for hybridization quality control.
RNA labeling and array hybridization
Sample labeling and array hybridization were
performed according to the Agilent One-Color Microarray-Based Gene
Expression Analysis protocol (Agilent Technology) with minor
modifications. Briefly, mRNA was purified from total RNA after
removal of rRNA (mRNA-ONLY™ Eukaryotic mRNA Isolation kit,
Epicentre). Then, each sample was amplified and transcribed into
fluorescent cRNA along the entire length of the transcripts without
3′ bias utilizing a random priming method (Arraystar Flash RNA
Labeling kit, Arraystar). Each labeled cRNA (1 μg) was fragmented
by adding 5 μl 10X blocking agent and 1 μl of 25X fragmentation
buffer, then heated the mixture at 60°C for 30 min, finally 25 μl
2X GE hybridization buffer was added to dilute the labeled cRNA.
Hybridization solution (50 μl) was dispensed into the gasket slide
and assembled to the lncRNA expression microarray slide. The slides
were incubated for 17 h at 65°C in an Agilent Hybridization Oven.
The hybridized arrays were washed, fixed and scanned with using the
Agilent DNA Microarray Scanner (part number G2505C).
Data analysis
Data analysis were performed by KangChen Biotech
(Shanghai, China). Agilent Feature Extraction software (version
11.0.1.1) was used to analyze acquired array images. Quantile
normalization and subsequent data processing were performed with
using the Gene Spring GX v12.1 software package (Agilent
Technologies). After quantile normalization of the raw data,
lncRNAs and mRNAs of ≥3 out of 6 samples have flags in Present or
Marginal (All Targets Value) were chosen for further data analysis.
Differentially expressed lncRNAs and mRNAs with statistical
significance between the two groups were identified through
P-value/FDR filtering. Differentially expressed lncRNAs and mRNAs
between the two samples were identified through fold-change
filtering. Hierarchical clustering and combined analysis were
performed using in-house scripts.
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using TRIzol reagent
(Invitrogen) and then reverse transcribed using
PrimeScript® RT Reagent kit with gDNA Eraser (Perfect
Real Time) (Takara, Dalian, China) according to the manufacturer's
instructions. The expression levels of seven upregulated and seven
downregulated lncRNAs in the 3 patients included in the microarray
study were measured by qRT-PCR using SYBR Green assays (Takara).
The expression levels of HOTAIRM1 in twelve PDAC specimens and
their paired adjacent non-cancerous tissues were also measured by
qRT-PCR. The lncRNA expression differences between the matched
cancer and non-cancerous samples were analyzed using Student's
paired t-test with the IBM SPSS Statistics version 20.0 (IBM Corp.,
New York, NY, USA). A probability value of P<0.05 was considered
statistically significant.
Results
Differentially expressed lncRNAs in
PDAC
The clinical parameters of all patients are shown in
Table I. The lncRNA expression
profile data from the microarray analysis contained a total of
21,558 lncRNAs that were expressed in PDAC tissue samples. To
determine the relationships among the specimens, hierarchical
clustering analysis was used to group the specimens according to
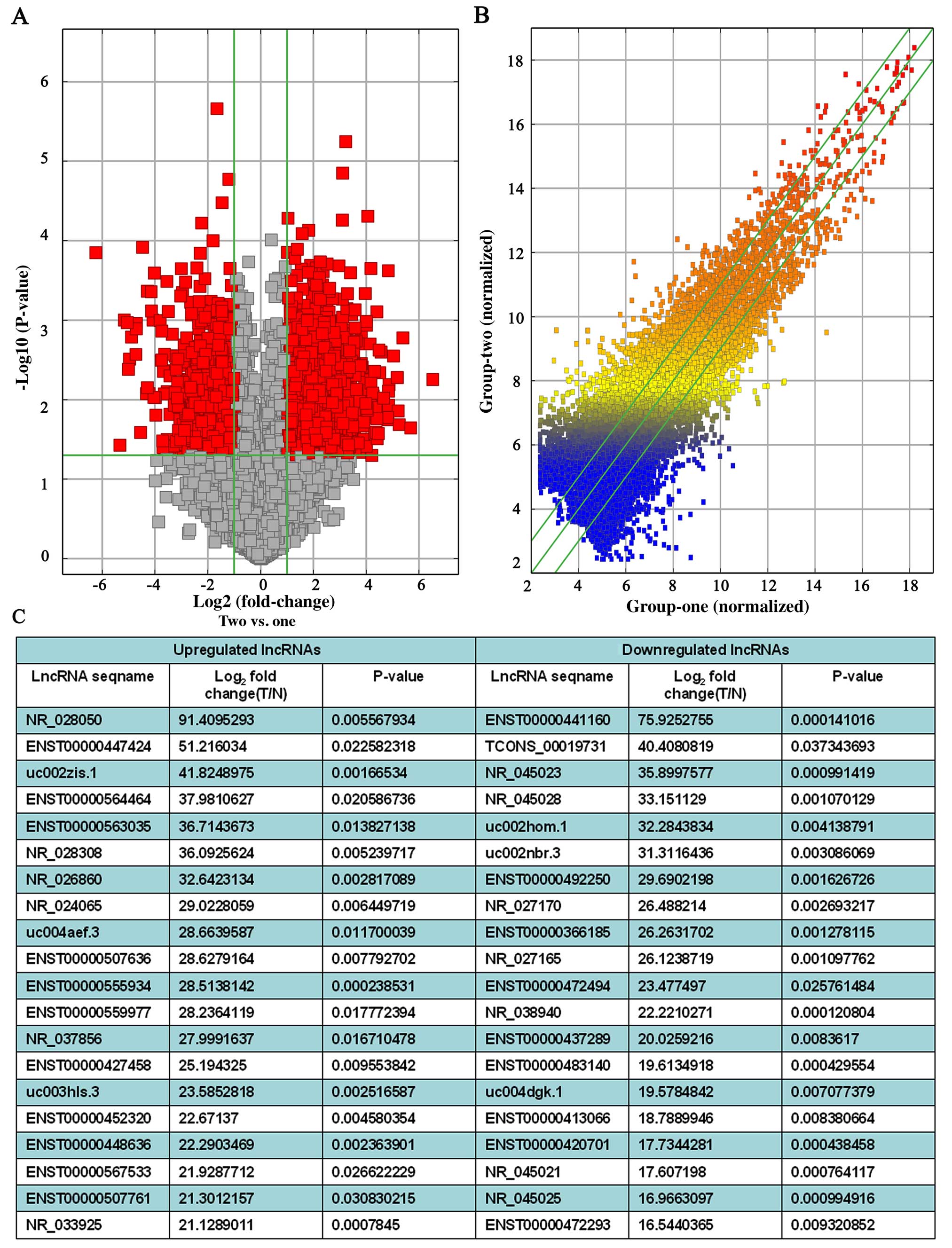
their expression levels (data not shown). Volcano Plots and the
scatterplot of lncRNA expression profile are useful for assessing
the variation or reproducibility (Fig.
1A and B). We identified hundreds of significantly
differentiated lncRNAs (fold change ≥2.0, P≤0.05) between 3 human
PDAC tissue samples and the matched adjacent non-tumor samples. In
total, there were 2,331 upregulated lncRNAs and 1,641 downregulated
lncRNAs found in the 3 PDAC patients (Fig. 1C). Upregulated lncR NAs were more
common than downregulated lncRNAs in our microarray data. Among
these lncRNAs, ASHGA5P050875 (fold change, 91.4095293) was the most
upregulated lncRNA, and ASHGA5P044551 (fold change, 75.9252755) was
the most downregulated lncRNA.
 | Table IClinical parameter of 15 PDAC
patients. |
Table I
Clinical parameter of 15 PDAC
patients.
| Sample nos. | Age (years) | Gender | Location of
tumor | Vascular
permeation | TNM stage |
Differentiation |
|---|
| 1 | 46 | Male | Head | Present | T2N1M0 | Poorly |
| 2 | 53 | Female | Body and tail | Absent | T3N1M0 | Moderately |
| 3 | 67 | Female | Head | Absent | T2N0M0 | Poorly |
| 4 | 72 | Male | Head | Absent | T3N1M0 | Moderately |
| 5 | 61 | Male | Body and tail | Present | T1N0M0 | Moderately |
| 6 | 42 | Female | Body and tail | Absent | T2N1M0 | Poorly |
| 7 | 55 | Male | Head | Absent | T1N0M0 | Well |
| 8 | 56 | Male | Head | Absent | T2N0M0 | Well |
| 9 | 75 | Male | Head | Absent | T4N1M0 | Moderately |
| 10 | 52 | Female | Head | Absent | T2N0M0 | Poorly |
| 11 | 60 | Male | Head | Absent | T1N1M0 | Moderately |
| 12 | 65 | Male | Body and tail | Absent | T3N1M0 | Well |
| 13 | 71 | Female | Head | Present | T2N1M0 | Poorly |
| 14 | 52 | Male | Body and tail | Absent | T1N0M0 | Moderately |
| 15 | 49 | Male | Body and tail | Present | T3N1M0 | Poorly |
Further analysis proceeded by classifying and
stratifying the lncRNAs into subgroups. Subgroups such as antisense
lncRNAs, enhancer lncRNAs and lincRNAs are thought to participate
in numerous diseases such as cancers. We found 69 antisense RNAs,
82 enhancer RNAs and 147 lincRNAs were upregulated in PDAC samples,
respectively, and 50 antisense RNAs, 70 enhancer RNAs and 236
lincRNAs were downregulated in the adjacent non-tumor samples,
respectively (data not shown). The changes of lncRNAs subgroup
between the PDAC samples and adjacent non-tumor samples play an
important role in the regulation of PDAC tumor progression and we
will focus on subgroup lncRNAs and their related mRNA in PDAC
samples in our further study.
Differentially expressed mRNAs in
PDAC
The mRNA expression profile data from the microarray
analysis contained a total of 14,609 mRNAs that were expressed in
the PDAC tissue samples. Volcano plots and the scatterplot of
lncRNA expression profile are useful for assessing the variation or
reproducibility (Fig. 2A and B).
Among them, 1,676 mRNAs were significantly upregulated and 1,981
mRNAs downregulated (fold change ≥2.0, P≤0.05) in the PDAC samples
(Fig. 2C). The most significantly
deregulated mRNAs were ASHGA5P012017 (upregulated, fold change,
89.8272773) and ASHGA5P033632 (downregulated, fold change,
463.3570246).
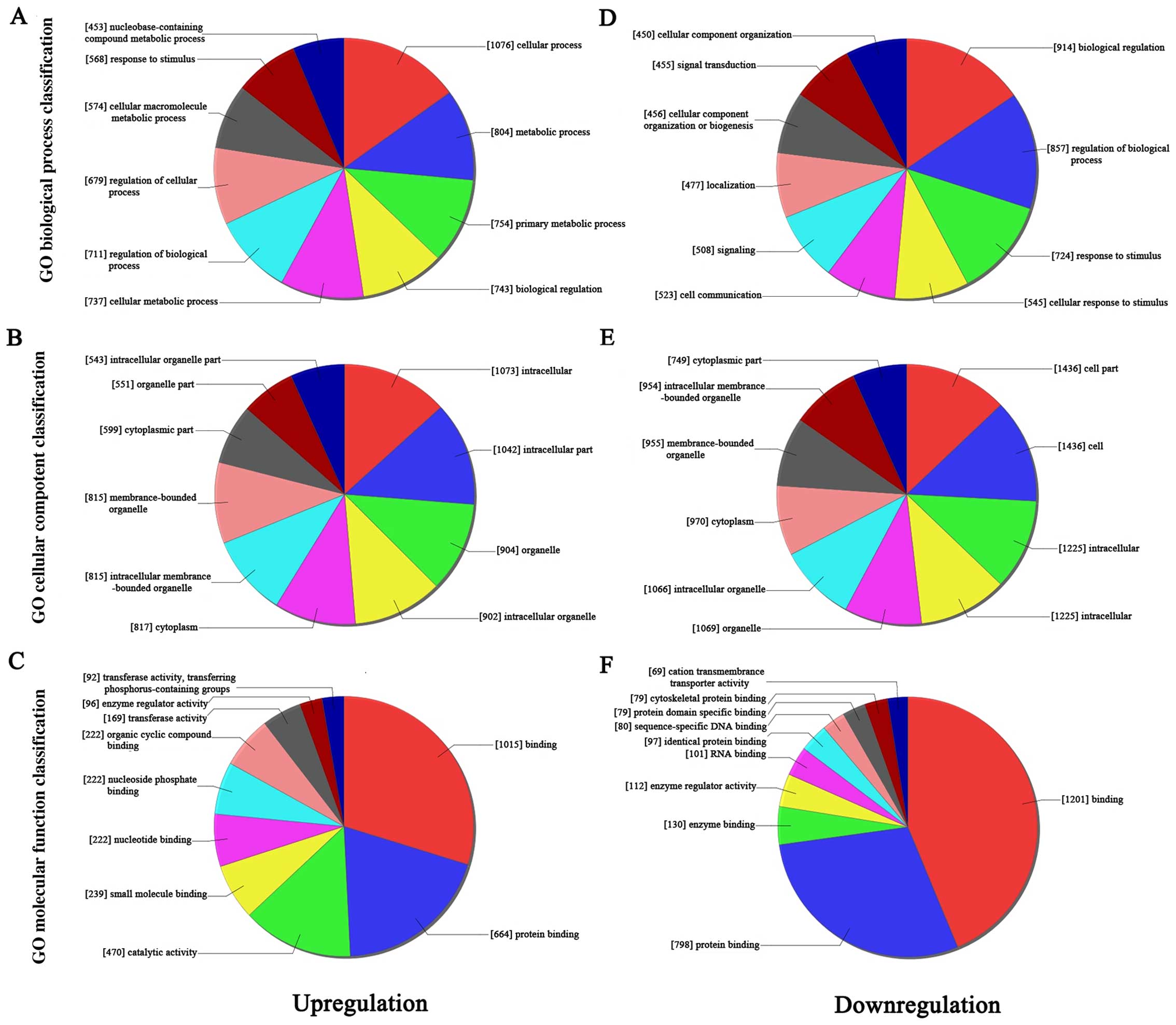
GO analysis
Gene Ontology (GO) analysis was performed to
determine the transcripts with terms under the biological process,
cellular component, and molecular function ontology in this study.
Fisher's exact test was applied to find if there were more overlap
between the differentially expressed list and the GO annotation
list than would be expected by chance. The P-values were used to
estimate the significance of GO terms enrichment in the
differentially expressed lncRNAs and mRNAs; the lower the P-value,
the more significant the GO term (P-values ≤0.05 is recommended).
We found that the highest enriched GO terms for the upregulated
transcripts were purine nucleoside catabolic process (Fig. 3A; GO:0006152 under biological
process, P=8.072E-06), cytoplasm (Fig.
3B; GO:0005737 under cellular component; P=1.158E-09), and
protein binding (Fig. 3C;
GO:0005515 under molecular function; P=1.014E-09). The most highly
enriched GO terms targeted by the downregulated transcripts were
establishment of localization (Fig.
3D, GO:0051234 under biological process; P=1.968E-05),
cytoplasmic part (Fig. 3E,
GO:0044444 under cellular component; P=1.394E-09), and protein
binding (Fig. 3F, GO:0005515 under
molecular function; P=9.546E-09).
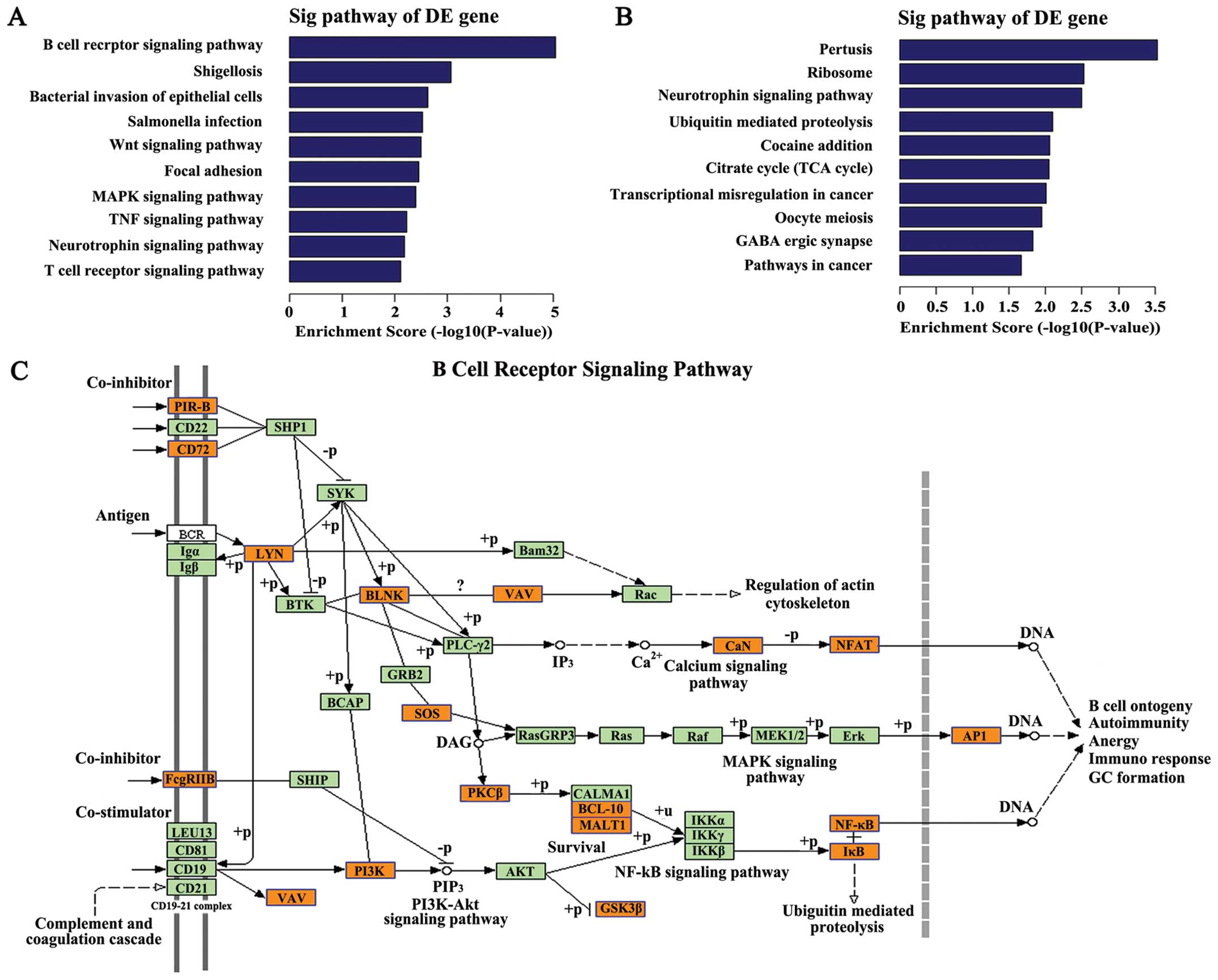
Pathway analysis
Pathway analysis indicated that 41 pathways
corresponded to the upregulated transcripts (Fig. 4A). The most enriched network was ‘B
cell receptor signaling pathway (human)’ (Fisher P=9.08198E-06,
Fig. 4C) with 18 transcripts
annotated with this term. Twenty-five pathways corresponded to the
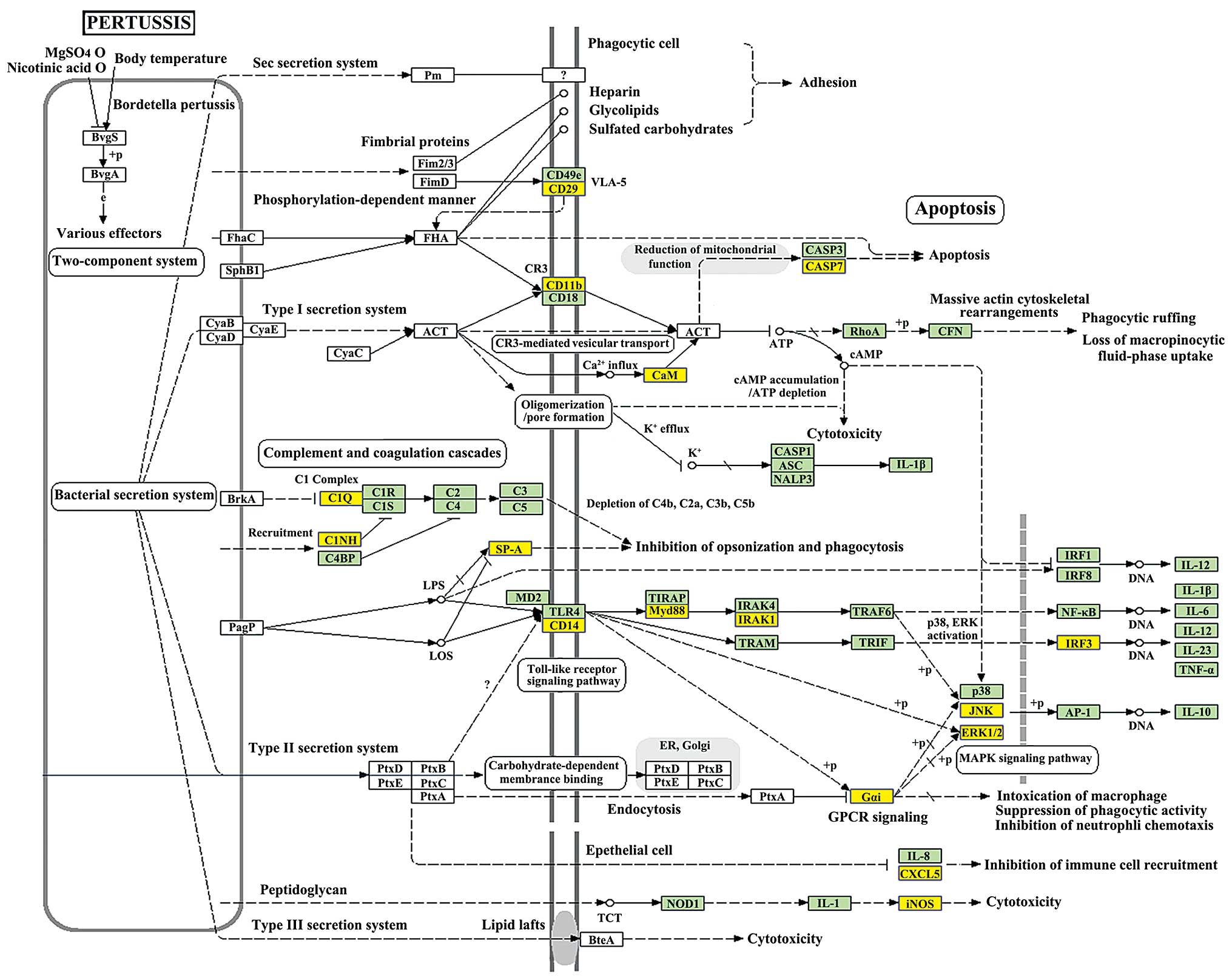
downregulated transcripts (Fig.
4B) and the most enriched network was ‘Pertussis-Homo
sapiens (human)’ (Fisher P=0.0002931875, Fig. 5) with 18 transcripts annotated with
this term. P-values ≤0.05 were taken as the cut-off. Among these
pathways, the gene category ‘Wnt signaling pathway’, has been
reported to be involved in metastasis of pancreatic carcinogenesis
(25), and the gene category ‘MAPK
signaling pathway’ has been shown to participate in the progression
of pancreatic cancer though multiple mechanisms (26–28).
The gene categories ‘FoxO signaling pathway’ have been reported to
suppress or activate pancreatic cancer progression by different
drugs or compound (29,30). The gene categories ‘Ubiquitin
mediated proteolysis’ participate in pancreatic cancer cell growth
in vitro and in vivo (31).
Quantitative real-time PCR
validation
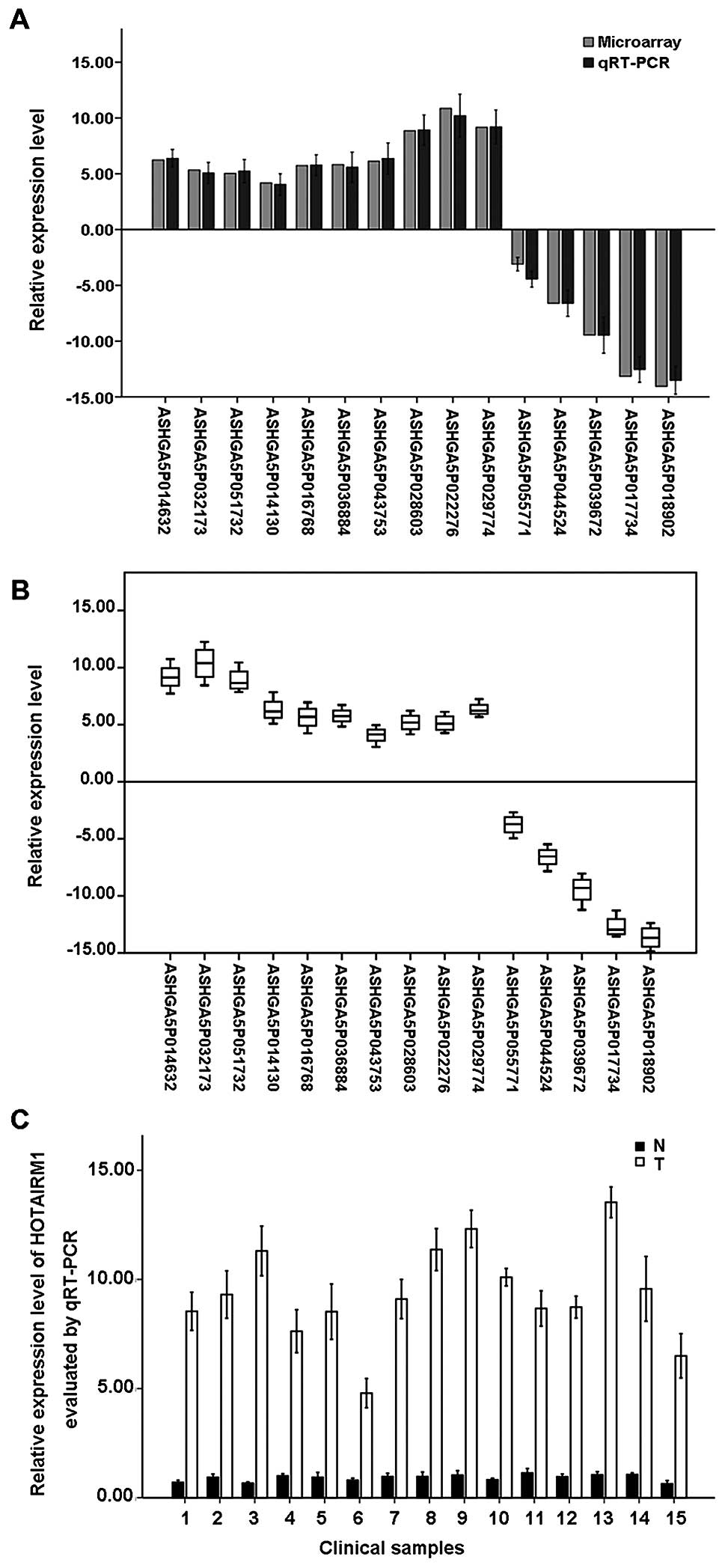
We used qRT-PCR to validate the expression levels of
the altered lncRNAs in the PDAC patients. We randomly selected ten
upregulated lncRNAs and five downregulated lncRNAs among the
differentially expressed lncRNAs. We found that ASHGA5P022276,
ASHGA5P029774, ASHGA5P028603, ASHGA5P014632, ASHGA5P043753,
ASHGA5P036884, ASHGA5P016768, ASHGA5P032173, ASHGA5P051732 and
ASHGA5P014130 were upregulated, and ASHGA5P055771, ASHGA5P044524,
ASHGA5P039672, ASHGA5P017734 and ASHGA5P018902 were downregulated
in the PDAC samples compared with adjacent non-tumor samples. Thus,
the results from the qRT-PCR analysis and the microarray data
analysis were consistent (P<0.05; Fig. 6A and B).
Moreover, we found a significant increase of the
expression level of HOTAIRM1 (fold change, 6.9263288, P=0.00282) in
PDAC samples compared with adjacent non-tumor samples via
microarray analysis. To examine whether upregulated expression of
HOTAIRM1 is pathologically specific, a total of 12 PDAC samples and
matched adjacent non-tumor samples were subjected to qRT-PCR. The
level of HOTAIRM1 expression was 2.92–8.53-fold higher in PDAC
samples than the mean level in matched adjacent non-tumor samples
(Fig. 6C). However, the sample
size of this study is limited and we will further collect more
samples and investigate the function of HOTAIRM1 in PDAC.
Discussion
In this study, we used a microarray to test the
lncRNAs expression profiles in PDAC tissues. The lncRNA expression
profiling data showed that there were lncRNAs that were
differentially expressed between the PDAC tissues and matched
adjacent non-tumor tissues. Previous studies showed that
dysregulation of lncRNAs expression such as HOTAIR (32,33),
HULC (34–36) and GAS5 (37,38),
is a potential molecular marker for diagnostic and therapeutic
purposes in several human cancers. There are still lncRNAs as
potential novel candidate molecular markers for clinical diagnosis
and therapy of PDAC that need to be further identified.
Although special lncRNAs as molecular markers in
other digestive tumors, such as hepatocellular carcinoma and
gastric cancer (39,40) have been reported, there is no
direct evidence shown that special lncRNAs are molecular markers
for PDAC. Moreover, several lncRNAs have been reported to be
significantly correlated with PDAC outcome and are involved in
cancer progression. HOTAIR is a negative prognostic factor for
breast, colon and liver cancer patient survival, and increased
HOTAIR expression in patients has been correlated with enhanced
breast and colon cancer metastasis (41–45).
Kyounghyun et al (46)
showed that HOTAIR expression was increased markedly in pancreatic
tumors compared to non-tumor tissues, and was associated with more
aggressive tumors. MALAT1 (47,48),
also known as nuclear-enriched abundant transcript 2 (NEAT2),
regulates gene expression and post-transcriptionally modifies
primary transcripts and is found to be upregulated in a variety of
human cancers of the breast, prostate, colon, liver, and uterus
(49). Recently, MALAT1 mRNA level
was found significantly higher in PADC tissues and some PC cell
lines. A high expression of MALAT1 was detected in PDAC tumors of
larger size, advanced tumor stage and deeper invasion. In addition,
the overexpression of MALAT1 was associated with poor prognosis of
PDAC patients (50). The HOTTIP
lncRNA, located at the 5′-end of the HOXA cluster, was
significantly expressed in anatomically distal human fibro-blasts
(51). Recent study demonstrates
that HOTTIP, which is significantly overexpressed in PDAC, plays a
significant role in PDAC progression and gemcitabine
chemoresistance (52). H19 was
characterized as an oncogenic lncRNA in some tumors and upregulated
remarkably in primary PDAC tumors that subsequently metastasized,
compared to those with non-metastasis. H19 also promoted PDAC cell
invasion and migration at least partially by increasing
HMGA2-mediated epithelial-mesenchymal transition (EMT) through
antagonizing let-7 (53). The
previous studies also reported several other lncRNAs related to
PDAC, such as HULC, PVTI, MAP3K14, PPP3Cb, DAPKI and LOC285194
(54–57). In the present study, we also
examined the expression of some most studied lncRNAs in PDAC, such
as MALAT1, HOTTIP, H19, HULC, PVTI, MAP3K14, PPP3Cb, DAPKI and
LOC285194 in the combination data set of 3 pairs of microarrays,
showing that MALAT1 and HOTTIP were significantly upregulated
16.22- and 23.48-fold in PDAC tissues compared with paired
non-tumor tissues respectively; however, other lncRNAs were not
significantly differentially expressed between PDAC tissues and
paired non-tumor tissues.
The microarray expression profiles revealed 21,558
lncRNAs that were expressed in those samples; 2,331 lncRNAs were
significantly upregulated and 1,641 lncRNAs were significantly
downregulated in 3 PDAC samples compared with the paired non-tumor
tissues. We then randomly selected 14 lncRNAs for validation by
qRT-PCR in other 12 PDAC samples and paired non-tumor tissues.
Additionally, the results from the qRT-PCR analysis and the
microarray data analysis were consistent. In these deregulated
lncRNAs, we then analyzed the subgroup lncRNAs, including the
antisense lncRNAs, the enhancer lncRNAs and the lincRNAs, and their
related mRNA that may play an important role in the regulation
mechanism of PDAC progression. Antisense lncRNAs have been
recognized to regulate expression of corresponding coding genes at
post-transcriptional level (58),
and therefore participate in carcinogenesis by regulation of
oncogenes as well as anti-oncogenes. Enhancer RNAs are required for
efficient transcriptional enhancement of interacting target genes
and also are required for p53-dependent enhancer activity and gene
transcription (59–62). LincRNAs play pivotal roles in
cancer-related gene regulatory system, and the disorder of their
gene expression is thought to promote cancer cell proliferation,
invasion and metastasis (63–67).
In the present study, our data showed that 69 antisense RNAs, 82
enhancer RNAs and 147 lincRNAs were upregulated in PDAC tissues,
and 50 antisense RNAs, 70 enhancer RNAs and 236 lincRNAs were
downregulated in adjacent non-tumor tissues. In our additional
study, we focused on the function and regulation mechanism of the
interesting subgroup lncRNAs in PDAC progression.
We performed GO and pathway analyses to predict the
biological functions and potential mechanisms of the differentially
expressed lncRNAs in PDAC progression. In this study, we found that
the highest enriched GO terms for the upregulated transcripts were
purine nucleoside catabolic process, cytoplasm, and protein binding
and the most highly enriched GO terms targeted by the downregulated
transcripts were establishment of localization, cytoplasmic part,
and protein binding. The GO project is a collaborative effort that
addresses the need for consistent descriptions of gene products in
terms of their ‘biology’ in a species-independent manner (68). To gain insight into the underlying
biology of the differentially expressed transcripts, we performed
pathway analysis and found that the upregulated transcripts were
associated with 41 pathways; the downregulated transcripts were
associated with 25 pathways. Among these pathways, the gene
category ‘B cell receptor signaling pathway (human)’ is involved in
the initiation and growth of human pancreatic ductal adenocarcinoma
(69). The gene category Wnt
signaling pathway, has been reported to be involved in metastasis
of pancreatic carcinogenesis (25), and the gene category MAPK signaling
pathway has been shown to participate in the progression of
pancreatic cancer though multiple mechanisms (26–28,70).
The gene categories FoxO signaling pathway have been reported to
suppress or activate pancreatic cancer progression by different
drugs or compound (29,30). The gene categories ‘Ubiquitin
mediated proteolysis’ participates in pancreatic cancer cell growth
in vitro and in vivo (31). The result of pathway analysis using
bioinformatics to find the specific regulation mechanisms in PDAC
progression is important for our further studies.
Moreover, we found a significant increase of the
expression level of HOTAIRM1 in PDAC samples comparing with the
non-tumor tissues via microarray analysis. To examine whether the
upregulated expression of HOTAIRM1 is pathologically specific, a
total of 12 PDAC samples and paired non-tumor tissues were
subjected to qRT-PCR. HOTAIRM1 expression level was higher in PDAC
samples than the mean level in paired non-tumor tissues. HOTAIRM1
is a long intergenic non-coding RNA located at the 3′-end of the
HOXA cluster, upregulated during myeloid maturation (71). HOTAIRM1 may affect cell fate by
regulating cell cycle progression and serving as a link in the
coordinated regulation of an extensive gene expression program.
Although, the expression of HOTAIRM1 was previously shown to be
specific to the myeloid lineage of hematopoietic cells (72), a recent study reported that the
HOTAIRM1 was overexpressed in the basal-like subtype of breast
cancer (73). Together with our
present study in PDAC tissues by microarray and qRT-PCR, the long
intergenic non-coding RNA HOTAIRM1 may participate in the
development and progression of several cancers. Thus, further
studies are needed to clarify its role in the regulation effect of
PDAC.
This study revealed differential expression patterns
of lncRNAs in 3 PDAC patients, in which 2,331 upregulated and 1,641
downregulated lncRNAs were found in PDAC tissues relative to paired
non-tumor tissues. In addition, the study helped us to understand
the potential mechanisms of the carcinogenesis of PDAC
preliminarily through ‘GO’ analysis, signaling pathway analysis and
lncRNA classification analysis. Furthermore, this study is the
first on the long intergenic non-coding RNA HOTAIRM1 in PDAC, which
may be used as a molecular marker in the future to predict response
to treatment as well as patient outcome of PDAC.
Acknowledgements
The authors thank all the patients who participated
in this study. This study was supported by grants from the Natural
Science Foundation of China (no. 81271007).
References
|
1
|
Kazanjian KK, Hines OJ, Duffy JP, Yoon DY,
Cortina G and Reber HA: Improved survival following
pancreaticoduodenectomy to treat adenocarcinoma of the pancreas:
The influence of operative blood loss. Arch Surg. 143:1166–1171.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Oettle H, Neuhaus P, Hochhaus A, Hartmann
JT, Gellert K, Ridwelski K, Niedergethmann M, Zülke C, Fahlke J,
Arning MB, et al: Adjuvant chemotherapy with gemcitabine and
long-term outcomes among patients with resected pancreatic cancer:
The CONKO-001 randomized trial. JAMA. 310:1473–1481. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang J, Li J, Zhu R, Zhang H, Zheng Y, Dai
W, Wang F, Shen M, Chen K, Cheng P, et al: K-ras mutational status
in cytohistological tissue as a molecular marker for the diagnosis
of pancreatic cancer: A systematic review and meta-analysis. Dis
Markers. 2014:5737832014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cowan RW and Maitra A: Genetic progression
of pancreatic cancer. Cancer J. 20:80–84. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dempe S, Stroh-Dege AY, Schwarz E,
Rommelaere J and Dinsart C: SMAD4: A predictive marker of PDAC cell
permissiveness for oncolytic infection with parvovirus H-1PV. Int J
Cancer. 126:2914–2927. 2010.
|
|
6
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ho AS, Huang X, Cao H, Christman-Skieller
C, Bennewith K, Le QT and Koong AC: Circulating miR-210 as a novel
hypoxia marker in pancreatic cancer. Transl Oncol. 3:109–113. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kawaguchi T, Komatsu S, Ichikawa D,
Morimura R, Tsujiura M, Konishi H, Takeshita H, Nagata H, Arita T,
Hirajima S, et al: Clinical impact of circulating miR-221 in plasma
of patients with pancreatic cancer. Br J Cancer. 108:361–369. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tay Y, Karreth FA and Pandolfi PP:
Aberrant ceRNA activity drives lung cancer. Cell Res. 24:259–260.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Beroukhim R, Mermel CH, Porter D, Wei G,
Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J,
Urashima M, et al: The landscape of somatic copy-number alteration
across human cancers. Nature. 463:899–905. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Futreal PA, Coin L, Marshall M, Down T,
Hubbard T, Wooster R, Rahman N and Stratton MR: A census of human
cancer genes. Nat Rev Cancer. 4:177–183. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stratton MR, Campbell PJ and Futreal PA:
The cancer genome. Nature. 458:719–724. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Flynn RA and Chang HY: Active chromatin
and noncoding RNAs: An intimate relationship. Curr Opin Genet Dev.
22:172–178. 2012. View Article : Google Scholar :
|
|
17
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hung T and Chang HY: Long noncoding RNA in
genome regulation: Prospects and mechanisms. RNA Biol. 7:582–585.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa
HJ, Cotton AM, Carone DM, Carone BR, Shivak DA, Guschin DY, et al:
Translating dosage compensation to trisomy 21. Nature. 500:296–300.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yildirim E, Kirby JE, Brown DE, Mercier
FE, Sadreyev RI, Scadden DT and Lee JT: Xist RNA is a potent
suppressor of hematologic cancer in mice. Cell. 152:727–742. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hajjari M and Salavaty A: HOTAIR: An
oncogenic long non-coding RNA in different cancers. Cancer Biol
Med. 12:1–9. 2015.PubMed/NCBI
|
|
23
|
Pang EJ, Yang R, Fu XB and Liu YF:
Overexpression of long non-coding RNA MALAT1 is correlated with
clinical progression and unfavorable prognosis in pancreatic
cancer. Tumour Biol. 36:2403–2407. 2015. View Article : Google Scholar
|
|
24
|
Ding C, Yang Z, Lv Z, Du C, Xiao H, Peng
C, Cheng S, Xie H, Zhou L, Wu J, et al: Long non-coding RNA PVT1 is
associated with tumor progression and predicts recurrence in
hepatocellular carcinoma patients. Oncol Lett. 9:955–963.
2015.PubMed/NCBI
|
|
25
|
Yu M, Ting DT, Stott SL, Wittner BS,
Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, et
al: RNA sequencing of pancreatic circulating tumour cells
implicates WNT signalling in metastasis. Nature. 487:510–513. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miyabayashi K, Ijichi H, Mohri D, Tada M,
Yamamoto K, Asaoka Y, Ikenoue T, Tateishi K, Nakai Y, Isayama H, et
al: Erlotinib prolongs survival in pancreatic cancer by blocking
gemcitabine-induced MAPK signals. Cancer Res. 73:2221–2234. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jun S, Lee S, Kim HC, Ng C, Schneider AM,
Ji H, Ying H, Wang H, DePinho RA and Park JI: PAF-mediated MAPK
signaling hyperactivation via LAMTOR3 induces pancreatic
tumorigenesis. Cell Rep. 5:314–322. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Yan W, Collins MA, Bednar F,
Rakshit S, Zetter BR, Stanger BZ, Chung I, Rhim AD and di Magliano
MP: Interleukin-6 is required for pancreatic cancer progression by
promoting MAPK signaling activation and oxidative stress
resistance. Cancer Res. 73:6359–6374. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Boreddy SR, Pramanik KC and Srivastava SK:
Pancreatic tumor suppression by benzyl isothiocyanate is associated
with inhibition of PI3K/AKT/FOXO pathway. Clin Cancer Res.
17:1784–1795. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Roy SK, Chen Q, Fu J, Shankar S and
Srivastava RK: Resveratrol inhibits growth of orthotopic pancreatic
tumors through activation of FOXO transcription factors. PLoS One.
6:e251662011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma Y, Gu Y, Zhang Q, Han Y, Yu S, Lu Z and
Chen J: Targeted degradation of KRAS by an engineered ubiquitin
ligase suppresses pancreatic cancer cell growth in vitro and in
vivo. Mol Cancer Ther. 12:286–294. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hajjari M and Khoshnevisan A: Potential
long non-coding RNAs to be considered as biomarkers or therapeutic
targets in gastric cancer. Front Genet. 4:2102013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang L, Liao LM, Liu AW, Wu JB, Cheng XL,
Lin JX and Zheng M: Overexpression of long noncoding RNA HOTAIR
predicts a poor prognosis in patients with cervical cancer. Arch
Gynecol Obstet. 290:717–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M, et al: Characterization of HULC, a novel gene with
striking up-regulation in hepatocellular carcinoma, as noncoding
RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Matouk IJ, Abbasi I, Hochberg A, Galun E,
Dweik H and Akkawi M: Highly upregulated in liver cancer noncoding
RNA is overexpressed in hepatic colorectal metastasis. Eur J
Gastroenterol Hepatol. 21:688–692. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J,
Zhang Y, Chen J, Shen H and Hu Z: A genetic variant in long
non-coding RNA HULC contributes to risk of HBV-related
hepatocellular carcinoma in a Chinese population. PLoS One.
7:e351452012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun M, Jin FY, Xia R, Kong R, Li JH, Xu
TP, Liu YW, Zhang EB, Liu XH and De W: Decreased expression of long
noncoding RNA GAS5 indicates a poor prognosis and promotes cell
proliferation in gastric cancer. BMC Cancer. 14:3192014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Renganathan A, Kresoja-Rakic J, Echeverry
N, Ziltener G, Vrugt B, Opitz I, Stahel RA and Felley-Bosco E: GAS5
long non-coding RNA in malignant pleural mesothelioma. Mol Cancer.
13:1192014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Arita T, Ichikawa D, Konishi H, Komatsu S,
Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T,
et al: Circulating long non-coding RNAs in plasma of patients with
gastric cancer. Anticancer Res. 33:3185–3193. 2013.PubMed/NCBI
|
|
40
|
Xie H, Ma H and Zhou D: Plasma HULC as a
promising novel biomarker for the detection of hepatocellular
carcinoma. BioMed Res Int. 2013:1361062013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et
al: Functional demarcation of active and silent chromatin domains
in human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar
|
|
47
|
Lin R, Maeda S, Liu C, Karin M and
Edgington TS: A large noncoding RNA is a marker for murine
hepatocellular carcinomas and a spectrum of human carcinomas.
Oncogene. 26:851–858. 2007. View Article : Google Scholar
|
|
48
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Schorderet P and Duboule D: Structural and
functional differences in the long non-coding RNA hotair in mouse
and human. PLoS Genet. 7:e10020712011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu JH, Chen G, Dang YW, Li CJ and Luo DZ:
Expression and prognostic significance of lncRNA MALAT1 in
pancreatic cancer tissues. Asian Pac J Cancer Prev. 15:2971–2977.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang KC, Yang YW, Liu B, Sanyal A,
Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta
RA, et al: A long noncoding RNA maintains active chromatin to
coordinate homeotic gene expression. Nature. 472:120–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H,
Wang Y, Zeng J, Song Y, Gao W, et al: The long non-coding RNA
HOTTIP promotes progression and gemcitabine resistance by
regulating HOXA13 in pancreatic cancer. J Transl Med. 13:842015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan
Z and Ai K: H19 promotes pancreatic cancer metastasis by
derepressing let-7's suppression on its target HMGA2-mediated EMT.
Tumour Biol. 35:9163–9169. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Peng W, Gao W and Feng J: Long noncoding
RNA HULC is a novel biomarker of poor prognosis in patients with
pancreatic cancer. Med Oncol. 31:3462014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
You L, Chang D, Du HZ and Zhao YP:
Genome-wide screen identifies PVT1 as a regulator of Gemcitabine
sensitivity in human pancreatic cancer cells. Biochem Biophys Res
Commun. 407:1–6. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tahira AC, Kubrusly MS, Faria MF, Dazzani
B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC
and Reis EM: Long noncoding intronic RNAs are differentially
expressed in primary and metastatic pancreatic cancer. Mol Cancer.
10:1412011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ding YC, Yu W, Ma C, Wang Q, Huang CS and
Huang T: Expression of long non-coding RNA LOC285194 and its
prognostic significance in human pancreatic ductal adenocarcinoma.
Int J Clin Exp Pathol. 7:8065–8070. 2014.
|
|
58
|
Katayama S, Tomaru Y, Kasukawa T, Waki K,
Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et
al: FANTOM Consortium: Antisense transcription in the mammalian
transcriptome. Science. 309:1564–1566. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang D, Garcia-Bassets I, Benner C, Li W,
Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al:
Reprogramming transcription by distinct classes of enhancers
functionally defined by eRNA. Nature. 474:390–394. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li W, Notani D, Ma Q, Tanasa B, Nunez E,
Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al: Functional
roles of enhancer RNAs for oestrogen-dependent transcriptional
activation. Nature. 498:516–520. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Hah N, Danko CG, Core L, Waterfall JJ,
Siepel A, Lis JT and Kraus WL: A rapid, extensive, and transient
transcriptional response to estrogen signaling in breast cancer
cells. Cell. 145:622–634. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Melo CA, Drost J, Wijchers PJ, van de
Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Léveillé N,
Kalluri R, et al: eRNAs are required for p53-dependent enhancer
activity and gene transcription. Mol Cell. 49:524–535. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Huarte M and Rinn JL: Large non-coding
RNAs: Missing links in cancer? Hum Mol Genet. 19R:R152–R161. 2010.
View Article : Google Scholar
|
|
64
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Maruyama R and Suzuki H: Long noncoding
RNA involvement in cancer. BMB Rep. 45:604–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Reference Genome Group of the Gene
Ontology Consortium. The Gene Ontology's Reference Genome Project:
A unified framework for functional annotation across species. PLoS
Comput Biol. 5:e10004312009. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Mizuma M, Rasheed ZA, Yabuuchi S, Omura N,
Campbell NR, de Wilde RF, De Oliveira E, Zhang Q, Puig O, Matsui W,
et al: The gamma secretase inhibitor MRK-003 attenuates pancreatic
cancer growth in preclinical models. Mol Cancer Ther. 11:1999–2009.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Feng J, Ma T, Ge Z, Lin J, Ding W, Chen H,
Zhu W, Zhou S and Tan Y: PKM2 gene regulates the behavior of
pancreatic cancer cells via mitogen-activated protein kinase
pathways. Mol Med Rep. 11:2111–2117. 2015.
|
|
71
|
Zhang X, Lian Z, Padden C, Gerstein MB,
Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM and
Newburger PE: A myelopoiesis-associated regulatory intergenic
noncoding RNA transcript within the human HOXA cluster. Blood.
113:2526–2534. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
McCarthy DJ and Smyth GK: Testing
significance relative to a fold-change threshold is a TREAT.
Bioinformatics. 25:765–771. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Su X, Malouf GG, Chen Y, Zhang J, Yao H,
Valero V, Weinstein JN, Spano JP, Meric-Bernstam F, Khayat D, et
al: Comprehensive analysis of long non-coding RNAs in human breast
cancer clinical subtypes. Oncotarget. 5:9864–9876. 2014. View Article : Google Scholar : PubMed/NCBI
|