Introduction
Colorectal cancer (CRC), one of the most common
malignancies, is the fourth leading cause of cancer-related death
globally (1). In China, there is
an increasing trend in both morbidity and mortality rates of CRC
(2). Although early diagnosis and
enhanced surgical resections have increased survival rates in
patients who are diagnosed with CRC, almost 50% of patients with
CRC will die due to complications associated with metastasis
(3). Understanding the mechanisms
of metastatic disease has important implications in formulating the
treatment and metastatic prevention strategies.
The junctional adhesion molecule (JAMs) family
belongs to an immunoglobulin subfamily involved in the formation of
tight junctions (TJ) in both endothelial and epithelial cells and
are characterized by two immunoglobulin-like domains (V-C2 type Ig
domains) (4,5). There are four main members of the JAM
protein family, which have been named; JAM-A, JAM-B, JAM-C and
JAM-L (6). Junctional adhesion
molecule-B (JAMB), also known as JAM-2, is specifically localized
to cell-cell contacts and enriched at TJ with homotypic
interactions. It is mainly expressed in the heart, endothelium,
trophoblasts of the placenta, high endothelial venules and in the
endothelium of arterioles (7,8).
JAM-2 has multiple functions involved in regulation of endothelial
and epithelial paracellular permeability, leukocyte recruitment
during inflammation, angiogenesis, cell proliferation and migration
(9).
The hallmarks of a metastatic cancer cells are their
ability to seperate from the original tumour and enter the
circulation via intravasation, leading to the development of tumour
metastasis, anaplasia of the primary tumour or lymphovascular
invasion (10). The metastatic and
invasive cascade consists of many complex cellular interactions and
pathways (11). Approximately 90%
of all cancer patients experience tumour metastasis (12). Due to the role JAM-2 plays in
integrity of junctions between cells, it has become an interesting
target for further research. It has been reported that JAM-2
interacts with JAM-3 in leukocytes involved in leukocyte
trafficking (13–15). It has been shown that JAM-2 can
regulate invasion and metastasis of melanoma with the expression of
JAM-3 (16). Also, JAM-2 increases
JAM-3-dependent gastric adenocarcinoma tumor metastasis (17). Moreover, JAM-2 interferes with the
signaling pathway of angiogenic VEGF/VEGFR2 (18). A previous study suggested that
JAM-2 plays an important role in regulating tumor growth and
angiogenesis (19). An interesting
finding was that JAM-2 has low expression in colorectal cancer due
to hypermethylation of its promotor (20,21).
In this study we used JAM-2 overexpression in colon cancer cells to
study its role in the development of colon tumour progression.
Materials and methods
Human colorectal specimens
A total of 169 patient tissues (94 were colorectal
cancer tissues; 75 were normal background tissues) were collected
immediately after surgery and snap-frozen in liquid nitrogen until
further use. Background normal mammary tissues were gained from the
same patients. The size of tumour tissues and normal tissues was
confirmed by a pathologist and the background tissues were free of
tumour deposits. All protocols were approved by the local ethics
committee. Full details of patient clinical data are shown in
Table I.
 | Table ICorrelation of mRNA of JAM-2 and
clinical parameters. |
Table I
Correlation of mRNA of JAM-2 and
clinical parameters.
| Category | No. | Median | IQR | P-value |
|---|
| T/N |
| Normal | 75 | 6.4 | <0.000001–616 | |
| Tumour | 94 | <0.000001 | <0.000001 | 0.042 |
| Paired T-N |
| Paired normal | 68 | 6.4 |
<0.000001–525 | |
| Paired tumour | 68 | <0.000001 |
<0.000001–0.01 | 0.03 |
| Location |
| Left colon | 22 | <0.000001 |
<0.000001–0.01 | |
| Right colon | 28 | <0.000001 | <0.000001 | |
| Trans-colon | 2 | 0.00175 | N/A | |
| Rectum | 22 | <0.000001 | <0.000001 | |
| Dukes' stage |
| A | 7 | <0.000001 |
<0.000001–0.0008 | |
| B | 33 | 3.19 |
<0.000001–0.01 | 0.2 |
| C | 32 | 12.8 | <0.000001 | 0.34 |
| BC | 65 | 7.93 |
<0.000001–0.01 | 0.17 |
| Tumour stage |
| T1 | 2 | <0.000001 | N/A | |
| T2 | 10 | <0.000001 |
<0.000001–0.00745 | 0.19 |
| T3 | 40 | <0.000001 |
<0.000001–0.03 | 0.25 |
| T4 | 18 | <0.000001 | <0.000001 | 0.36 |
| T23 | 50 | <0.000001 |
<0.000001–0.02 | 0.09 |
| T34 | 58 | <0.000001 |
<0.000001–0.01 | 0.27 |
| Lymph node
involvement stage |
| N0 | 39 | <0.000001 |
<0.000001–0.01 | |
| N1 | 16 | <0.000001 |
<0.000001–0.6 | 0.33 |
| N2 | 15 | <0.000001 |
<0.000001–0.028 | 0.19 |
| TNM stage |
| I | 9 | <0.000001 |
<0.000001–0.01 | |
| II | 30 | <0.000001 |
<0.000001–0.01 | 0.54 |
| III&IV | 32 | <0.000001 | <0.000001 | 0.32 |
| Clinical
outcome |
| No invasion | 50 | <0.000001 |
<0.000001–0.01 | |
| Invasion | 26 | <0.000001 | <0.000001 | |
| Disease free | 35 | 0.001 |
<0.000001–0.012 | |
| Incidence | 23 | <0.000001 |
<0.000001–0.32 | |
| No metastasis | 50 | <0.000001 |
<0.000001–0.008 | |
| Alive | 36 | <0.000001 |
<0.000001–0.009 | |
| Died | 22 | <0.000001 |
<0.000001–0.17 | |
Cell culture
RKO and HT115 human cancer cell lines were acquired
from the European Collection of Animal Cell Cultures (ECACC;
Salisbury, UK). These two wild-type cells were routinely cultured
in DMEM/F12 HAM (Dulbecco's modified Eagle's medium; Sigma-Aldrich)
supplemented with 10% fetal calf serum (FCS; PAA Laboratories,
Somerset, UK), penicillin and streptomycin (Sigma-Aldrich), in an
incubator at 37.0°C, 95% humidity and 5% CO2.
Construction of JAM-2 expression vectors
and transfection
The JAM-2-GFP and pCMV-entry plasmids were purchased
from OriGene Technoloqies (Rockville, MD, USA). The JAM-2 cDNA
sequences which were obtained from the JAM-2-GFP plasmid were
individually cloned into a mammalian expression pCMV-entry plasmid
vector. Purified JAM-2 transgenes and control plasmid vectors were
transfected into RKO and HT115 cells respectively using an Easjet
Plus electroporator (EquiBio Ltd., Kent, UK).
RNA isolation and reverse transcription
PCR
RNA was isolated from wild-type and selected cells
using total-RNA isolation reagent (Sigma-Aldrich, Dorset, UK).
Following the manufacturer's protocol, cDNA was generated from the
total RNA using GoScript™ Reverse Transcription System kit
(Promega, Madison, WI, USA). Subsequently, PCR was conducted using
a REDTaq™ ReadyMix PCR reaction mix (primer sequences shown in
Table II). Reaction conditions
were: 94°C for 5 min (initial denaturation), followed by 35 cycles
of 94°C for 30 sec, 55°C for 30 sec and 72°C for 40 sec. This was
followed by a final 10-min extension period at 72°C. The products
were visualized on 2% agarose gel stained with ethidium bromide.
Quantitative analysis of the JAM-2 transcript was carried out using
real-time quantitative PCR with on the Amplifluor™ technology.
Q-PCR primers (Table II) were
designed using Beacon Design software (Premier Biosoft, Palo Alto,
CA, USA), and one of the primers carried a Z-sequence. Colorectal
cDNA samples were synchronously examined for JAM-2 and GAPDH along
with a set of internal control. Real-time PCR was carried out using
iCycler iQ™ (Bio-Rad Laboratories, Hemel Hempstead, UK) following
the cycling conditions: 94°C for 5 min, 70 cycles of: 94°C for 10
sec, 55°C for 35 sec and 72°C for 20 sec.
 | Table IIPrimer sequences. |
Table II
Primer sequences.
| Molecule | Sense primers
(5′-3′) | Antisense primers
(5′-3′) |
|---|
| JAM-2 (Q-PCR) |
TGATAGGGGCTGTAAATCT |
ACTGAACCTGACCGTACATAATGATGCAAGACAGTTCC |
| GAPDH (Q-PCR) |
CTGAGTACGTCGTGGAGTC |
ACTGAACCTGACCGTACACAGAGATGATGACCCTTTTG |
| JAM-2 |
GAACTGTGGTAGAGCTACGATGTC |
TTTCACTCATTGTCGTGGCTTTAG |
| GAPDH |
GGCTGCTTTTAACTCTGGTA |
GACTGTGGTCATGAGTCCTT |
Western blot analysis
Total protein concentrations were determined with
the DC Protein Assay kit (Bio-Rad Laboratories, Hertfordshire, UK)
and an ELx800 spectrophotometer (Elx800 ™; Bio-Tek, Swindon, UK).
Equal amounts of protein sample were separated using 10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
blotted onto PVDF membrane. Following protein transfer, the
membrane was blocked with 5% skimmed milk for 1 h. Proteins were
then separately probed with the polyclonal goat anti-human JAM-2
antibody (R&D Systems, Inc., Minneapolis, MN, USA), monoclonal
mouse anti-human GAPDH antibody (BD Biosciences, San Diego, CA,
USA) and corresponding peroxidase-conjugated secondary antibody.
Protein bands were visualized and analysed using Luminata Forte
(Merck Millipore, Hertfordshire, UK) and an UVITech imager
(UVITech, Inc., Cambridge, UK).
In vitro cell function assays
In vitro cell growth assay
Cell suspensions were added into a 96-well plate
(3,000 cells/200 μl/well). Cell growth was assessed after a period
of incubation (up to 5 days) in quadruplicate (overnight, day 3,
day 4 and day 5). Cells were fixed in 4% formalin and stained with
0.5% crystal violet. Crystal violet was extracted with 10% (v/v)
acetic acid and the absorbance of the dissolved dye determined
using an ELx800 spectrophotometer (ELx800; Bio-Tek) at a wavelength
of 540 nm.
In vitro cell migration assay
Cells (700,000/well) were seeded in a 24-well plate
and cultured in the incubation overnight, then scratched with a
10-μl pipette tip to create a wound and washed twice with PBS to
remove floating cells. The cells were photographed at intervals
using an inverted microscope; the size of the wounds were
subsequently measured with the ImageJ software.
In vitro cell adhesion assay
Plates (96-well) were pre-coated with Matrigel (BD
Matrigel™ Basement Membrane Matrix) (5 μg/100 μl/well) diluted with
serum free media and dried. Following rehydration with serum-free
media 40,000 cells were added into each well. After incubation for
40 min, the wells were washed with BSS to remove non-adherent cells
and the adherent cells were fixed with 4% formalin and stained with
0.5% crystal violet. Crystal violet staining was dissolved with 10%
acetic acid and measured using a spectrophotometer (Elx800;
Bio-Tek).
In vitro cell invasion assay
Transwell inserts with an 8 μm pore size were coated
with 50 μg Matrigel/100 μl (BD Matrigel™ Basement Membrane Matrix)
and air-dried. Following rehydration, 30,000 cells/200 μl/well were
added with 0.5% FCS and 1 ml with 10% FCS medium in the bottom
well. After 48 h, the cells that migrated through the matrix and
pores were fixed with 4% formalin, stained in crystal violet and
analyzed.
Transepithelial resistance (TER)
TER was measured with an EVOM Volt-Ohm meter (EVOL,
World Precision Instruments, Aston, Herts, UK) with STX2 electrode
(World Precision Instruments, Inc., Sarasota, FL, USA). Briefly,
cells were added into the 0.4-μm pore size insert (Greiner Bio-One
Ltd., Stonehouse, UK) and allowed to reach full confluency.
Electrodes were placed at the upper and lower chambers, then
resistance was measured with the Volt-Ohm meter. Fluorescein
isothiocyanate (FITC)-dextran 10 kDa and TRITC dextran 40 kDa were
applied to cells apically. Basolateral dextran passage was analyzed
with a GloMax®-Multi Microplate Multimode reader
(Promega UK Ltd., Southampton, UK).
Gelatin zymography assay
Cells (1×106) were seeded into a tissue
culture flask and cultured in serum-free medium. After 6 h, samples
were centrifuged (4,000 rmp for 10 min) to remove cell debris. The
supernatant was collected and centrifuged for 15 min (14,000 × g)
in Amicon Ultra-0.5 ml centrifugal filters (Merck Millipore, East
Midlands, UK). Thereafter, the sample was separated via 10%
SDS-PAGE containing 0.1% gelatin. Following electrophoresis, gels
were washed and incubated overnight at 37°C. After incubation, gels
were stained with Coomassie blue and ubsequently scanned on digital
scanner images (SNP-id 2.0 machine; Millipore UK Ltd., Watford, UK)
and digital data were saved for analysis.
Statistical analysis
Experimental procedures were repeated independently
at least 3 times. Statistical analysis was performed using the
Minitab statistical software package (version 14). Data with a
non-parametric distribution were assessed with the Mann-Whitney U
test, whilst a two sample t-test was used for normally distributed
data. Differences were considered to be statistically significant
at P<0.05.
Results
JAM-2 expression in colorectal cancer
cells and tissues
The expression of JAM-2 was examined in colon cancer
cell lines and a cohort of colon tumour tissues using reverse
transcription-PCR. We found that JAM-2 exhibited very low
expression in all four colon cancer cell lines and in colon cancer
tissues which were selected randomly (Fig. 1A). JAM-2 transcript levels were
quantified in the colon specimens (tumour, n=94; background, n=75)
using real-time quantitative PCR (all values displayed as mean
JAM-2 transcript copies/μl of cDNA from 50 ng total RNA). In
comparison with normal colon tissues, a significantly decreased
expression (P=0.042) of JAM-2 was observed in tumour tissues
(Fig. 1B).
Correlation of JAM-2 expression in
colorectal adenocarcinoma and of the clinical characteristics of
the disease
JAM-2 transcripts were analysed in colorectal cancer
tissues and adjacent normal tissues using Q-RT-PCR. Decreased
levels of JAM-2 demonstrated a lower level in tumours than in
normal background tissues (P=0.03). The expression level of JAM-2
decreased as TNM stage (P=0.32) and nodal stage (P=0.19) and levels
of JAM-2 transcript were lower in node-positive tumours than in
node-negative tumours, lower in tumours with distant metastasis
than those without metastasis, and much lower in the patients who
had died of colon cancer than in those who remained alive and well,
but this did not reach statistical significance. Medical notes and
histology reports were used to extract clinico-pathological data
(Table I).
Expression of JAM-2 in colon cancer cell
lines reduces cell growth, adhesion, migration and invasion
To study the role of JAM-2 in colon cancer
metastasis and progression, we over-expressed JAM-2 in RKO and
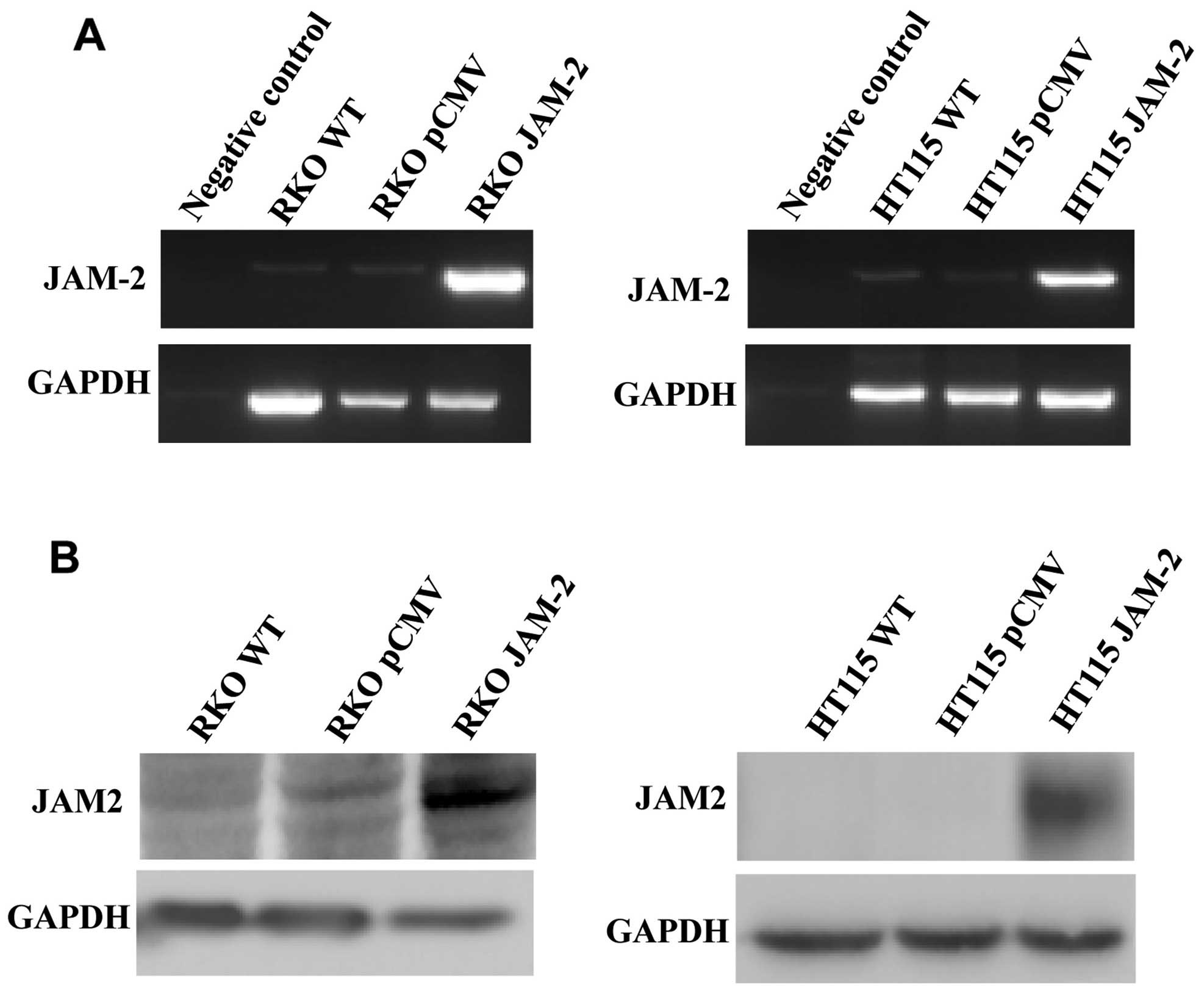
HT115 cells. We confirmed that JAM-2 plasmid had successfully
expressed JAM-2 within the RKO and HT115 colon cancer cell lines
(Fig. 2). RT-PCR and western blot
data demonstrated that JAM-2 was expressed at mRNA and protein
level in comparison to the level of expression in the empty plasmid
control cells (RKOpCMV-entry and
HT115pCMV-entry) and in wild-type cells
(RKOWT and HT115WT) (Fig. 2).
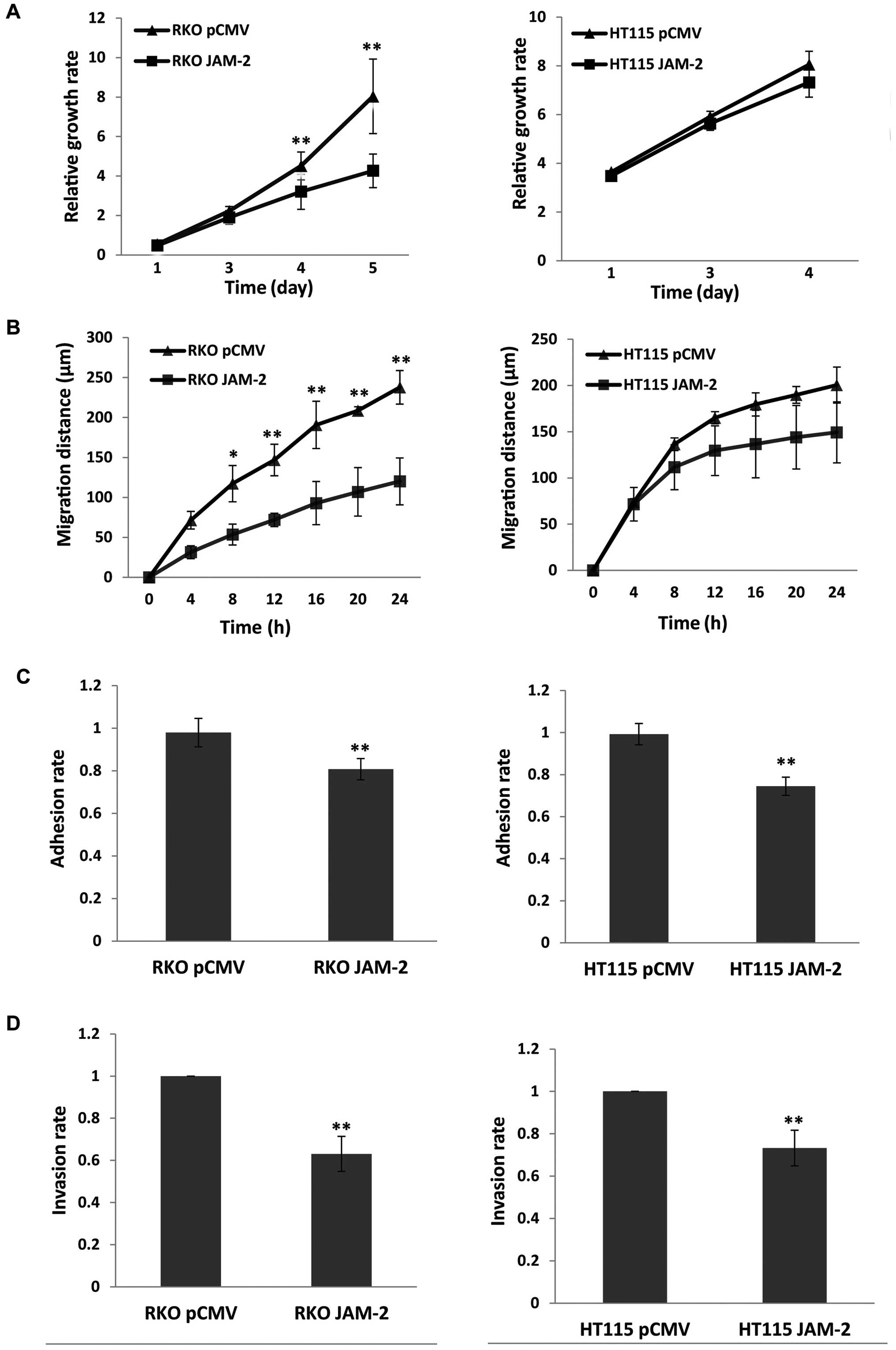
Compared with the pCMV-entry control, JAM-2
expression caused a significant reduction of RKO cell growth
(P<0.01) but not in HT115 (P>0.05) cells, however, the growth
trend of the two cell lines was similar (Fig. 3A). The migration assay showed that
expression of JAM-2 in RKO cells resulted in a strong reduction in
the degree of migration (P<0.01 vs. pCMV-entry control)
(Fig. 3B) but did not result in a
significant change on HT115 cells. We also determined the influence
on cell adhesion. Expression of JAM-2 decreased adhesion of the
colon cancer cell lines and produced a significant change in both
RKO and HT115 cells (P<0.01 vs. pCMV-entry control) (Fig. 3C). Moreover, invasion data suggest
that increasing JAM-2 expression depressed colon cancer cells
invasion (P<0.01 vs. pCMV-entry control) (Fig. 3D).
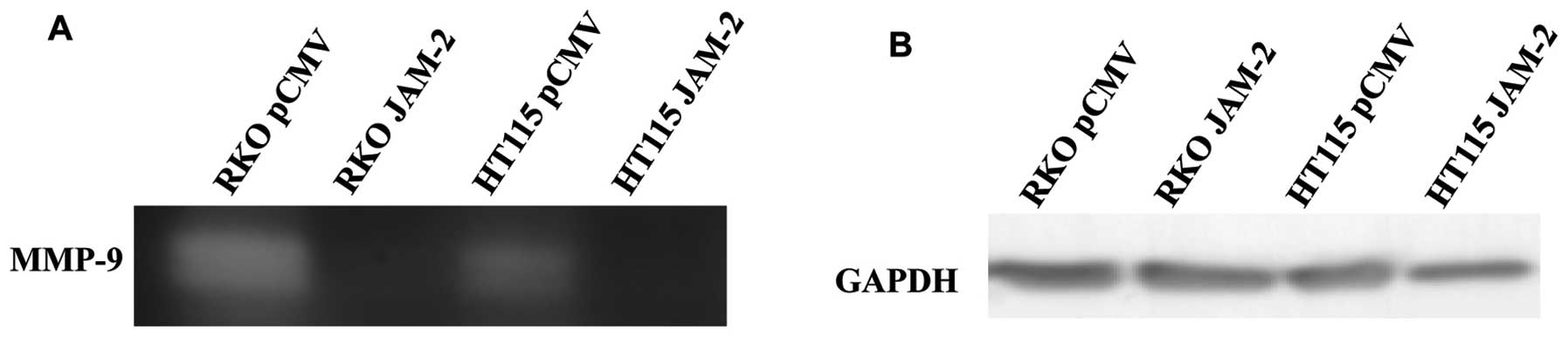
Expression of JAM-2 influences MMP
activity in colon cancer cells
Invasive potential relates to the ability of tumour
cells to degrade the extracellular matrix. We used gelatin
zymography on supernatants from RKO and HT115 JAM-2 expression
cells, which showed a decrease in MMP9 activity compared with
pCMV-entry control cells (Fig.
4).
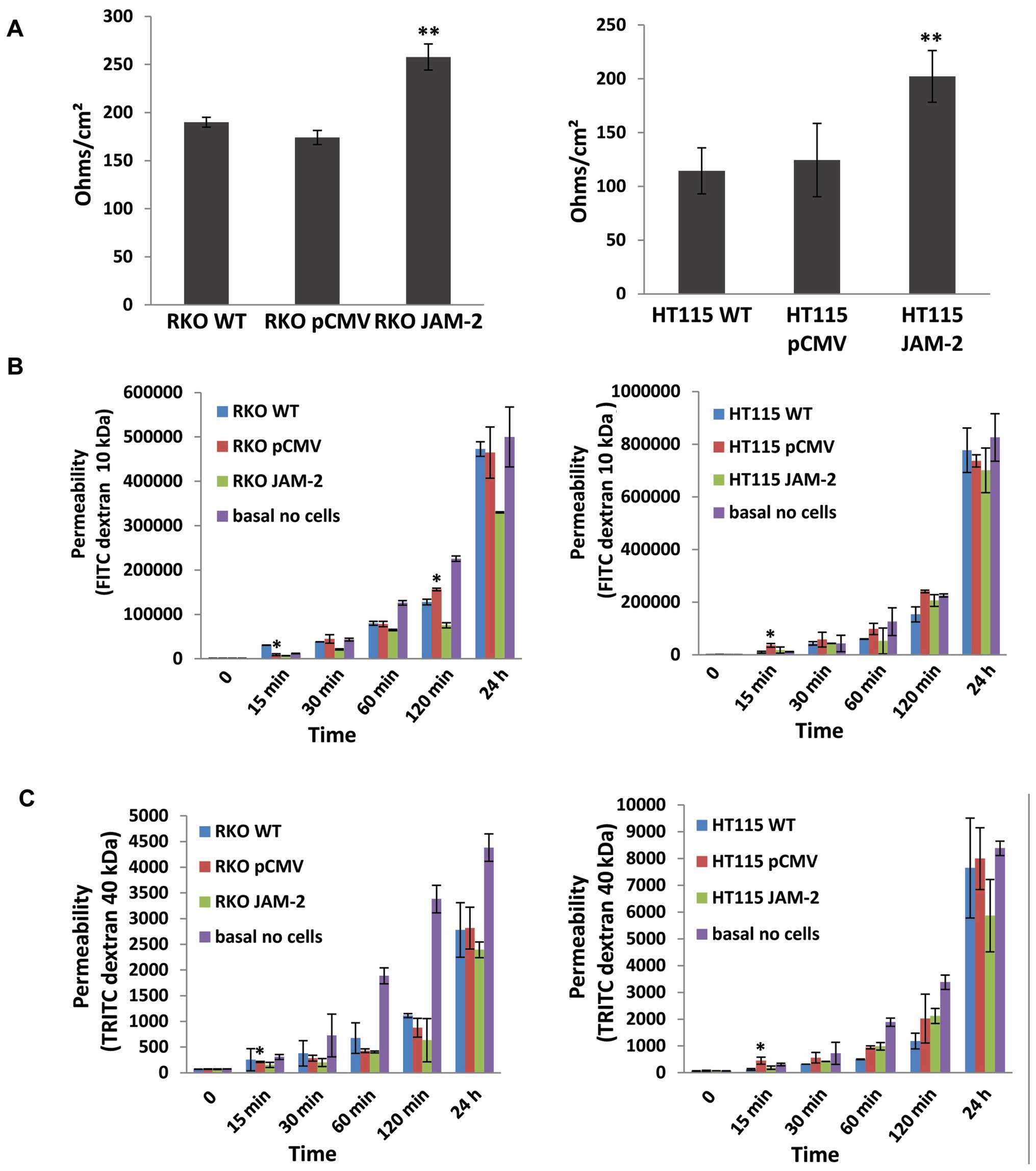
Expression of JAM-2 decreases the
trans-epithelial resistance (TER) and the permeability of polarized
monolayers
We next examined the effect of JAM-2 overexpression
on TJ barrier function. JAM-2 had a significant effect on the TJ
function of colon cancer cells. TER in both RKO and HT115 with
JAM-2 expression cells were reduced, in comparison to that in empty
plasmid control cells (RKOpCMV-entry and
HT115pCMV-entry) and in wild-type cells
(RKOWT and HT115WT) (P<0.01; Fig. 5A). In order to verify the TER
results, we also detected the flux between polarized cells
monolayers using paracellular perm-ability. This demonstrated that
the higher the TER, the lower the permeability of polarized
monolayers was with both FITC dextran 10 kDa and TRITC dextran 40
kDa (Fig. 5B and C).
Discussion
In the present study, we demonstrated that JAM-2 has
low expression in colon cancer which is consistent with previous
studies, where it was shown that JAM-2 is downregulated due to the
JAM-2 gene having a hyper-methylated promoter at the CpG islands
(20,22). We have also shown that JAM-2
expression exerts a significant effect on tumour metastasis and
invasion. JAM-2 expression decreases the invasive properties of RKO
and HT115 colon cancer in vitro, leading to reduced MMP9
activity.
Regarding to the role of JAM integrity at the
junctions between cells, some studies have focused on the
expression analysis of the JAM-2 gene in different cancers.
Recently, a report showed that the mRNA level of JAM-2 was
deregulated in gastric cancer (17). Another study found that JAM-2
occurred in primary cancer compared to chronic gastritis and
metastatic cancer (23). Taken
together, JAM-2 expression in glioma suggests that JAM-2 may play a
role in cancer metastasis by decreasing the expression of actin
filament-associated protein through interacting with JAM-3.
Arcangeli et al (16) have
shown that JAM-2 expression in endothelial cells contributed to
murine B16 melanoma cell metastasis through interacting with JAM-C
on tumour cells. The present study also detected the expression of
JAM-2 in relation to colorectal cancer patient clinical data in a
cohort of human colorectal cancer specimens through quantitative
PCR. Reduced transcript expression of JAM-2 was observed in the
colon cancer tissue sections in comparison to normal background
mammary tissues (P=0.042). This indicated that a loss of JAM-2 may
occur as cells and normal tissues progress to a cancerous
state.
Following overexpression of JAM-2 in colon cancer
cells, analysis of functional studies revealed a statistically
significant reduction in JAM-2 expression in RKO cells as compared
to controls in growth, migration, adhesion and invasion. This
occurred in HT115 cells, although the effect on proliferation and
migration was not as substantial. Cell-matrix adhesion plays a key
step in cancer metastasis and is essential for invasion through
matrix so as to progress the metastatic process.
Numerous studies suggest matrix metalloproteases
(MMP), a family of multidomain, zinc-containing neutral
endopeptidases, to contribute to form a microenvironment that
promotes tumour metastasis during early stages of tumourigenesis
(24,25). Degradation of extracellular matrix
components containing laminin, proteoglycans, collagen and other
glycoproteins by MMPs facilitates proliferation, migration and
metastasis of cancer cells, via blood and lymphatic routes
(26). MMP-9-induced release of
biological mediators from the extracellular matrix surrounding a
cancer may compose a system by which stromal and neoplastic cells
communicate. MMP-2 and MMP-9 are important members of MMPs family
and their role has been studied in colon cancer (27). MMP-9 has been related to tumour
progression in numerous studies (28–32).
MMP-9 is regarded as an extreme enzyme for damage of the basement
membrane, the first barrier for tumour invasion. Some studies have
demonstrated that it is associated with tumour metastasis (33,34).
Our study shows that MMP-9 is downregulated in JAM-2 overexpressing
cells, which is consistent with the decreased invasive capability
of JAM-2 expression in colon cancer cells.
JAM-2 is involved in the formation of TJs, with
increased TER and decreased permeability in several cell types. In
this study, resistance in both RKO and HT115 monolayer was reduced
with JAM-2 overexpression in comparison to the empty plasmid
control cells. Overall, JAM-2 decreased the permeability of
polarized monolayers.
In conclusion, we characterized a pattern of low
expression of JAM-2 in colon cancers which was correlated with
metastasis of colorectal cancer. With cells overexpressing JAM-2,
we found that JAM-2 can inhibit tumour metastasis by reducing
invasion. This study also showed that overexpression of JAM-2
inhibits the invasive potential of cancer cells by regulating the
transcription of MMP-9. JAM-2 provides a new research direction for
the diagnosis and treatment of relevant diseases.
Acknowledgements
The present study was supported by the Beijing
Municipal Science & Technology Commission (no.
Z151100001615039). H.S.Z. was a recipient of the Cardiff University
China Medical Scholarship. The authors wish to thank Cancer
Research Wales and the Welsh Network of Life Science.
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lei T, Chen WQ, Zhang SW, Lei TH, Ying Q,
He ZY and Wang XH: Prevalence trend of colorectal cancer in 10
cities and counties in China from 1988 to 2002. Zhonghua Zhong Liu
Za Zhi. 31:428–433. 2009.in Chinese. PubMed/NCBI
|
|
3
|
de Krijger I, Mekenkamp LJ, Punt CJ and
Nagtegaal ID: MicroRNAs in colorectal cancer metastasis. J Pathol.
224:438–447. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Martìn-Padura I, Lostaglio S, Schneemann
M, Williams L, Romano M, Fruscella P, Panzeri C, Stoppacciaro A,
Ruco L, Villa A, et al: Junctional adhesion molecule, a novel
member of the immunoglobulin superfamily that distributes at
intercellular junctions and modulates monocyte transmigration. J
Cell Biol. 142:117–127. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Aurrand-Lions M, Johnson-Leger C, Wong C,
Du Pasquier L and Imhof BA: Heterogeneity of endothelial junctions
is reflected by differential expression and specific subcellular
localization of the three JAM family members. Blood. 98:3699–3707.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shin K, Fogg VC and Margolis B: Tight
junctions and cell polarity. Annu Rev Cell Dev Biol. 22:207–235.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Palmeri D, van Zante A, Huang CC,
Hemmerich S and Rosen SD: Vascular endothelial junction-associated
molecule, a novel member of the immunoglobulin superfamily, is
localized to intercellular boundaries of endothelial cells. J Biol
Chem. 275:19139–19145. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aurrand-Lions M, Duncan L, Ballestrem C
and Imhof BA: JAM-2, a novel immunoglobulin superfamily molecule,
expressed by endothelial and lymphatic cells. J Biol Chem.
276:2733–2741. 2001. View Article : Google Scholar
|
|
9
|
Luissint AC, Nusrat A and Parkos CA:
JAM-related proteins in mucosal homeostasis and inflammation. Semin
Immunopathol. 36:211–226. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brambilla D and Fais S: The Janus-faced
role of ezrin in ‘linking’ cells to either normal or metastatic
phenotype. Int J Cancer. 125:2239–2245. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Steeg PS: Tumor metastasis: Mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Crnic I and Christofori G: Novel
technologies and recent advances in metastasis research. Int J Dev
Biol. 48:573–581. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Doñate C, Ody C, McKee T, Ruault-Jungblut
S, Fischer N, Ropraz P, Imhof BA and Matthes T: Homing of human B
cells to lymphoid organs and B-cell lymphoma engraftment are
controlled by cell adhesion molecule JAM-C. Cancer Res. 73:640–651.
2013. View Article : Google Scholar
|
|
14
|
Liang TW, Chiu HH, Gurney A, Sidle A,
Tumas DB, Schow P, Foster J, Klassen T, Dennis K, DeMarco RA, et
al: Vascular endothelial-junctional adhesion molecule (VE-JAM)/JAM
2 interacts with T, NK, and dendritic cells through JAM 3. J
Immunol. 168:1618–1626. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ludwig RJ, Zollner TM, Santoso S, Hardt K,
Gille J, Baatz H, Johann PS, Pfeffer J, Radeke HH, Schön MP, et al:
Junctional adhesion molecules (JAM)-B and -C contribute to
leukocyte extravasation to the skin and mediate cutaneous
inflammation. J Invest Dermatol. 125:969–976. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arcangeli ML, Frontera V, Bardin F,
Thomassin J, Chetaille B, Adams S, Adams RH and Aurrand-Lions M:
The Junctional Adhesion Molecule-B regulates JAM-C-dependent
melanoma cell metastasis. FEBS Lett. 586:4046–4051. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hajjari M, Behmanesh M, Sadeghizadeh M and
Zeinoddini M: Junctional adhesion molecules 2 and 3 may potentially
be involved in progression of gastric adenocarcinoma tumors. Med
Oncol. 30:3802013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meguenani M, Miljkovic-Licina M, Fagiani
E, Ropraz P, Hammel P, Aurrand-Lions M, Adams RH, Christofori G,
Imhof BA and Garrido-Urbani S: Junctional adhesion molecule B
interferes with angiogenic VEGF/VEGFR2 signaling. FASEB J.
29:3411–3425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reynolds LE, Watson AR, Baker M, Jones TA,
D'Amico G, Robinson SD, Joffre C, Garrido-Urbani S,
Rodriguez-Manzaneque JC, Martino-Echarri E, et al: Tumour
angiogenesis is reduced in the Tc1 mouse model of Down's syndrome.
Nature. 466:3982010. View Article : Google Scholar
|
|
20
|
Kok-Sin T, Mokhtar NM, Ali Hassan NZ,
Sagap I, Rose IM, Harun R and Jamal R: Identification of diagnostic
markers in colorectal cancer via integrative epigenomics and
genomics data. Oncol Rep. 34:22–32. 2015.PubMed/NCBI
|
|
21
|
Bujko M, Kober P, Mikula M, Ligaj M,
Ostrowski J and Siedlecki JA: Expression changes of cell-cell
adhesion-related genes in colorectal tumors. Oncol Lett.
9:2463–2470. 2015.PubMed/NCBI
|
|
22
|
Bujko M, Kober P, Mikula M, Ligaj M,
Ostrowski J and Siedlecki JA: Expression changes of cell-cell
adhesion-related genes in colorectal tumors. Oncol Lett.
9:2463–2470. 2015.PubMed/NCBI
|
|
23
|
Zhang J, Huang JY, Chen YN, Yuan F, Zhang
H, Yan FH, Wang MJ, Wang G, Su M, Lu G, et al: Whole genome and
transcriptome sequencing of matched primary and peritoneal
metastatic gastric carcinoma. Sci Rep. 5:137502015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Overall CM and López-Otín C: Strategies
for MMP inhibition in cancer: Innovations for the post-trial era.
Nat Rev Cancer. 2:657–672. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chakraborti S, Mandal M, Das S, Mandal A
and Chakraborti T: Regulation of matrix metalloproteinases: An
overview. Mol Cell Biochem. 253:269–285. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Roomi MW, Monterrey JC, Kalinovsky T, Rath
M and Niedzwiecki A: Patterns of MMP-2 and MMP-9 expression in
human cancer cell lines. Oncol Rep. 21:1323–1333. 2009.PubMed/NCBI
|
|
27
|
Frewer KA, Sanders AJ, Owen S, Frewer NC,
Hargest R and Jiang WG: A role for WISP2 in colorectal cancer cell
invasion and motility. Cancer Genomics Proteomics. 10:187–196.
2013.PubMed/NCBI
|
|
28
|
Arnold S, Mira E, Muneer S, Korpanty G,
Beck AW, Holloway SE, Mañes S and Brekken RA: Forced expression of
MMP9 rescues the loss of angiogenesis and abrogates metastasis of
pancreatic tumors triggered by the absence of host SPARC. Exp Biol
Med (Maywood). 233:860–873. 2008. View Article : Google Scholar
|
|
29
|
Belotti D, Paganoni P, Manenti L, Garofalo
A, Marchini S, Taraboletti G and Giavazzi R: Matrix
metalloproteinases (MMP9 and MMP2) induce the release of vascular
endothelial growth factor (VEGF) by ovarian carcinoma cells:
Implications for ascites formation. Cancer Res. 63:5224–5229.
2003.PubMed/NCBI
|
|
30
|
Bergers G, Brekken R, McMahon G, Vu TH,
Itoh T, Tamaki K, Tanzawa K, Thorpe P, Itohara S, Werb Z, et al:
Matrix metalloproteinase-9 triggers the angiogenic switch during
carcinogenesis. Nat Cell Biol. 2:737–744. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim HJ, Park CI, Park BW, Lee HD and Jung
WH: Expression of MT-1 MMP, MMP2, MMP9 and TIMP2 mRNAs in ductal
carcinoma in situ and invasive ductal carcinoma of the breast.
Yonsei Med J. 47:333–342. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kondratiev S, Gnepp DR, Yakirevich E, Sabo
E, Annino DJ, Rebeiz E and Laver NV: Expression and prognostic role
of MMP2, MMP9, MMP13, and MMP14 matrix metalloproteinases in
sinonasal and oral malignant melanomas. Hum Pathol. 39:337–343.
2008. View Article : Google Scholar
|
|
33
|
Coussens LM, Fingleton B and Matrisian LM:
Matrix metalloproteinase inhibitors and cancer: Trials and
tribulations. Science. 295:2387–2392. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Egeblad M and Werb Z: New functions for
the matrix metalloproteinases in cancer progression. Nat Rev
Cancer. 2:161–174. 2002. View
Article : Google Scholar : PubMed/NCBI
|