Introduction
Hepatocellular carcinoma (HCC) occurs mostly on the
basis of pre-existing chronic liver disease and cirrhosis (1) and is a major health issue worldwide
as the sixth most common cancer and second leading etiology of
cancer-related deaths due to its poor prognosis associated with
high recurrence rate and limited treatment options (2–4).
Further investigations show that HCC is a genetic disease
developing from a multi-step process. Gene aberrance linked to
growth control, invasion and metastasis is frequent and provides
molecular genetic basis of malignant transformation and tumor
progression (5,6). Therefore, to find key genes related
to tumorigenesis is of great importance for the diagnosis, targeted
therapy, disease monitoring and clinical outcomes in HCC
patients.
Long noncoding RNAs (lncRNAs) have no open reading
frame and map to intronic and intergenic regions involved in
regulating several biological processes such as transcription,
translation, cellular differentiation, cell cycle regulation, and
chromatin modification (7–9). lncRNAs (1772) have been found
differentially expressed between HCC tissues and normal liver
tissues (10), of which lncRNA
GAS5 is downregulated in HCC indicating an independent prognostic
factor for HCC patients (11), and
lncRNA MEG3 functions as a growth suppressor via activation of p53
protein (12,13). Inhibition of cellular lncRNA-DREH
by Hepatitis B virus X protein (HBx) promotes HCC cell
proliferation in vivo and in vivo (14). In addition, overexpression of
lncRNA HOTAIR and MALAT-1 may be candidate biomarkers for
predicting tumor recurrence in HCC patients (15,16).
Enforced expression of lncRNA HEIH facilitates HCC growth through
enhancer of zeste homolog 2 (EZH2) (17) and lncRNA MVIH promotes
tumor-inducing angiogenesis through inhibiting the secretion of
phosphoglycerate kinase 1 (PGK1) (18). In HBV-related HCC, lncRNA HULC
decreases p18 expression and boosts growth (19). Hence, lncRNAs play an important
role in hepatocarcinogenesis, invasion, and metastasis.
Moreover, investigations have revealed that lncRNA
AFAP1-AS1 has been implicated in tumorigenesis of various cancers.
Increased expression of AFAP1-AS1 is found in Barrett esophagus,
esophageal adenocarcinoma (20)
and pancreatic ductal adenocarcinoma (21). Upregulation of AFAP1-AS1 promotes
cell invasion and metastasis via regulation of the actin filament
integrity, suggesting a poor prognosis and survival for
nasopharyngeal carcinoma (22) and
lung cancer (23).
However, to our knowledge, few studies have been
reported regarding the expression and functions of AFAP1-AS1 in
HCC. In the present study, we showed that AFAP1-AS1 was remarkably
increased in HCC tissues compared with the adjacent non-tumor
tissues and served as an independent predictor for overall survival
in HCC. In addition, knockdown of AFAP1-AS1 by si-AFAP1-AS1
inhibited cell growth in vitro and in vivo and cell
invasion and induced cell apoptosis and cycle arrest in S phase,
associated with regulating the transduction of the RhoA/Rac2
signaling, indicating that AFAP1-AS1 plays a critical role in the
progression of HCC.
Materials and methods
Materials
Human HCC cell lines (SMCC7721 and HepG2) were from
Institute of Biochemistry and Cell Biology (Shanghai, China).
Lentivirus-mediated si-AFAP1-AS1 was purchased from Genechem
Biotech Co., Ltd. (Shanghai, China). All antibodies including RhoA,
Rac2, PCNA, MMP-9, CyclinD1 and Bax were from Santa Cruz
Biotechnology (Santa Cruz, CA, USA).
Drugs and reagents
Dulbecco's modified Eagle's medium (DMEM) and fetal
bovine serum (FBS) were purchased from Gibco BRL (Gaithersburg, MD,
USA); TRIzol Reagent and Lipofectamine 2000 were from Invitrogen
(Carlsbad, CA, USA); M-MLV Reverse Transcriptase was from Promega
(Madison, WI, USA); SYBR Green Master Mixture was from Takara
(Otsu, Japan). ECL-PLUS/kit was obtained from Beyotime (Hainan,
China).
Clinical samples
HCC tissues and the adjacent non-tumor tissues were
acquired from Shanghai First People's hospital from May 2010 to Dec
2014. Our present study was approved by Medical Ethics Committee of
Shanghai Jiaotong University School of Medicine and written
informed consent was received from the HCC patients or their
parents before sample collection. Two pathologists decided and
checked the HCC cases.
Cell culture and infection
HCC cells, placed in a humidified atmosphere
containing 5% CO2 at 37°C, were cultured in DMEM medium
supplemented with 10% heat-inactivated FBS, 100 U/ml of penicillin
and 100 μg/ml of streptomycin. When the cells reached more than 50%
confluence, they were infected with lentivirus vector si-AFAP1-AS1
or negative control virus, and cultured at 37°C and 5%
CO2. The clone infected with si-AFAP1-AS1 was defined as
si-AFAP1-AS1 group, and that infected with negative control vectors
was considered as s-i-NC group. si-AFAP1-AS1 forward, 5′-CCG
GAACACCAATCCCAAGAGGTGACTCGAGTCACCTCTTGGGATTGGTGTTTTTTTG-3′ and
reverse,
5′-AATTCAAAAAAACACCAATCCCAAGAGGTGACTCGAGTCACCTCTTGGGATTGGTGTT-3′;
si-NC, forward
5′-CCGGTTTCTCCGAACGTGTCACGTCTCGAGACGTGACACGTTCGGAGAATTTTTG-3′ and
reverse,
5′-AATTCAAAAAGTTCTCCGAACGTGTCACGTCTCGAGACGTGACACGTTCGGAGAA-3′.
Quantitative real-time PCR
To quantitatively examine the RNA expression of
AFAP1-AS1 in HCC cells, real-time PCR was carried out. Total RNA of
each clone was extracted with TRIzol according to the
manufacturer's protocol. Reverse-transcription was performed using
M-MLV and cDNA amplification was done using SYBR Green Master Mix
kit. The AFAP1-AS1 gene was amplified using a specific
oligonucleotide primer: sense 5′-ACTGAAGAGGAACCAGGGACAG-3′ and
antisense 5′-GGGGAAACTGAAATGAATGAAG-3′. Human
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as
an endogenous control.
Western blot assay
HCC cells were harvested and extracted using lysis
buffer (Tris-HCl, SDS, mercaptoethanol, glycerol). Cell extracts
were boiled for 5 min in loading buffer and then equal amount of
cell extracts were separated on 15% SDS-PAGE gels. Separated
protein bands were transferred into polyvinylidene fluoride (PVDF)
membranes and the membranes were blocked in 5% skim milk powder.
The primary antibodies against RhoA, Rac2, PCNA, MMP-9, CyclinD1
and Bax were diluted according to the instructions of antibodies
and incubated overnight at 4°C. Horseradish peroxidase-linked
secondary antibodies were added at a dilution ratio of 1:1000, and
incubated at room temperature for 2 h. The membranes were washed
with PBS three times and the immunoreactive bands were visualized
using ECL-PLUS kit according to the kit instructions.
Cell proliferation assay
HCC cells infected with si-AFAP1-AS1 were incubated
in 96-well-plates with DEME medium supplemented with 10% FBS. HCC
cells were treated with 20 μl MTT dye and incubated with 150 μl of
DMSO for 5 min. The color reaction was measured at 570 nm with
enzyme immunoassay analyzer (Bio-Rad, Berkeley, CA, USA).
Transwell invasion assay
Transwell assay was performed by using a Transwell
chamber (Qiagen, Hilden, Germany) with pore size of 8.0 μm. The
Transwell chamber was coated with Matrigel. Total 1×106
cells were suspended in 200 μl serum-free medium and seeded in the
upper compartment of the chamber. The lower compartment was loaded
with 750 μl full culture medium containing 10% FBS. After being
incubated at 37°C for 12 h, the membrane was fixed with
formaldehyde, and stained with hematoxylin. Then the trans-membrane
cells were counted.
Flow cytometric analysis
To detect cell apoptosis, HCC cells were
trypsinized, washed with cold PBS and resuspended in binding buffer
according to the instruction of the apoptosis kit. FITC-AnnexinV
and PI were added to the fixed cells for 20 min in the dark, at
room temperature. Then, Annexin V binding buffer was added to the
mixture before the fluorescence was measured on FAC sort flow
cytometer. The cell apoptosis was analyzed using Cell Quest
software (Becton Dickinson, Mountain View, CA, USA). Three separate
experiments were performed for each clone.
After PBS washing, the fixed cells were stained with
PI in the presence of RNase A for 30 min at room temperature in the
dark. Each sample was filtered through a 50 μm nylon filter to
obtain single-cell suspension. The samples were then analyzed on
FACsort flow cytometer (Becton Dickinson). ModFit3.0 software
(Verity Software House, Topsham, ME, USA) was used for cell cycle
analysis. Three separate experiments were performed for each
clone.
In vivo tumor xenograft studies
Six-week-old female immune-deficient nude mice
(BALB/c-nu) were bred at the laboratory animal facility (Institute
of Chinese Academy of Sciences, Shanghai), and were housed
individually in microisolator ventilated cages with free access to
water and food. All experimental procedures were performed
according to the regulations and internal biosafety and bioethics
guidelines of Shanghai Jiaotong University and the Shanghai
Municipal Science and Technology Commission. Two mice were injected
subcutaneously with 1×106 HCC cells in 50 μl of PBS
pre-mixed with an equal volume of matrigel matrix (Becton
Dickinson). Mice were monitored daily and developed a subcutaneous
tumor. When the tumor size reached approximately 5 mm in length,
they were surgically removed, cut into 1–2 mm3 pieces,
and reseeded individually into other mice. When tumor size reached
approximately 5 mm in length, the mice were randomly assigned as
si-NC group (n=5) and si-AFAP1-AS1 group (n=5). In si-AFAP1-AS1
treatment group, 15 μl of lentivirus was injected into subcutaneous
tumors using a multi-site injection format. Injections were
repeated every other day after initial treatment. The tumor volume
was measured with a caliper, using the formula volume = (length ×
width)2/2.
Statistical analysis
The result of each experiment was shown as mean ± SD
when applicable. Statistically significant difference in each assay
was determined by SPSS version 20.0. Difference in each group was
tested for significance using Kruskal-Wallis H test and ANOVA
analysis of variance. P<0.05 was considered significant.
Results
Expression of AFAP1-AS1 is increased in
human HCC tissues and correlates with poor prognosis
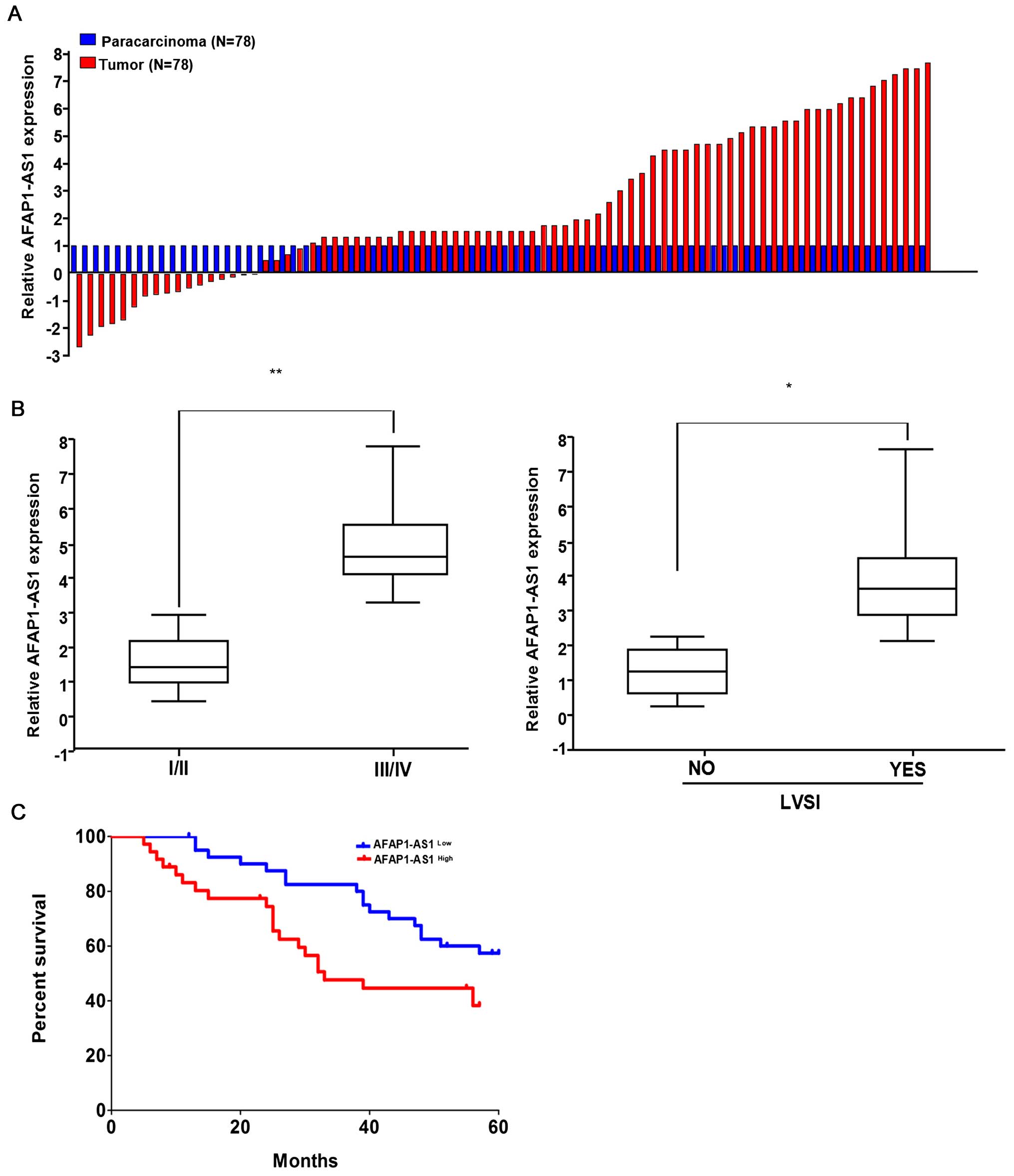
To observe the expression of AFAP1-AS1 in HCC, we
examined the AFAP1-AS1 expression levels in 78 paired HCC tissues
and corresponding non-tumor tissues by using qRT-PCR. The
transcript levels of AFAP1-AS1 were significantly increased in
71.25% (57 of 78) cancerous tissues compared with their
corresponding adjacent non-tumor tissues (P<0.01) (Fig. 1A). Then, we analyzed the
correlation of AFAP1-AS1 expression level with the clinical
features in HCC patients. As shown in Fig. 1B and Table I, high expression of AFAP1-AS1 was
associated with pathological staging (P=0.024) and lymph-vascular
space invasion (LVSI) (P=0.007). However, other clinical parameters
were not found correlated with AFAP1-AS1 expression.
 | Table ICorrelation of lncRNA AFAP1-AS1
expression with clinicopathological features in HCC patients. |
Table I
Correlation of lncRNA AFAP1-AS1
expression with clinicopathological features in HCC patients.
| | AFAP1-AS1 | | |
|---|
| |
| | |
|---|
| Variables | Cases no. | Low
21 | High
57 | χ2 | P-value |
|---|
| Age (years) |
| <60 | 51 | 14 | 37 | | |
| ≥60 | 27 | 7 | 20 | 0.021 | 0.896 |
| Gender |
| Male | 58 | 16 | 42 | | |
| Female | 19 | 5 | 14 | 0.011 | 0.915 |
| Liver
cirrhosis |
| No | 49 | 15 | 34 | | |
| Yes | 29 | 6 | 23 | 0.900 | 0.343 |
| Pathological
staging |
| I–II | 32 | 13 | 19 | | |
| III–IV | 46 | 8 | 38 | 5.111 | 0.024 |
| Tumor size
(cm) |
| <5 | 45 | 15 | 30 | | |
| ≥5 | 33 | 6 | 27 | 2.193 | 0.139 |
| TNM staging |
| T1+ T2 | 41 | 10 | 31 | | |
| T3+ T4 | 37 | 11 | 26 | 0.278 | 0.598 |
| Lymph-vascular
space invasion (LVSI) |
| No | 26 | 12 | 14 | | |
| Yes | 52 | 9 | 43 | 7.237 | 0.007 |
Kaplan-Meier analysis using the log-rank test
indicated that HCC patients with high AFAP1-AS1 expression had a
shorter median survival time of 33.7 months, while those with low
AFAP1-AS1expression had a median survival time of 59.3 months
(P=0.0378; Fig. 1C). Multivariate
analysis showed that, AFAP1-AS1 expression might serve as an
independent prognostic factor for overall survival (OS) in HCC
patients (P=0.029, Table II).
 | Table IISummary of univariate and
multivariate Cox regression analysis of overall survival
duration. |
Table II
Summary of univariate and
multivariate Cox regression analysis of overall survival
duration.
| | Multivariate
analysis |
|---|
| |
|
|---|
| Parameter | Univariate P | P | HR | 95% CI |
|---|
| Age (≥60 vs. <60
years) | 0.137 | NA | | |
| Gender (Male vs.
Female) | 0.269 | NA | | |
| Liver cirrhosis
(Positive vs. Negative) | 0.371 | NA | | |
| Pathological stage
(I/II vs. III/IV) | 0.217 | NA | | |
| Tumor size (≥5 vs.
<5 cm) | 0.016 | NS | 1.175 | 0.914–1.939 |
| TNM classification
(T1/T2 vs. T3/T4) | 0.067 | NA | | |
| LVSI (Positive vs.
Negative) | 0.017 | NS | 2.013 | 1.237–2.514 |
| AFAP1-AS1
expression (High vs. Low) | 0.0012 | 0.029 | 1.471 | 0.987–2.626 |
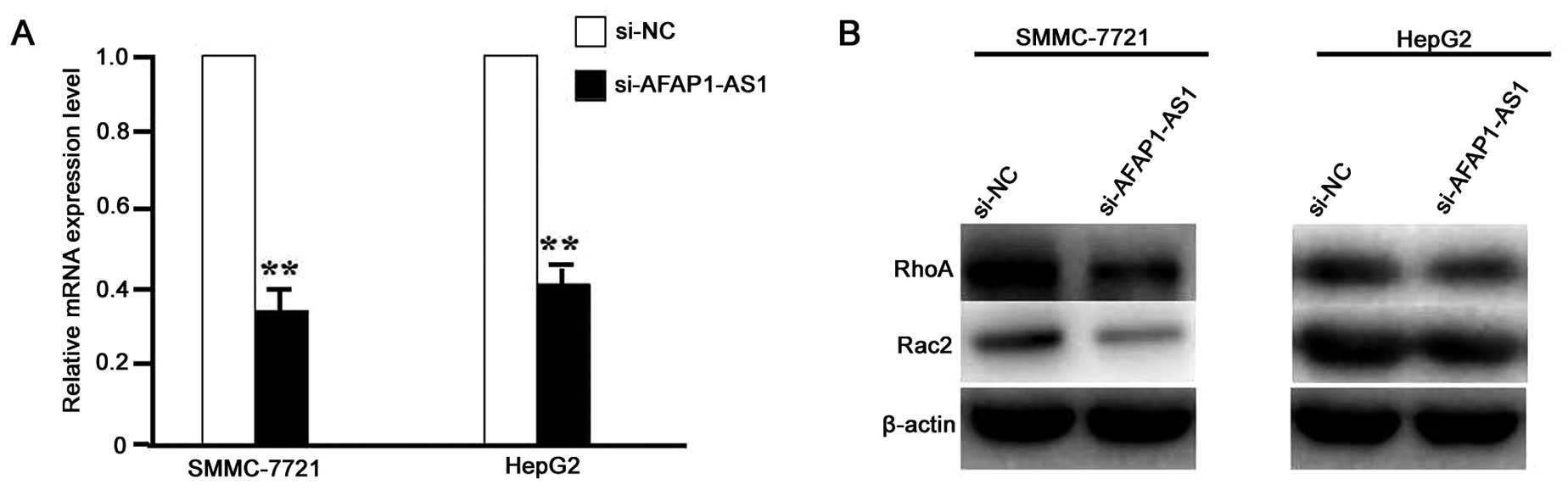
AFAP1-AS1 knockdown downregulated the
transduction of RhoA/Rac2 signaling
After HCC cell lines (SMCC-721 and HepG2) were
infected with lentivirus-mediated si-AFAP1-AS1 for 24 h, the RNA
expression level of AFAP1-AS1 (Fig.
2A) and protein expression levels of RhoA and Rac2 (Fig. 2B) were detected by real-time PCR
and western blot assays, which indicated the decreased expression
levels of AFAP1-AS1, RhoA and Rac2 in si-AFAP1-AS1 group compared
with the si-NC group (P<0.01).
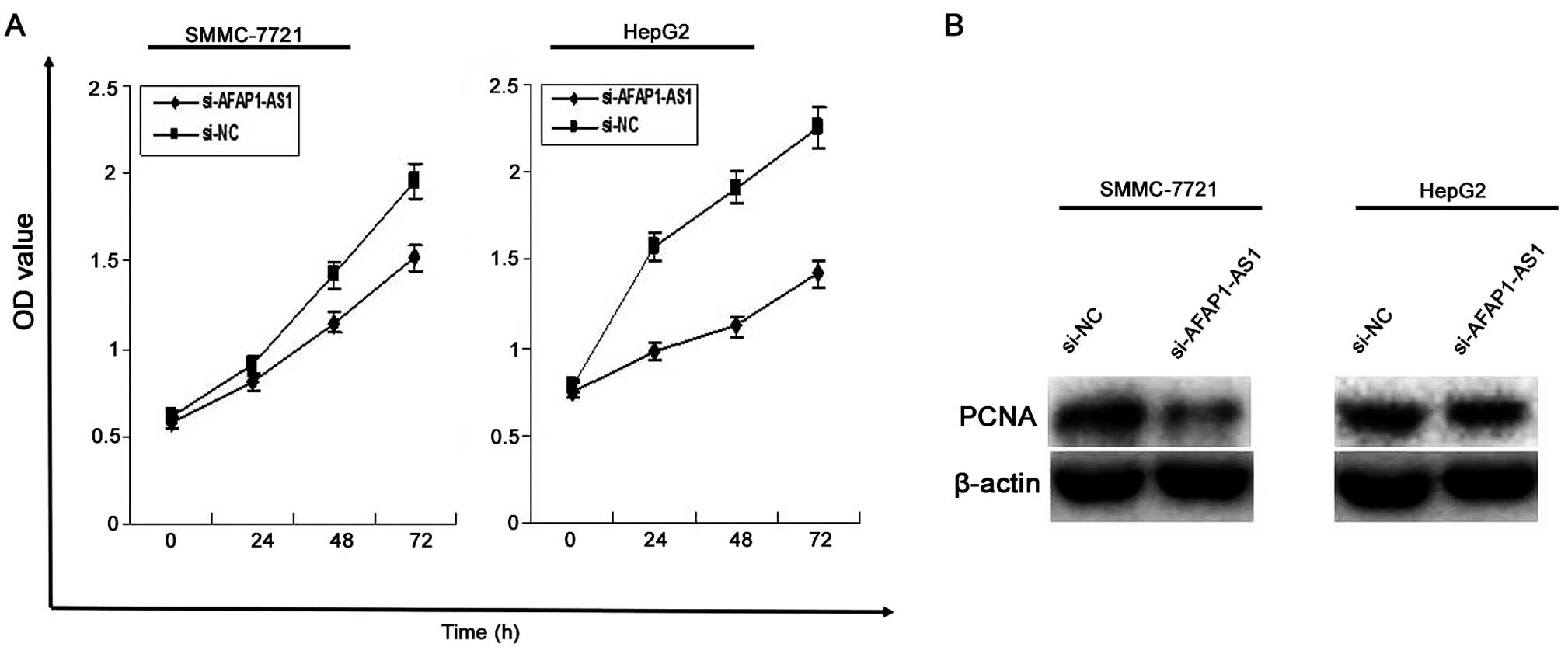
AFAP1-AS1 knockdown inhibits cell
proliferation
To investigate the effect of AFAP1-AS1 on HCC cell
proliferation, MTT assay was used to evaluate cell proliferative
activity, indicating that cell proliferation activity of HCC cells
was significantly reduced in si-AFAP1-AS1 group compared to those
in si-NC group (P<0.01, Fig.
3A). In addition, the protein expression level of PCNA examined
by western blotting (Fig. 3B)
assay, was decreased in si-AFAP1-AS1 group compared to the si-NC
group (P<0.01).
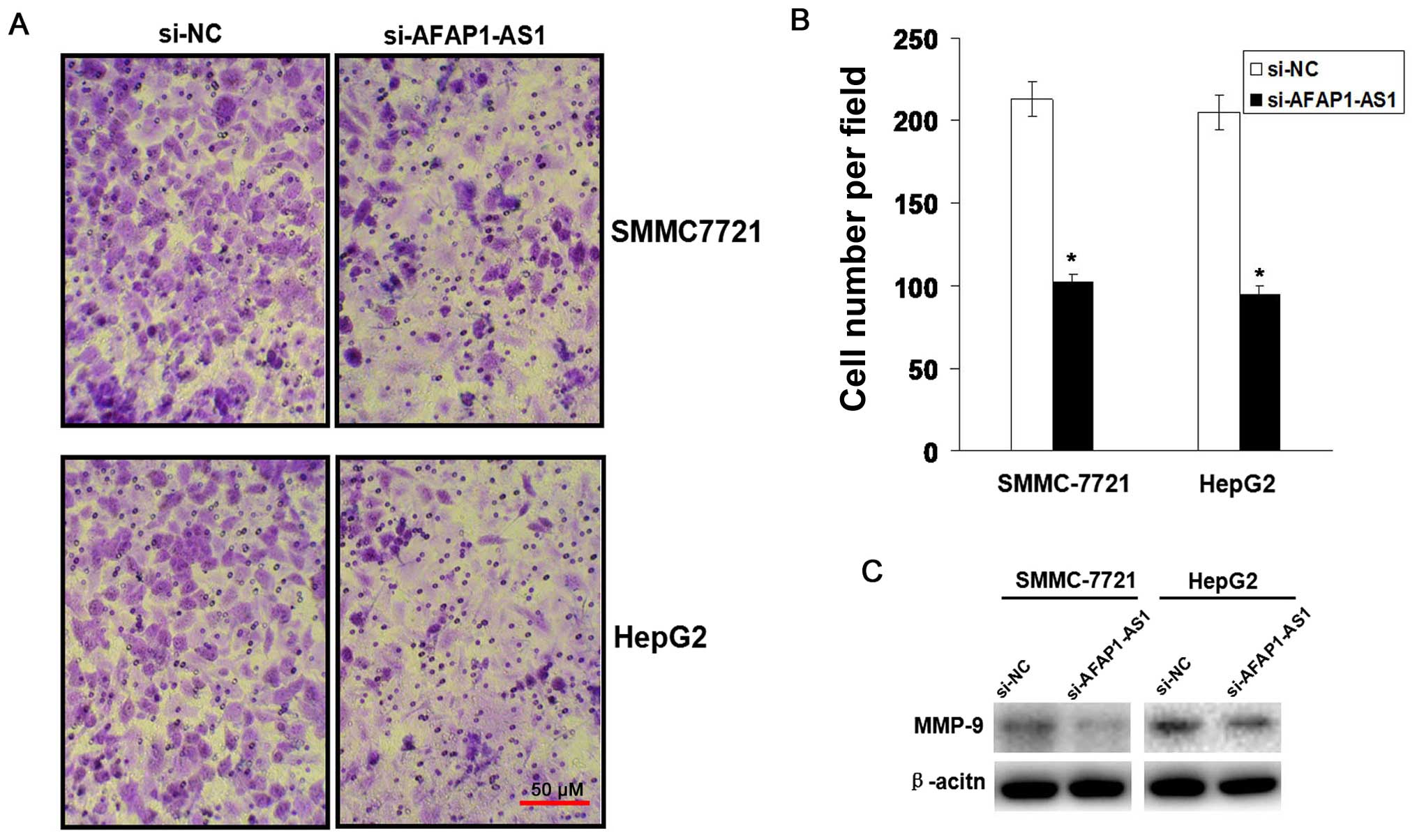
AFAP1-AS1 knockdown inhibits cell
invasion
To observe the effect of AFAP1-AS1 on cell invasive
potential in HCC cells, Transwell assay was performed. We found
that the invasive potential of HCC cells was lower in si-AFAP1-AS1
group compared to those in si-NC group (P<0.01, Fig. 4A and B). The protein expression
level of MMP-9 examined by western blot (Fig. 4C) assay was downregulated in
si-AFAP1-AS1 group compared to the si-NC group.
AFAP1-AS1 knockdown induces cell
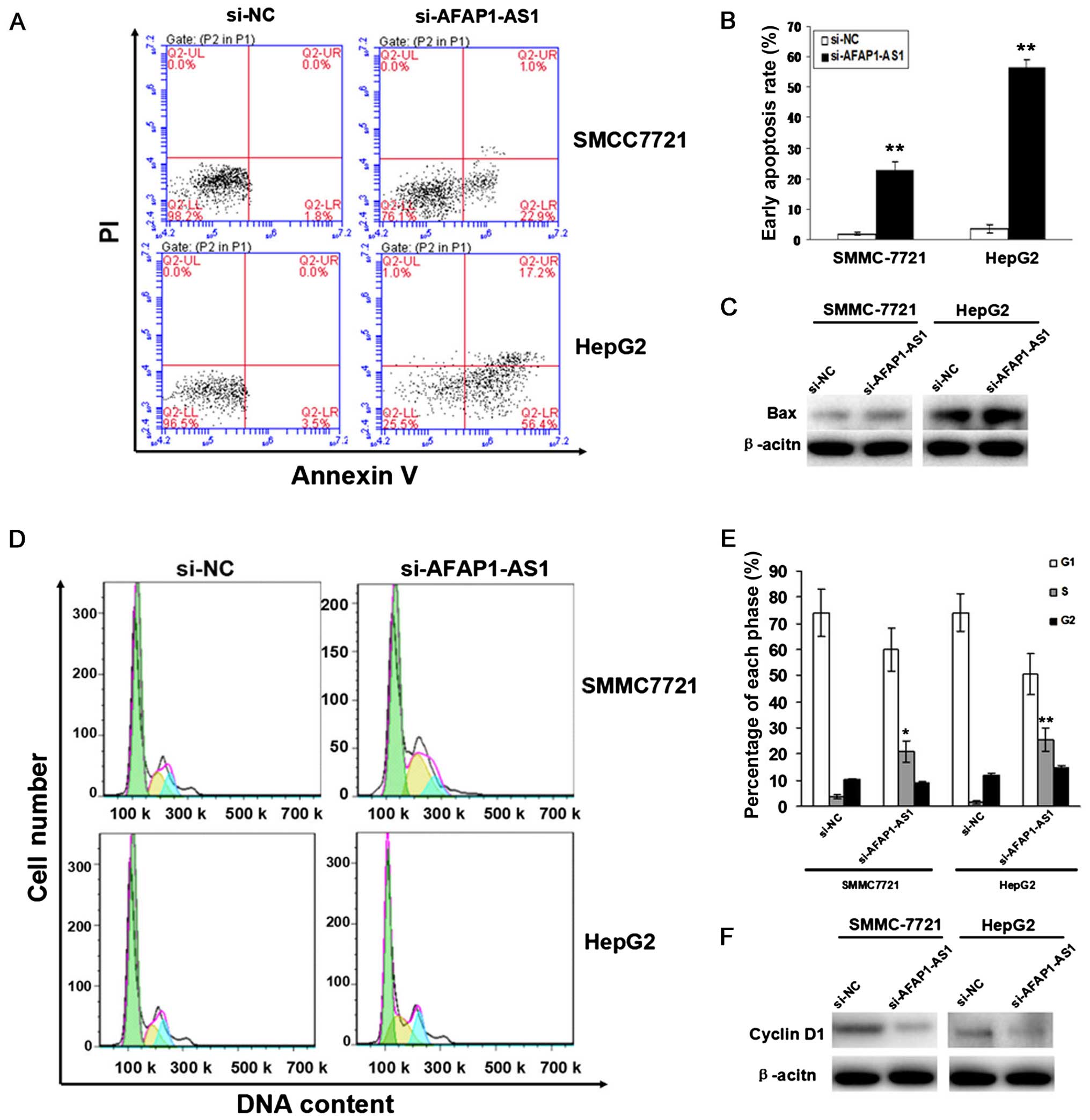
apoptosis and cycle arrest
To evaluate the effect of AFAP1-AS1 on cell
apoptosis and cycle distribution in HCC cells, flow cytometric
analysis was performed. We found that the apoptotic indexes of HCC
cells were elevated in si-AFAP1-AS1 group compared to those in NC
group (P<0.01, Fig. 5A and B).
The number of HCC cells was significantly increased in S phase in
si-AFAP1-AS1 group compared to those in the si-NC group, and cell
cycle was arrested in S phase (P<0.05, P<0.01, Fig. 5D and E). The protein expression
levels of Bax examined by western blot assay were upregulated while
those of and cyclinD1 were downregulated in si-AFAP1-AS1 group
compared to the si-NC group (Fig. 5C
and F).
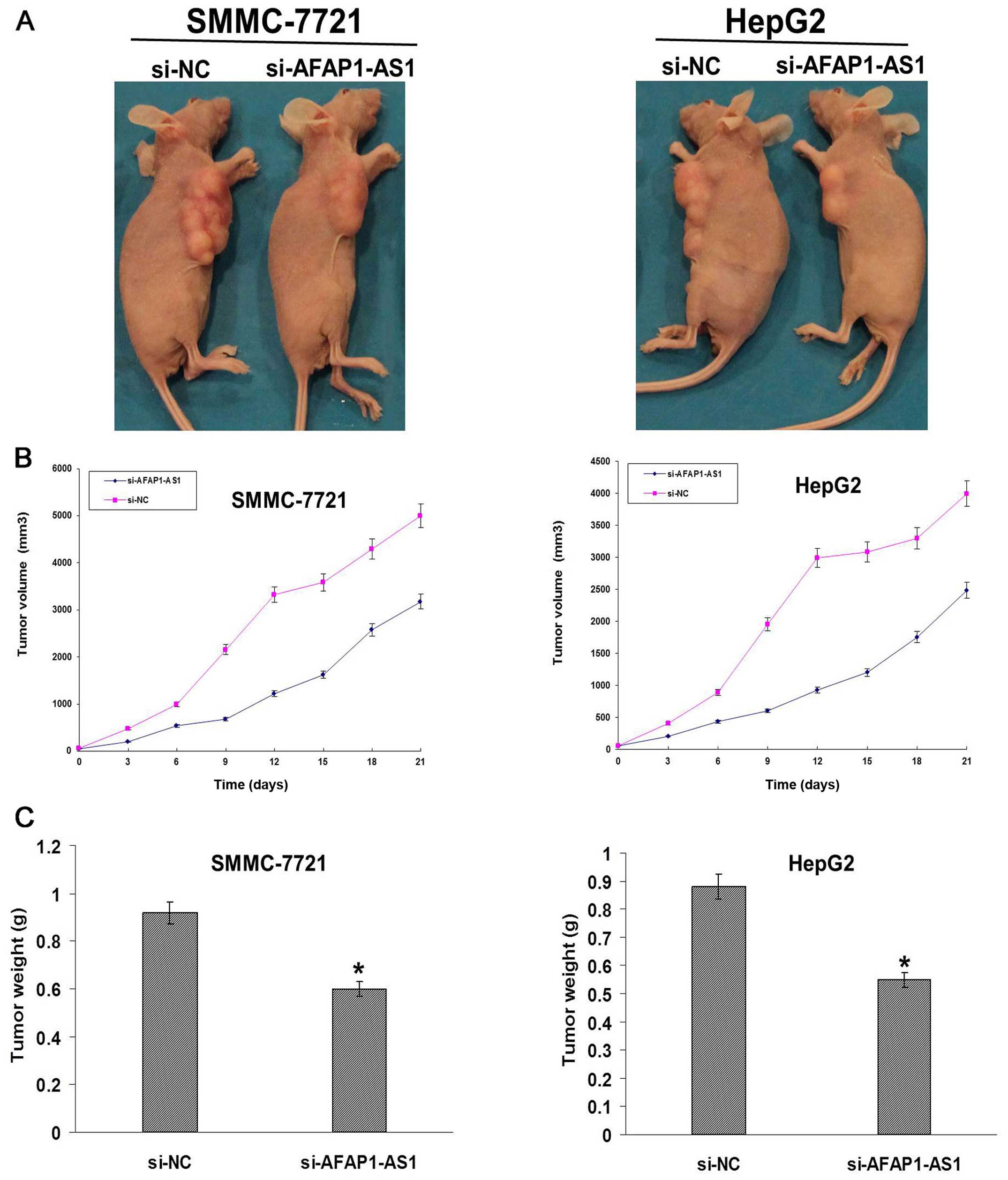
AFAP1-AS1 knockdown inhibits xenograft
tumor growth in vivo
Xenograft tumor models were established to assess
the tumor growth in vivo. During the whole tumor growth
period, the tumor growth activity was measured. The tumors grew
substantially slowly in si-AFAP1-AS1 group compared to the si-NC
group (Fig. 6A and B). When the
tumors were harvested, the average weight of the tumors in
si-AFAP1-AS1 group was significantly smaller than that in si-NC
group (P<0.05, Fig. 6B).
Discussion
Molecular targeting therapy is of particular
significance for treatment of malignancies because of the lack of
effective systemic therapies and options. Tremendous evidence shows
that lncRNAs over 200 nucleotides (nt) in length are emerging as
important regulatory molecules at the transcriptional and
post-transcriptional levels, and play essential roles in a variety
of cancer development and progression and provide potential
therapeutic biomarkers for cancer diagnosis and prognosis such as
H19, HOTAIR, MALAT1, MEG3, and XIST (24–26).
The combination of lncRNAs SOX2OT, PTPRG-AS1, ANRASSF1, ANRIL and
RP11-397D12.4, AC007403.1, ERICH1-AS1 may be helpful for early
detection and evaluation of prognosis in breast cancer (27) and non-small cell lung cancer
(NSCLC) (28). To confirm the
expression and clinical significance of AFAP1-AS1 in HCC, in the
present study, we found that AFAP1-AS1 was highly expressed in HCC
tissues and was correlated with the LVSI in HCC patients.
Multivariate analysis showed that AFAP1-AS1 might serve as an
independent prognostic factor for overall survival in HCC
patients.
Many studies have confirmed that lncRNAs are
involved in cell proliferation, angiogenesis, invasion and
metastasis invarious types of cancers (29–32).
LncRNA Hh maintains the mammosphere-formation efficiency (MFE) and
self-renewal capacity of cancer stem cells in Twist-positive breast
cancer (29), and HOTAIR induces
androgen-independent androgen receptor (AR) activation, drives the
AR-mediated transcriptional program and facilitates
castration-resistant prostate cancer progression (30). Silencing of lncRNA MALAT1 or
HOXA-AS2 inhibits epithelial-mesenchymal transition and malignant
transformation by inducing G1 arrest and promoting apoptosis in
gastric cancer (31,32). Depletion of lncRNA ANRIL leads to
cell cycle arrest at the G2/M phase in NSCLC and cervical cancer
(33). LncRNA ODRUL increases
doxorubicin-resistance molecule via ABCB1 gene in osteosarcoma
cells (34). To demonstrate the
function of AFAP1-AS1 in HCC, we found that knockdown of AFAP1-AS1
by si-AFAP1-AS1 decreased the proliferation and invasion in
vitro and in vivo, induced cell apoptosis and blocked
cell cycle in S phase.
RhoA activation has been confirmed to regulate many
molecular events including cell proliferation, differentiation,
inflammation response and angiogenesis (35). Activation of RhoA contributes to a
poor prognosis and mediates cell migration in HCC (36–38).
However, inhibition of RhoA by miR-200b/200c/429 counteracts the
metastatic capacity of HCC cells (39). Rac2 is frequently mutated and have
a high transcript level in HCC (40,41).
However, the relationship between AFAP1-AS1 expression and
RhoA/Rac2 signaling is not comprehensively understood. Our present
studies showed that knockdown of AFAP1-AS1 decreased the expression
of RhoA and Rac2 in HCC cells, suggesting that AFAP1-AS1 might
promote the HCC progression via upregulation of RhoA/Rac2
signaling.
In conclusion, our findings indicate that AFAP1-AS1
may promote the HCC progression and invasion through upregulation
of RhoA/Rac2 signaling. Our studies may provide a novel and
potential therapeutic target for treatment of HCC.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (81200328), the Shanghai Natural
Science Foundation (12ZR1424100) and the Scientific Research
Project of Shanghai Science and Technology Committee
(15411967200).
References
|
1
|
Rinella ME: Nonalcoholic fatty liver
disease: A systematic review. JAMA. 313:2263–2273. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mancuso A and Perricone G: Hepatocellular
Carcinoma and Liver Transplantation: State of the Art. J Clin
Transl Hepatol. 2:176–181. 2014. View Article : Google Scholar
|
|
3
|
Khan FZ, Perumpail RB, Wong RJ and Ahmed
A: Advances in hepatocellular carcinoma: Nonalcoholic
steatohepatitis-related hepatocellular carcinoma. World J Hepatol.
7:2155–2161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
She WH and Chok KS: Strategies to increase
the resectability of hepatocellular carcinoma. World J Hepatol.
7:2147–2154. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meng X, Franklin DA, Dong J and Zhang Y:
MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res.
74:7161–7167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Watson ME, Diepeveen LA, Stubbs KA and
Yeoh GC: Glycosylation-related diagnostic and therapeutic drug
target markers in hepatocellular carcinoma. J Gastrointestin Liver
Dis. 24:349–357. 2015.PubMed/NCBI
|
|
7
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim ED and Sung S: Long noncoding RNA:
Unveiling hidden layer of gene regulatory networks. Trends Plant
Sci. 17:16–21. 2012. View Article : Google Scholar
|
|
9
|
Yoon JH, Abdelmohsen K and Gorospe M:
Posttranscriptional gene regulation by long noncoding RNA. J Mol
Biol. 425:3723–3730. 2013. View Article : Google Scholar :
|
|
10
|
Yu TT, Xu XM, Hu Y, Deng JJ, Ge W, Han NN
and Zhang MX: Long noncoding RNAs in hepatitis B virus-related
hepatocellular carcinoma. World J Gastroenterol. 21:7208–7217.
2015.PubMed/NCBI
|
|
11
|
Tu ZQ, Li RJ, Mei JZ and Li XH:
Down-regulation of long non-coding RNA GAS5 is associated with the
prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol.
7:4303–4309. 2014.PubMed/NCBI
|
|
12
|
Zhang X, Rice K, Wang Y, Chen W, Zhong Y,
Nakayama Y, Zhou Y and Klibanski A: Maternally expressed gene 3
(MEG3) noncoding ribonucleic acid: Isoform structure, expression,
and functions. Endocrinology. 151:939–947. 2010. View Article : Google Scholar :
|
|
13
|
Zhuo H, Tang J, Lin Z, Jiang R, Zhang X,
Ji J, Wang P and Sun B: The aberrant expression of MEG3 regulated
by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol
Carcinog. Epub: Jan 16. 2015.(Epub ahead of print). View Article : Google Scholar
|
|
14
|
Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang
Y, Tang GN, Zhou WP and Sun SH: Hepatitis B virus X protein
(HBx)-related long noncoding RNA (lncRNA) down-regulated expression
by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by
targeting the intermediate filament protein vimentin. Hepatology.
57:1882–1892. 2013. View Article : Google Scholar
|
|
15
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar
|
|
17
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long noncoding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yuan SX, Yang F, Yang Y, Tao QF, Zhang J,
Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al: Long noncoding RNA
associated with microvascular invasion in hepatocellular carcinoma
promotes angiogenesis and serves as a predictor for hepatocellular
carcinoma patients' poor recurrence-free survival after
hepatectomy. Hepatology. 56:2231–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Du Y, Kong G, You X, Zhang S, Zhang T, Gao
Y, Ye L and Zhang X: Elevation of highly up-regulated in liver
cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell
proliferation via down-regulating p18. J Biol Chem.
287:26302–26311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu W, Bhagat TD, Yang X, Song JH, Cheng Y,
Agarwal R, Abraham JM, Ibrahim S, Bartenstein M, Hussain Z, et al:
Hypomethylation of noncoding DNA regions and overexpression of the
long noncoding RNA, AFAP1-AS1, in Barrett's esophagus and
esophageal adenocarcinoma. Gastroenterology. 144:956–966.e4. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ye Y, Chen J, Zhou Y, Fu Z, Zhou Q, Wang
Y, Gao W, Zheng S, Zhao X, Chen T, et al: High expression of
AFAP1-AS1 is associated with poor survival and short-term
recurrence in pancreatic ductal adenocarcinoma. J Transl Med.
13:1372015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao
Q, Chen P, Shi L, Lian Y, Jing Y, et al: Upregulated long
non-coding RNA AFAP1-AS1 expression is associated with progression
and poor prognosis of nasopharyngeal carcinoma. Oncotarget.
6:20404–20418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zeng Z, Bo H, Gong Z, Lian Y, Li X, Li X,
Zhang W, Deng H, Zhou M, Peng S, et al: AFAP1-AS1, a long noncoding
RNA upregulated in lung cancer and promotes invasion and
metastasis. Tumour Biol. Aug 6–2015.(Epub ahead of print).
|
|
24
|
Sun J, Bie B, Zhang S, Yang J and Li Z:
Long non-coding RNAs: Critical players in hepatocellular carcinoma.
Int J Mol Sci. 15:20434–20448. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li CH and Chen Y: Targeting long
non-coding RNAs in cancers: Progress and prospects. Int J Biochem
Cell Biol. 45:1895–1910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Iranpour M, Soudyab M, Geranpayeh L,
Mirfakhraie R, Azargashb E, Movafagh A and Ghafouri-Fard S:
Expression analysis of four long noncoding RNAs in breast cancer.
Tumour Biol. Sep 27–2015.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Q, Ni Z, Cheng Z, Xu J, Yu H and Yin
P: Three circulating long non-coding RNAs act as biomarkers for
predicting NSCLC. Cell Physiol Biochem. 37:1002–1009. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou M, Hou Y, Yang G, Zhang H, Tu G, Du
YE, Wen S, Xu L, Tang X, Tang S, et al: LncRNA-Hh strengthen cancer
stem cells generation in twist-positive breast cancer via
activation of Hedgehog signaling pathway. Stem Cells. Sept
29–2015.(Epub ahead of print). PubMed/NCBI
|
|
30
|
Zhang A, Zhao JC, Kim J, Fong KW, Yang YA,
Chakravarti D, Mo YY and Yu J: LncRNA HOTAIR enhances the
androgen-receptor-mediated transcriptional program and drives
castration-resistant prostate cancer. Cell Rep. 13:209–221. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu L, Luo F, Liu Y, Liu X, Shi L, Lu X and
Liu Q: Post-transcriptional silencing of the lncRNA MALAT1 by
miR-217 inhibits the epithelial-mesenchymal transition viaenhancer
of zeste homolog 2 in the malignant transformation of HBE cells
induced by cigarette smoke extract. Toxicol Appl Pharmacol.
289:276–285. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xie M, Sun M, Zhu YN, Xia R, Liu YW, Ding
J, Ma HW, He XZ, Zhang ZH, Liu ZJ, et al: Long noncoding RNA
HOXA-AS2 promotes gastric cancer proliferation by epigenetically
silencing P21/PLK3/DDTT3 expression. Oncotarget. 6:33587–33601.
2015.PubMed/NCBI
|
|
33
|
Naemura M, Murasaki C, Inoue Y, Okamoto H
and Kotake Y: Long noncoding RNA ANRIL regulates proliferation of
non-small cell lung cancer and cervical cancer cells. Anticancer
Res. 35:5377–5382. 2015.PubMed/NCBI
|
|
34
|
Zhang CL, Zhu KP, Shen GQ and Zhu ZS: A
long non-coding RNA contributes to doxorubicin resistance of
osteosarcoma. Tumour Biol. Sep 25–2015.(Epub ahead of print).
|
|
35
|
Yu OM and Brown JH: G protein-coupled
receptor and RhoA-stimulated transcriptional responses: Links to
inflammation, differentiation, and cell proliferation. Mol
Pharmacol. 88:171–180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yin K, Zhao G, Huang X, Gao G, Sun H, Wei
Q, Liu Q, Li M, Xu C, Zhu S, et al: Inhibition of RhoA expression
by adenovirus-mediated siRNA combined with TNF-α induced apoptosis
of hepatocarcinoma cells. Biomed Mater Eng. 26(Suppl 1):
S2055–S2067. 2015.
|
|
37
|
Serizawa N, Tian J, Fukada H, Baghy K,
Scott F, Chen X, Kiss Z, Olson K, Hsu D, Liu FT, et al: Galectin 3
regulates HCC cell invasion by RhoA and MLCK activation. Lab
Invest. 95:1145–1156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lin L, Yang XM, Li J, Zhang YL, Qin W and
Zhang ZG: Microfilament regulatory protein MENA increases activity
of RhoA and promotes metastasis of hepatocellular carcinoma. Exp
Cell Res. 327:113–122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wong CM, Wei L, Au SL, Fan DN, Zhou Y,
Tsang FH, Law CT, Lee JM, He X, Shi J, et al: MiR-200b/200c/429
subfamily negatively regulates Rho/ROCK signaling pathway to
suppress hepatocellular carcinoma metastasis. Oncotarget.
6:13658–13670. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gu DL, Chen YH, Shih JH, Lin CH, Jou YS
and Chen CF: Target genes discovery through copy number alteration
analysis in human hepatocellular carcinoma. World J Gastroenterol.
19:8873–8879. 2013. View Article : Google Scholar :
|
|
41
|
Cleary SP, Jeck WR, Zhao X, Chen K,
Selitsky SR, Savich GL, Tan TX, Wu MC, Getz G, Lawrence MS, et al:
Identification of driver genes in hepatocellular carcinoma by exome
sequencing. Hepatology. 58:1693–1702. 2013. View Article : Google Scholar : PubMed/NCBI
|