Introduction
Breast cancer ranks as the second cause of cancer
death among women (1–4). Clinical cases have indicated that
most of the breast cancer deaths are caused by metastasis (5,6), and
as a distinct metastatic pattern, breast cancer cells are more
inclined to migrate into lymph nodes, bone marrow, lung, liver and
brain (7). Extensive efforts have
been made towards a better understanding of human breast cancer
oncogenic events, and many biomarkers have been reported to be an
indicator for initiation and invasion of breast cancer. These may
include among others; proline rich inositol polyphosphate
5-phosphatase (PIPP), which when depleted inhibits PI3K/AKT
signaling, enhanced the transformation ability of breast cancer
cells, but reduced cell migration and invasion thus indicating that
PIPP is a potential suppressor of oncogenic PI3K/AKT signaling in
breast cancer (8); bone marrow
stromal antigen 2 which is expressed in cancer cells and
facilitates the emergence of neoplasia and malignant progression of
breast cancer (9) and others such
as oncoprotein hepatitis B X-interacting protein which when
mediated by general control non-derepressible 5 promotes the
migration of breast cancer cells and therapeutically could act as a
novel target in breast cancer (10).
PEG10 (paternally expressed imprinted gene 10)
located at human chromosome 7q21 was first reported to a gene from
a newly defined imprinted region in 2001 (11). Akamatsu et al have reported
that PEG10 was highly expressed in neuroendocrine prostate cancer
(NEPC), and distinct isoforms of PEG10 promoted proliferation and
invasion of NEPC cells, which could be recognized as a specific
therapeutic target for NEPC (12).
Abnormal overexpression of PEG10 was found in most hepatocellular
carcinoma cells, and regenerating mouse liver cells (13). PEG10 expression was reported to be
associated with worse survival and recurrence in hepatocellular
carcinoma, which could indicate that PEG10 as a potential target
predicting early recurrence and recurrence-free survival after
curative resection (14).
Long-term inhibition of PEG10 was found to lead to induction of
apoptosis in B-cell chronic lymphocytic leukemia (B-CLL) cells
(15). The PEG10 knockout mice
showed early embryonic lethality owing to defects in the placenta,
which indicated that PEG10 played an important role in placental
development (16). PEG10
expression was closely associated with the clinical, pathological
and biological behaviors, and a poor prognosis in gallbladder
cancer (17). In breast cancer
transgenic mouse models with c-MYC overexpression, most of the
mammary carcinomas developed by overexpressing c-MYC had increased
PEG10 mRNA expression compared with normal mammary gland (18). Though PEG10 has been implicated in
many types of cancers, the function of PEG10 in human breast cancer
still remains unclear.
In recent years, we have focused on PEG10 in several
cancers, including hematological malignancies and lung cancer. We
have observed that in CD19+CD34+ B cells from
patients with B-cell lineage acute (B-ALL) and B-CLL, PEG10 was
activated by CXCR5 and CCR7 co-simulation thus contributing to
apoptosis resistance (19).
Similarly in B-ALL CD23+CD5+ B cells, PEG10
expression level was upregulated by CCL19 and CXCR13, which also
enhanced the anti-apoptosis ability (20). PEG10 promoted Raji cell migration
capacity by up-regulating matrix metalloproteinase-2 (MMP-2) and
MMP-9 expression (21). Recently,
we observed that the expression of PEG10 was closely related to
clinical TNM grade and patient prognosis in lung cancer through
publicly available datasets. Amplifying this finding, in
vitro experiments in A549 cells, indicated that PEG10
facilitated cell proliferation and promoted tumor cell migration
and invasion by upregulating the expression of β-catenin, MMP-2 and
MMP-9 (22). Therefore using Gene
Expression Omnibus (GEO) datasets, we analyzed the relationship
between PEG10 and breast cancer, and found that the high expression
of PEG10 was positively correlated with the poor grade of breast
cancer and closely related with overall survival of the patients.
Thus, we hypothesize that the elevation of PEG10 also enhances
breast cancer cells proliferation and invasion.
Materials and methods
Cell culture
Human breast cancer cell line MDA-MB-231 and human
hepatoma cell line HepG2 were obtained from American type culture
collection (ATCC) and maintained in Dulbecco's modified Eagle's
medium (DMEM) with high glucose (Gibco, Carlsbad, CA, USA)
supplemented with 10% fetal bovine serum (FBS; Hyclone Corp.,
Logan, UT, USA), containing 100 U/ml penicillin and 100 mg/ml
streptomycin. Cells were cultured in a 5% CO2 air
incubator at 37°C and passaged using 0.25% trypsin-EDTA (Gibco)
when they reached confluence.
Plasmid and transfection
Full-length PEG10 was isolated from HepG2 cells, and
then subcloned into pEGFP-c1 plasmid between HindIII and
SalI restriction sites. The orientation and correct frame of
the recombinant vector pEGFP-c1/PEG10 (PEG10-EGFP) and the empty
vector pEGFP-c1 plasmid used as control (Control-EGFP) were
confirmed by sequencing.
For transient transfection, cells were first seeded
in plate at 60% confluency (5×105 cells per well)
overnight. The next day plasmids were diluted with an appropriate
concentration of Opti-MEM according to manufacturer's instruction
of Lipofectamin™ 3000 to form complexes. The cells were transiently
transfected with PEG10-EGFP plasmid or Control-EGFP plasmid
complexes for 48 h. The viability of the cells was tested by trypan
blue staining.
Cell proliferation assay
MDA-MB-231 cells were seeded in a 96-well plate at
the density of 1×104 cells per well overnight for
adherence. The next day cells were transfected with the above
mentioned plasmids. After 48 h of transfection, 10 μl CCK-8
solutions was added to each well at indicated times and incubated
for another 3 h. The absorbance of each well was obtained by
PerkinElmer 2030 VICTOR X Multilabel Plate Reader (Perkin-Elmer,
Waltham, MA, USA) at 450 nm.
Cell cycle
Cells were transfected for 48 h and digested by
trypsin. After incubation with 70% ethanol overnight, cells were
stained with propidium iodide (PI) for 30 min. The cells were then
washed with ice cold PBS twice and PI intensity was detected by
flow cytometry. The results were analyzed using ModFit software as
directed by manufacturer's specifications.
Wound-healing assay
Confluent monolayers of MDA-MB-231 cells (60%) were
triple plated in a 24-well plate, and transfection was done as
described above. After transfection for 48 h, cells were starved
with DMEM solution containing 1% FBS for 24 h. A wound was then
scratched in each well using a sterile P10 micropipette tip. Cells
were allowed to migrate for 24 h and images were obtained by
microscopy using a charge coupled device (CCD) camera. All analyses
were repeated in triplicate.
Transwell migration and Matrigel invasion
assay
Cells were transfected with plasmids for 48 h as
described above. The cells were then digested, centrifuged and
suspended into DMEM medium with 1% FBS. A total of 5×104
PEG10-EGFP and Control-EGFP cells were plated in upper chamber,
which was a 6.5-mm-diameter, polycarbonate membrane with an 8-μm
pore size filter, coated (invasion) or not coated (migration)
Matrigel (Corning, NY, USA). The lower chamber was filled with 500
μl DMEM solution containing 20% FBS, which acted as a
chemoattractant. After 24 h of incubation at 37°C, non-migrated
cells were removed with cotton swabs and migrated cells were fixed
in methanol and stained with 0.1% crystal violet. Images were
obtained by light microscope with charge coupled device camera.
Cells from four different fields were counted.
Clone formation assay
Transfected cells (500) were seeded to each well of
6-well plate and allowed to grow for 10 days in 37°C incubator. The
cells were then washed twice with ice cold PBS, fixed by methanol
for 10 min and stained with crystal violate for 10 min. The images
of stained and clone spheres of cells were obtained by a charge
coupled device camera.
RNA extraction and real-time quantitative
RT-PCR
Total RNA was extracted from MDA-MB-231 cells using
TRIzol (Invitrogen, USA) according to the manufacturer's
specifications and quantified by NanoDrop 2000 (Thermo Scientific,
Waltham, MA, USA). RNA (2 μg) was reverse-transcribed to cDNA with
random primers using the reverse transcriptase kit (Promega,
Madison, WI, USA) according to manufacturer's protocol. The
sequences of specific primers were as follows: PEG10: Forward:
5′-GCTAAGCTTCGATGACCGAACGAAGAAGGGACGAG-3′, Reverse:
5′-ATATAGTCGACCTACAGCGGGGCCGGGGAGTTT-3′; MMP-1: Forward:
5′-GGGGCTTTGATGTACCCTAGC-3′, Reverse: 5′-TGTCACACGCTTTTGGGGTTT-3′;
MMP-2: Forward: 5′-TGACATCAAGGGCATTTCAGGAGC-3′, Reverse:
5′-GTCCGCCAAATGAACCGGTCCTTG-3′; MMP-9: Forward:
5′-GAGGTTCGACGTGAAGGCGCAGATG-3′, Reverse:
5′-CATAGGTCACGTAGCCCACTTGGTC-3′; TIMP-1: Forward:
5′-CTTCTGCAATTCCGA CCTCGT-3′, Reverse: 5′-ACGCTGGTATAAGGTGGTCTG-3′;
TIMP-2: Forward: 5′-GCTGCGAGTGCAAGATCAC-3′, Reverse:
5′-TGGTGCCCGTTGATGTTCTTC-3′; E-cadherin: Forward:
5′-ATTTTTCCCTCGACACCCGAT-3′, Reverse: 5′-TCCCAGGCGTAGACCAAGA-3′;
GAPDH: Forward: 5′-CTGGGCTACACTGAGCACC-3′, Reverse:
5′-AAGTGGTCGTTGAGGGCAATG-3′;
Protein extraction and western
blotting
Transfected cells were washed with ice cold PBS and
lysed by RIPA supplemented with protease inhibitor PMSF on ice.
Concentrations of total protein were measured by BCA kit (Thermo
Scientific) and absorbance was obtained by the PerkinElmer 2030
VICTOR X Multilabel Plate Reader. The extracted proteins were
separated by 10% SDS-polyacrylamide gel electrophoresis and
transferred to PVDF membranes (Millipore, Billerica, MA, USA). The
membrane were blocked with 5% non-fat milk TBS-T (0.1% Tween-20,
100 nM Tris-HCl, 0.9% NaCl) and incubated with primary antibodies
with gentle shaking at 4°C overnight. After washing four times, the
membranes were incubated with HRP-conjugated secondary antibodies
for 2 h. The signals were detected using an enhanced
chemiluminescence detection kit (Thermo Scientific).
Bioinformatics analysis
Several GEO datasets were used to analyze the
correlations of PEG10 and clinical pathological features. PEG10
expression was traced by probe 212092 and 212094 in the platform.
The PEG10 expression of different groups were obtained and compared
by two-tail t-test. In order to observe the overall survival,
patients were divided into PEG10-high-expression (top 25%) and
PEG10-low-expression group (remaining 75%) and survival curves were
obtained by Kaplan-Meier and Log-rank tests which could also be
obtained using www.kmplot.com website with auto select
best cutoff for dividing group. In addition, for Gene Set
Enrichment Analysis (GSEA), GSE3494 dataset was analyzed with GMT
file C5 (GO gene set) as directed by the manufacturer's
specifications.
Statistics
Statistical analysis of significant differences were
acquired using the GraphPad Prism 5 software. Significance was
assessed by t-test followed by unpaired comparisons. ANOVA test was
used to evaluate PEG10 expression variation in different tumor
grades. Kaplan-Meier plots were constructed and log-rank tests were
used to observe overall survival rates related to PEG10 expression.
A probability (P) value of ≤0.05 was considered to indicate a
statistically significant difference.
Results
PEG10 was highly expressed in breast
cancer and closely correlated with clinicopathological
features
From GEO database, we applied the t-test to analyze
several datasets containing breast cancer samples with or without
normal breast tissues and corresponding clinical information. PEG10
expression was measured as log2 (probe intensities)
using Affymetrix microarrays. As shown in Fig. 1A, PEG10 expression was notably
elevated in breast carcinoma compared to normal breast samples
(GSE42568, p=0.0049). Similar results were obtained in GSE10780
dataset indicating that PEG10 was expressed highly in breast cancer
compared to normal tissues (Fig.
1B, p=0.0002). High PEG10 expression was related to high grade
of breast cancer (Fig. 1C,
GSE42568, p=0.006). From GSE4922 dataset, PEG10 high expression was
correlated with tumor size and presented more heterogeneity than
low-level group (Fig. 1D,
p=0.0333). In addition, patients with invasive ductal carcinomas
(IDC) (GSE21422, Fig. 1E, p=0.039)
and lymph node metastasis (GSE42568, Fig. 1F, p=0.023) were likely to show
PEG10 high expression.
Upregulation of PEG10 led to a poor
outcome of breast cancer patients
Consistent with above pathological observations, we
analyzed the GEO datasets GSE3494 and GSE42568 to investigate the
relationship between PEG10 expression and overall survival rate.
According to PEG10 expression, top 25% of samples were considered
as the PEG10 high-expression group, and 75% of the samples were
included in the PEG10 low-expression group. The Kaplan-Meier method
and log-rank test were used to compare the survival rate of
patients with breast cancers in the two groups described above. The
data indicated that patients in high-expression group present
poorer survival rates than the low-expression group in GSE3494
(Fig. 2A, p=0.0077) and GSE42568
(Fig. 2B, p=0.0496). Similar
results were observed using www.kmplot.com with
auto selected best cutoff in which PEG10 high group have lower
survival rates with p value of 0.016 (Fig. 2C). In addition, survival rate of
patients with high PEG10 expression was even lower in patients with
lymph node metastasis (Fig. 2D,
p=0.0029), while there was no significant difference in PEG10 high
and low group in non-metastatic patients (Fig. 2E, p=0.24). These results indicated
that PEG10 is negatively correlated with clinical outcome.
PEG10 enhanced cell cycle processes in
breast cancer
GSE3494 dataset was used to analyze PEG10
relationship with biological processes by GSEA. The gene profile of
high PEG10 expression group (top 25%) and low PEG10 expression
group (75%) were entered to the GSEA software and GMT file C5 (GO
gene sets) was selected to process the analysis. The first twenty
relevant biological processes that had p<0.05 and false
discovery rate p<0.08 were selected and are shown in Table I with enrichment score, normalized
enrichment score, normal p-value and false discovery rate values.
Fig. 3 shows gene set differences
in PEG10 high vs. low patients, indicating that PEG10 regulates
gene sets mainly associated with cell cycle progression.
 | Table IBiological processes enriched by
PEG10 elevation. |
Table I
Biological processes enriched by
PEG10 elevation.
| No. | GS Details | Size | ES | NES | NOM p-val | FDR q-val |
|---|
| 1 |
MRNA_METABOLIC_PROCESS | 56 | 0.58 | 2.13 | 0 | 0.016 |
| 2 |
MRNA_PROCESSING_GO_0006397 | 45 | 0.62 | 2.12 | 0 | 0.009 |
| 3 | RNA_BINDING | 196 | 0.38 | 2.04 | 0.002 | 0.017 |
| 4 |
REGULATION_OF_CELL_CYCLE | 153 | 0.55 | 1.97 | 0 | 0.036 |
| 5 |
CELLULAR_PROTEIN_COMPLEX_ASSEMBLY | 28 | 0.53 | 1.92 | 0.002 | 0.059 |
| 6 |
CELL_CYCLE_GO_0007049 | 260 | 0.56 | 1.91 | 0.002 | 0.059 |
| 7 |
RIBONUCLEOPROTEIN_COMPLEX | 105 | 0.49 | 1.91 | 0.002 | 0.05 |
| 8 | RNA_PROCESSING | 117 | 0.46 | 1.89 | 0.022 | 0.055 |
| 9 |
MITOTIC_CELL_CYCLE | 124 | 0.65 | 1.87 | 0.004 | 0.065 |
| 10 | CHROMOSOME | 104 | 0.6 | 1.85 | 0.004 | 0.071 |
| 11 |
REGULATION_OF_MITOSIS | 34 | 0.75 | 1.84 | 0.002 | 0.075 |
| 12 |
CHROMOSOMAL_PART | 80 | 0.62 | 1.84 | 0.004 | 0.069 |
| 13 |
GUANYL_NUCLEOTIDE_BINDING | 42 | 0.49 | 1.83 | 0.004 | 0.073 |
| 14 | GTP_BINDING | 42 | 0.49 | 1.83 | 0.004 | 0.068 |
| 15 |
CELL_CYCLE_PROCESS | 156 | 0.62 | 1.82 | 0.004 | 0.069 |
| 16 |
CELL_CYCLE_PHASE | 140 | 0.61 | 1.82 | 0.004 | 0.065 |
| 17 |
REGULATION_OF_CYCLIN_DEPENDENT
PROTEIN_KINASE_ACTIVITY | 39 | 0.63 | 1.81 | 0.002 | 0.067 |
| 18 |
RIBONUCLEASE_ACTIVITY | 21 | 0.64 | 1.81 | 0 | 0.065 |
| 19 | INTERPHASE | 58 | 0.57 | 1.81 | 0.004 | 0.063 |
| 20 |
REGULATION_OF_MITOTIC_CELL_CYCLE | 16 | 0.69 | 1.81 | 0.004 | 0.06 |
PEG10 accelerates breast cancer cell
proliferation and clone formation
First, we constructed PEG10 overexpression plasmid
PEG10-EGFP. MDA-MB-231 cells were transfected with PEG10-EGFP
plasmid or Control-EGFP plasmid, and mRNA and protein were
collected after 48 h of transfection. Real-time quantitative PCR
and western blotting were performed for PEG10 expression at mRNA
level (Fig. 4A) and protein level
(Fig. 4B), respectively. To assess
the effect of proliferation by PEG10, MDA-MB-231 cells were
transfected with plasmids for 48 h and then seeded in culture
plates. Cell viability was measured by CCK-8 assay, and results
showed that overexpressed PEG10 promoted the proliferation rates of
MDA-MB-231 cells compared with the control group in a
time-dependent manner (Fig. 4C).
In addition, clone formation assay also presented similar results
(Fig. 4D) in which clone spheres
were enlarged after the PEG10 elevation (Fig. 4E). All results gathered illustrated
that PEG10 could enhance breast cancer cell proliferation and clone
formation.
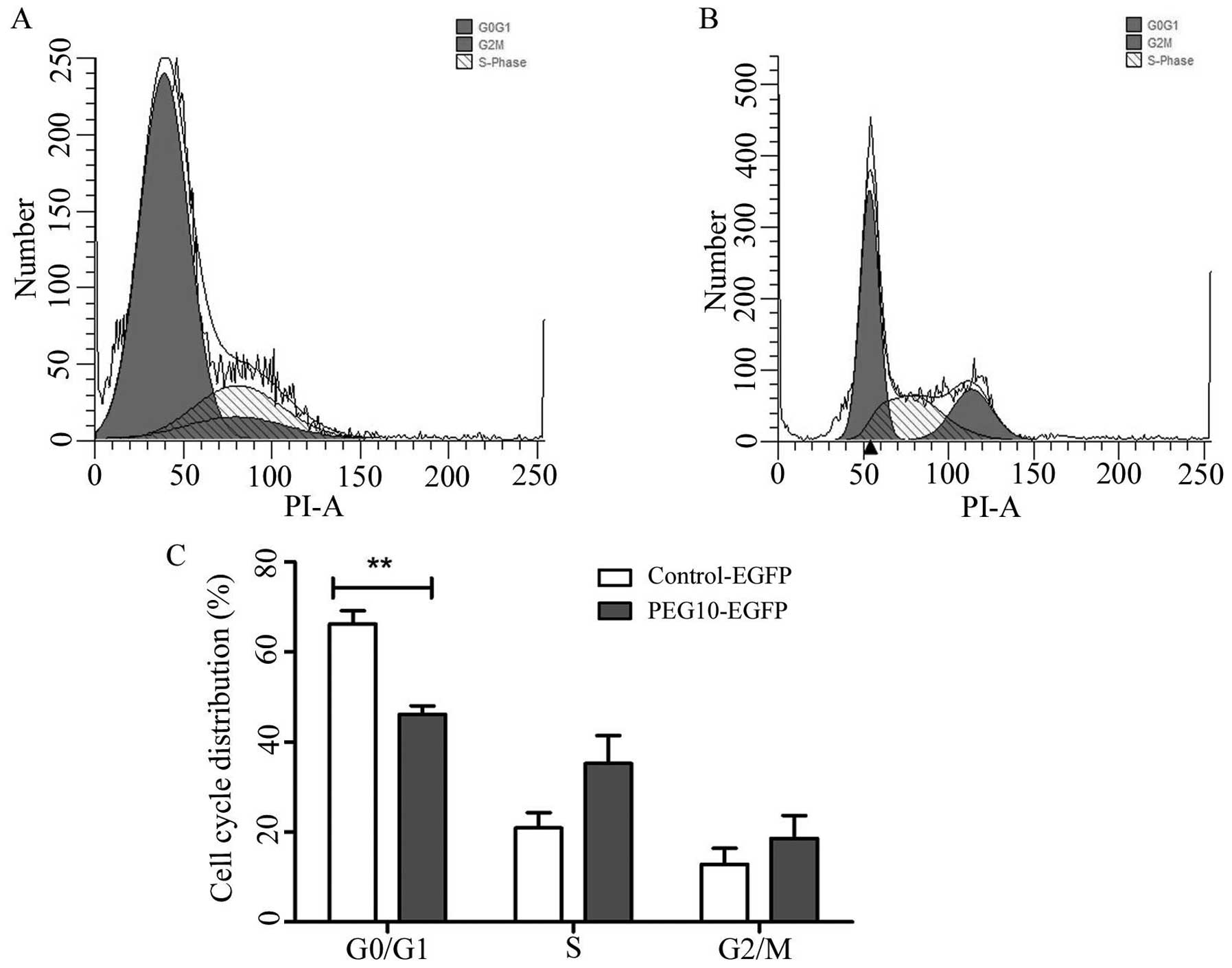
PEG10 facilitated MDA-MB-231 cells into S
and G2/M phase
Flow cytometry analysis revealed that the percentage
of MDA-MB-231/PEG10 transfect cells was higher in the S phase and
G2/M phase, while the proportion was lower in the G0/G1 phase of
the MDA-MB-231/PEG10 transfected cells as compared to the control
group, as shown in Fig. 5A and B.
These data provided evidence that PEG10 facilitated the cell cycle
progression of MDA-MB-231 cells. Fig.
5C shows the quantitative statistical data of the cell
cycle.
Overexpression of PEG10 promotes breast
cancer cell migration and invasion
To investigate the migration ability initiated by
PEG10, would healing assay were performed in MDA-MB-231 cells.
Fig. 6A demonstrates that PEG10
promoted MDA-MB-231 cells to migrate. Transwell assay (Fig. 6B) showed a similar result that
PEG10 overexpressing cells migrated more than the control group.
Furthermore, Matrigel invasion assay (Fig. 6C) also suggested that PEG10 was
involved in breast cancer cells invasion. After overexpressing
PEG10 levels in breast cancer, MMP-1, MMP-2 and MMP-9, which
function as matrix decomposers, were evidently increased, while
TIMP-1 and TIMP-2 decreased (Fig.
6D). Importantly, the vital cell-to-cell junction molecule
E-cadherin was downregulated after PEG10 overexpression (Fig. 6D), suggesting that PEG10 might
influence the detachment and movement of cancer cells from primary
sites to invade secondary sites.
Discussion
Breast cancer is frequently lethal with the second
highest mortality rate (15%) among women in America (1,23,24).
There were 64,640 new diagnosed in situ breast cancer cases,
232,340 invasive cases and 39,620 deaths due to breast cancer among
all ages in the United States in 2013. One in every eight women
will develop breast cancer, and invasive incidence is much higher
in ages over 50, and so is the mortality incidence (25). Breast cancer with metastasis is
devastating and effective treatments options are facing numerous
challenges. Hence, efficient specific tumor-related genes, which
could be used in the early diagnosis, prognosis and treatment of
breast cancer are urgently required.
PEG10 was initially identified in 2001, and since
then, it has been continuously positively associated with many
types of cancers, including leukemia (15,19,20),
lymphoma (21), prostate cancer
(12), liver cancer (13,14),
and lung cancer (22). Our recent
report unveiled the role of PEG10 in Raji cell apoptosis
resistance, proliferation, adhesion, migration and invasion
(21). In addition, we
demonstrated that PEG10 is associated with lung cancer progression
and enhanced proliferation, carcinogenesis, migration and invasion
ability of A549 cells (22). In
this study, we investigated the relationship between PEG10 and
breast cancer progression by bioinformatics analysis and the
function of PEG10 in breast cancer cells proliferation, cell cycle,
clone formation, migration and invasion by in vitro
experiments.
With the development of genomics and computer
sciences, bioinformatics analysis is emerging as a tool to evaluate
and predict diseases and it is widely applied in research (26–28).
For bioinformatics analysis, a large cohorts of samples (n>100)
with sufficient clinical information and follow-up records are used
as the selecting criteria and GSE42568 (n=121), GSE3494 (n=251),
GSE4922 (n=289), GSE10780 (n=185) were chosen in this study. Our
results showed that PEG10 was highly expressed in breast cancer
tissues compared to normal breast tissues and highly expressed in
metastasis tissues versus the non-metastatic tissues. At the same
time, PEG10 expression was also linked with tumor grades and tumor
size. Univariate survival analysis indicated that patients with
PEG10 high-expression were more vulnerable to breast cancer. GSEA
results showed that elevated PEG10 expression was closely related
with cell cycle processes. These results confirmed that PEG10 may
be important to breast cancer development and could be used as a
prognosis indicator. In addition, the GSEA table results not only
demonstrated that PEG10 was highly related to cell cycle, but also
revealed that PEG10 may influence the mRNA metabolic process,
cellular protein complex assembling and other biological
processes.
For better understanding the role of PEG10 in breast
cancer, we performed cytology experiments to verify the results we
obtained from GEO datasets. We constructed a PEG10 full-length
plasmid and transfected it to MDA-MB-231 cells. Overexpression of
PEG10 was observed to strongly enhance cell proliferation, clone
formation ability, and the clone size was much bigger than in the
control group. PEG10 also promoted cells into S and G2/M phases,
which was consisted with GSEA results. In addition, PEG10
overexpressing cells exhibited enhanced migration and invasion
ability based on wound healing assay and invasion assay.
Altogether, we propose that PEG10 is strongly associated with
breast cancer development and progression.
As tumors progress to higher pathological grades of
malignancy, they exhibit invasive properties, such as decreased
attachment to other cells. E-cadherin has been seen as a key
cell-to-cell adhesion molecule, and its loss has been associated
with weakened maintenance of adherens junctions (29,30).
In our study, we observed that E-cadherin expression was reduced by
PEG10 overexpression, indicating the occurrence of weakened cell
junction and the possibility of migration. The process of cancer
invasion requires extracellular matrix degradation, which is
orchestrated by MMPs and TIMPs (31–33).
It has been widely reported that MMP-2 exhibited a vital role in
invasion of breast cancer (34–37).
MMP-2 is highly expressed in breast cancer and is also positively
related to shortened survival (38). Siegel et al reported that in
breast cancer invasion, not only MMP-2 and MMP-9 expression was
enhanced, but also MMP-1 and MMP-13 were upregulated in a
CCR9-dependent fashion (1).
Therefore, in this study we also showed that MMP-1, MMP-2 and MMP-9
were upregulated while TIMP-1 and TIMP-2 were downregulated due to
PEG10 overexpression. These results confirmed that PEG10 could
enhance the migration and invasion ability of breast cancer.
However, the expression of Twist, one of the most important
molecules regulating cancer metastasis (39,40),
was not significant (data not shown), suggesting that PEG10 effect
on migration may not be influenced by Twist signaling.
Taken together, this study elucidated that PEG10 was
highly expressed in breast cancer and positively correlated with
clinicopathological features. Furthermore, we verified that
overexpressing PEG10 could promote breast cancer cell
proliferation, clone formation, cell cycle, migration and invasion.
These findings may be beneficial to a better understanding of
breast cancer and to shed light on potential targets for breast
cancer diagnosis, and therapeutics.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (nos. 81400121, 81270607, 81541027 and
81501352).
Abbreviations:
|
PEG10
|
paternally expressed imprinted gene
10
|
|
GEO
|
Gene Expression Omnibus
|
|
GSEA
|
gene set enrichment analysis
|
|
MMPs
|
matrix metalloproteinases
|
|
TIMPs
|
tissue inhibitor of
metalloproteinases
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ricceri F, Fasanelli F, Giraudo MT, Sieri
S, Tumino R, Mattiello A, Vagliano L, Masala G, Quirós JR, Travier
N, et al: Risk of second primary malignancies in women with breast
cancer: Results from the European prospective investigation into
cancer and nutrition (EPIC). Int J Cancer. 137:940–948. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cappello P, Blaser H, Gorrini C, Lin DC,
Elia AJ, Wakeham A, Haider S, Boutros PC, Mason JM, Miller NA, et
al: Role of Nek2 on centrosome duplication and aneuploidy in breast
cancer cells. Oncogene. 33:2375–2384. 2014. View Article : Google Scholar
|
|
4
|
Bush TL, Payton M, Heller S, Chung G,
Hanestad K, Rottman JB, Loberg R, Friberg G, Kendall RL, Saffran D,
et al: AMG 900, a small-molecule inhibitor of aurora kinases,
potentiates the activity of microtubule-targeting agents in human
metastatic breast cancer models. Mol Cancer Ther. 12:2356–2366.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Q, Wei J, Su P and Gao P: Histone
demethylase JARID1C promotes breast cancer metastasis cells via
down regulating BRMS1 expression. Biochem Biophys Res Commun.
464:659–666. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee M, Beggs SM, Gildea D, Bupp S,
Lichtenberg J, Trivedi NS, Hu Y, Bodine DM and Crawford NP; NISC
Comparative Sequencing Program. Necdin is a breast cancer
metastasis suppressor that regulates the transcription of c-Myc.
Oncotarget. 6:31557–31568. 2015.PubMed/NCBI
|
|
7
|
Johnson-Holiday C, Singh R, Johnson E,
Singh S, Stockard CR, Grizzle WE and Lillard JW Jr: CCL25 mediates
migration, invasion and matrix metalloproteinase expression by
breast cancer cells in a CCR9-dependent fashion. Int J Oncol.
38:1279–1285. 2011.PubMed/NCBI
|
|
8
|
Ooms LM, Binge LC, Davies EM, Rahman P,
Conway JR, Gurung R, Ferguson DT, Papa A, Fedele CG, Vieusseux JL,
et al: The inositol polyphosphate 5-phosphatase PIPP regulates
AKT1-dependent breast cancer growth and metastasis. Cancer Cell.
28:155–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mahauad-Fernandez WD, DeMali KA, Olivier
AK and Okeoma CM: Bone marrow stromal antigen 2 expressed in cancer
cells promotes mammary tumor growth and metastasis. Breast Cancer
Res. 16:4932014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li L, Liu B, Zhang X and Ye L: The
oncoprotein HBXIP promotes migration of breast cancer cells via
GCN5-mediated microtubule acetylation. Biochem Biophys Res Commun.
458:720–725. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ono R, Kobayashi S, Wagatsuma H, Aisaka K,
Kohda T, Kaneko-Ishino T and Ishino F: A retrotransposon-derived
gene, PEG10, is a novel imprinted gene located on human chromosome
7q21. Genomics. 73:232–237. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Akamatsu S, Wyatt AW, Lin D, Lysakowski S,
Zhang F, Kim S, Tse C, Wang K, Mo F, Haegert A, et al: The
placental gene PEG10 promotes progression of neuroendocrine
prostate cancer. Cell Rep. 12:922–936. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tsou AP, Chuang YC, Su JY, Yang CW, Liao
YL, Liu WK, Chiu JH and Chou CK: Overexpression of a novel
imprinted gene, PEG10, in human hepatocellular carcinoma and in
regenerating mouse livers. J Biomed Sci. 10:625–635.
2003.PubMed/NCBI
|
|
14
|
Bang H, Ha SY, Hwang SH and Park CK:
Expression of PEG10 is associated with poor survival and tumor
recurrence in hepatocellular carcinoma. Cancer Res Treat.
47:844–852. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kainz B, Shehata M, Bilban M, Kienle D,
Heintel D, Krömer-Holzinger E, Le T, Kröber A, Heller G,
Schwarzinger I, et al: Overexpression of the paternally expressed
gene 10 (PEG10) from the imprinted locus on chromosome 7q21 in
high-risk B-cell chronic lymphocytic leukemia. Int J Cancer.
121:1984–1993. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ono R, Nakamura K, Inoue K, Naruse M,
Usami T, Wakisaka-Saito N, Hino T, Suzuki-Migishima R, Ogonuki N,
Miki H, et al: Deletion of Peg10, an imprinted gene acquired from a
retrotransposon, causes early embryonic lethality. Nat Genet.
38:101–106. 2006. View
Article : Google Scholar
|
|
17
|
Liu Z, Yang Z, Liu D, Li D, Zou Q, Yuan Y,
Li J, Liang L, Chen M and Chen S: TSG101 and PEG10 are prognostic
markers in squamous cell/adenosquamous carcinomas and
adenocarcinoma of the gallbladder. Oncol Lett. 7:1128–1138.
2014.PubMed/NCBI
|
|
18
|
Li CM, Margolin AA, Salas M, Memeo L,
Mansukhani M, Hibshoosh H, Szabolcs M, Klinakis A and Tycko B:
PEG10 is a c-MYC target gene in cancer cells. Cancer Res.
66:665–672. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu C, Xiong J, Zhang L, Huang B, Zhang Q,
Li Q, Yang M, Wu Y, Wu Q, Shen Q, et al: PEG10 activation by
co-stimulation of CXCR5 and CCR7 essentially contributes to
resistance to apoptosis in CD19+CD34+ B cells
from patients with B cell lineage acute and chronic lymphocytic
leukemia. Cell Mol Immunol. 1:280–294. 2004.
|
|
20
|
Chunsong H, Yuling H, Li W, Jie X, Gang Z,
Qiuping Z, Qingping G, Kejian Z, Li Q, Chang AE, et al: CXC
chemokine ligand 13 and CC chemokine ligand 19 cooperatively render
resistance to apoptosis in B cell lineage acute and chronic
lymphocytic leukemia CD23+CD5+ B cells. J
Immunol. 177:6713–6722. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiong J, Qin J, Zheng Y, Peng X, Luo Y and
Meng X: PEG10 promotes the migration of human Burkitt's lymphoma
cells by up-regulating the expression of matrix metalloproteinase-2
and -9. Clin Invest Med. 35:E117–E125. 2012.PubMed/NCBI
|
|
22
|
Deng X, Hu Y, Ding Q, Han R, Guo Q, Qin J,
Li J, Xiao R, Tian S, Hu W, et al: PEG10 plays a crucial role in
human lung cancer proliferation, progression, prognosis and
metastasis. Oncol Rep. 32:2159–2167. 2014.PubMed/NCBI
|
|
23
|
Vila J, Gandini S and Gentilini O: Overall
survival according to type of surgery in young (≤40 years) early
breast cancer patients: A systematic meta-analysis comparing
breast-conserving surgery versus mastectomy. Breast. 24:175–181.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mariz K, Ingolf JB, Daniel H, Teresa NJ
and Erich-Franz S: The Wnt inhibitor dickkopf-1: A link between
breast cancer and bone metastases. Clin Exp Metastasis. 32:857–866.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar
|
|
26
|
Wu W, Morrissey CS, Keller CA, Mishra T,
Pimkin M, Blobel GA, Weiss MJ and Hardison RC: Dynamic shifts in
occupancy by TAL1 are guided by GATA factors and drive large-scale
reprogramming of gene expression during hematopoiesis. Genome Res.
24:1945–1962. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wilson NK, Kent DG, Buettner F, Shehata M,
Macaulay IC, Calero-Nieto FJ, Sánchez Castillo M, Oedekoven CA,
Diamanti E, Schulte R, et al: Combined single-cell functional and
gene expression analysis resolves heterogeneity within stem cell
populations. Cell Stem Cell. 16:712–724. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shirai CL, Ley JN, White BS, Kim S,
Tibbitts J, Shao J, Ndonwi M, Wadugu B, Duncavage EJ, Okeyo-Owuor
T, et al: Mutant U2AF1 expression alters hematopoiesis and pre-mRNA
splicing in vivo. Cancer Cell. 27:631–643. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Piao HL, Yuan Y, Wang M, Sun Y, Liang H
and Ma L: α-catenin acts as a tumour suppressor in
E-cadherin-negative basal-like breast cancer by inhibiting NF-κB
signalling. Nat Cell Biol. 16:245–254. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Waleh NS, Murphy BJ and Zaveri NT:
Increase in tissue inhibitor of metalloproteinase-2 (TIMP-2) levels
and inhibition of MMP-2 activity in a metastatic breast cancer cell
line by an anti-invasive small molecule SR13179. Cancer Lett.
289:111–118. 2010. View Article : Google Scholar
|
|
32
|
Lee S, Desai KK, Iczkowski KA, Newcomer
RG, Wu KJ, Zhao YG, Tan WW, Roycik MD and Sang QX: Coordinated peak
expression of MMP-26 and TIMP-4 in preinvasive human prostate
tumor. Cell Res. 16:750–758. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nagase H: Cell surface activation of
progelatinase A (proMMP-2) and cell migration. Cell Res. 8:179–186.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vizoso FJ, González LO, Corte MD,
Rodríguez JC, Vázquez J, Lamelas ML, Junquera S, Merino AM and
García-Muñiz JL: Study of matrix metalloproteinases and their
inhibitors in breast cancer. Br J Cancer. 96:903–911. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jacob A, Jing J, Lee J, Schedin P, Gilbert
SM, Peden AA, Junutula JR and Prekeris R: Rab40b regulates
trafficking of MMP2 and MMP9 during invadopodia formation and
invasion of breast cancer cells. J Cell Sci. 126:4647–4658. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kambach DM, Sodi VL, Lelkes PI,
Azizkhan-Clifford J and Reginato MJ: ErbB2, FoxM1 and 14-3-3ζ prime
breast cancer cells for invasion in response to ionizing radiation.
Oncogene. 33:589–598. 2014. View Article : Google Scholar :
|
|
37
|
Addison JB, Koontz C, Fugett JH, Creighton
CJ, Chen D, Farrugia MK, Padon RR, Voronkova MA, McLaughlin SL,
Livengood RH, et al: KAP1 promotes proliferation and metastatic
progression of breast cancer cells. Cancer Res. 75:344–355. 2015.
View Article : Google Scholar :
|
|
38
|
Talvensaari-Mattila A, Pääkkö P, Höyhtyä
M, Blanco-Sequeiros G and Turpeenniemi-Hujanen T: Matrix
metalloproteinase-2 immunoreactive protein: A marker of
aggressiveness in breast carcinoma. Cancer. 83:1153–1162. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang
Q, Lin Y, Li J, Kang T, Tao M, et al: Disrupting the interaction of
BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like
breast cancer. Cancer Cell. 25:210–225. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fu J, Qin L, He T, Qin J, Hong J, Wong J,
Liao L and Xu J: The TWIST/Mi2/NuRD protein complex and its
essential role in cancer metastasis. Cell Res. 21:275–289. 2011.
View Article : Google Scholar
|