Introduction
The epithelial-mesenchymal transition (EMT) is a
normal process of early embryonic development and also contributes
pathologically to multiple conditions such as renal fibrosis and
cancer progression. In this process, the epithelial cells undergo
both structural and phenotypic changes, including changes in
adhesion, cytoskeletal and polarity structures, which result in
loss of cell polarity, enhanced migration and invasion, and
increased resistance to apoptosis (1). EMT plays important roles in
tumorigenesis, invasion and metastasis of cancers (2), and has consequently been studied
extensively in the cancer research field (3).
The Sp1 transcription factor is critical for many
biological processes, including cellular growth and differentiation
(4). In cancer growth and
metastasis, Sp1 influences a variety of cancer cell types through
the regulation of oncogenes, tumor suppressor genes, cell cycle
regulators, growth related signal transduction pathways, angiogenic
factors and apoptosis (5,6). Sp1 is expressed at high levels in
many cancers (7) and is closely
associated with EMT (8,9). New therapies targeting Sp1 could,
therefore, represent beneficial additions to current clinical
chemotherapy strategies.
EMT behavior increases invasiveness and mobility of
gastric cancer cells and makes them more resistant to apoptosis
(10). A previous study suggests
Sp1 is an important target in the study of gastric cancer (11). Sp1 can affect invasive ability of
gastric cancer cells through regulating transcription of MTA2 gene
(12). miRNA-22 also regulates
invasion and migration of gastric cancer via its target gene Sp1
(13). Since Sp1 is closely
involved in invasion and migration of gastric cancer, it is worthy
to study whether it plays a role in EMT of gastric cancer cells. In
addition, whether miRNAs are involved in this process is a focus of
this study.
In this study, we immunohistochemically assessed Sp1
expression in selected specimens from primary and metastatic
tissues of gastric cancers and adjacent tissues, and the results
showed that Sp1 was higher in metastatic and primary cancer tissues
than in normal adjacent tissues, suggesting close association of
high Sp1 expression and invasion and metastasis of gastric cancer.
Further study showed that the high expression of Sp1 was attributed
to inactivation of translation mechanism and low miRNA-223 level
was the direct cause for increased Sp1 expression. Therefore, we
reasoned that miRNA-223/Sp1/EMT is a pathway regulating metastasis
and invasion, and low expression of miRNA-223 increases Sp1 gene
expression in gastric cancer, induces EMT, and consequently
increases its ability to invade, proliferate and resist apoptosis.
Afterwards, we focused our study on verification of this pathway
regulating EMT and investigated whether overexpressing miRNA-223
can inhibit EMT in gastric cancer cells.
Materials and methods
Detection of Sp1 protein in gastric
cancer tissues and liver metastatic tissues
Gastric cancers, their associated liver meta-static
tissues and para-carcinoma tissues from 20 cases were collected
from the General Surgery Department of Shanghai Changzheng
Hospital, China. Sp1 expression was detected by
immunohistochemistry. Tissues were collected and fixed with 4%
paraformaldehyde and soaked in 20% saccharose solution at 4ºC
overnight. The specimens were embedded in paraffin, sliced and
subjected to conventional immunohistochemical detection of Sp1.
Briefly, each slice was washed three times with 0.01 M potassium
phosphate-buffered saline (PBS) for 5 min each, treated with a 0.3%
hydrogen peroxide-methanol solution for 30 min, and then washed
with PBS an additional three times. Next, the samples were treated
with 0.3% Triton X-100 in PBS for 30 min, washed with PBS three
times, and incubated with the primary antibody (Anti-Sp1, 1:200;
sc-420, Santa Cruz Biotechnology, USA) in serum diluent solution
(1% bovine serum albumin, 0.08% sodium azide in PBS) at 4ºC for 24
h. The slice was then washed with PBS, and incubated with
horseradish peroxidase-conjugated secondary antibody (anti-mouse,
1:3,000; #7076, Cell Signaling Technology, USA) at room temperature
for 2 h, before washing again in PBS. Avidin-biotin complex was
then added and the slice incubated at room temperature for 2 h,
after which it was washed three times with PBS followed by an
additional three washes with distilled water before addition of
developing solution for 15 min. The slice was then rinsed with
distilled water and PBS to terminate the reaction. Finally, the
slice was dehydrated through a graded series of alcohol washes,
sealed and photographed.
Detection of miRNA-223, Sp1 mRNA and
protein, EMT-marker proteins in tumor specimens
Tissue samples of ~100 mg each were cut from gastric
cancers, liver metastases and adjacent para-carcinoma tissues of 20
cases and stored in liquid nitrogen. For sample processing, tissues
were treated with 1 ml TRIzol (Invitrogen, Carlsbad, CA, USA) or
protein lysis buffer (Pierce, USA) for the subsequent extraction of
RNA and proteins, respectively. After homogenization, total RNA and
protein was extracted in accordance with the manufacturer's
instructions. The purity of total RNA was measured by ultraviolet
spectrophotometry, and miRNA-223 and Sp1 mRNA levels were detected
by real-time RT-PCR (for experimental methods, see the the details
below). The concentration of each protein sample was measured using
the BCA protein assay (Pierce), and the protein expression was
determined by western blotting.
Detection of mature miRNA-223
Total RNA (2 μg) was reverse-transcribed into cDNA
using primers specific for human U6 snRNA
(5′-TACCTTGCGAAGTGCTTAAAC-3′), and miRNA-223
(5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGAATGGGGT-3′). The
reaction product (2 μl) was used as the template for fluorogenic
quantitative PCR to measure miRNA-223. The results were analyzed by
the 2−ΔΔCt method using U6 snRNA (NM_004394.1) as the
internal reference. The forward and reverse PCR primers for U6
snRNA were 5′-GTGCTCGCTTCGGCAGCACAT-3′ and
5′-TACCTTGCGAAGTGCTTAAAC-3′, respectively. The forward and reverse
primers for miRNA-223 were 5′-GCCGGCGCCCGAGCTCTGGCTC-3′ and
5′-TGTCAGTTTGTCAAATACCCCA-3′, respectively. The PCR reaction mix
(20 μl) included 10 μl of SYBR Premix Ex Taq (Takara, Japan), 0.2
μl each of forward and reverse primers (20 μM stock), 2 μl of the
cDNA template, and 7.6 μl dH2O. The PCR reaction
conditions involved 40 cycles of a 95ºC denaturing step, a 58ºC
annealing step, and a 72ºC elongation step, each of which was
performed for 10 sec.
Detection of Sp1 mRNA
Analysis of Sp1 mRNA expression was performed using
real-time PCR and the 2−ΔΔCt method. Briefly, total RNA
of patient tissues was reverse-transcribed into cDNA using oligo
dT(18) primers (Takara, Japan)
and this was used as a template for the PCR reaction with
amplification of β-actin providing the internal reference. Forward
and reverse primers for the detection of Sp1 mRNA (NM_138473.2)
were 5′-AACAGATTATCACAAATCGAGG-3′ and 5′-AAGGTAGCTGCAGAAACGCTG-3′,
respectively. Forward and reverse primers for β-actin mRNA
(NM_001101.3) were 5′-GTGCTCGCTTCGGCAGCACAT-3′ and
5′-TACCTTGCGAAGTGCTTAAAC-3′, respectively. The PCR reaction mix and
cycle parameters were as above with the exception that the
annealing step was performed at 56ºC.
Western blotting
Protein samples were separated by SDS-PAGE using an
11% polyacrylamide gel and transferred to PVDF membranes. Membranes
were blocked at room temperature for 2 h in Tris-buffered saline
containing 0.1% Tween-20 (TBST) and 5% nonfat milk and then
incubated overnight at 4ºC with primary antibodies (Sp1, sc-420,
1:300; Vimentin, sc-53464, 1:200; Fibronectin, sc-81769, 1:400;
N-cadherin, sc-393933, 1:600; E-cadherin, sc-21791, B actin,
sc-130300, 1:1,000; Santa Cruz Biotechnology). Membranes were then
rinsed with TBST three times and incubated with horseradish
peroxidase-conjugated secondary antibody (rabbit anti-mouse IgG,
#7076, 1:3,000; Cell Signaling Technology) for 2 h. ECL
chemiluminescence substrate (Pierce) and X-ray film were used to
detect protein bands. Relative protein concentration was quantified
by densitometry analysis using image processing software by
normalizing the intensity of the target band to that of the
corresponding β-actin control band.
miRNA-223 binding site prediction and
luciferase reporter assays
To predict the miRNA-223 binding site, we used
Targetscan (http://www.targetscan.org/) to predict whether a
miRNA-223 binding site exists within the 3′-UTR of human Sp1 mRNA
(NM_138473.2). The results showed that a seven-base miRNA-223 seed
sequence is present in the 3′-UTR of Sp1 mRNA.
To construct luciferase reporter vectors, total RNA
was extracted from 293 cells, reverse-transcribed into cDNA, and 2
μl of the reaction product subsequently used as a template for PCR.
Primers were designed that targeted the 3′-UTR of the Sp1 gene such
that flanking XbaI restriction sites were introduced into
the 261 base-pair (bp) PCR product containing the 5′-AACTGAC-3′
miRNA-223 target site. The forward and reverse primer sequences
were 5′-GCTCTAGAGCTAACAGAAATTAATTTAACTG-3′ and
5′-GCTCTAGAAAGACGGTGTGGGTTGTTAC-3′, respectively. PCR reaction
conditions were as follows: 35 cycles of a 94ºC denaturing step for
30 sec, a 55ºC annealing step for 30 sec and a 72ºC elongation step
for 10 sec. The PCR product was digested with XbaI and
cloned into the pGL3-promoter luciferase reporter vector (Promega,
USA) to generate the vector pGL3-Pro-WT-Sp1. The miRNA-223 target
site in the pGL3-Pro-WT-AP1 vector was mutated from 5′-AACTGAC-3′
to 5′-ACCTATC-3′ to construct the mutated reporter vector,
pGL3-Pro-MT-Sp1. The products of all cloning and mutagenesis
reactions were confirmed by DNA sequencing. Endotoxin free DNA was
prepared in all cases. The miRNA-223 mimic
(5′-UGUCAGUUUGUCAAAUACCCCA-3′), the miRNA-223 inhibitor
(5′-UGGGGUAUUUGACAAACUGACA-3), and negative control miRNA
(miRNA-NC, 5′-CUCAUGUCACUGUGACAUAUAC-3′) were all chemically
synthesized.
A suspension of 293 (SIBCB, China) cells in
logarithmic phase growth was prepared and the number of viable
cells counted using a hemocytometer in conjunction with trypan blue
staining. The cells were seeded into 6-well plates at a
concentration of 2×105 cells per well and maintained in
Dulbecco's modified Eagle's medium (Invitrogen) supplemented with
10% fetal calf serum at 37ºC for 24 h in a 5% CO2
atmosphere. The transfection of plasmid DNA and RNA was performed
using Lipofectamine 2000 (Invitrogen). Transfection of cells with
pGL-TK (100 ng) (Invitrogen) served as a reference for luciferase
detection. Luciferase activity was measured using the dual
luciferase reporter assay system (Promega) 48 h after
transfection.
Generation of recombinant lentivirus for
miRNA-223 and Sp1 overexpression studies
Genome DNA was extracted from para-carcinoma tissues
and the 465-bp DNA sequence containing the miRNA-223 precursor
sequence was amplified by PCR. The forward and reverse primer
sequences were 5′-CGGAATTCGCCACCTTCTGGTGCTTTGGTTGGTC-3′ and
5′-CGGGATCCTTCCATTCCTGGACACTTTATAC-3′, respectively. EcoRI
and BamHI restriction sites were introduced into the forward
and reverse primers, respectively, to enable subsequent cloning of
the PCR product into the pcDNA lentiviral expression vector
resulting in the generation of pcDNA-miRNA-223. To clone human Sp1,
total RNA was extracted from human gastric cancer samples using
TRIzol, and reverse-transcribed into cDNA. This reaction product
was then used for the amplification of Sp1 cDNA by PCR using
primers that incorporated an EcoRI site and a Kozak sequence
(5′-GCCACC-3′) upstream, and a BamHI site downstream, of the
target sequence. Forward and reverse primers were
5′-CGGAATTCGCCACCATGAGCGACCAAGATCACTCC-3′ and
5′-CGGGATCCTCAGAAGCCATTGCCACTGAT-3′, respectively. The Sp1 PCR
product was cloned into the pcDNA lentiviral expression vector
(SystemBiosciences, USA) to generate pcDNA-Sp1. In all cases,
successful cloning was confirmed by DNA sequencing. For the
generation of lentiviral particles, expression vectors were
co-transfected into 293T producer cells (SystemBiosciences) with a
lentiviral packaging plasmid mix (SystemBiosciences) using
Lipofectamine 2000 (Invitrogen). Culture supernatants were
collected 48 h later, clarified by centrifugation, and passed
through a 0.45-μm filter (Millipore, USA). Viral titer was
evaluated by the gradient dilution method. Lentiviral particles
generated using the pcDNA-miRNA-223 and pcDNA-Sp1 vectors were
designated Lv-miRNA-223 and Lv-Sp1, respectively. Cells were
infected with lentivirus 1 day after seeding using a multiplicity
of infection (MOI) of 20.
Measurement of correlation between
miRNA-223 and Sp1 protein and effect of overexpression of miRNA-223
on Sp1 protein in gastric cancer cells
GES-1, a normal human gastric mucosal cell line and
three gastric cancer cell lines, MGC-803, BGC-823 and SGC-7901,
were maintained in DMEM+10% FBS at 37ºC and 5% CO2.
Total RNA and protein were extracted and miRNA-223 and Sp1 contents
were examined to determine whether there was abnormal expression or
correlation between miRNA-223 and Sp1. MGC-803, BGC-823 and
SGC-7901 cells were seeded to 6-well plates, incubated overnight
and infected with Lv-miRNA223 or Lv-NC. Cells were collected 72 h
after infection, and total RNA and protein were extracted and
measured for miRNA-223 and Sp1, to evaluate the effect of
overexpression of miRNA-223 on Sp1 protein level.
TGF-β1-dependent in vitro model of
gastric cancer EMT
TGF-β1 is commonly used to induce EMT in a variety
of cancer cell types (14,15). We therefore established a
TGF-β1-dependent model of EMT for gastric cancer cell lines.
MGC-803, BGC-823 and SGC-7901 cells were incubated with TGF-β1 for
72 h and subsequently the expression of EMT related markers was
evaluated to determine whether the model had been established
successfully. Cells in logarithmic phase growth were seeded into
6-well plates at a density of 2×105 cells/well, and then
treated with 100 ng/ml TGF-β1 (Pierce) for 72 h. At the same time,
the cells treated with or without TGF-β1 were infected with
Lv-miRNA-223 (MOI of 20) and then maintained for 48 h under normal
culture conditions. The cells were then collected and total RNA and
protein extracted and analyzed by real-time PCR and western
blotting to measure miRNA-223 and Sp1 protein levels,
respectively.
To determine whether EMT could successfully be
induced following TGF-β1 treatment, we examined the expression of a
group of EMT markers. SGC-7901 cells were stimulated with TGF-β1 at
a final concentration of 100 ng/ml for 72 h. The cells were
infected with recombinant lentiviruses when appropriate and
collected 48 h later for protein extraction and western blotting to
determine Vimentin, Fibronectin, N-cadherin and E-cadherin
expression. Based on this model, we carried out experiments of gene
intervention, i.e., 24 h after lentiviral infection, the cells were
incubated with TGF-β1 for 72 h and EMT protein markers were
measured. Moreover, we analyzed EMT progress by evaluating
proliferation, apoptosis and invasion to determine the effect of
miRNA-223 on TGF-β1 induced EMT in gastric cancer cells.
Cellular proliferation assay
Increased proliferation is one indicator of EMT and
so we measured changes in proliferation to determine whether
miRNA-223 overexpression could inhibit EMT. MGC-803 cells infected
with recombinant lentiviruses were stimulated with 100 ng/ml TGF-β1
for 72 h, trypsinized, and seeded into 96-well plates at a density
of 1×104 cells per well. The cells were cultured under
normal conditions and cell viability was examined using CCK-8 at
24-, 48-, and 72-h time-points. Briefly, 10 μl CCK-8 solution was
added, and the cells then cultured under normal conditions for an
additional 4 h before measurement of absorbance at 450 nm.
Apoptosis assay
Increased apoptosis is a marker of EMT inhibition.
BGC-823 cells in logarithmic phase growth were seeded into 6-well
plates at a density of 2×105 cells/well, and incubated
with 100 ng/ml TGF-β1 for 72 h. At the same time, the cells treated
with or without TGF-β1 were infected with Lv-miRNA-223 or Lv-Sp1
(MOI of 10) and then maintained for 48 h under normal conditions.
The cells were collected and apoptosis assessed using the Annexin
V-FITC Apoptosis Detection Kit II (BD Biosciences, USA). The cells
were washed twice by centrifugation in PBS (2,000 × g, 5 min),
resuspended in 500 μl binding buffer containing 5 μl of Annexin
V-FITC, and incubated for 10 min. The cells were then stained with
5 μl propidium iodide for 5 min. Apoptosis was analyzed at an
excitation wavelength of 488 nm using the FL1 channel for Annexin
V-FITC and the FL2 channel for PI.
Cell invasion assay
The EMT model was established using SGC-7902 cells
which were subsequently infected with lentiviruses. Forty-eight
hours later, the cells were seeded into Transwell chambers
(Millipore) for the analysis of cell invasion according to the
manufacturer's instructions. For quantitative analysis, we
conducted DAPI staining and α-SMA immunolabeling to fluorescently
stain nuclei and the cell body, respectively. This enabled the
number of cells that had transversed the chamber filter to be
observed and counted using epifluorescence microscopy (IX81,
Olympus, Japan). In addition, in accordance with the kit's
instructions, the cells were detached and counted after
staining.
Analysis of specificity of
miRNA-223/Sp1/EMT pathway
There may be several target genes of miRNA-223, so
we had to verify whether the Sp1 effect is critical to miRNA-223 on
EMT. We designed the following experiment: we cloned Sp1 coding
sequence to construct a recombinant vector pcDNA-Sp1 to express Sp1
in cancer cells via our lentiviral system. In contrast with
endogenous Sp1, it lacks the binding site of miRNA-223 due to no
wild-type 3′-UTR. Therefore, in the presence of exogenous Sp1, we
could determine the specificity of miRNA-223/Sp1/EMT according to
the effect of miRNA-223 on EMT indicators.
Statistical analysis
The data, expressed as means ± SD, were analyzed
using SPSS software (version 13.0; SPSS). Statistical analysis was
determined by one-way ANOVA followed by multiple comparisons with
the least significant difference (LSD). P-values <0.05 were
considered statistically significant.
Results
Analysis of Sp1 protein expression in
clinical gastric cancer tissues and associated liver metastatic
tissues
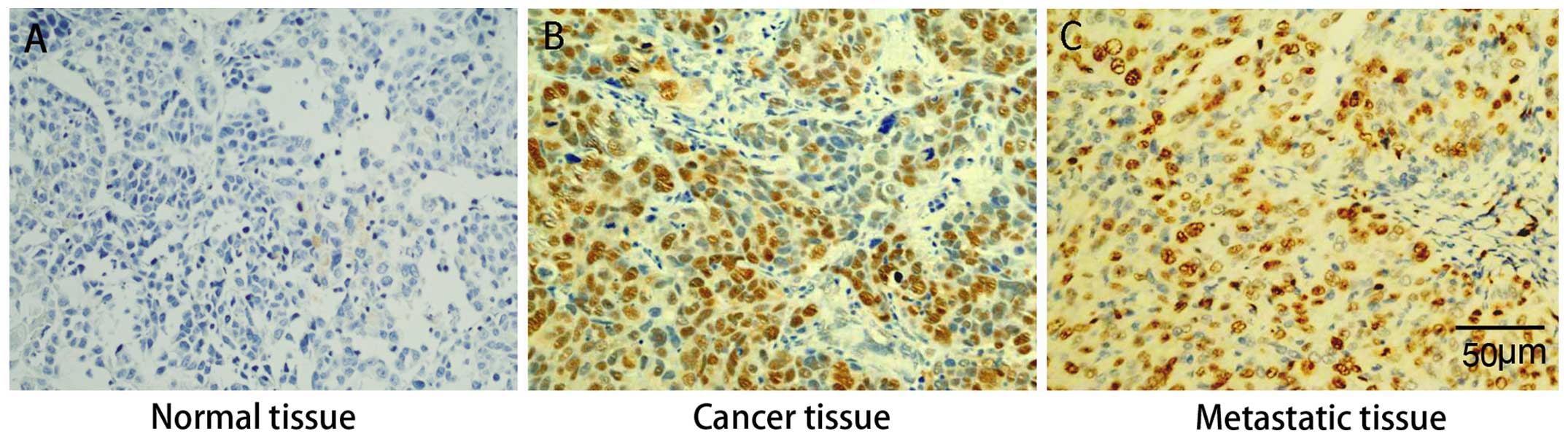
Immunohistochemical analysis revealed that Sp1
protein was expressed at higher levels in primary gastric cancer
and liver metastatic tissues than in para-carcinoma tissues. There
was no observable difference in Sp1 expression between primary
gastric cancer and liver metastatic tissues, suggesting that high
expression of Sp1 is associated with both tumorigenesis and
metastasis of gastric cancer (Fig.
1).
Detection of miRNA-223, Sp1 mRNA and
protein, EMT markers in tumor specimens
Expression of miRNA-223 in normal adjacent tissues,
gastric cancer tissues and metastatic tissues negatively correlated
with Sp1 protein levels and was lower in the gastric cancer and
metastatic tissues than in the adjacent tissues (P<0.05;
Fig. 2A). Sp1 mRNA and protein
expression data (Fig. 2B and C)
demonstrate that, in comparison with normal para-carcinoma tissues,
Sp1 protein was elevated in gastric cancer and its metastatic
tissues in the liver (P<0.05). However, no significant
differences in mRNA levels were observed between the three tissue
groups (P>0.05). These results suggest that the high expression
of Sp1 is due to an inactivation of post-transcriptional
regulation. We also examined the expression of EMT-related proteins
in these tissues, and found an increase in Vimentin, Fibronectin,
N-cadherin and E-cadherin in gastric cancers and their metastatic
tissues when compared with the adjacent tissues (P<0.05;
Fig. 2D). These results suggest
that EMT is closely associated with the invasion and metastasis of
gastric cancer cells and that Sp1 expression positively correlates
with EMT and gastric cancer spread.
 | Figure 2Expression analysis of miRNA-223, Sp1
mRNA, Sp1 protein and EMT-associated proteins in normal adjacent
tissue, gastric cancer tissue, and the tissue of liver metastases.
(A) miRNA-223 levels in normal adjacent tissue, gastric cancer
tissue, and the tissue of liver metastases. (B) Sp1 mRNA expression
levels in the same tissues, as measured by quantitative PCR. (C)
Sp1 protein levels in normal adjacent tissue, gastric cancer
tissue, and the tissue of liver metastases as determined by western
blotting. Upper panel, representative western blotting; lower
panel, quantification of relative Sp1 protein expression (the
intensity of each Sp1 protein band was normalized to that of the
corresponding β-actin control and the data then normalized to that
of the control tissue group). Data are expressed as means ± SD of
at least three independent experiments. (D) Protein levels of
Vimentin, Fibronectin, and N-cadherin in normal adjacent tissue,
gastric cancer tissue, and the tissue of liver metastases as
determined by western blotting. Upper panel, representative western
blots; lower panel, quantification of relative protein expression
(normalization of data for the comparison of treatments was
performed as in (C). Data are expressed as means ± SD of at least
three independent experiments. *P<0.05 and
**P<0.01, when compared with the normal adjacent
group. |
Prediction and verification of a
miRNA-223 binding site within the 3′-UTR of Sp1
The measurements of Sp1 protein and mRNA levels, and
the levels of miRNA-223 suggested that miRNA regulation may
underlie the increase in Sp1 expression observed in gastric cancers
and their metastases. Consequently, to further explore the
relationship between Sp1 expression, miRNA regulation and gastric
cancer dissemination via the EMT pathway, we next examined whether
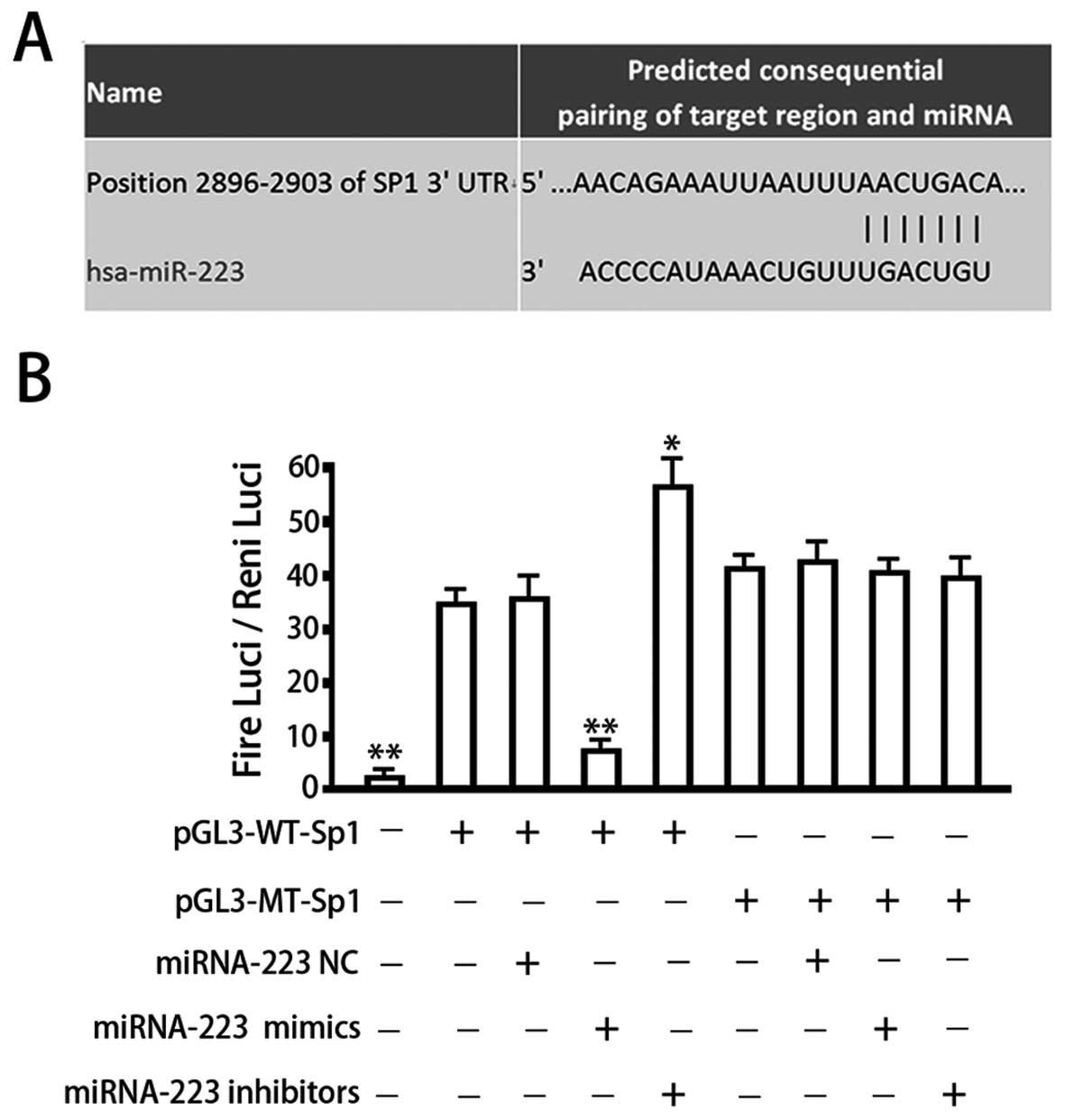
Sp1 represents one of the target genes of miRNA-223. Our
bioinformatics analysis identified a seven-base hsa-miRNA-223 seed
sequence in the 3′-UTR of Sp1 mRNA (Fig. 3A). We therefore constructed
luciferase reporter vectors to verify whether this site represents
a valid miRNA-223 target. Reporter vectors were generated that
contained the wild-type Sp1 3′-UTR or a variant in which the
miRNA-223 target site within the 3′-UTR had been mutated. Both
reporter constructs expressed luciferase at a high level (Fig. 3B). However, the miRNA-223 mimic
significantly inhibited luciferase activity in cells transfected
with the reporter vector encoding the wild-type 3′-UTR (34.25±3.69
vs. 8.11±1.24; P<0.05), while the miRNA-223 inhibitor
significantly increased luciferase activity in these cells
(34.25±3.69 vs. 55.71±6.68; P<0.05). Conversely, in cells
transfected with the reporter vector encoding the mutated miRNA-223
target site, neither the miRNA-223 mimic nor the
miRNA-223-inhibitor had any observable effect on luciferase
activity (P>0.05). Co-transfection of miRNA-143-NC
(non-targeting control) had no effect on the luciferase activity of
either of the vectors (P>0.05). These results verified the
presence of a hsa-miRNA-223 target site in the 3′-UTR of Sp1 mRNA
and demonstrated that binding of miRNA-223 to this target site
downregulates Sp1 expression.
Detection of correlation between
miRNA-223 and Sp1 protein in normal cells and gastric cancer cells
and effect of over-expression of miRNA-223 via lentiviral approach
on Sp1 expression in gastric cancer cells
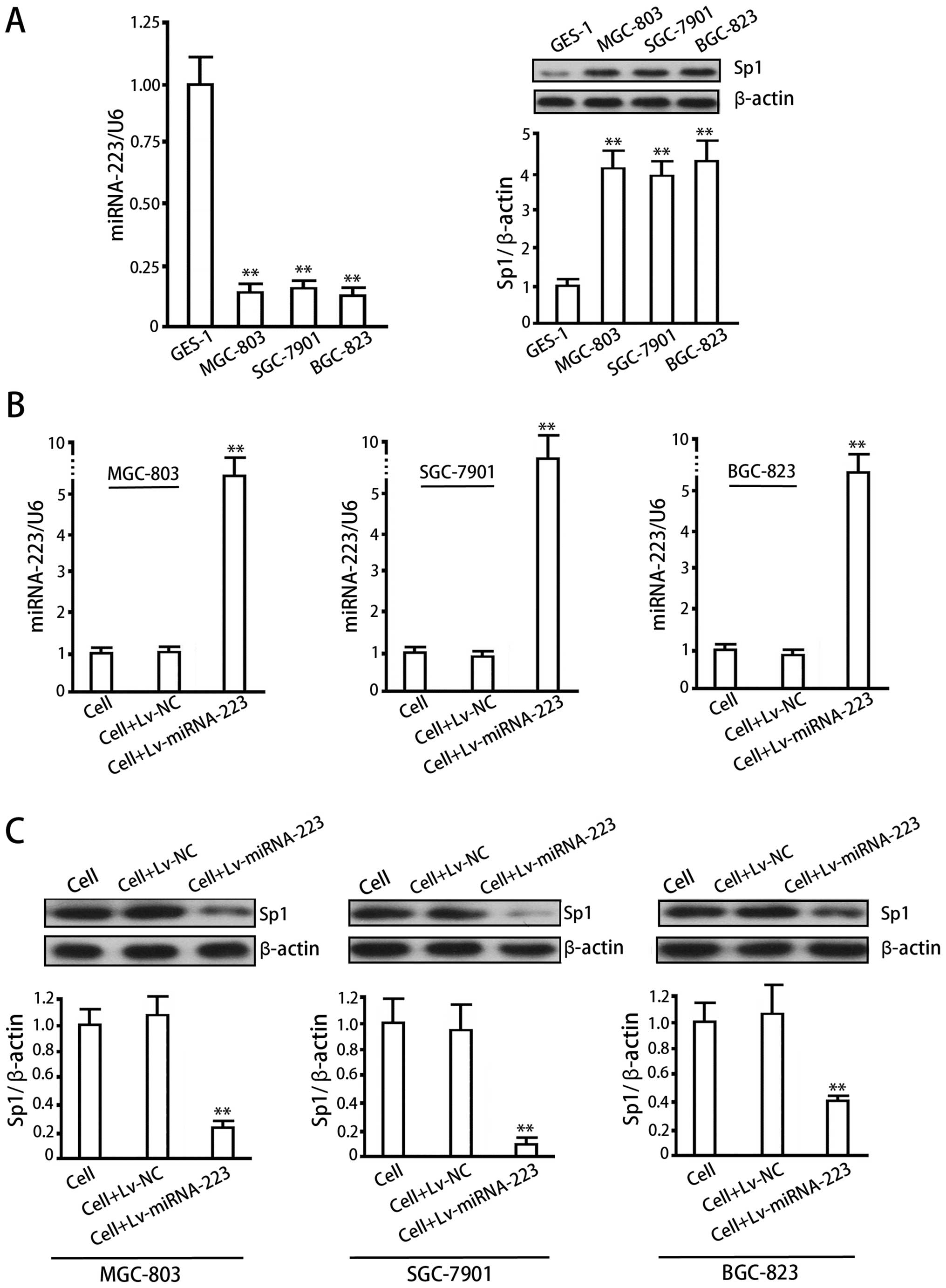
As compared with normal gastric mucosa cells,
MGC-803, BGC-823 and SGC-7901 cells showed lower expression levels
of miRNA-223 (P<0.05, vs GES-1) and higher levels of Sp1 protein
(P<0.05, vs GES-1), with a significant negative correlation
between miRNA-223 and Sp1 (Fig.
4A). The lentiviral system effectively delivered miRNA-223 in
MGC-803, BGC-823 and SGC-7901 cells (Fig. 4B) (P<0.01, vs cell control or
Lv-NC), which inhibited Sp1 protein in turn (Fig. 4C) (P<0.01, vs cell control or
Lv-NC).
Verification of EMT model established by
TGF-β1 induction in gastric cancer cells
We measured EMT markers 72 h after incubating
MGC-803 cells with 100 ng/ml TGF-β1. The results (Fig. 5A) showed that TGF-β1 significantly
increased Vimentin, Fibronectin, cadherin and E-cadherin
(P<0.05, vs control), indicating the successful establishment of
the EMT model.
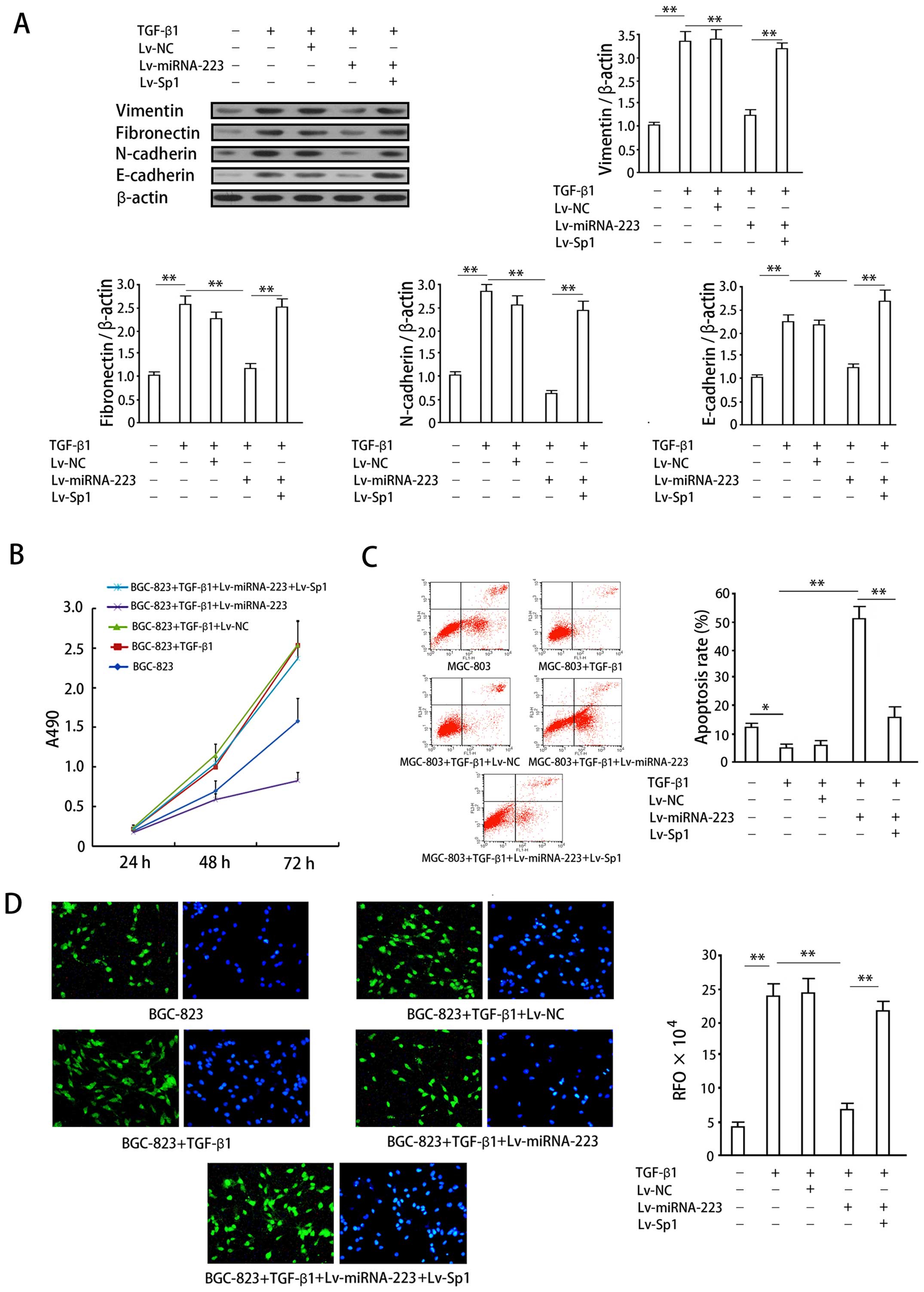 | Figure 5Effects of miRNA-223 overexpression
on cellular proliferation, apoptosis, invasion and protein
expression in an in vitro gastric cancer model of EMT. (A)
Vimentin, fibronectin, and N-cadherin protein expression in
SGC-7091 cells was determined by western blotting. Upper panel,
quantification of protein expression as determined by densitometric
analysis. The intensity of each target protein band was normalized
to that of the corresponding β-actin control band and the data then
normalized to that of the no-treatment control group. Lower panel,
representative western blots. (B) Growth curves for BGC-823 cells
exposed to the indicated treatments. Data are expressed as means ±
SD of at least three independent experiments. (C) Apoptosis in
MGC-803 cells. Left panel, representative plots of MGC-803 cell in
the presence of the indicated treatments. Right panel,
quantification of apoptosis for the indicated treatments. Data
represent means ± SD of at least three separate experiments.
*P<0.05; **P<0.01. (D) Invasion assay
of BGC-823 cells in the presence of the indicated treatments.
Epifluorescence images of migrated cells after DAPI staining (left
panel) and α-SMA immunofluorescence staining (right panel). Right
hand graph, quantification of cell invasion (RFU = relative
fluorescence units). Data represent means ± SD of relative
fluorescence units from at least three separate experiments,
**P<0.01. |
Effects of miRNA-223 overexpression on
cellular proliferation, apoptosis, and invasion in a gastric cancer
model of EMT
TGF-β1 (100 ng) significantly increased vimentin,
fibronectin, and N-cadherin protein expression in SGC-7901 gastric
cancer cell lines (P<0.05; Fig.
5A), indicating the successful induction of EMT. Overexpression
of miRNA-223 significantly impaired the increase in expression of
these proteins induced by TGF-β1 (P<0.05, when compared with the
induction group). Conversely, co-expression of exogenous Sp1
reversed the inhibition of vimentin, fibronectin, and N-cadherin
expression by miRNA-223 (P<0.05).
Stimulation with 100 ng/ml TGF-β1 increased BGC-823
cell proliferation (P<0.05, when compared with the control
group), which is an expected response associated with the induction
of EMT (Fig. 5B). Overexpression
of miRNA-223 significantly inhibited the increase in proliferation
induced by TGF-β1 (P<0.05), whereas co-expression of exogenous
Sp1 with miRNA-223 reversed this inhibitory effect (P<0.05).
A decrease in apoptosis was also observed in BGC-823
cells following 72-h stimulation with 100 ng/ml TGF-β1 (P<0.05;
Fig. 5C), which again is an
expected response associated with the induction of EMT.
Overexpression of miRNA-223 significantly increased BGC-823 cell
apoptosis (P<0.05, when compared with the group treated with
TGF-β1 alone). Co-expression of Sp1 with miRNA-223 reversed the
increase in apoptosis observed following the overexpression of
miRNA-223 alone (P<0.05).
Cell invasion assays were performed in combination
with fluorescence imaging to observe and quantify the effects of
the TGF-β1-dependent induction of EMT on the invasive properties of
gastric cancer cells. Fluorescence imaging enabled the direct
observation of cells penetrating through the matrix membrane of the
invasion chamber (Fig. 5D, left,
DAPI staining; right, α-SMA immunofluorescence). Induction of
SGC-7901 cells with 100 ng/ml TGF-β1 resulted in a significant
increase in cellular invasion (P<0.05, when compared with the
non-induced group), a finding that was in agreement with the
decrease in cellular apoptosis observed with the induction of EMT.
Overexpression of miRNA-223 significantly inhibited cellular
invasion (P<0.05), while co-expression of exogenous Sp1 with
miRNA-223 reversed this inhibitory effect (P<0.05). The data
obtained from cell counts were in agreement with those obtained
using the fluorescent plate reader.
Analysis of specificity of
miRNA-223/Sp1/EMT pathway
Based on that miRNA-223 inhibits EMT in gastric
cancer cells, we then verified the specificity of miRNA-223/Sp1/EMT
pathway. We overexpressed Sp1 protein by using the lentiviral
system, which would not be regulated by miRNA-223 due to lack of
wild 3′-UTR. The results showed that exogenous expression of Sp1
significantly reversed inhibition of EMT by miRNA-223: exogenous
Sp1 revered decrease of EMT markers (Fig. 5A), proliferation inhibition
(Fig. 5B), apoptosis (Fig. 5C), and invasion inhibition
(Fig. 5D) induced by miRNA-223 in
the EMT model. We hence considered that exogenous Sp1 counteracts
miRNA-223 in respect of EMT inhibition in gastric cancer cells,
i.e., the inhibition of EMT by miRNA-223 specifically mediated by
its target gene Sp1.
In summary we have found that TGF-β1 induction
decreased miRNA-223 in cancer cells, resulting in an increase in
Sp1 protein (a target gene of miRNA-223) and the promotion of EMT.
Overexpression of miRNA-223 effectively inhibited EMT-associated
processes, and this inhibition could be reversed by the
co-expression of exogenous Sp1. Analysis of EMT was performed by
observing changes in EMT-associated markers such as the expression
of the EMT-related proteins vimentin, fibronectin, and N-cadherin,
and behavioral processes including cellular proliferation,
apoptosis and invasion.
Discussion
Highly evolutionarily conserved, miRNAs represent
endogenous non-coding RNAs of ~20–25 bases in length that regulate
gene expression at the post-transcriptional level through binding
to the 3′-UTR of target genes (16). miRNAs have been implicated in
tumorigenesis and tumor development, and their roles in these
processes have been the subject of extensive research (17). Studies show that dysfunction of
intrinsic miRNA regulatory mechanisms results in abnormal
expression of various oncogenes and tumor suppressor genes in a
variety of tumor types (18);
therefore, miRNAs may act as both oncogenes and tumor suppressor
genes. miRNAs have been implicated in the biology of gastric cancer
(19,20): miRNA array analysis has shown that
expression of miR-21, miR-191, miR-223, miR-24, miR-107, miR-92 and
miR-221 is elevated in gastric cancer (21), while that of miR-128b, miR-129 and
miR-148 is decreased (22).
Gastric cancer is a disease involving the complex activation and
inactivation of multiple genes; miRNAs may therefore assume the
role of both oncogene and tumor suppressor in the onset and
development of this disease. With progress now being made in our
understanding of the relationship between gastric cancer and
miRNAs, the search for the target proteins of miRNAs, as well as
the molecular mechanisms regulating miRNA expression, remains an
important area of study in this field, especially considering the
potential implications such research has for the development of
gene therapies. miRNAs are also involved in the regulation of EMT:
curcumin suppresses EMT in chemo-resistant colorectal cancer by
upregulating EMT-suppressive miRNAs and mediating
chemosensitization to 5-fluorouracil (23). Moreover, miRNA-153 has been shown
to be a marker of gastric cancer metastasis associated with EMT
(24). The involvement of
miRNA-223 in gastric cancer (including EMT associated with this
disease) has not been reported previously.
The invasion and metastasis of cancer refers to the
dissemination of tumor cells from the primary tumor to discrete
target tissues, often of remote organs, where these cells then
proliferate into cancers of a similar nature (25). This process depends on the
interaction between cancer cells and the local tumor environment
which promotes their survival, growth, invasion, and ultimately
metastasis (26). EMT, which
drives many of these processes, is therefore a major factor
contributing to the invasion and metastasis of cancer cells
(27–29).
In recent years, an increasing number of studies
have shown that EMT is related not only to cancer invasion and
metastasis, but also to the acquisition of cancer stem cell
characteristics (30,31). Gastric cancer is the second most
common cancer in the world. Studies have shown that EMT is closely
related to gastric cancer metastasis (including post-operative
metastasis). Zhang et al reported that Wnt5a, through
stimulation of the non-classical Wnt signaling pathway, mediates
EMT during the metastasis of cancer cells, and may serve as a
target for the inhibition of EMT and gastric cancer metastasis
(32). Lu et al have shown
that miRNA-19a, which is associated with cancer invasion and
metastasis and is expressed highly in gastric cancer cell lines,
induces EMT in gastric cancer cells by activating the PI3K/AKT
signal pathway (33).
Sp1, a member of the SP/Kruppel-like factor
super-family (Sp/KLF family) of transcription factors (34), contains four domains: a DNA binding
domain, an Sp1 activation domain, a Btd box and an SP box (4). Sp1 initiates gene transcription by
forming a tetramer and binding the gene promoter. Once bound, the
tetramer can recruit and promote the binding of additional
tetramers to DNA, driving gene transcription via a positive
feedback loop. Sp1 is highly expressed in gastric cancer, lung
cancer, cervical cancer, breast cancer, and thyroid tumors
(35–38). Qiu et al have shown that
inhibition of Sp1 expression in gastric cancer cells suppresses
proliferation, migration and invasion (39). Zhou et al demonstrated that
high expression of Sp1 could enhance gastric cancer metastasis
(12). Previous study by Nam and
colleagues on cancer cell EMT has shown that Sp1 plays crucial
roles in the integrin α5-dependent induction of EMT and metastasis
(8). Given these findings, we
sought to determine whether Sp1 is involved in the EMT of gastric
cancer, and whether abnormal expression of Sp1 in gastric cancer
EMT, invasion and metastasis is regulated in a post-transcriptional
manner. We additionally sought to determine whether miRNAs are key
players in this regulation.
We found that Sp1 expression is closely associated
with gastric cancer metastasis, and that abnormal expression of Sp1
is regulated at a post-transcriptional level. Consequently we
explored the possibility that miRNA regulates Sp1 expression. We
identified miRNA-223 as a putative regulator of Sp1 using a
bioinformatics approach, and verified experimentally that miRNA-223
can negatively regulate Sp1 expression by targeting its 3′-UTR. Sp1
expression negatively correlated with that of miRNA-223. We
selected three gastric cancer cell lines and established a model of
gastric cell EMT using TGF-β induction. This model was used in
conjunction with miRNA-223 overexpression studies to observe
possible changes in the expression of EMT-related markers in
gastric cancer cells. Our results demonstrated that overexpression
of miRNA-223 in BGC823 cells effectively suppressed proliferation.
Moreover, high expression of miRNA-223 induced apoptosis and
inhibited the invasion of gastric cancer cells. Subsequent analysis
of EMT-related proteins verified our conclusion. We also
investigated whether miRNA-223 affects EMT through Sp1 by
expressing exogenous Sp1 in gastric cancer cells. We found that
overexpression of Sp1 impaired the effects of miRNA-223 on
EMT-related indicators suggest that there is a specific
miRNA-223/Sp1/EMT signaling axis regulating gastric cancer
metastasis.
In this study, we found that the direct cause for
TGF-β1 to induce EMT in gastric cancer cells is that it further
reduces miRNA-223 expression. It is worth additional study to
clarify how TGF-β1 inhibits miRNA-223, which will be our future
direction. A reasonable hypothesis is that TGF-β1 may change some
transcription factors responsible for miRNA-223 regulation. We,
therefore, are screening differentially expressed transcription
factors in gastric cancer and EMT model focusing on sequence
analysis of the promotor region of miRNA-223, which has made some
preliminary progress.
Our study demonstrates that low expression of
miRNA-223 in gastric cancer cells gives rise to high expression of
Sp1, and consequently the invasion and metastasis of cancer cells
through EMT. Overexpression of miRNA-223 in gastric cancer cells
inhibits expression of Sp1 protein and the invasion and metastasis
of tumor cells via EMT. These findings provide new possibilities
for gene therapy strategies designed as adjuvant postoperative
treatments for gastric cancer and for the development of drugs
targeting new genes.
Acknowledgements
This study was supported by Heilongjiang Province
Science Funds for Distinguished Young Scientists (JC2015019,
H2015079), National Science Foundation of China (81372293,
81273161), Heilongjiang Province Graduate Innovation and Creativity
Funds (YJSCX2011-321HLJ), New Century Excellent Talents in
Heilongjiang Province University (1254-NECT-023).
References
|
1
|
Sakamoto S and Ichikawa T: Mechanism of
prostate cancer invasion and metastasis. Nihon Rinsho.
72:2086–2089. 2014.(In Japanese). PubMed/NCBI
|
|
2
|
Ombrato L and Malanchi I: The EMT
universe: Space between cancer cell dissemination and metastasis
initiation. Crit Rev Oncog. 19:349–361. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu H, Ren G, Wang T, Chen Y, Gong C, Bai
Y, Wang B, Qi H, Shen J, Zhu L, et al: Aberrantly expressed Fra-1
by IL-6/STAT3 transactivation promotes colorectal cancer
aggressiveness through epithelial-mesenchymal transition.
Carcinogenesis. 36:459–468. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Safe S and Abdelrahim M: Sp transcription
factor family and its role in cancer. Eur J Cancer. 41:2438–2448.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Beishline K and Azizkhan-Clifford J: Sp1
and the ‘hallmarks of cancer’. FEBS J. 282:224–258. 2015.
View Article : Google Scholar
|
|
6
|
Li S, Wang Q, Qiang Q, Shan H, Shi M, Chen
B, Zhao S and Yuan L: Sp1-mediated transcriptional regulation of
MALAT1 plays a critical role in tumor. J Cancer Res Clin Oncol.
141:1909–1920. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sankpal UT, Goodison S, Abdelrahim M and
Basha R: Targeting Sp1 transcription factors in prostate cancer
therapy. Med Chem. 7:518–525. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nam EH, Lee Y, Park YK, Lee JW and Kim S:
ZEB2 upregulates integrin α5 expression through cooperation with
Sp1 to induce invasion during epithelial-mesenchymal transition of
human cancer cells. Carcinogenesis. 33:563–571. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karkhanis M and Park JI: Sp1 regulates
Raf/MEK/ERK-induced p21(CIP1) transcription in TP53-mutated cancer
cells. Cell Signal. 27:479–486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Katoh M: Epithelial-mesenchymal transition
in gastric cancer (Review). Int J Oncol. 27:1677–1683.
2005.PubMed/NCBI
|
|
11
|
Jiang W, Jin Z, Zhou F, Cui J and Wang L
and Wang L: High co-expression of Sp1 and HER-2 is correlated with
poor prognosis of gastric cancer patients. Surg Oncol. 24:220–225.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou C, Ji J, Cai Q, Shi M, Chen X, Yu Y,
Liu B, Zhu Z and Zhang J: MTA2 promotes gastric cancer cells
invasion and is transcriptionally regulated by Sp1. Mol Cancer.
12:1022013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo MM, Hu LH, Wang YQ, Chen P, Huang JG,
Lu N, He JH and Liao CG: miR-22 is down-regulated in gastric
cancer, and its overexpression inhibits cell migration and invasion
via targeting transcription factor Sp1. Med Oncol. 30:5422013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jungert K, Buck A, von Wichert G, Adler G,
König A, Buchholz M, Gress TM and Ellenrieder V: Sp1 is required
for transforming growth factor-beta-induced mesenchymal transition
and migration in pancreatic cancer cells. Cancer Res. 67:1563–1570.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jia L, Ma X, Gui B, Ge H, Wang L, Ou Y,
Tian L, Chen Z, Duan Z, Han J, et al: Sorafenib ameliorates renal
fibrosis through inhibition of TGF-β-induced epithelial-mesenchymal
transition. PLoS One. 10:e01177572015. View Article : Google Scholar
|
|
16
|
Chen XH, Liu ZC, Zhang G, Wei W, Wang XX,
Wang H, Ke HP, Zhang F, Wang HS, Cai SH, et al: TGF-β and EGF
induced HLA-I downregulation is associated with
epithelial-mesenchymal transition (EMT) through upregulation of
snail in prostate cancer cells. Mol Immunol. 65:34–42. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li PF, Chen SC, Xia T, Jiang XM, Shao YF,
Xiao BX and Guo JM: Non-coding RNAs and gastric cancer. World J
Gastroenterol. 20:5411–5419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang M, Shan X, Zhou X, Qiu T, Zhu W, Ding
Y, Shu Y and Liu P: miR-1271 regulates cisplatin resistance of
human gastric cancer cell lines by targeting IGF1R, IRS1, mTOR, and
BCL2. Anticancer Agents Med Chem. 14:884–891. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jin Z, Jiang W and Wang L: Biomarkers for
gastric cancer: Progression in early diagnosis and prognosis
(Review). Oncol Lett. 9:1502–1508. 2015.PubMed/NCBI
|
|
21
|
Hu J, Fang Y, Cao Y, Qin R and Chen Q:
miR-449a regulates proliferation and chemosensitivity to cisplatin
by targeting cyclin D1 and BCL2 in SGC7901 cells. Dig Dis Sci.
59:336–345. 2014. View Article : Google Scholar
|
|
22
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Toden S, Okugawa Y, Jascur T, Wodarz D,
Komarova NL, Buhrmann C, Shakibaei M, Boland CR and Goel A:
Curcumin mediates chemosensitization to 5-fluorouracil through
miRNA-induced suppression of epithelial-to-mesenchymal transition
in chemoresistant colorectal cancer. Carcinogenesis. 36:355–367.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Z, Sun J, Bai Z, Li H, He S, Chen R
and Che X: MicroRNA-153 acts as a prognostic marker in gastric
cancer and its role in cell migration and invasion. Onco Targets
Ther. 8:357–364. 2015.PubMed/NCBI
|
|
25
|
Kulesa PM and McLennan R: Neural crest
migration: Trailblazing ahead. F1000Prime Rep. 7:022015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Maritzen T, Schachtner H and Legler DF: On
the move: Endocytic trafficking in cell migration. Cell Mol Life
Sci. 72:2119–2134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Deng B, Zhang S, Miao Y, Zhang Y, Wen F
and Guo K: Down-regulation of Frizzled-7 expression inhibits
migration, invasion, and epithelial-mesenchymal transition of
cervical cancer cell lines. Med Oncol. 32:1022015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Greening DW, Gopal SK, Mathias RA, Liu L,
Sheng J, Zhu HJ and Simpson RJ: Emerging roles of exosomes during
epithelial-mesenchymal transition and cancer progression. Semin
Cell Dev Biol. 40:60–71. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuan W, Yuan Y, Zhang T and Wu S: Role of
Bmi-1 in regulation of ionizing irradiation-induced
epithelial-mesenchymal transition and migration of breast cancer
cells. PLoS One. 10:e01187992015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Choi YJ, Kim N, Chang H, Lee HS, Park SM,
Park JH, Shin CM, Kim JM, Kim JS, Lee DH, et al: Helicobacter
pylori-induced epithelial-mesenchymal transition, a potential role
of gastric cancer initiation and an emergence of stem cells.
Carcinogenesis. 36:553–563. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lin J, Liu X and Ding D: Evidence for
epithelial-mesenchymal transition in cancer stem-like cells derived
from carcinoma cell lines of the cervix uteri. Int J Clin Exp
Pathol. 8:847–855. 2015.PubMed/NCBI
|
|
32
|
Zhang Y, Du J, Zheng J, Liu J, Xu R, Shen
T, Zhu Y, Chang J, Wang H, Zhang Z, et al: EGF-reduced Wnt5a
transcription induces epithelial-mesenchymal transition via
Arf6-ERK signaling in gastric cancer cells. Oncotarget.
6:7244–7261. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu W, Xu Z, Zhang M and Zuo Y: MiR-19a
promotes epithelial-mesenchymal transition through PI3K/AKT pathway
in gastric cancer. Int J Clin Exp Pathol. 7:7286–7296.
2014.PubMed/NCBI
|
|
34
|
Black AR, Black JD and Azizkhan-Clifford
J: Sp1 and krüppel-like factor family of transcription factors in
cell growth regulation and cancer. J Cell Physiol. 188:143–160.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Choi ES, Nam JS, Jung JY, Cho NP and Cho
SD: Modulation of specificity protein 1 by mithramycin A as a novel
therapeutic strategy for cervical cancer. Sci Rep. 4:71622014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen
WM, Huang MD and Shu YQ: SP1-induced upregulation of the long
noncoding RNA TINCR regulates cell proliferation and apoptosis by
affecting KLF2 mRNA stability in gastric cancer. Oncogene.
34:5648–5661. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yao Y, Hu J, Shen Z, Yao R, Liu S, Li Y,
Cong H, Wang X, Qiu W and Yue L: MiR-200b expression in breast
cancer: A prognostic marker and act on cell proliferation and
apoptosis by targeting Sp1. J Cell Mol Med. 19:760–769. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao S, Wu J, Zheng F, Tang Q, Yang L, Li
L, Wu W and Hann SS: β-elemene inhibited expression of DNA
methyltransferase 1 through activation of ERK1/2 and AMPKα
signalling pathways in human lung cancer cells: The role of Sp1. J
Cell Mol Med. 19:630–641. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Qiu T, Zhou X, Wang J, Du Y, Xu J, Huang
Z, Zhu W, Shu Y and Liu P: MiR-145, miR-133a and miR-133b inhibit
proliferation, migration, invasion and cell cycle progression via
targeting transcription factor Sp1 in gastric cancer. FEBS Lett.
588:1168–1177. 2014. View Article : Google Scholar : PubMed/NCBI
|