Introduction
Recent studies have shown that a very small
percentage of cells (0.1–1%) present in a variety of solid tumors
possess characteristics similar to stem cells, such stem-like tumor
cells have been designated cancer stem cells (CSCs) by cancer
researchers. CSCs are a critical factor in the sensitivity of
tumors to treatments and tumor recurrence (1). Glioma is the most common type of
primary central nervous system (CNS) tumor. The isolation and
identification of glioma stem cells are achieved mainly by virtue
of functional distinctions and molecular markers. Currently
recognized markers specific for glioma stem cells include CD133,
integrin α6, CD171, CD15 and nestin (2). Among these markers, CD133 is the most
widely recognized marker for glioma stem cells. The subset of
glioma cells expressing high levels of CD133 can be isolated using
flow cytometry-based cell sorting techniques and serially passaged
in a serum-free culture system, allowing investigation of the
functional status of these cells in vitro and the
development of therapeutic strategies specifically targeting such
cells (3).
Long non-coding RNAs (lncRNAs) are a class of
non-coding RNA transcripts with a length of >200 nucleotides.
They regulate gene expression at various levels in the form of RNA
(4). HOX transcript antisense RNA
(HOTAIR) was the first trans-acting lncRNA gene to be discovered.
The expression of HOTAIR is upregulated in a variety of tumor
cells, which is related to poor prognosis of the tumors (5,6). The
5′ terminus of HOTAIR binds to chromatin-modifying complexes such
as polycomb repressive complex 2 (PRC2) which mediates the
trimethylation of lysine 27 on histone H3 (H3K27), thereby
silencing the transcription of specific genes. EZH2 plays a major
role in the trimethylation of H3K27 (5). The 3′ terminus of HOTAIR binds to the
histone deacetylase 1 (HDAC1)/lysine-specific demethylase 1 (LSD1)/
REST corepressor 1 (CoREST)/RE1-silencing transcription factor
(REST) repressor complex. The complex mediates the demethylation of
dimethylated histone H3 lysine 4 (H3K4me2), thereby silencing the
transcription of target genes (7,8). In
a previous study, we showed that interference with HOTAIR
expression inhibits the proliferation and in vitro
tumorigenicity of renal cancer cells (9). Research has also demonstrated that
interference with HOTAIR inhibits the cell cycle progression and
invasive capability of glioma cells; however, adenoviral vectors
were used in that study, which lack clinical applicability
(10). In this study, a non-viral
carrier was used, which further allowed the possibility of
employing HOTAIR as a molecular target to achieve effective
treatment in vivo.
Magnetic nanomaterials are a class of nanomaterials
that generally refers to materials with a grain size of <100 nm,
consisting of iron, cobalt, nickel and their alloys. Among a series
of magnetic nanoparticles, iron oxide nanoparticles have been
widely studied due to their high saturation magnetization value,
low toxicity, easily obtainable raw material and high surface
reactivity. In recent years, iron oxide nanoparticles have received
increasing attention as gene carriers. When the size of a magnetic
nanoparticle is <20 nm, it often exhibits superparamagnetic
properties. Superparamagnetic iron oxide nanoparticles (SPIONs)
have currently become a hotspot of gene carrier research because of
their controllable properties, high stability and susceptibility to
modification. After binding to plasmid DNA or small interfering RNA
(siRNA), SPIONs deliver the bound nucleic acids into mammalian
cells under the influence of an external magnetic field. SPIONs
overcome the intracellular and extracellular barriers impeding gene
delivery via magnetic absorption, thereby increasing the local DNA
concentration and improving transfection efficiency (11–13).
Based on the findings described above, a HOTAIR
siRNA sequence was constructed in this study. In addition, SPIONs
were used to mediate the transfection and expression of siRNAs
targeting HOTAIR in CD133+ GSCs. The goal of this study
was to evaluate whether si-HOTAIR is capable of reducing the
proliferation, invasion and tumorigenicity of GSCs through
targeting the expression of the programmed cell death 4
(PDCD4).
Materials and methods
Isolation and culture of
CD133+ human glioma stem cells
CD133+ human glioma stem cells were
isolated from the human glioma cell line SHG44 (Shanghai Institutes
for Biological Sciences, Chinese Academy of Sciences). Briefly,
after digestion with 0.25% trypsin, the detached SHG44 cells were
collected via centrifugation and then resuspended in 0.5 ml of
ice-cold sterile phosphate-buffered saline (PBS, Hyclone).
Subsequently, 5 μl of a fluorescein isothiocyanate
(FITC)-conjugated anti-human CD133 antibody (eBioscience) was added
to the cell suspension to achieve a final concentration of 0.01
mg/ml. The cells were then incubated for 30 min at 4°C in the dark.
Upon the completion of antigen-antibody interactions, the cells
were washed twice with ice-cold PBS. Then, the isolation and
enrichment of CD133+ human glioma stem cells were
achieved using a flow cytometer (BD FACS Aria, BD Bioscience, CA,
USA). CD133+ human glioma stem cells were subsequently
seeded at a density of 1,000 cells/ml in Dulbecco's modified
Eagle's medium (DMEM)-F12 medium (Hyclone) containing 10 ng/ml
basic fibroblast growth factor (bFGF), 10 ng/ml epidermal growth
factor (EGF), 5 μg/ml insulin and 0.5% bovine serum albumin (BSA)
(all from Sigma-Aldrich, St. Louis, MO, USA) to generate
non-adherent spherical clusters. The cells were cultured until
passage 3.
SPION-induced transfection of microRNA
plasmid DNA into cells
SPIONs were purchased from Novobio (Novobio
Biotechnology Co., Ltd., Shanghai, China). According to the
procedure specified in the instruction manual and the methods
described in previous studies, 5 μl of SPIONs (0.2 mM) was mixed
thoroughly with 5 μl of a HOTAIR-siRNA or mock-siRNA plasmid (10
μM). The mixture was then vortexed for 10 sec and allowed to stand
at room temperature (RT) for 20 min. Subsequently, the SPION-siRNA
plasmid DNA conjugates (10 μl) were mixed with 90 μl of serum-free
DMEM and added to the cells (1×104 cells/ml), and the
cells were cultured in the presence of the SPION-siRNA plasmid DNA
conjugates for 72 h at 37°C in an atmosphere of 5%
CO2.
CCK-8 assay
Briefly, various groups of cells were seeded into
96-well cell culture plates at a density of 2×103
cells/ml. At 72 h after transfection, 10 μl of CCK-8 solution
(Sigma-Aldrich Chemical) was added to each group of cells. The
cells were then incubated at 37°C for 3 h, after which the cell
culture plates were placed on a microplate reader, and the
absorbance at a wavelength of 450 nm was recorded. The cell
proliferation inhibition rate (%) was calculated using the
following formula: (1 - OD value of cells in the experimental group
- blank / OD value of cells in the control group - blank) x
100%.
RNA extraction and analysis via
quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from each group of cells
using the TRIzol reagent (Invitrogen) in accordance with the
manufacturer's instructions. Total RNA was treated with DNAse I
(Sigma-Aldrich), quantified and then reverse transcribed into
complementary DNA (cDNA) using the ReverTra Ace-α First-Strand cDNA
Synthesis kit (Toyobo). qRT-PCR was performed on the RealPlex4
real-time PCR detection system (Eppendorf Co. Ltd., Germany), and
SYBR Green Real-Time PCR Master Mix (Toyobo) was used as the
fluorescent dye to label the amplified nucleic acids. qRT-PCR
amplification was performed over 40 cycles with denaturation at
95°C for 15 sec annealing at 58°C for 45 sec. Target cDNA was
quantified using the relative quantification method. A comparative
threshold cycle (Ct) was used to determine gene expression relative
to a control (calibrator) and steady-state mRNA levels are reported
as an n-fold difference relative to the calibrator. For each
sample, the maker genes Ct values were normalized using the formula
ΔCt = Ct_markers - Ct_18 sRNA. To determine the relative expression
levels, the following formula was used ΔΔCt = ΔCt_si-HOTAIR -
ΔCt_Mock. The values used to plot relative expressions of markers
were calculated using the expression 2-ΔΔCt. The mRNA levels were
calibrated based on levels of 18 sec RNA. The cDNA of each gene was
amplified with primers as previously described (Table I).
 | Table IThe sequences of qRT-PCR primers. |
Table I
The sequences of qRT-PCR primers.
| Gene product | Forward (F) and
reverse (R) primers |
|---|
| 18s rRNA | F:
CAGCCACCCGAGATTGAGCA
R: TAGTAGCGACGGGCGGTGTG |
| Cd44 | F:
GACAAGTTTTGGTGGCACG
R: CACGTGGAATACACCTGCAA |
| Cd133 | F:
CCATTGGCATTCTCTTTGAA
R: TTTGGATTCATATGCCTTCTGT |
| Nestin | F:
AAGATGTCCCTCAGCCTGG
R: GAGGGAAGTCTTGGAGCCAC |
| Bdnf | F:
GTCTCTGGGGATGCAGAG
R: AGCCTTCATGCAACCAAAGT |
| Pdcd4 | F:
GTGCAAGCGAAATTAAGGGA
R: TTCATCACCGGAAAAGAGAGA |
| Ccnd1 | F:
TCCTCTCCAAAATGCCAGAG
R: GGCGGATTGGAAATGAACTT |
| Cdk4 | F:
TGCAGTCCACATATGCAACA
R: GTCGGCTTCAGAGTTTCCAC |
PI stain and FCM assay
PI staining and flow cytometric (FCM) assays were
performed according to procedures that have been described
previously (14,15). Briefly, cells (5×105
cells/ml) were harvested and fixed in 1 ml of ice-cold 70% ethanol
for 48 h. After fixation, the cells were centrifuged at 1,500 r/min
and 4°C for 5 min, and the cell pellets were collected.
Subsequently, the cells were incubated in PI staining solution
(Sigma Chemicals) for 30 min at 4°C in the dark. After PI staining,
the cell cycle distribution was examined in each group of cells via
flow cytometry (BD FACSAria), and the results were analyzed using
CellQuest software.
Transwell migration assay
Briefly, cells were seeded into the upper chambers
of Transwell plates (membrane pore size: 8.0 μm) at a density of
2×103 cells/ml in 200 μl of serum-free cell culture
medium. A 600-μl volume of complete medium containing 10% fetal
bovine serum (FBS) was added to the lower chambers of the Transwell
plates. The cells were then cultured for 48 h at 37°C in an
atmosphere of 5% CO2. Subsequently, cells attached to
the lower surface of membrane were fixed with 4% paraformaldehyde
at RT for 30 min and stained with 4,6-diamidino-2-phenylindole
(DAPI, Sigma-Aldrich Chemical) for 10 min. Three non-overlapping
fields of view were selected under a microscope, and the total
number of cells was counted.
Northern blotting
Northern blot analysis was performed according to a
procedure described previously (14,15).
Briefly, total RNA was extracted from all groups of cells using the
TRIzol kit and then analyzed and quantified. Next, 20 μg of
high-quality total RNA was subjected to gel electrophoresis on a
7.5 M urea-12% formaldehyde [polyacrylamide (PAA)] denaturing gel.
After electrophoresis, the RNA was transferred to Hybond
N+ nylon membranes (Amersham, Freiburg, Germany) and
then cross-linked to the membranes via exposure to ultraviolet (UV)
radiation for 30 sec (UV dose:1,200 mJ/cm2). To examine
the expression status of miR-374a, a labeled antisense DNA probe
against miR-374a was allowed to hybridize with the immobilized RNA.
After hybridization and extensive washing of the membranes, the
membranes were exposed to Kodak XAR-5 film (Sigma-Aldrich Chemical)
for 20–40 h. As a positive control, the human U6 small nuclear RNA
(snRNA) probe (5′-GCAGGGGCCATGCTAATCTTCTCTGTATCG-3′) was hybridized
to the RNA on the membranes. The exposure time for the U6 snRNA
probe was controlled between 15 and 30 min.
Western blotting
Western blot analysis was performed according to a
procedure described previously (14,15).
Briefly, total protein was extracted from each group of cells and
subjected to sodium dodecyl sulfate polyacrylamide gel
electrophoresis (SDS-PAGE) on a 12% denaturing gel. After
electrophoresis, total protein was transferred to a polyvinylidene
difluoride (PVDF) membrane (Millipore). The membrane was blocked,
washed and then incubated with primary antibodies at 37°C for 45
min (Table II). After extensive
washing, the membrane was incubated with secondary antibodies at
37°C for 45 min, followed by washing 4 times for 14 min each with
Tris-buffered saline-Tween-20 (TBST) at RT. The blot was developed
with the enhanced chemiluminescence (ECL) kit (Pierce
Biotechnology) and film exposed to light (Sigma-Aldrich Chemical)
for visualization.
 | Table IIPrimary antibodies, their source and
dilutions. |
Table II
Primary antibodies, their source and
dilutions.
| Antibodies | Companies | Applications |
|---|
| Rabbit anti-human
PDCD4 (#9535) | Cell Signaling
Technology, Inc. (Danvers, MA, USA) | IF
(1:100)
WB (1:1,000) |
| Rabbit anti-human
BDNF (#3897) | Cell Signaling
Technology, Inc. (Danvers, MA, USA) | IF
(1:100)
WB (1:1,000) |
| Rabbit anti-human
CCND1 (#2978) | Cell Signaling
Technology, Inc. (Danvers, MA, USA) | WB (1:1,000) |
| Rabbit anti-human
CDK4 (#12790) | Cell Signaling
Technology, Inc. (Danvers, MA, USA) | WB (1:1,000) |
| Rabbit anti-human
Ki67 (#9129) | Cell Signaling
Technology, Inc. (Danvers, MA, USA) | IF (1:100) |
| Rabbit anti-human
H3K4Me2 (#9725) | Cell Signaling
Technology, Inc. (Danvers, MA, USA) | IP (1:100) |
| Rabbit anti-human
H3K4Me3 (#9751) | Cell Signaling
Technology, Inc. (Danvers, MA, USA) | IP (1:100) |
| Rabbit anti-human
EZH2 (#5246) | Cell Signaling
Technology, Inc. (Danvers, MA, USA) | IP (1:100) |
| Rabbit anti-human
LSD1 (#2184) | Cell Signaling
Technology, Inc. (Danvers, MA, USA) | IP (1:100) |
| Rabbit anti-human
GAPDH (#5174) | Cell Signaling
Technology, Inc. (Danvers, MA, USA) | WB (1:1,000) |
CHIP
The antibodies used in the experiments included an
anti-trimethylated H3K27 antibody, anti-dimethylated H3K4 antibody
and normal rabbit IgG (negative control). Briefly, the cells were
fixed with 1% paraformaldehyde at 37°C for 30 min and then
incubated with 125 mM glycine at RT for 10 min to terminate the
cross-linking reaction. Subsequently, the cells were sonicated on
ice until the DNA had been sheared into chromatin fragments of
~200–1,000 bp in size. The DNA fragments were then incubated with
primary antibodies at 4°C overnight. Immunoprecipitates were
ultimately obtained via adsorption to proteinA/G plus-agarose beads
and subjected to PCR amplification. The PCR conditions were as
follows: denaturation at 95°C for 30 sec; annealing at 55°C for 30
sec; elongation at 72°C for 30 sec; and a total of 33 amplification
cycles. The amplification products were examined via agarose gel
electrophoresis.
In vivo xenograft experiments
The experiments were conducted according to a
procedure described previously (14,15).
Briefly, various groups of cells were transfected with different
plasmids. Logarithmically growing cells (1×105 cells/ml)
were collected and inoculated subcutaneously into
BALB/Cnu/nu mice. Each experimental group contained 4
mice (6–8-week-old male BALB/Cnu/nu mice, provided by
the Experimental Animal Center of Fudan University). Animal
experiments were performed in accordance with the National
Institutes of Health Guide for the Care and Use of Laboratory
Animals (NIH publication no. 80–23) and approved by the ethics
committee for the use of experimental animals in Fudan University
and every attempt was made to limit animal numbers and suffering.
After 8 weeks of continuous monitoring, the mice were sacrificed,
and their tumors were removed. The tumors were subsequently
weighed, and the tumor volume was calculated according to the
following formula: tumor volume (mm3) =
(ab2)/2 [a, the longest axis (mm); b, the shortest axis
(mm)].
Histopathology assay
Briefly, fresh tissues were fixed via immersion in
4% paraformaldehyde (Sigma-Aldrich) at RT for 30 min. The tissues
were then subjected to gradient ethanol dehydration, embedded in
paraffin, sectioned (thickness, 6 μm), and dewaxed in xylene.
Tissue sections were stained with hematoxylin and eosin (H&E,
Sigma-Aldrich), permeabilized with xylene (Sigma-Aldrich) and
mounted in neutral resin (Sigma-Aldrich).
Immunohistochemical staining assay
Briefly, fresh tissues were fixed via immersion in
4% paraformaldehyde (Sigma-Aldrich) at RT for 30 min. The tissues
were then subjected to gradient ethanol dehydration, embedded in
paraffin, sectioned (thickness, 6 μm), and dewaxed in xylene. Next,
the tissue sections were blocked at 37°C for 30 min in
immunohistochemistry (IHC) blocking solution (Beyotime
Biotechnology Co., Ltd., Zhejiang, China). After removal of the
blocking solution, the tissue sections were washed 3 times at RT
for 5 min each using IHC wash buffer (Beyotime Biotechnology Co.,
Ltd.). Subsequently, the tissue sections were incubated with
primary antibodies (Table II) at
37°C for 45 min. After removal of the primary antibodies, the
tissue sections were washed 3 times at RT for 5 min each using IHC
wash buffer (Beyotime Biotechnology Co., Ltd.). The tissue sections
were then incubated with secondary antibodies (Table I) at 37°C for 45 min. The secondary
antibodies were subsequently discarded, and the tissue sections
were washed 3 additional times at RT for 5 min each using IHC wash
buffer (Beyotime Biotechnology Co., Ltd.). Finally, the tissue
sections were mounted using neutral resin (Sigma-Aldrich) or
fluorescence mounting medium (Sigma-Aldrich).
Transmission electron microscopy (TEM)
analysis
Tissue samples were fixed and embedded according to
the procedure described in a previous study. The tissue samples
were first fixed with 1% glutaraldehyde (Sigma-Aldrich) for 4 h and
then with 1% osmium tetroxide (Sigma-Aldrich) for 1 h.
Subsequently, the tissue samples were dehydrated in acetone and
embedded in resin 12 (Ted Pella, USA). Ultrathin sections of the
embedded tissues (cross-sectional thickness, 70 nm) were attached
to a copper grid, stained with 1% uranyl acetate (Sigma-Aldrich)
and 1% lead citrate (Sigma-Aldrich), and imaged using a JEM-1230
transmission electron microscope (Jeol, Japan).
Statistical analysis
Each experiment was performed as least three times,
and data are presented as the mean ± SE where applicable.
Differences were evaluated using Student's t-test, and a
probability of <0.05 was considered to be statistically
significant.
Results
CD133+ human glioma stem cells
express a high level of HOTAIR
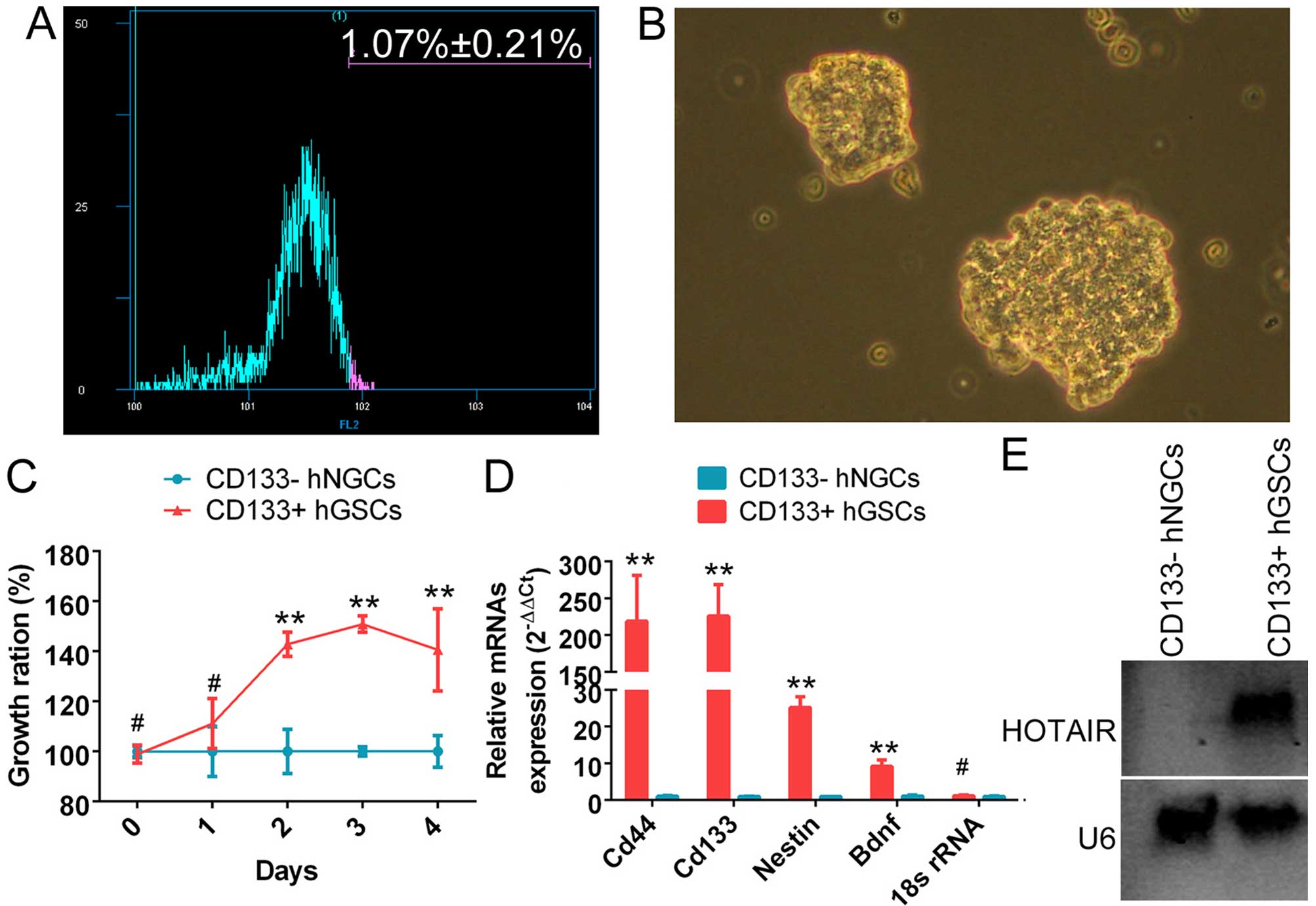
The CD133+ cell subset (1.07±0.21%) was
isolated using a flow cytometry-based cell sorting technique
(Fig. 1A). It was observed that
the CD133+ cells grew in suspension. The cells were
densely packed, and intercellular spaces were not clearly
detectable. The colonies of CD133+ cells appeared full
and highly refractive (Fig. 1B).
Moreover, the CD133+ cell subset exhibited a
significantly higher proliferation rate. It reached the maximum
growth rate after ~4 days in suspension culture (Fig. 1C). qRT-PCR revealed that the mRNA
levels of CD133, CD44, nestin and brain-derived neurotrophic factor
(Bdnf) were significantly higher in the subset of CD133+
cells compared with the control group (Fig. 1D). The above experiments
demonstrated that the subset of CD133+ cells was indeed
glioma stem cells. Northern blot analysis results revealed a
significantly stronger endogenous HOTAIR hybridization signal in
CD133+ human glioma stem cells compared with normal
CD133− glioma cells (Fig.
1E).
SPION-mediated si-HOTAIR transfection of
human glioma stem cells inhibits cell proliferation and
invasion
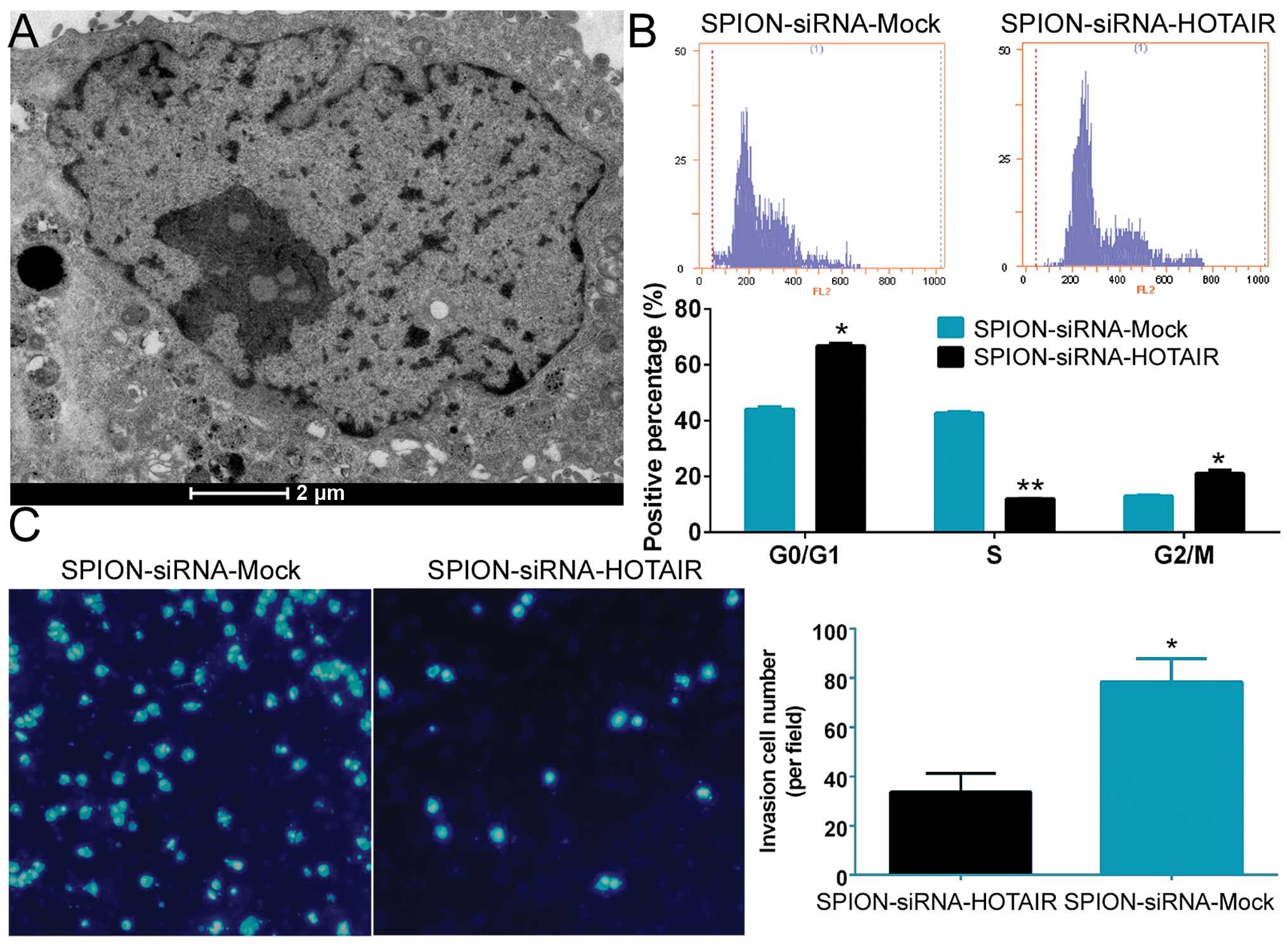
Si-HOTAIR was added to cultured CD133+
human glioma stem cells after being crosslinked to SPION. The
glioma stem cells were observed under a projection electron
microscope at 72 h. Multiple circular-shaped, dense electron clouds
of ~80–100 nm in size were detected in both the cytoplasm and the
nucleus of the glioma stem cells, indicating that SPION
successfully entered the cells (Fig.
2A). Flow cytometric analysis of the cell cycle showed that
SPION-mediated si-HOTAIR transfection markedly reduced the number
of human glioma stem cells in the S phase (12.06±0.08%) of the cell
cycle, while significantly increasing the number of cells in both
the G0/G1 phase (66.78±0.96%) and the G2/M phase (21.17±1.04%,
Fig. 2B). In addition, Transwell
assays found that the number of cells invading through the
Transwell insert membrane was significantly lower in the group that
was nanomagnetically transfected with si-HOTAIR compared with the
group subjected to SPION-mediated transfection of the si-mock
control (Fig. 2C). The above
results demonstrated that SPION effectively mediated the entry of
exogenous si-RNA sequences into the cells and that interference
with HOTAIR expression effectively suppressed the proliferation and
invasion of CD133+ human glioma stem cells in
vitro.
Si-HOTAIR-mediated interference with
HOTAIR expression in human glioma stem cells promotes the
expression of PDCD4 and inhibits the expression of cell cycle
regulatory proteins
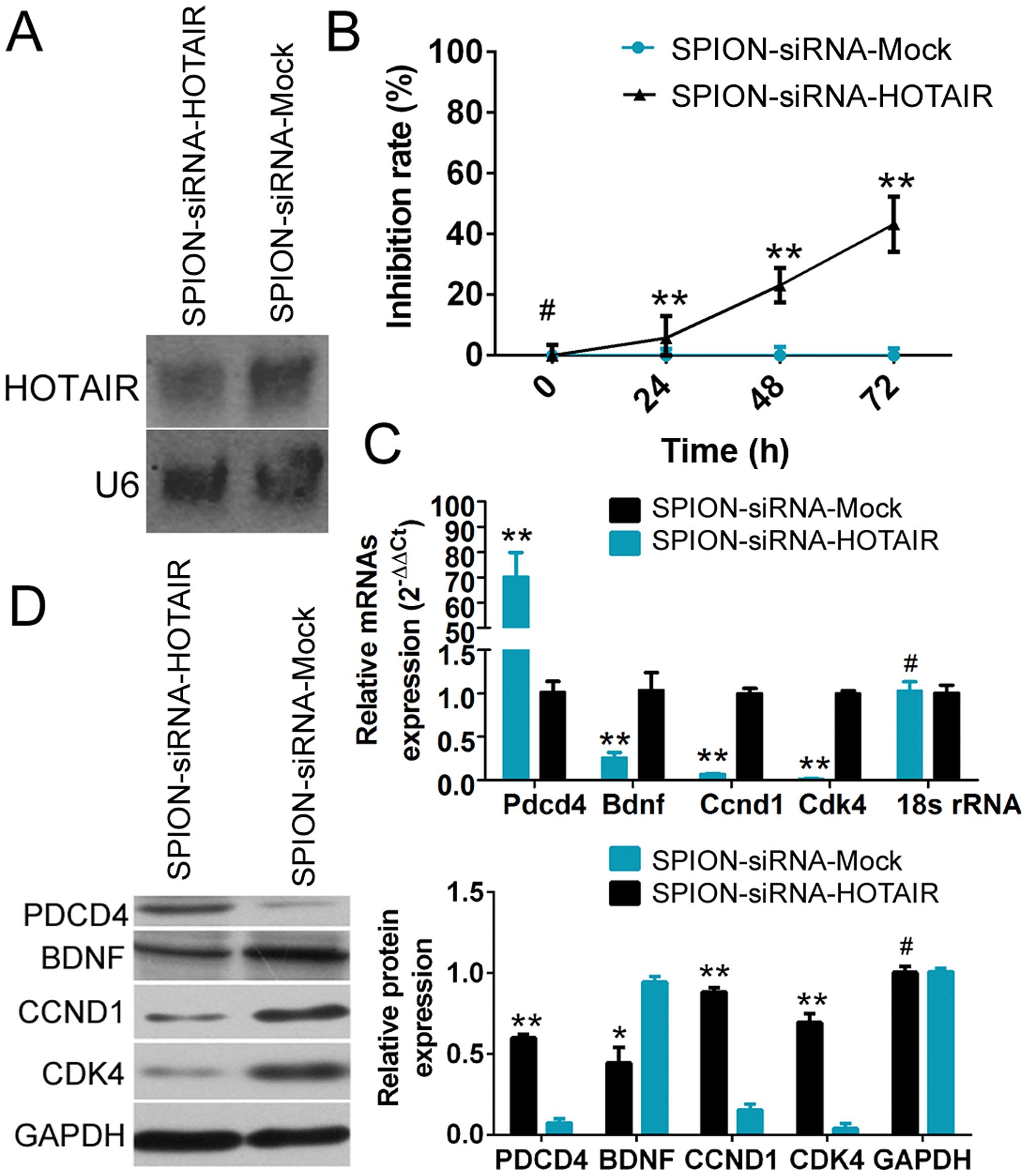
Northern blot analysis showed that the HOTAIR probe
yielded a significantly weaker hybridization signal in
si-HOTAIR-transfected human glioma stem cells compared with the
si-mock-transfected group (Fig.
3A). The CCK-8 cell proliferation assay demonstrated that
SPION-mediated low HOTAIR expression in CD133+ human
glioma stem cells effectively enhanced the rate of inhibition of
cell proliferation in vitro (Fig.
3B). The qRT-PCR results showed that compared with the control
group, the expression of PDCD4 was significantly increased, while
the expression of BDNF (which is related to cell proliferation) and
the cell cycle regulatory factors cyclin D1 (CCND1) and
cyclin-dependent kinase 4 (CDK4) was markedly reduced in
SPION-si-HOTAIR-transfected glioma stem cells expressing a low
level of HOTAIR (Fig. 3C).
Moreover, the results of western blot analysis also demonstrated
that compared with the control group, a low level of HOTAIR
expression led to significantly increased PDCD4 expression and
dramatically reduced levels of the BDNF, CCND1 and CDK4 proteins in
human glioma stem cells. The above results demonstrated that
SPION-mediated low HOTAIR expression in human glioma stem cells
significantly promoted the expression of PDCD4 and inhibited the
expression of cell cycle regulatory factors.
HOTAIR regulates the expression of PDCD4
at the transcriptional level by recruiting EZH2 and LSD1
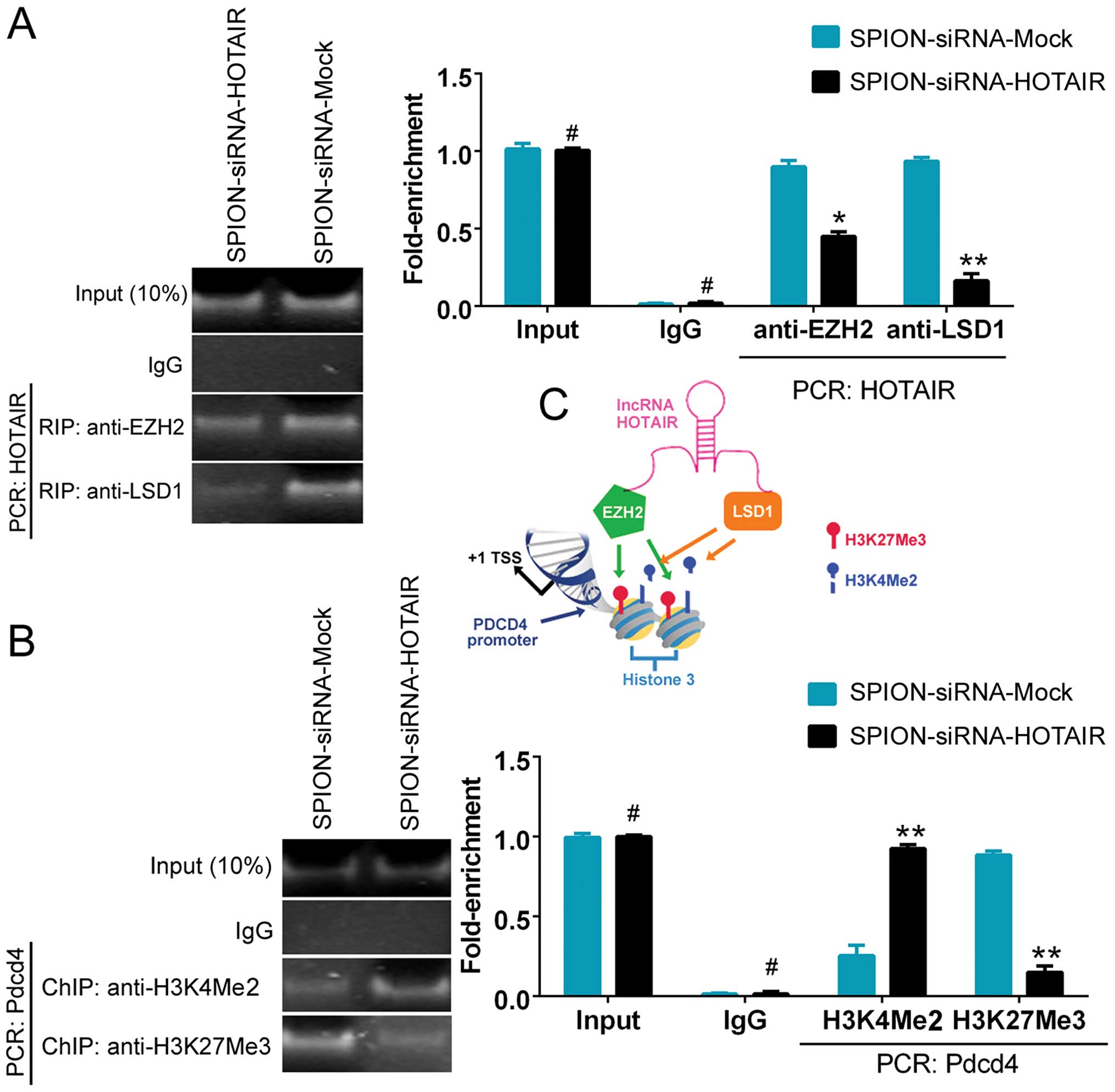
The ability of HOTAIR to recruit and bind to the
transcriptional corepressors EZH2 and LSD1 was assessed using the
RNA-binding protein immunoprecipitation (RIP) method. Fig. 4A shows that the ability of HOTAIR
to bind to EZH2 and LSD1 was significantly reduced in the
si-HOTAIR-transfected group compared with the control group. The
results indicated that the low level of HOTAIR expression affected
the recruitment of the EZH2 and LSD1 proteins. To further
investigate the effect of HOTAIR on PDCD4, a ChIP assay was
conducted. A decreased level of H3K27me3 and an elevated level of
H3K4me2 were detected in the promoter region of PDCD4 after
interference with the expression of HOTAIR. The results indicated
that si-HOTAIR regulated PDCD4 promoter-binding histones and,
consequently, maintained the transcriptional activation of the
PDCD4 gene by affecting HOTAIR expression (Fig. 4B and C).
A low level of HOTAIR expression
suppresses tumorigenicity of CD133+ human glioma stem
cells in nude mice
Human glioma stem cells that were transfected with
SPION/si-HOTAIR or SPION/si-mock conjugates were inoculated
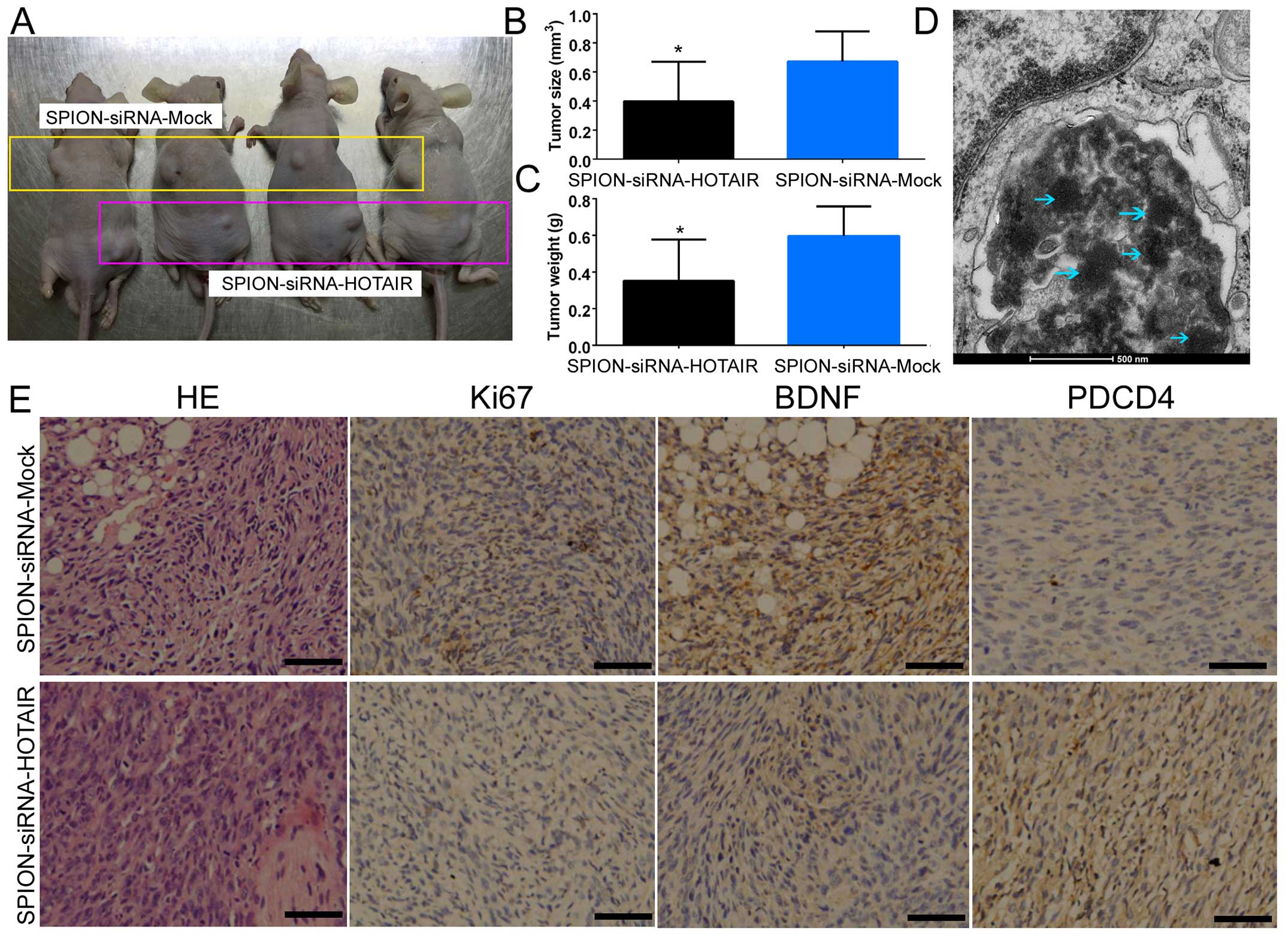
subcutaneously into BABL/Cnu/nu mice (Fig. 5A). Statistical analysis showed that
human glioma stem cells expressing a low level of HOTAIR exhibited
a dramatically reduced tumorigenic rate and produced significantly
smaller xenograft tumors compared with the control group (Fig. 5B). Moreover, the tumor weight was
significantly reduced in the group of mice inoculated with human
glioma stem cells expressing a low level of HOTAIR in comparison
with the control group (Fig. 5C).
In addition, multiple circular-shaped high-density electron clouds
of ~80–100 nm in size were observed in the xenograft tumor tissues
derived from the si-HOTAIR and si-mock-transfected groups (Fig. 5D). It was speculated that the
(intercellular) substance corresponding to the electron clouds was
SPION. The results of H&E staining indicated that both groups
of cells were capable of generating human glioma xenografts in nude
mice. However, xenograft tumors derived from glioma stem cells
expressing a low level of HOTAIR exhibited a significantly lower
degree of malignancy compared with the control group (Fig. 5E). The results of
immunohistochemical staining also indicated that the expression of
PDCD4 was significantly increased in the si-HOTAIR-transfected
group, while the expression of BDNF and Ki67 (proteins capable of
reflecting the proliferative status of the cells) was dramatically
decreased (Fig. 5E). The above
results indicated that low HOTAIR expression inhibited the growth
and tumorigenic capability of CD133+ human glioma stem
cells in nude mice.
Discussion
This study showed that inhibition of HOTAIR
expression targeted by SPION-mediated siRNA transfection promoted
the expression of PDCD4 at the transcriptional level, thereby
reducing the proliferation, invasion and tumorigenicity of human
glioma stem cells.
PDCD4 has been widely recognized as a tumor
suppressor gene and a target for antitumor therapy. It is closely
related to the development, progression and prognosis of tumors
(16). It has been demonstrated
that multiple signaling pathways are involved in the regulation of
PDCD4 expression. For example, PDCD4 is a downstream target of
microRNA 21 (miR21). Inhibition of miR21 in tumor cells increases
the expression of endogenous PDCD4 and promotes the apoptosis of
tumor cells (17). The
relationship between transforming growth factor-β (TGF-β) and PDCD4
has not been clearly defined, although it has been found that
treatment of the human hepatoma cell line Huh7 with TGF-β increases
the expression of the PDCD4 gene. However, it has also been shown
that TGF-β downregulates the expression of PDCD4 through inducing
the expression of miR-21 (18,19).
In addition to regulating PDCD4 protein expression at the
translational level, Akt and s6 kinase 1 (S6K1) induce the
phosphorylation of the PDCD4 protein and consequently promote PDCD4
degradation through the ubiquitin-proteasome pathway, thereby
decreasing the expression of PDCD4. Drugs targeting the above
pathway enhance the level of PDCD4 (16). A study conducted at the
transcriptional level demonstrated that the transcription factor
v-Myb and DNA demethylation induce the expression of PDCD4
(16). This study revealed, for
the first time, that HOTAIR was capable of affecting the expression
of PDCD4 through regulating the methylation levels of histone H3K4
and H3K27. Our findings showed that inhibition of HOTAIR expression
by si-HOTAIR significantly enhanced the mRNA and protein levels of
PDCD4.
Histone methylation is a type of epigenetic
modification involving the methylation of arginine or lysine
residues in the N-terminal domain of histones H3 and H4. The
effects of histone methylation are mainly reflected in
heterochromatin formation, gene imprinting, X-chromosome
inactivation and transcriptional regulation (20). To date, 24 histone methylation
sites (mainly at lysines) have been discovered. Histone methylation
can occur in the form of mono-, di-, or trimethylation, which is
catalyzed by histone methyltransferase and histone demethylase.
Methylation of different lysine residues may produce different
effects on gene transcription. Methylation at H3K4, H3K36 and H3K79
activates gene transcription, whereas methylation at H3K9, H3K27,
H3K79 and H4K20 inhibits gene transcription (21). A previous study by our group on
renal cancer cells showed that HOTAIR affects the expression of the
cell cycle-related proteins p53, p21 and p16 at the transcriptional
level through binding to EZH2 and histone H3 trimethyl Lys27
(H3K27me3) (9). This study further
investigated the mechanisms by which HOTAIR regulates PDCD4. It was
found that HOTAIR recruited and enriched the EZH2 and LSD1
proteins. EZH2 maintained the trimethylation state of histone H3K27
in the promoter region of PDCD4, while LSD1 functioned as a histone
deacetylase to mediate the demethylation of H3K4me2. The
synergistic effect of EZH2 and LSD1 led to the inhibition of PDCD4
transcription. Inhibition of HOTAIR expression by si-HOTAIR reduced
the recruitment of the EZH2 and LSD1 proteins, thereby upregulating
the expression of PDCD4 at the epigenetic level.
Taking into account the phospholipid bilayer
structure of the eukaryotic cell membrane, past studies have
generally employed lipid carriers as the medium for plasmid DNA
transfection. Although liposomes have been used for many years, the
transfection efficiencies of liposomes are not particularly high.
With the development of nanotechnology research, certain studies
have reported the use of nanoparticles as transfection media for
intracellular delivery of drugs or nucleic acids. In this study,
SPIONs were employed as a transfection vehicle to deliver siRNA
into cells. The transfected cells were analyzed via TEM, and SPION
signals were detected in the transfected cells. Northern blot
analysis showed that the HOTAIR hybridization signal was
significantly reduced in si-HOTAIR-transfected human glioma stem
cells. The results indicated that SPION successfully facilitated
the passage of si-HOTAIR through the cell membrane and interfered
with the amplification of the target genes. Both in vivo and
in vitro experiments demonstrated that the in vivo
and in vitro proliferation and invasion of glioma cells were
inhibited, and the tumorigenic capacity of glioma cells was
significantly reduced after suppressing the expression of HOTAIR by
the method described above.
In conclusion, SPIONs exhibited great potential in
mediating the transfection of nucleic acids into mammalian cells.
SPIONs effectively mediated the expression of si-HOTAIR in human
glioma stem cells. Si-HOTAIR inhibited the downstream expression of
PDCD4 at the transcriptional level and ultimately suppressed the
proliferation, invasion and tumorigenicity of glioma stem
cells.
Acknowledgements
This study was supported by grants from National
Natural Science Foundation of China (no. 81202811, 81371410), and
Project funded by China Postdoctoral Science Foundation (no.
2014M550250, 2015T80455), and Shanghai Natural Science Foundation
(no. 16ZR1434000) to Te Liu. It was also partially supported by the
Biomedical Multidisciplinary Program of Shanghai Jiao Tong
University (no. YG2014MS31) to Yuncheng Wu.
References
|
1
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
He J, Liu Y and Lubman DM: Targeting
glioblastoma stem cells: Cell surface markers. Curr Med Chem.
19:6050–6055. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Galli R, Binda E, Orfanelli U, Cipelletti
B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F and Vescovi
A: Isolation and characterization of tumorigenic, stem-like neural
precursors from human glioblastoma. Cancer Res. 64:7011–7021. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rinn JL, Ker tesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et
al: Functional demarcation of active and silent chromatin domains
in human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shah N and Sukumar S: The Hox genes and
their roles in oncogenesis. Nat Rev Cancer. 10:361–371. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hayami S, Kelly JD, Cho HS, Yoshimatsu M,
Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BA, et al:
Overexpression of LSD1 contributes to human carcinogenesis through
chromatin regulation in various cancers. Int J Cancer. 128:574–586.
2011. View Article : Google Scholar
|
|
9
|
Wu Y, Liu J, Zheng Y, You L, Kuang D and
Liu T: Suppressed expression of long non-coding RNA HOTAIR inhibits
proliferation and tumourigenicity of renal carcinoma cells. Tumour
Biol. 35:11887–11894. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou X, Ren Y, Zhang J, Zhang C, Zhang K,
Han L, Kong L, Wei J, Chen L, Yang J, et al: HOTAIR is a
therapeutic target in glioblastoma. Oncotarget. 6:8353–8365. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, Cui H, Li K, Sun C, Du W, Cui J,
Zhao X and Chen W: A magnetic nanoparticle-based multiple-gene
delivery system for transfection of porcine kidney cells. PLoS One.
9:e1028862014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Delyagina E, Schade A, Scharfenberg D,
Skorska A, Lux C, Li W and Steinhoff G: Improved transfection in
human mesenchymal stem cells: Effective intracellular release of
pDNA by magnetic polyplexes. Nanomedicine (Lond). 9:999–1017. 2014.
View Article : Google Scholar
|
|
13
|
Wang Y, Cui H, Sun C, Du W, Cui J and Zhao
X: Study on performance of magnetic fluorescent nanoparticles as
gene carrier and location in pig kidney cells. Nanoscale Res Lett.
8:1272013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao Y, Liu T and Huang Y: MicroRNA-134
suppresses endometrial cancer stem cells by targeting POGLUT1 and
Notch pathway proteins. FEBS Lett. 589:207–214. 2015. View Article : Google Scholar
|
|
15
|
Cheng W, Liu T, Wan X, Gao Y and Wang H:
MicroRNA-199a targets CD44 to suppress the tumorigenicity and
multidrug resistance of ovarian cancer-initiating cells. FEBS J.
279:2047–2059. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lankat-Buttgereit B and Göke R: The tumour
suppressor Pdcd4: Recent advances in the elucidation of function
and regulation. Biol Cell. 101:309–317. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Poria DK, Guha A, Nandi I and Ray PS:
RNA-binding protein HuR sequesters microRNA-21 to prevent
translation repression of proinflammatory tumor suppressor gene
programmed cell death 4. Oncogene. 35:1703–1715. 2016. View Article : Google Scholar :
|
|
18
|
Yao Q, Cao S, Li C, Mengesha A, Kong B and
Wei M: Micro-RNA-21 regulates TGF-beta-induced myofibroblast
differentiation by targeting PDCD4 in tumor-stroma interaction. Int
J Cancer. 128:1783–1792. 2011. View Article : Google Scholar
|
|
19
|
Zhang H, Ozaki I, Mizuta T, Hamajima H,
Yasutake T, Eguchi Y, Ideguchi H, Yamamoto K and Matsuhashi S:
Involvement of programmed cell death 4 in transforming growth
factor-beta1-induced apoptosis in human hepatocellular carcinoma.
Oncogene. 25:6101–6112. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ning B, Li W, Zhao W and Wang R: Targeting
epigenetic regulations in cancer. Acta Biochim Biophys Sin
(Shanghai). 48:97–109. 2016.
|
|
21
|
Barski A, Cuddapah S, Cui K, Roh TY,
Schones DE, Wang Z, Wei G, Chepelev I and Zhao K: High-resolution
profiling of histone methylations in the human genome. Cell.
129:823–837. 2007. View Article : Google Scholar : PubMed/NCBI
|