Introduction
Renal cell carcinoma (RCC) is the most common renal
neoplasm in adults, and clear cell renal cell carcinoma (ccRCC) is
the most common pathological type, accounting for approximately
75–80% of the cases of renal tumors (1). Although most patients can be treated
by surgical resection in early stage, there are still many patients
who will develop recurrence or the localized tumor will occur
distant metastasis. The traditional therapeutic options are still
not effective enough on renal cell carcinoma (2). Thus, there is an urgent need for
novel diagnosis and treatment of RCC.
MicroRNAs are endogenously expressed single-stranded
non-coding RNAs. Their length is approximately 18-25 nucleotides,
which can bind to the 3′untranslated region (3′UTR) of target
mRNAs, causing degradation of these mRNAs or inhibition of their
translation to functional proteins (3). Post-transcriptional regulatory
factors, play an important role in the proliferation,
differentiation and other biological processes including
tumorigenesis (4). MicroRNA-195 is
downregulated in a variety of cancers, such as human tongue
squamous cell carcinoma (5),
gastric (6), breast (7) and ovarian cancer (8). These studies showed that miR-195
acted as an anti-oncogene in these types of cancer. Although Slaby
et al (9) have found that
the expression level of miR-195 in recurrent tumor is lower, the
functional role and mechanism by which miR-195 exerts its
activities in clear cell renal cell carcinoma remain to be
elucidated. In the present study, we confirmed that microRNA-195
was significantly decreased in renal cell carcinoma tissues
compared with non-cancerous renal tissues suggesting that it may
participate in the occurrence and development of ccRCC. At the same
time, it also suggests that miR-195 may function as a tumor
suppressor gene in ccRCC.
Materials and methods
Human tissue samples
A total of 30 clear cell renal cell carcinoma tissue
samples and 30 corresponding non-cancerous renal tissue samples
were collected from the patients diagnosed with clear cell renal
cell carcinoma (n=30) by the Department of Urology (the Forth
Affiliated Hospital of Harbin Medicine University, Harbin, China).
Tissue samples were immediately frozen in liquid nitrogen after
surgical removal and classified according to the criteria provided
by AJCC (American Joint Committee on Cancer). None of the patients
recruited in the present study had undergone preoperative
chemotherapy or radiotherapy. Informed written consent was obtained
from all patients. The Medical Association Ethics Committee of the
Fourth Affiliated Hospital of Harbin Medical University approved
all aspects of the present study in accordance with the Helsinki
Declaration. Correlation between miR-195 expression and
clinicopathological variables of ccRCC are shown in Table I.
 | Table IThe correlation between miR-195
expression and clinicopathological variables of ccRCC. |
Table I
The correlation between miR-195
expression and clinicopathological variables of ccRCC.
| Variables | No. of
patients | Low expression | High
expression | P-value |
|---|
| Age (years) |
| <60 | 10 | 6 | 4 | 0.794 |
| ≥60 | 20 | 11 | 9 | |
| Gender |
| Male | 15 | 8 | 7 | 0.712 |
| Female | 15 | 9 | 6 | |
| Size (cm) |
| <3 | 16 | 9 | 7 | 0.961 |
| ≥3 | 14 | 8 | 6 | |
| Location |
| Right | 18 | 10 | 8 | 0.925 |
| Left | 12 | 7 | 5 | |
| Histological |
| I, II | 24 | 14 | 10 | 0.713 |
| III, IV | 6 | 3 | 3 | |
| Metastasis |
| Yes | 16 | 12 | 4 | 0.028 |
| No | 14 | 5 | 9 | |
RNA isolation and quantitative real-time
polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissues using TRIzol
reagent (Invitrogen, Grand Island, NY, USA) according to the
manufacturer’s protocol. RNA concentrations were measured using the
SpectraMax microplate spectrophotometer (Molecular Devices,
Sunnyvale, CA, USA). For reverse transcription, 1.0 μl of cDNA and
SYBR-Green real-time PCR Master Mix (Takara Co., Ltd., Tokyo,
Japan) were used according to the manufacturer’s protocol. The PCR
amplifications were performed in a 96-well plate for 1 cycle of
94°C for 30 sec, 40 cycles of 95°C for 5 sec, and 60°C for 30 sec
on Applied Biosystems 7900HT. The expression level of miR-195 was
analyzed by SDS2.4 software (Applied Biosystems) and internally
normalized to that of U6 with 2−ΔΔCt method. The primers
of miR-195 and U6 used for qRT-PCR are listed as Table II.
 | Table IISequences of primers for qRT-PCR and
miR-195 related sequence. |
Table II
Sequences of primers for qRT-PCR and
miR-195 related sequence.
| Name | Sequence
(5′-3′) |
|---|
| Primers used for
mRNA detection |
| miR-195
(Forward) |
GGGGTAGCAGCACAGAAAT |
| miR-195
(Reverse) |
TCCAGTGCGTGTCGTGGA |
| miR-195 (RT) |
(GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGCCAAT) |
| U6 (Forward) |
GCTTCGGCAGCACATATACTAAAAT |
| U6 (Reverse) |
CGCTTCACGAATTTGCGTGTCAT |
| U6 (RT) |
(GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTACATAC) |
| Gene names |
| miR-195
mimics |
UAGCAGCACAGAAAUAUUGGC |
| Mimics NC |
UUCUCCGAACGUGUCACGUTT |
| miR-195
inhibitor |
GCCAAUAUUUCUGUGCUGCUA |
| Inhibitor NC |
CAAUAUUUCUGUGCUGCUAUU |
Cell culture and transfection
The human clear cell renal cell carcinoma cell line
(ACHN) was purchased from the American Type Culture Collection
(ATCC; Manassas, VA, USA) and maintained in Dulbecco’s modified
Eagle’s medium (DMEM; HyClone Laboratories, Logan, UT, USA)
supplemented with 10% fetal bovine serum (FBS; Gibco), 100 μg/ml
streptomycin, 100 μg/ml penicillin and incubated at 37°C in a
humidified incubator at 5% CO2. ACHN cells were seeded
in antibiotic-free medium for 24 h for transfection. The cells were
then transfected with miR-195 mimics (50 nM), miR-195 inhibitor
(100 nM) and their respective negative controls (Shanghai
GenePharma Co., Ltd., Shanghai, China) using X-treme (Invitrogen,
Carlsbad, CA, USA) in serum-free Opti-MEM (Invitrogen) according to
the manufacturer’s instruction. The negative control (NC) scrambled
oligonucleotide does not encode for any known miRNAs. Transfection
efficiency was confirmed by SYBR-Green real-time PCR. Cells were
collected 48 h later. All miRNA sequences are listed in Table II.
Cell viability assay
ACHN cells (2×103) were seeded in 96-well
plates in 100 μl culture medium and allowed to attach for 24 h.
Then cells were transfected with miR-195 mimics, miR-195 inhibitor,
and their corresponding negative controls for 48 h as we mentioned
before. After the treatment, the medium in each well were removed
and replaced with PBS solution with 5 mg/ml
3-′4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetra-zolium bromide
(MTT) (Sigma-Aldrich, St. Louis, MO, USA) and then the plates were
further incubated at 37°C for 4 h. The remaining supernatant was
then removed and 150 μl dimethyl sulfoxide (DMSO; Sigma-Aldrich)
was added for 15 min to dissolve the formed crystal formazan.
Finally, the absorbance was measured at 490 nm using enzyme-labeled
analyzer. Three independent experiments were performed.
Cell migration and invasion assays
For migration assay, 5×104 cells
resuspended in serum-free DMEM after transfection were placed in
the upper chamber with 8 μm pores (Corning, Corning NY, USA) and
incubated at 37°C in 5% CO2 continuously for 24 h. The
bottom chamber contained 10% FBS as a chemoattractant. After the
incubation, the non-migration cells on the upper membrane surface
were removed with cotton swab. Cells that had migrated to the
bottom of the insert were washed twice with PBS and fixed in 100%
methanol stained with 0.2% crystal violet. The migrated cells were
photographed under a microscope (magnification, x200) and counted
in five random fields. Matrigel-precoated Transwell inserts (BD
Biosciences, San Jose, CA, USA) were used for the cell invasion
assay. Similarly, the lower chamber was filled with 10% FBS as a
chemoattractant. After 24 h of incubation at 37°C in 5%
CO2, the cells that had invaded through the membrane
were fixed, stained and counted. Each experiment was performed in
triplicate.
Cell cycle analysis
The influence of the change of cell cycle was
analyzed by flow cytometry. The transfected ACHN cells
(1×106 cells) were washed with PBS and fixed in 70%
ethanol overnight at 4°C. The cells were then labeled with
propidium iodide (50 μg/ml) and RNase (100 μg/ml) (Sigma-Aldrich)
for 30 min and were analyzed using a fluorescence-activated cell
sorter scanning (Becton-Dickinson, San Jose, CA, USA). The
percentages of G0/G1, S and G2/M
cells were counted and compared. Each experiment was performed in
triplicate.
TUNEL assay and DAPI staining
Cell apoptosis was measured by TUNEL/DAPI double
staining assay as the manual recommended. The ACHN cells were
seeded in coverslips in 6-well plates with the same cell number per
well (1×105). Cells harvested 48 h after transfection
were washed with PBS/1% BSA and fixed in paraformaldehyde solution
(4% in PBS) for 20 min at room temperature. After washing with PBS
three times, the cells were permeabilized in a solution containing
0.1% Triton X-100 in 0.1% sodium citrate for 2 min on ice, followed
by incubation in freshly prepared TUNEL reaction mixture terminal
deoxynucleotidyl transferase (TdT) and TUNEL dilution buffer at the
ratio of 1:10 for 60 min at 37°C in the dark. Then anti-digoxigenin
peroxidase conjugate was added, and incubation continued in a humid
box for 30 min at room temperature. After washing with PBS three
times, DAPI (Roche Molecular Biochemicals) was added for 10 min at
room temperature to stained nuclei. The coverslips were then washed
with PBS three times and manipulated with anti-fading solution.
Finally, the cells were observed on slides by a confocal microscope
(Olympus FluoView™ FV1000).
Western blot analysis
Total protein samples were extracted from
transfected ACHN cells using lysis buffer containing protease
inhibitor. The protein concentrations were measured using the
Bio-Rad protein assay system (Bio-Rad Laboratories, Hercules, CA,
USA). After boiling the samples for 5 min, the protein samples were
run on SDS-PAGE (8–15% polyacryl-amide gels). The lysates were
resolved by electrophoresis (70 V for 25 min and 120 V for 1.5 h)
and transferred onto NC membranes (nitrocellulose membrane; Bio-Rad
Laboratories). After blocking in 5% non-fat milk in Tris-buffered
saline with Tween (TBST) for 2 h, the NC membranes were treated
overnight at 4°C with the following primary antibodies: Bax (1:200
dilution; Cell Signaling Technology, Danvers, MA, USA), caspase-3
(1:200 dilution; Cell Signaling Technology), Bcl-2 (1:200 dilution;
Cell Signaling Technology), VEGFR2 (1:200 dilution; Abcam,
Cambridge, MA, USA), total-Akt (1:200 dilution; Santa Cruz
Biotechnology, Santa Cruz, CA USA), p-Akt (1:500 dilution; Cell
Signaling Technology), Raf (1:500 dilution; Cell Signaling
Technology), MEK (1:500 dilution; Cell Signaling Technology) ERK
(1:500 dilution; Cell Signaling Technology), p-ERK1/2 (p44/p42)
(1:500 dilution; Cell Signaling Technology). The next day, the NC
membranes were washed in PBS three times and incubated with
secondary antibody: Alexa Fluor® 800 goat anti-mouse or
anti-rabbit IgG (Invitrogen) diluted at 1:4,000 at room temperature
for 1.5 h. Western blot bands were quantified using Odyssey v1.2
software by measuring the band intensity (Area x OD; Optical
Density) for each group and normalized to GAPDH
(glyceraldehyde-3-phosphate). All the presented results are
representative of at least three independent experiments.
Luciferase assay
To generate reporter vectors bearing miRNA-binding
sites, the 3′-untranslated region (3′-UTR) of VEGFR2 and its
mutation type were synthesized. The construct was inserted into
multiple cloning sites downstream of the luciferase gene
(SacI and HindIII sites) in the pMIR-REPORT
luciferase miRNA expression reporter vector (Ambion, Austin, TX,
USA). For the luciferase assay, 0.1 μg of luciferase reporters
containing 3′-UTR were cotransfected with miR-195 or miR-195
inhibitor into HEK-293 cells using Lipofectamine 2000 (Invitrogen).
As an internal control, 10 ng of Renilla luciferase
reporters were also included. Forty-eight hours after transfection,
the cells were collected and Dual-Luciferase activities were
measured by a luminometer.
Data analysis
Data were analyzed using the GraphPad Prism 5.0 and
the SPSS 14.0. The data were presented as mean ± SEM. Two-tailed
unpaired Student’s t-tests and one-way ANOVA were used for the
statistical evaluation of the data. The Chi-squared test was used
to investigate the significance of miR-195 expression as correlated
with clinicopathological features in renal cell carcinoma.
P<0.05 was considered as indicating statistically significant
differences.
Results
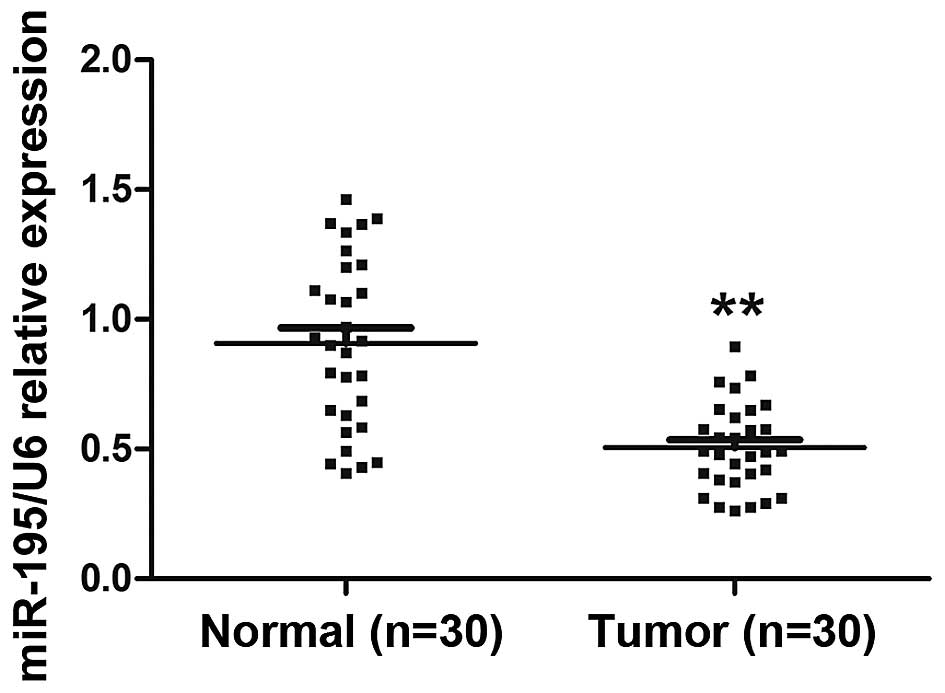
Expression of miR-195 is decreased in
ccRCC tissues and associated with lymph node metastasis
To detect the expression and significance of miR-195
in ccRCC tumorigenesis, we compared miR-195 expression profiles
between ccRCC tissues and paired adjacent non-cancerous renal
tissues from 30 individual patients using quantitative RT-PCR. The
results showed that miR-195 expression was significantly decreased
in ccRCC tissues compared with adjacent non-cancerous renal tissues
(Fig. 1; P<0.01). Next we
analyzed the association between miR-195 expression and
clinicopathological features in the 30 ccRCC patients. The
relationship between miR-195 levels and patients’ clinical features
suggests that expression of miR-195 was correlated with metastasis,
while no obvious correlation was observed in the patient gender,
age, tumor size or location (Table
I). These results suggest that the change of miR-195 expression
level may affect the occurrence and development of ccRCC. miR-195
may act as a tumor suppressor gene in ccRCC.
miR-195 inhibits ACHN cell viability in
vitro
To explore the effect of miR-195 on the biological
behavior of ccRCC, we transfected ACHN cells with miR-195 mimics,
miR-195 inhibitor and their corresponding negative controls as
mentioned above. The expression of miR-195 in ACHN cells was
detected in order to guarantee the efficiency of transfection. Each
group of transfected cells was compared with the NC. In the miR-195
mimics group, miR-195 expression was significantly higher than the
mimic NC group. There was also a notable decrease in miR-195
expression in the miR-195 inhibitor group as compared to the mimics
group (Fig. 2A; P<0.01). The
transfected cells were then used to do the viability assay. The
experimental results showed that the viability of ACHN cells was
suppressed by overexpressing miR-195 compared with that of control
(Fig. 2B; P<0.01).
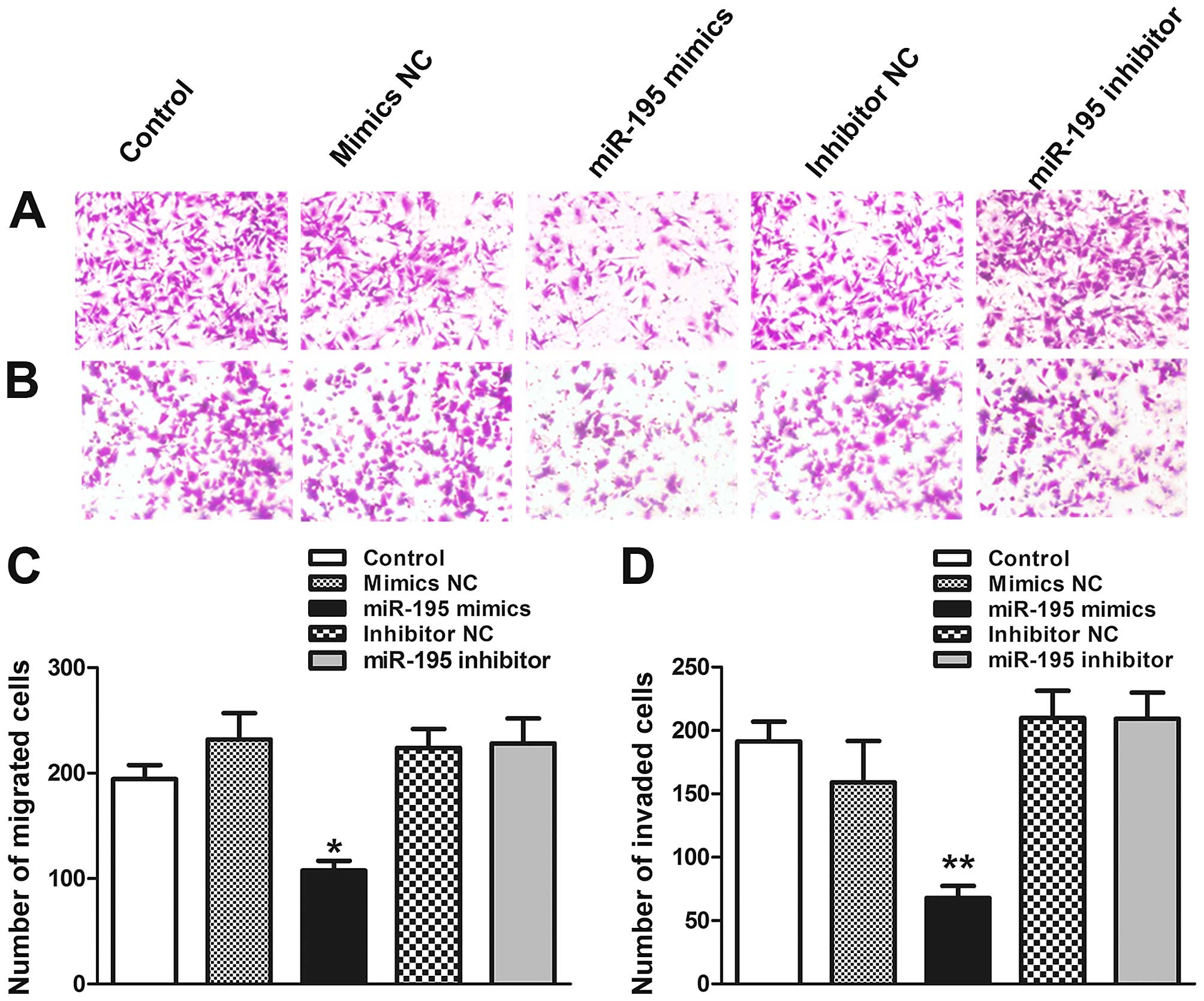
miR-195 attenuates migration and invasion
potential of ACHN cells in vitro
Invasiveness and migratory capacity of the tumor
cells are essential for tumor progression. To test whether miR-195
worked on migratory and invasive capabilities of ACHN cells,
Transwell and Matrigel invasion assays were performed in
vitro. The results showed that the migratory capability of ACHN
cells transfected with miR-195 was reduced by 45.3% in x200
magnification (Fig. 3A and C;
P<0.05). In order to further verify the effect of miR-195 on
ACHN cell metastasis, Matrigel invasion assays was performed.
Similarly, the Matrigel invasion assay showed that the invasiveness
of ACHN cells was reduced by 41.6% compared to control group
(Fig. 3B and D; P<0.01). In
conclusion, these results suggest that overexpression of miR-195
can suppress ACHN cells migration and invasion.
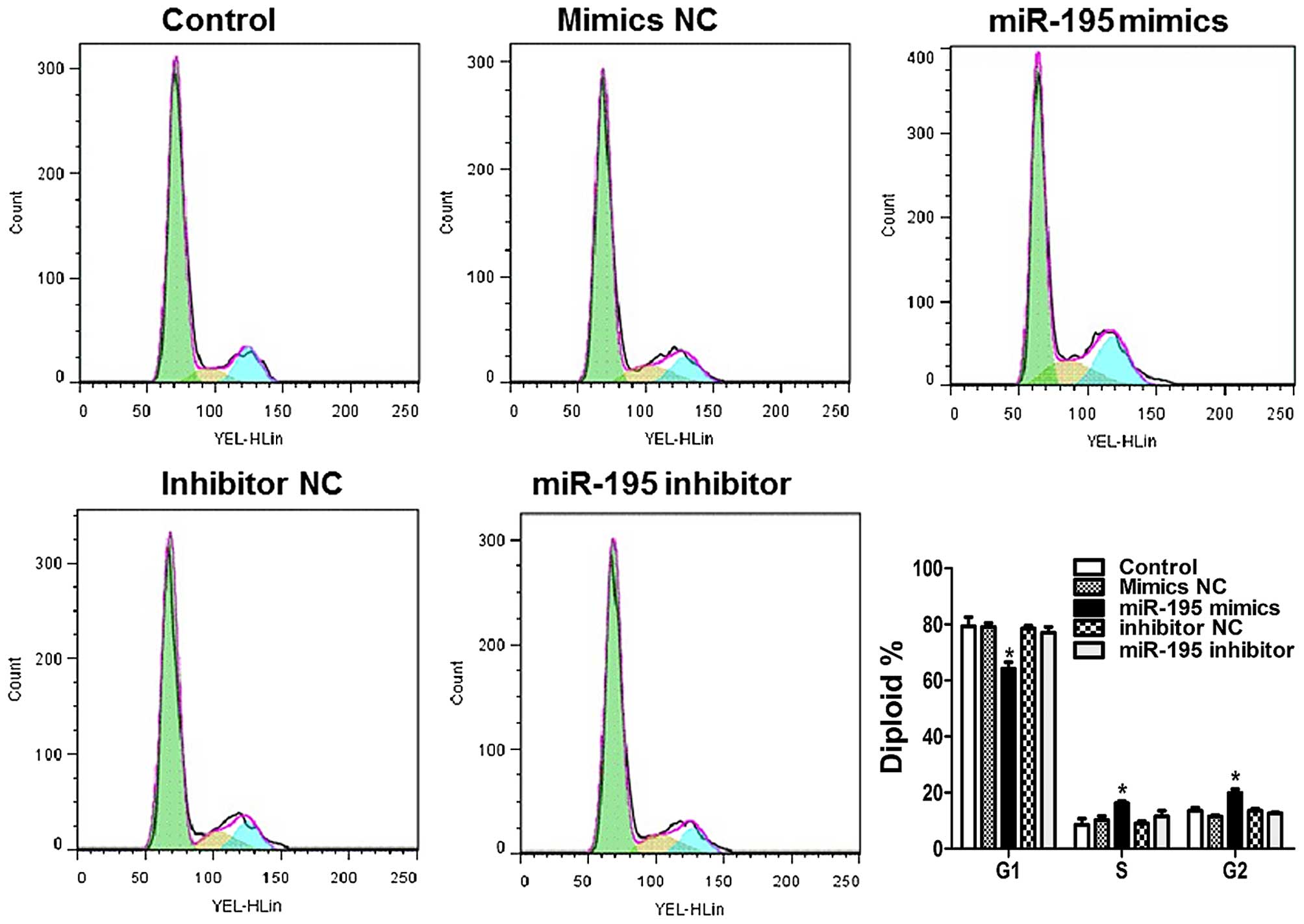
miR-195 induces cell cycle arrest in ACHN
cells
By exploring the effects of miR-195 on cell cycle
distribution, we can learn more about the intrinsic mechanism of
miR-195 on cell proliferation. As compared with control in Fig. 4, the percentage of cells in S and
G2 phase was significantly increased upon treatment with
miR-195 mimics.
miR-195 induces apoptosis in ACHN
cells
To investigate whether miR-195 can induce ACHN cells
to apoptosis, TUNEL assays were used to confirm apoptosis changes.
DAPI positive cells showed that total number of each group were
almost the same (Fig. 5A, upper
panel). In the TUNEL staining, the merged pictures showed the
double labeled cells which were the ACHN cells occurring apoptosis
(Fig. 5A, lower panel). From the
statistical graph, we found that the apoptotic rate of ACHN cells
increased when the cells were treated by miR-195 mimics (P<0.01;
Fig. 5B). Taken together, these
data indicated that miR-195 can induce apoptosis in ACHN cells
compared with normal groups.
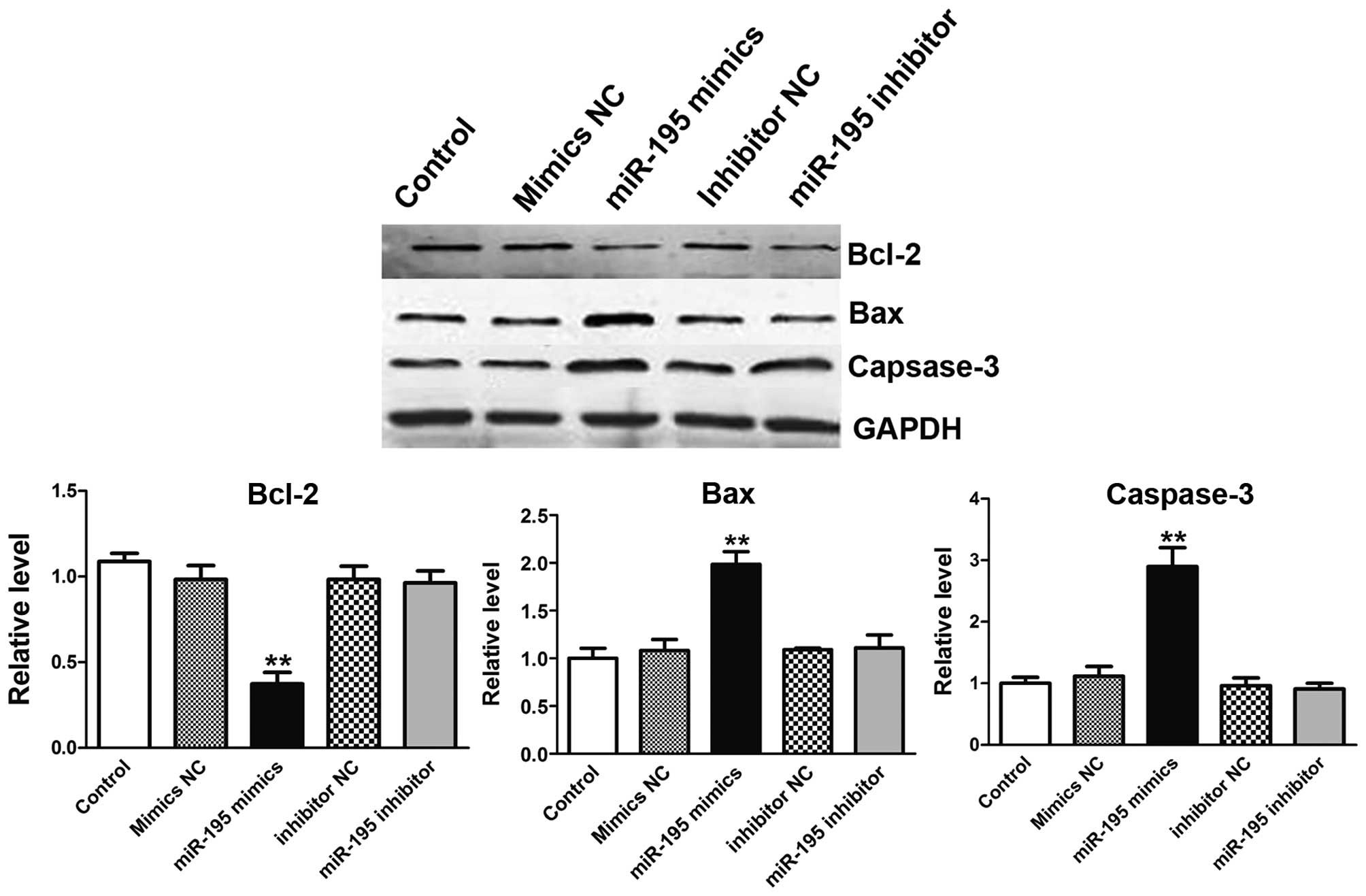
miR-195 overexpression inhibits Bcl-2
expression, and boosts Bax, caspase-3 proteins expression in the
process of apoptosis
Apoptosis is a complex process which involves a
series of proteins. Bcl-2 is a member of the human inhibitors of
apoptosis protein family (10).
Bcl-2 expression in cancer has been implicated not only in
inhibition of apoptosis, but also related to promotion of
proliferation (11). On the
country, Bax and caspase-3 are symbols for apoptosis (12–14).
To further confirm the apoptosis effect induced by miR-195, western
blot analysis was used to detect the apoptosis-related proteins as
mentioned before. In treatment of ACHN cells with miR-195 mimics,
the expression of Bcl-2 expression was depressed (Fig. 6A). Interestingly, Bax in cell
extracts was increased (Fig. 6B).
Furthermore, cells transfected with miR-195 resulted in the
significant increase in expression level of relative caspase-3
suggesting the involvement of cell apoptosis prompted by miR-195
(Fig. 6C) (15). When ACHN cells were transfected
with miR-195 inhibitor, no obvious phenomenon was observed.
miR-195 can regulate PI3K/Akt and
Raf/MEK/ERK signaling pathway via targeting VEGFR2
It has been reported that VEGFR2 is not only a vital
signal receptor, but also involved in the regulation of cellular
processes, including survival, growth, migration, invasion,
apoptosis and angiogenesis (16–21).
Furthermore, its downstream signaling pathways phosphatidylinositol
3-kinase (PI3K/Akt) occupied a central position in the pathogenesis
of renal cell carcinoma (22). In
addition, the downstream signaling pathway: Raf/MEK/ERK pathway is
involved in the proliferation and inhibition of apoptosis (23–25).
Thus, we investigated whether VEGFR2 could be regulated by miR-195,
which had an inhibitory effect on ACHN cells through PI3K/Akt and
Raf/MEK/ERK signaling pathways. We used qRT-PCR and western blot
analysis to detect the expression levels of VEGFR-2 and its
downstream signal molecules: Akt, MEK and ERK in different groups,
tyrosine phosphorylation of these proteins is essential for cancer
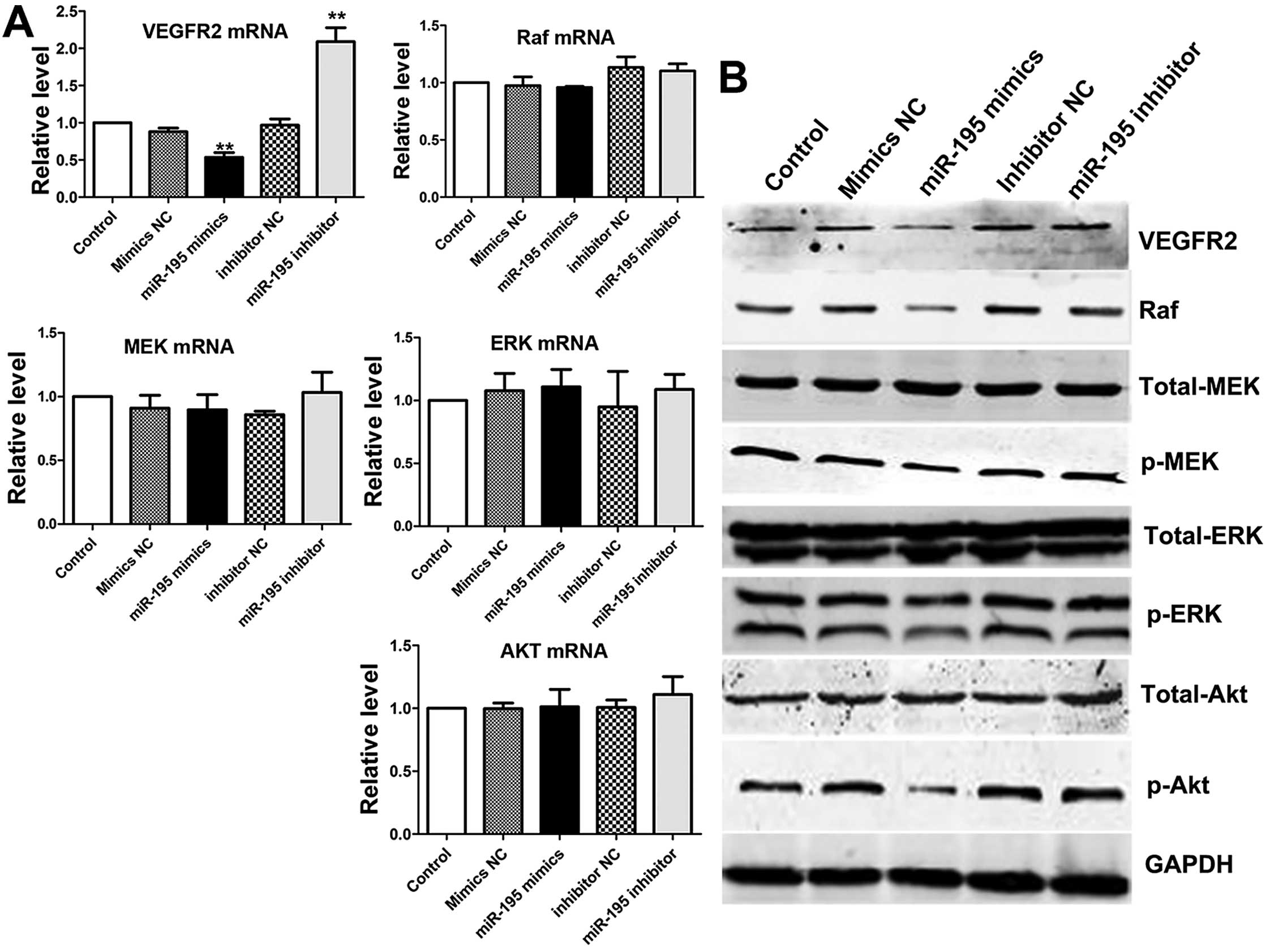
cell proliferation (26,27). The results indicated miR-195
suppressed the VEGFR2 mRNA, but had no effect on mRNA of MEK, ERK
or Akt. Then, western blot analysis was performed to analyze the
effect on proteins induced by miR-195 (Fig. 7A). The expression level of VEGFR2
was significantly reduced and its active downstream factors: Raf,
p-MEK and p-ERK1/2 were dramatically decreased following
downregulation of VEGFR2 in ACHN cells. However, the expression of
total MEK and total ERK1/2 remained only slightly changed after
transfection in ACHN cells compared with control groups. We noted
that the level of total-Akt was almost the same in overexpression
miR-195 ACHN cells compared with control groups. As expected, the
level of p-Akt was decreased in overexpression miR-195 ACHN cells
compared with control groups (Fig. 7B
and C). Based on these results, we confirmed that
overexpression of miR-195 in ACHN cells suppressed the
phosphorylation of MEK, ERK and Akt, which are the downstream
targets of VEGFR2. These processes could come through PI3K/Akt and
RAF/MEK/ERK two signal pathways.
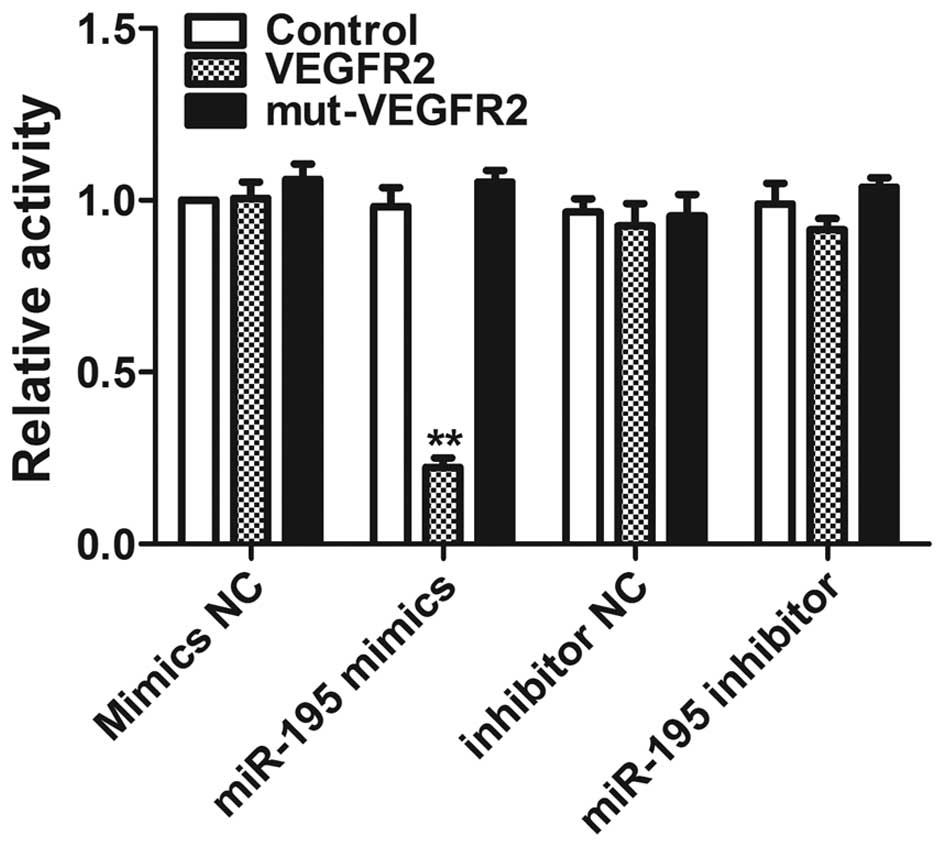
VEGFR2 is a direct target of miR-195
To further validate whether VEGFR2 is a direct
target of miR-195, luciferase reporter constructs containing a
segment of the 3′-UTR of VEGFR2 (VEGFR2) or a mutated 3′-UTR of
VEGFR2, which contained a mutated seed sequence (mut-VEGFR2), were
generated (Fig. 8). The constructs
were co-transfected with miR-195 into HEK293 cells. Co-transfection
of miR-195 strongly inhibited the luciferase activity of the
reporter construct containing the 3′-UTR.
Discussion
The formation of renal cell carcinoma is a complex
process which involves changes of various genes (28,29).
A class of endogenous, single stranded, small non-coding RNAs, are
known as miRNAs which can regulate multiple function gene
expression at the post-transcriptional level and play important
roles in biological processes such as cell differentiation,
proliferation and apoptosis (3,4).
Emerging evidence show that aberrant expression of microRNAs may
contribute to kidney tumorigenesis (30,31).
Recently, it has been shown that some microRNAs inhibit cancer via
suppressing certain cytokines. For example, hepatocellular
carcinoma was inhibited by miR-195 through VEGF inhibition and
colorectal cancer was inhibited by miR-497 via regulating IGF1
(32,33). Slaby et al (9) showed that miR-195 was decreased in
tumors from patients who developed relapse and in primary
metastasis. In the present study, we confirmed that by qRT-PCR and
considered miR-195 may play an important role in ccRCC progression.
As expected, the viability of ACHN cells was suppressed by miR-195
mimics, the migratory and invasive activities were inhibited
compared to that of the control. In addition, bioinformatic
analysis shows that VEGFR2 is a potential target of miR-195. We
first identified that miR-195 targets VEGFR2 in ACHN cells and then
investigated the downstream signaling moleculars further in this
study. Renal cell carcinoma (RCC), is the second most common form
of urologic tumor associated with an alteration of multiple
signaling pathways (34). In
addition, the PI3K/Akt pathway plays a critical role in kidney
cancer pathogenesis. The p-Akt then activates a series of
cancer-related functions such as cell growth, proliferation,
survival and motility, which drive tumor progression (35,36).
We found that overexpression of miR-195 significantly
down-regulated p-Akt protein expression in ACHN cells. Likewise,
the expression of p-Raf and its downstream proteins p-MEK, p-ERK
were decreased. The expression level of total Akt, total MEK, and
total ERK had little change before and after transfection.
Furthermore, we found that increased Bax and caspase-3 proteins,
and attenuated expression of Bcl-2 protein level in transfected
miR-195 mimics ACHN cells. In summary, overexpression miR-195 could
inhibit activation of PI3K/Akt and Raf/MEK/ERK signaling pathways,
which play critical roles in cell proliferation, migration and
apoptosis. A schema of the role of miR-195 in ACHN cells is shown
in Fig. 7. Our findings provided a
theoretical basis for further research on the mechanism of miR-195
in renal clear cell carcinoma, and provide a direction for gene
therapy for renal cell carcinoma.
To the best of our knowledge, VEGFR2 is an important
receptor of VEGF (vascular endothelium derived factor) reported to
be powerful in promoting angiogenesis function (37). Angiogenesis plays an important role
in the process of renal cell carcinoma (38). Hence we intend to focus on the
anti-angiogenesis role of miR-195 in renal cell carcinoma in future
research.
In conclusion, miR-195 suppresses ACHN proliferation
and potentiates apoptosis by inhibiting both the Raf/MEK/ERK and
the PI3K/Akt pathways via targeting VEGFR2. This may provide
promising targets for ccRCC which remains an incurable disease.
Overexpression of miR-195 could be considered as a potential
therapeutic strategy for ccRCC therapy. More research will be
required before applying it in the clinic.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (30900354).
References
|
1
|
Rasmussen F: Metastatic renal cell cancer.
Cancer Imaging. 13:374–380. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ljungberg B, Cowan NC, Hanbury DC, Hora M,
Kuczyk MA, Merseburger AS, Patard JJ, Mulders PF and Sinescu IC;
European Association of Urology Guideline Group. EAU guidelines on
renal cell carcinoma: The 2010 update. Eur Urol. 58:398–406. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saxena S, Jónsson ZO and Dutta A: Small
RNAs with imperfect match to endogenous mRNA repress translation.
Implications for off-target activity of small inhibitory RNA in
mammalian cells. J Biol Chem. 278:44312–44319. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jia LF, Wei SB, Gong K, Gan YH and Yu GY:
Prognostic implications of micoRNA miR-195 expression in human
tongue squamous cell carcinoma. PLoS One. 8:e566342013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Deng H, Guo Y, Song H, Xiao B, Sun W, Liu
Z, Yu X, Xia T, Cui L and Guo J: MicroRNA-195 and microRNA-378
mediate tumor growth suppression by epigenetical regulation in
gastric cancer. Gene. 518:351–359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Heneghan HM, Miller N, Kelly R, Newell J
and Kerin MJ: Systemic miRNA-195 differentiates breast cancer from
other malignancies and is a potential biomarker for detecting
noninvasive and early stage disease. Oncologist. 15:673–682. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim YW, Kim EY, Jeon D, Liu JL, Kim HS,
Choi JW and Ahn WS: Differential microRNA expression signatures and
cell type-specific association with Taxol resistance in ovarian
cancer cells. Drug Des Devel Ther. 8:293–314. 2014.PubMed/NCBI
|
|
9
|
Slaby O, Redova M, Poprach A, Nekvindova
J, Iliev R, Radova L, Lakomy R, Svoboda M and Vyzula R:
Identification of MicroRNAs associated with early relapse after
nephrectomy in renal cell carcinoma patients. Genes Chromosomes
Cancer. 51:707–716. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hardwick JM and Soane L: Multiple
functions of BCL-2 family proteins. Cold Spring Harb Perspect Biol.
5:a0087222013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Davids MS and Letai A: Targeting the
B-cell lymphoma/leukemia 2 family in cancer. J Clin Oncol.
30:3127–3135. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yip KW and Reed JC: Bcl-2 family proteins
and cancer. Oncogene. 27:6398–6406. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Antonsson B: Bax and other pro-apoptotic
Bcl-2 family ‘killer-proteins’ and their victim the mitochondrion.
Cell Tissue Res. 306:347–361. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li CL, Chang L, Guo L, Zhao D, Liu HB,
Wang QS, Zhang P, Du WZ, Liu X, Zhang HT, et al: β-elemene induces
caspase-dependent apoptosis in human glioma cells in vitro through
the upregulation of Bax and Fas/FasL and downregulation of Bcl-2.
Asian Pac J Cancer Prev. 15:10407–10412. 2014. View Article : Google Scholar
|
|
15
|
Zhang W, Ha M, Gong Y, Xu Y, Dong N and
Yuan Y: Allicin induces apoptosis in gastric cancer cells through
activation of both extrinsic and intrinsic pathways. Oncol Rep.
24:1585–1592. 2010.PubMed/NCBI
|
|
16
|
Lang SA, Schachtschneider P, Moser C, Mori
A, Hackl C, Gaumann A, Batt D, Schlitt HJ, Geissler EK and
Stoeltzing O: Dual targeting of Raf and VEGF receptor 2 reduces
growth and metastasis of pancreatic cancer through direct effects
on tumor cells, endothelial cells, and pericytes. Mol Cancer Ther.
7:3509–3518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen CH, Lai JM, Chou TY, Chen CY, Su LJ,
Lee YC, Cheng TS, Hong YR, Chou CK, Whang-Peng J, et al: VEGFA
upregulates FLJ10540 and modulates migration and invasion of lung
cancer via PI3K/AKT pathway. PLoS One. 4:e50522009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pal HC1, Sharma S, Strickland LR, Agarwal
J, Athar M, Elmets CA and Afaq F: Delphinidin reduces cell
proliferation and induces apoptosis of non-small-cell lung cancer
cells by targeting EGFR/VEGFR2 signaling pathways. PLoS One.
8:e772702013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sakurai Y, Ohgimoto K, Kataoka Y, Yoshida
N and Shibuya M: Essential role of Flk-1 (VEGF receptor 2) tyrosine
residue 1173 in vasculogenesis in mice. Proc Natl Acad Sci USA.
102:1076–1081. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fontanella C, Ongaro E, Bolzonello S,
Guardascione M, Fasola G and Aprile G: Clinical advances in the
development of novel VEGFR2 inhibitors. Ann Transl Med.
2:1232014.
|
|
21
|
Claesson-Welsh L and Welsh M: VEGFA and
tumour angiogenesis. J Intern Med. 273:114–127. 2013. View Article : Google Scholar
|
|
22
|
Conti A, Santoni M, Amantini C, Burattini
L, Berardi R, Santoni G, Cascinu S and Muzzonigro G: Progress of
molecular targeted therapies for advanced renal cell carcinoma.
BioMed Res Int. 2013:4191762013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Troppmair J and Rapp UR: Raf and the road
to cell survival: A tale of bad spells, ring bearers and detours.
Biochem Pharmacol. 66:1341–1345. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stadler WM: Targeted agents for the
treatment of advanced renal cell carcinoma. Cancer. 104:2323–2333.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sridhar SS, Hedley D and Siu LL: Raf
kinase as a target for anticancer therapeutics. Mol Cancer Ther.
4:677–685. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chan CH, Jo U, Kohrman A, Rezaeian AH,
Chou PC, Logothetis C and Lin HK: Posttranslational regulation of
Akt in human cancer. Cell Biosci. 4:592014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shaul YD and Seger R: The MEK/ERK cascade:
From signaling specificity to diverse functions. Biochim Biophys
Acta. 1773:1213–1226. 2007. View Article : Google Scholar
|
|
28
|
Walker C: Molecular genetics of renal
carcinogenesis. Toxicol Pathol. 26:113–120. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bussolati B, Satolli MA and Camussi G: The
role of angiogenesis in renal carcinoma. G Ital Nefrol. 25:297–305.
2008.(In Italian). PubMed/NCBI
|
|
30
|
Redova M, Svoboda M and Slaby O: MicroRNAs
and their target gene networks in renal cell carcinoma. Biochem
Biophys Res Commun. 405:153–156. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma L and Qu L: The function of microRNAs
in renal development and pathophysiology. J Genet Genomics.
40:143–152. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang R, Zhao N, Li S, Fang JH, Chen MX,
Yang J, Jia WH, Yuan Y and Zhuang SM: MicroRNA-195 suppresses
angiogenesis and metastasis of hepatocellular carcinoma by
inhibiting the expression of VEGF, VAV2, and CDC42. Hepatology.
58:642–653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo ST, Jiang CC, Wang GP, Li YP, Wang CY,
Guo XY, Yang RH, Feng Y, Wang FH, Tseng HY, et al: MicroRNA-497
targets insulin-like growth factor 1 receptor and has a tumour
suppressive role in human colorectal cancer. Oncogene.
32:1910–1920. 2013. View Article : Google Scholar :
|
|
34
|
Kim WY and Kaelin WG Jr: Molecular
pathways in renal cell carcinoma - rationale for targeted
treatment. Semin Oncol. 33:588–595. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li F, Ambrosini G, Chu EY, Plescia J,
Tognin S, Marchisio PC and Altieri DC: Control of apoptosis and
mitotic spindle checkpoint by survivin. Nature. 396:580–584. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Reuter CW, Morgan MA, Grünwald V, Herrmann
TR, Burchardt M and Ganser A: Targeting vascular endothelial growth
factor (VEGF)-receptor-signaling in renal cell carcinoma. World J
Urol. 25:59–72. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rini BI and Rathmell WK: Biological
aspects and binding strategies of vascular endothelial growth
factor in renal cell carcinoma. Clin Cancer Res. 13:741s–746s.
2007. View Article : Google Scholar : PubMed/NCBI
|