Introduction
Thyroid carcinoma (TC) is the most frequent
endocrine system tumor and a malignancy with fast growing
prevalence rate in recent years. Papillary thyroid carcinoma (PTC)
remains the most common pathological type of thyroid malignancy,
comprising ≥80% of all thyroid carcinomas (1,2). To
date, surgical resection combined with radio iodine and
levothyroxine treatment are the primary mode of therapy for
patients with PTC (3). Although
most PTCs have a good prognosis, regional recurrence is observed in
5–20% of patients who have undergone total thyroidectomy and lymph
node metastasis in the neck occurs in 20–50% of all patients
(4–7). Advanced age at diagnosis, larger
primary tumor, extrathyroidal invasion, lymph node metastasis and
advanced tumor-node-metastasis (TNM) stage are demonstrated to be
associated with a poor prognosis (8). Given this, there is an urgent need to
explore the mechanisms involved in the development and progression
of thyroid cancers.
MicroRNAs (miRNAs) are a group of endogenous, 19–25
nucleotides non-coding RNAs which can regulate gene expression
through binding to the 3′-untranslated region (3′-UTR) of target
genes to promote mRNA degradation or protein translation inhibition
(9). Recent evidence has shown
that miRNAs are involved in many important physiological and
pathological processes, such as cell proliferation, development,
differentiation, virus infection and tumorigenesis, and are widely
dyregulated in various cancers (10). Nearly half of the human miRNAs are
located in cancer-associated genomic regions which are frequently
amplified, deleted, or rearranged in cancer, suggesting that miRNAs
may function as either tumor-suppresor genes or oncogenes (11). In PTC, miRNA analysis was reported
as a potential diagnostic tool, and the miRNA signature was shown
to be indicative of the degree of aggressiveness (12). MicroRNA-7 (miR-7) was found to be
significantly downregulated in various tumors, including breast
cancer (13), lung cancer
(14), glioma (15), hepatocellular carcinoma (16). In our previous study, miR-7 is
downregulated in PTC tissue relative to adjacent normal tissue by
miRNA-microarray analysis conducted in 10 pairs of PTC specimens
and normal thyroid tissues. Until now, little was known on the role
of miR-7 in PTC, let alone their functions and mechanisms of
action.
CKS2, firstly identified in 1990, is located in
chromosome 9q22 (17). Accumulated
evidence suggests that CKS2 is an oncoprotein that promotes cancer
tumorigenesis and progress. It has abnormal expression in a variety
of malignant tumor tissues and is closely associated with various
biological behavior, such as tumor development, progression and
metastasis (18). Previous
evidence has demonstrated that CKS2 was regulated by miR-26a in PTC
and CKS2 levels were associated with expression of its downstream
genes, including cyclin B1 and cdk1 (19). However, there are scarce related
research on the function of CKS2 in PTC, and deeper understanding
of the post-transcriptional control of the CKS2 gene in PTC remains
indefinable.
In this study, we investigated the function of miR-7
in PTC. Interestingly, our experiments demonstrate that
upregulation of miR-7 inhibited cellular growth, suppressed
cellular migration and invasion, caused a G0/G1 arrest in PTC
cells, likely by targeting CKS2. Our results indicate that
upregulation of miR-7 level may be a novel therapeutic target for
the treatments of thyroid cancer.
Materials and methods
MiRNA-microarray analysis
In this study, ten paired thyroid papillary cancer
specimens and adjacent normal thyroid tissues were collected from
the Department of Breast and Thyroid Surgery of Shanghai Tenth
People’s Hospital, Shanghai, China. All the samples were
immediately snap-frozen in liquid nitrogen and confirmed as thyroid
papillary cancer by at least two pathologists. None of the patients
had received any chemotherapy or radiotherapy before surgery. All
the patients participating in the study gave their informed consent
and protocols were approved by Institutional Ethics Committees of
Tongji University (approval no. SHSY-IEC-pap3.0/13-3).
The ten paired thyroid papillary cancer specimens
and adjacent normal thyroid tissues were handled using TRIzol
(Invitrogen) and miRNeasy mini kit (Qiagen) according to the
manufacturer’s instructions. Total RNA (10 μg) was size
fractionated (<200 nucleotides) by using a mirVana kit (Ambion
Inc., Austin, TX, USA) and labeled using the miRCURY™ Hy3™/Hy5™
Power labeling kit and hybridized on the miRCURY LNA array
(v.16.0). The slides were scanned using the Axon GenePix 4000B
microarray scanner. Scanned images were then imported into GenePix
Pro 6.0 software (Axon) for grid alignment and data extraction.
Data adjustments included data filtering, log2 transformation, and
gene centering and normalization. The t-test analysis was conducted
between thyroid papillary cancer specimens and adjacent normal
thyroid tissues, and miRNA with P-values <0.05 were considered
statistically significant.
PTC specimens and cell culture
In our study, 20 pairs of thyroid papillary cancer
and adjacent normal specimens were collected from the Department of
Breast and Thyroid Surgery of Shanghai Tenth People’s Hospital,
Shanghai, China. The samples were immediately snap-frozen in liquid
nitrogen. Both tumor and normal tissues were histologically
confirmed by more than one experienced pathologist according to the
World Health Organization (WHO) using H&E (hematoxylin and
eosin) staining, and none of these patients had received any
chemotherapy or radiotherapy prior to surgery. Human Nthy-ori 3-1
normal thyroid follicular epithelial cells, human TPC1 thyroid
papillary cancer cells, human K1 thyroid papillary cancer cells and
human B-CPAP thyroid papillary cancer cells were obtained from the
Chinese Science Institute (Shanghai, China). The Nthy-ori 3-1
cells, TPC1 cells and B-CPAP cells were cultured in RPMI-1640
medium (RPMI-1640; Gibco, USA) supplemented with 100 U/ml
penicillin and 100 μg/ml streptomycin (Enpromise, Hangzhou, China),
10% fetal bovine serum (FBS; Gibco). The K1 cells were cultured in
Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with
100 U/ml penicillin and 100 μg/ml streptomycin (Enpromise), 10%
fetal bovine serum (FBS; Gibco). Cells were incubated at 37°C in a
humidified chamber containing 5% CO2. Cells at ~90%
confluence were split at 1:3 ratio every 2–3 days.
Transfection assay
All miRNA mimics were chemically synthesized and
purified by GenePharma (Shanghai, China) based on the following
sequences: hsa-miR-7 mimics: 5′-UGG AAGACUAGUGAUUUUGUUGU-3′,
miR-negative control (miR-NC): 5′-UUCUCCGAACGUGUCCGGAGAATT-3′.
CKS2-siRNA and its NC were chemosynthesized by Guangzhou RiboBio
Co., Ltd. (Guangzhou, China). Cells (1×106) were added
into each well of a 6-well plate and cultured with RPMI-1640 or
DMEM medium without serum and antibiotics. When the confluency of
breast cancer cells reached 30–50%, miR-7 mimics, CKS2-siRNA and
their NC were transfected at working concentrations using
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer’s instructions. After 4–5 h of incubation, RPMI-1640
or DMEM medium was replaced by RPMI-1640 or DMEM with 10% FBS, and
all the cells were incubated at 37°C in a CO2 incubator
for 48–72 h prior to further testing.
RNA extraction and quantitative
reverse-transcription PCR (qRT-PCR)
According to the manufacturer’s protocol, total RNA
was extracted from the cells or tissues using TRIzol (Invitrogen)
and stored at −80°C. For detection of miR-7 expression, primer
design and qRT-PCR were carried out according to the manufacturer’s
instructions. The primers of miR-7 and U6 were purchased from
GenePharma, and U6 was used for normalization. cDNA was generated
by reverse transcription using the PrimeScript™ RT-PCR kit
according to the manufacturer’s instructions (Takara, Tokyo,
Japan). Real-time PCR was performed on a 7900HT Fast RT-PCR
instrument (Applied Biosystems, Singapore). The amplification
procedure was as follows: 5 min at 95°C, followed by 40 cycles at
95°C for 30 sec and 65°C for 45 sec. The relative expression was
evaluated following the relative quantification equation,
2−ΔΔCT. Each sample was tested in triplicate.
Quantitative detection of CKS2 was implemented using the same
strategy. The primers used were as followed: CKS2 forward,
5′-TTCGACGAACACTACGAGTACC-3′; reverse, 5′-GGACACCAAGTCTCCTCCAC-3′.
β-actin forward, 5′-AGCGAGCATCCCCCAAAGTT-3′; reverse,
5′-GGGCACGAAGGCTCATCATT-3′. The PCR parameters for relative
quantification were as follows: 2 min at 95°C, followed by 40
cycles of 45 sec at 57°C and 45 sec at 72°C. The relative
expression was evaluated following the relative quantification
equation, 2−ΔΔCT. All qRT-PCRs were performed in
triplicates.
Cell proliferation assay (MTT assay)
Cell proliferation was determined using the MTT
method. Briefly, 24 h after transfection, the TPC1 and K1 cells
were seeded into 96-well culture plates (BD Biosciences, Franklin
Lakes, NJ, USA) and incubated overnight at 37°C in 5%
CO2. Cell proliferation was assessed at 24, 48, 72, 96
and 120 h following addition of 0.5 mg/ml MTT (Sigma, USA)
solution. After a 4-h incubation, The reaction was stopped by
addition of 150 μl DMSO (Sigma). After 10 min of agitation (100
rpm), the optical density (OD) at 490 nm was determined with
microplate reader (BioTek). Each sample was tested with six
replicates. All experiments were performed in biological
triplicate.
Plate colony formation assay
Twenty-four hours after transfection, Five hundred
TPC1 and K1 cells were seeded in 6-well plates and were incubated
at 37°C. The medium was replaced every 3 days. After 7–10 days, or
when the colonies were visible, the culture was terminated. The
colonies were washed twice with PBS, fixed with 95% ethanol for 10
min and stained with 0.1% crystal violet for 10 min, then the plate
was slowly washed three times with water. When the plate was dried,
the number of visible colonies was counted and representative
colonies were captured. The number of colonies was counted only if
the colony contained >50 cells. Each experiment was performed in
triplicate.
Wound healing assay
In the in vitro wound healing assay, TPC1 and
K1 cells were cultured in 6-well plates until the cell confluence
reached ~90%. Then the plates were washed in PBS after making a
scratch in each well using a sterile pipette tip. Wound healing was
observed under a light microscope and images were captured at the
same view at 0, 12, 24 and 48 h after scratching to observe the
process of wound healing. The experiments were repeated twice and
representative images are shown.
Transwell invasion assay
A Transwell invasion assay was performed by using
Chemicon Cell Invasion assay kit (Chemicon, USA). Cells
(5×104 cells/Transwell) were plated in the top chamber
of Transwells with a Matrigel (2 mg/ml)-coated membrane with 8-μm
diameter pores in 200 μl serum-free DMEM. The lower chambers were
filled with 500 μl of DMEM containing 10% FBS. After 48 h of
incubation, the membrane was stained with 0.1% crystal violet and
observed under a microscope after removing the Matrigel and cells
in the upper chambers. Five fields were randomly selected from each
membrane, and the number of cells penetrating the membrane was
counted at a magnification of ×200. The invasion ability was
described as the number of invading cells. Each experiment was
carried out in triplicate. Membrane-binding crystal violet was
dissolved with 400 μl 33% glacial acetic acid, and then absorbance
at 573 nm was measured using a microplate reader.
Cell cycle assay and apoptosis assay
For all groups, 106 cells were collected
in PBS and resuspended in 70% ethanol to fix overnight at 4°C.
Cells were pelleted at 1,000 rpm/min for 5 min, washed in PBS, and
then pelleted at 1,000 rpm/min for 5 min. A total of 250 μl 0.05
g/l propidium iodide (PI) staining solution was added into each
sample and incubated for 30 min at room temperature, and cell cycle
distribution was analyzed using flow cytometry (FACSCanto™ II, BD
Biosciences). For cell apoptosis assay, cells transfected with
miR-7 and negative control were incubated in 6-well plates for 24
h. Cells were subsequently stained with fluorescein
(FITC)-conjugated Annexin V and propidium iodide (FITC-Annexin
V/PI) (BD Biosciences, San Diego, CA, USA), the rate of apoptosis
was detected by flow cytometry (FACSCanto II, BD Biosciences).
Dual-luciferase reporter assay
293T cells were seeded in 12-well plates (BD, USA)
and cultured until the cells reached 80–90% confluence. The CKS2
3′-UTR was cloned into the psiCHECK-2 vector containing the RL
gene, and co-transfected into cells together with miR-7 mimics or
negative control (100 nM) using Lipofectamine 2000 (Invitrogen)
according to the manufacturer’s instructions. Thirty-six hours
after transfection, luciferase activity was measured using the
dual-luciferase reporter assay kit (Promega, Madison, WI, USA).
Briefly, the cells were washed twice with PBS and lysed by
incubation on ice for 30 min with passive lysis buffer (PLB, 250
μl/well). The supernatants were collected, and 20 μl of the
aliquots were added to 96-well plates. The firefly luciferase (FL)
reporter was measured by a microplate spectrophotometer immediately
after adding 50 μl of Luciferase Assay Reagent II. Next, Stop &
Glo® reagent (50 μl/well) was added to each well to
initiate the Renilla luciferase (RL). RL activity was
normalized to FL activity. All experiments were performed three
times.
Western blot analysis
Cells were washed in ice-cold PBS and resuspended in
RIPA lysis buffer (100 μl/well, Beyotime). Then the cells were
collected and centrifuged for 30 min at 4°C (Eppendorf 5804R,
Eppendorf Biotech, Germany). Supernatants were collected and the
protein concentrations were quantified using a BCA protein assay
kit (Beyotime). Protein samples were denatured with 5X SDS loading
buffer (Beyotime) at 100°C for 10 min. Next, whole protein samples
were separated by 8% SDS-polyacrylamide gel electrophoresis
(SDS-PAGE) and transferred onto 0.45-μm nitrocellulose membranes
(Beyotime). After 1 h of blocking in 5% fat-free milk, the
membranes were incubated with the CKS2 (1:1,000), cyclin B1
(1:1,000), cdk1 (1:1,000), cyclin A (1:1,000) and the β-actin
(1:1,500) antibody (all from Cell Signaling Technology, USA)
overnight at 4°C. Protein blots were washed and then incubated for
1 h with specific secondary antibodies. After washing by PBST 3
times, immunoreactive protein bands were detected using the Odyssey
scanning system (LI-COR, Lincoln, NE, USA).
Statistical analysis
Data were presented as the means ± standard
deviation (SD) from at least three independent experiments. The
Students t-test was used to evaluate the differences between each
group in SPSS 20.0 software. Differences were considered
significant for P-values <0.05.
Results
miR-7 is downregulated in both human
thyroid papillary cancer specimens and cell lines
In our experiment, miRNA-microarray was first
performed to analyze the miRNA expression between thyroid papillary
cancer tissues and para-cancer tissues. As shown in Table I, expression of 83 miRNAs was
upregulated and 75 miRNAs were downregulated with a P-value
<0.05. We also found that the expression of miR-7 was
significantly decreased in cancer tissues compared to para-cancer
tissues, nearly 5.64-fold change. Our results were partly
consistent with the findings of Pallante et al (20). To explore the role of miR-7 in
human thyroid papillary carcinomas, we further analyzed 20 pairs of
thyroid papillary cancer and adjacent normal specimens in this
study. Total RNAs were isolated from excised tumor tissues and
benign tissues of patients with thyroid papillary cancer. Detection
of miR-7 by qRT-PCR indicated that miR-7 levels were obviously
downregulated in thyroid papillary cancer tissues compared with
benign tissues, which was consistent with our finding in
miRNA-microarray (P<0.001, Fig.
1A). Moreover, expression of miR-7 was also demontrated to be
downregulated in three collected thyroid papillary cancer cell
lines compared to Nthy-ori 3-1, a normal thyroid follicular
epithelial cell line (P<0.05, Fig.
1B). In this experiment, we also explored the relationship
between clinical features and miR-7 expression levels, in our case,
a negative correlation was found between tumor size and miR-7
expression levels (r=−0.249, P<0.01, Fig. 1C), as well as number of metastatic
lymph nodes and miR-7 expression levels (r=−0.515, P<0.001,
Fig. 1D). These findings imply a
potential role in tumor growth and metastasis of miR-7 in thyroid
papillary cancer.
 | Table IDifferential expression of miRNAs in
thyroid papillary cancer specimens compared with para-cancer
tissues.a |
Table I
Differential expression of miRNAs in
thyroid papillary cancer specimens compared with para-cancer
tissues.a
| MicroRNA | Fold change | P-value |
|---|
| Upregulated | hsa-miR-135b | 7.89 | 0.011 |
| hsa-miR-142 | 6.76 | 0.044 |
| hsa-miR-146b | 11.70 |
3.12e−5 |
| hsa-miR-15a | 9.11 | 0.002 |
| hsa-miR-19a-3p | 5.85 | 0.043 |
| hsa-miR-200a | 7.90 | 0.036 |
| hsa-miR-200b | 9.62 | 0.011 |
| hsa-miR-21 | 5.64 |
4.34e−6 |
| hsa-miR-221 | 8.16 |
3.11e−6 |
| hsa-miR-222 | 8.33 |
1.49e−5 |
| hsa-miR-301a | 3.86 | 0.002 |
| hsa-miR-31 | 5.76 |
1.55e−5 |
| hsa-miR-429 | 6.86 | 0.009 |
| hsa-miR-96 | 5.45 | 0.008 |
| Downregulated | hsa-miR-144 | 3.04 |
7.36e−4 |
|
hsa-miR-193b-5p | 3.73 |
2.84e−4 |
| hsa-miR-197 | 7.30 | 0.004 |
| hsa-miR-204 | 3.62 | 0.005 |
| hsa-miR-424 | 3.04 | 0.001 |
| hsa-miR-451a | 9.60 |
4.10e−7 |
| hsa-miR-486 | 3.57 |
7.90e−4 |
| hsa-miR-494 | 6.07 | 0.001 |
| hsa-miR-7 | 5.64 | 0.002 |
| hsa-miR-718 | 3.21 | 0.020 |
| hsa-miR-887 | 3.04 |
1.86e−5 |
miR-7 inhibits the proliferation of
thyroid papillary cancer cells in vitro
As two of the most representative thyroid papillary
cancer cell lines, TPC1 and K1 cells were selected in our following
experiments. To explore the role of endogenous miR-7 in thyroid
papillary cancer, miR-7 was overexpressed in the TPC1 and K1 cells
by transiently transfecting with miR-7 mimics, and scrambled miRNA
sequences (miR-NC) were used as negative control. Following
transfection, increased miR-7 expression was confirmed by qRT-PCR
(P<0.01, Fig. 2). Cell
proliferation assay (MTT assay) indicated that overexpression of
miR-7 resulted in significant inhibition of cell proliferation in
both TPC1 and K1 cells (P<0.05, P<0.01, P<0.001, Fig. 3A and B). Colony formation assays
also showed less colony formation in the group transfected with
miR-7 mimics compared with the miR-NC group (P<0.01, Fig. 3C and D). Our data indicated that
miR-7 can inhibit cell proliferation in thyroid papillary cancer
cells in vitro.
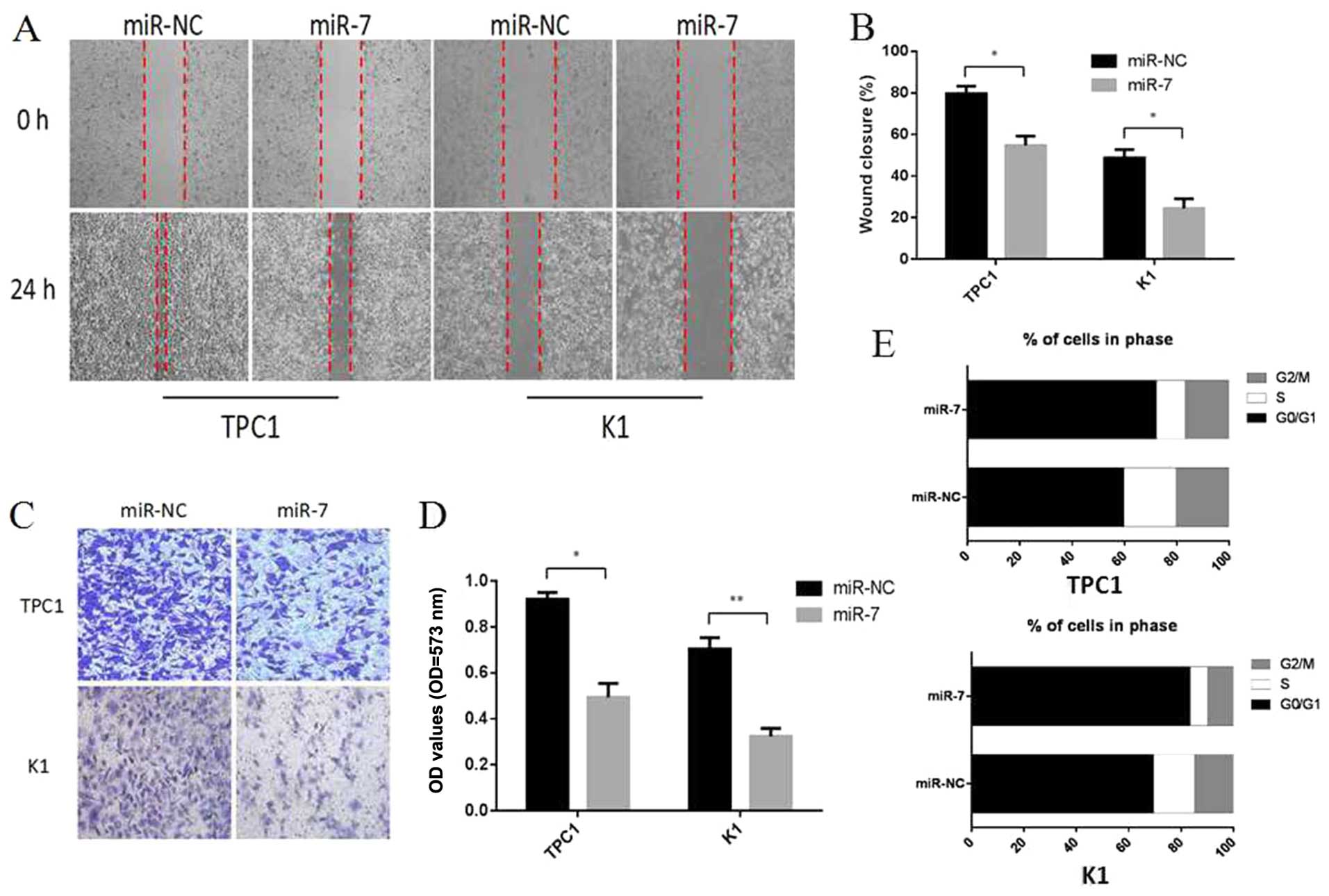
miR-7 inhibits thyroid papillary cancer
cell migration and invasion in vitro
To further elucidate the functional role of miR-7 in
thyroid papillary cancer, we assessed the impact of its
overexpression on metastatic processes, wound healing assays and
Transwell assays were performed in TPC1 and K1 cells. Cells were
transiently transfected with miR-7 mimics or miR-NC. As shown in
Fig. 4A and B, 24 h after drawing
the ‘scratch’ line on the monolayer TPC1 cells, the miR-NC group
nearly filled in the gap, while the miR-7 group still showed a
clear gap in the scratched region, and the experiments carrying out
in K1 cells also showed a similar trend. Percentage wound closure
was calculated using edge detection and area calculation with
ImageJ software (P<0.05). The results indicate that
overexpression of miR-7 in TPC1 and K1 cells can inhibit cellular
migration. The Transwell invasion assay revealed that the number of
TPC1 and K1 cells penetrating the membrane significantly decreased
at 48 h after miR-7 mimics transfection as compared to the miR-NC
group (P<0.05, P<0.01, Fig. 4C
and D). Taken together, these results showed that
overexpression of miR-7 can inhibit cellular migration and invasion
in vitro.
miR-7 regulates the cell cycle of thyroid
papillary cancer cells
The reason of the reduction in cell proliferation
following miR-7 overexpression was explored by using cell division
and cell apoptosis analysis. Cell division analysis detected by
flow cytometry revealed that TPC1 and K1 cells transfected with
miR-7 had a significantly reduced percentage in S and G2/M phase
and an increase in G0/G1 phase compared to miR-NC transfected cells
(P<0.05, Fig. 4E). Cell death
analysis using apoptosis assays showed no significant changes in
the cell population transfected with miR-7 compared to miR-NC group
(P>0.05, data not shown). These findings revealed that miR-7 can
lead to upregulation of G0/G1 phase cells and we propose that miR-7
acts to reduce cell proliferation partly by inducing G0/G1 cell
cycle arrest in thyroid papillary cancer.
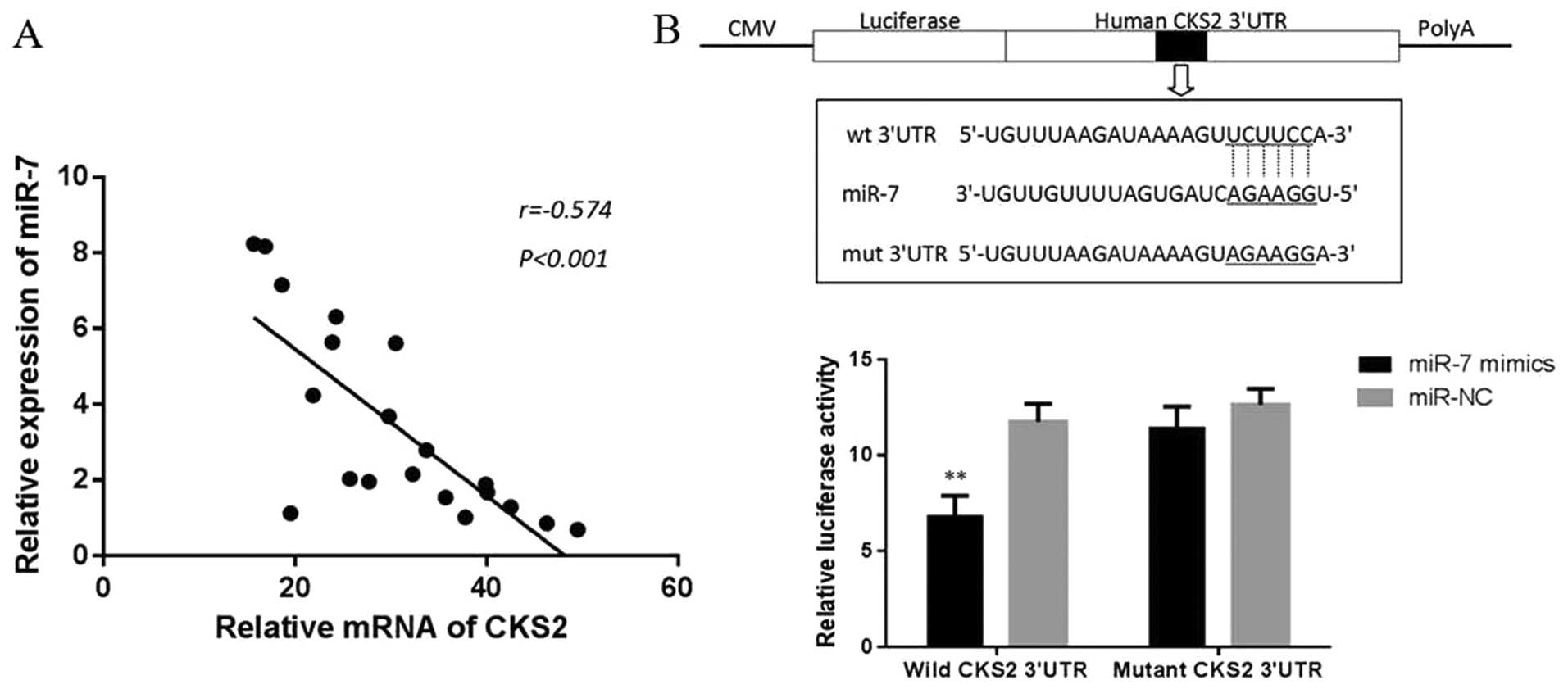
miR-7 regulates the expression of CKS2
and its downstream genes
To investigate the mechanism of miR-7 in thyroid
papillary cancer, we screened the target genes of miR-7 by using
Targetscan and microRNA.org (http://www.targetscan.org/ and http://www.microrna.org). CKS2 (also referred to as
CDC28 protein kinase regulatory subuinit2, or Cks2), a member of
cell cycle dependent protein kinase subunits family, was identified
as a candidate. Then, we explore the correlation between the
expression of miR-7 and CKS2 in 20 collected clinical thyroid
papillary cancer specimens. Interestingly, miR-7 levels were found
to be markedly inversely correlated with CKS2 expression (r=−0.574,
P<0.001, Fig. 5A), suggesting
that miR-7 might target CKS2 mRNA in thyroid papillary cancer. To
verify whether miR-7 can bind to the predicted site of CKS2, we
performed luciferase reporter assay in the 293T cell line. As shown
in Fig. 5B, the luciferase
activity significantly decreased after co-transfection with
psi-CHECK-2/CKS2 3′-UTR and miR-7 mimics in comparison with control
cells, indicating that miR-7 specifically binds to the 3′-UTR of
CKS2 mRNA. In addition, we analyzed the level of CKS2 expression in
TPC1 and K1 cells transfected with miR-7 mimic or miR-NC by western
blotting. The results showed that miR-7 overexpression
significantly reduced the CKS2 expression at protein levels. We
also found that the downstream gene of CKS2, such as cyclin B1 and
cdk1, can also be inhibited by miR-7 and CKS2 axis at the protein
level (Fig. 6). At last, we
investigated the role of CKS2 in thyroid papillary cancer.
Silencing of CKS2 by CKS2-siRNA can inhibit proliferation,
invasion, migration and induce G0/G1 cell cycle arrest in TPC1 and
K1 thyroid papillary cancer cells (Fig. 7). Thus, our data indicated that
miR-7 can regulate CKS2 and its downstream genes in thyroid
papillary cancer.
Discussion
Thyroid papillary carcinoma is the most common form
of follicular-cell derived carcinomas and comprises almost 75% of
all newly-diagnosed thyroid cancers (21). Accumulating evidence indicates that
the aberrant expression of miRNAs contributes to thyroid papillary
carcinoma tumorigenesis and metastasis through repression of their
target genes, suggesting that miRNAs may serve as molecular
biomarkers for the prediction and prognosis of PTC, and as novel
targets for disease treatment (22). Dysregulation of miRNAs is connected
with initiation and progression of thyroid cancer, since they may
serve as oncogenes or tumor suppressors. For example, upregulation
of miR-146b significantly promoted cell migration and invasiveness
and increased resistance to chemotherapy-induced apoptosis in
thyroid papillary carcinoma (23).
miR-34a can regulate growth arrest-specific 1 (GAS1) expression to
promote proliferation and suppress apoptosis in thyroid papillary
cancer cells via the PI3K/Akt/Bad pathway (24). In this study, we are interested in
the role of miR-7 in thyroid papillary carcinoma and speculate CKS2
as one of its possible targets.
CKS2 is the member of cell cycle dependent protein
kinase subunits family, which participates in cell cycle regulation
(25–27). CKS2 plays an important role in the
process of somatic cell division, it also exhibits certain
functions in tumor development (28–30).
It is reported that CKS2 is upregulated in many types of tumors,
including prostate cancer, bladder cancer, breast cancer and liver
cancer (31,32). As the downstream genes of CKS2,
cdk1 and cyclin B1 are known to be important players in the cell
cycle. Many studies have demonstrated that cyclin B1-cdk1 protein
kinase, also known as mitosis promoting factor (MPF), is essential
for mitosis and that in its absence, cells are unable to progress
past the G2 phase of the cell cycle, CKS2 binds to cdk1 via
interaction with the catalytic subunit of cdk1, and subsequently
affects cell cycle (33). Many
human malignancies are characterized by CKS2 overexpression, and it
is generally known as an oncogene. However, the underlying cellular
functions of CKS2 in thyroid papillary carcinoma and related
mechanisms remain largely unexplored.
In our experiment, miRNA-microarray was firstly
performed in thyroid papillary cancer specimens, miR-7 was
significantly decreased in cancer tissues compared to para-cancer
tissues. To validate the result in miRNA-microarray, we
investigated the expression levels of miR-7 in 20-paired thyroid
papillary cancer and adjacent normal specimens. Interestingly, we
observed that the expression levels of miR-7 were also remarkably
decreased in cancer tissues relative to paired non-tumor tissues.
Since miR-7 has been described as a tumor suppressor gene in
several human cancers including glioblastoma (34), hepatocellular carcinoma (16) and lung cancer (35), we hypothesized that miR-7 might be
a novel tumor-suppressor miRNA in thyroid papillary cancer. Then we
investigated the specific role of miR-7 in two typical thyroid
papillary cancer cell lines, TPC1 and K1. Cells were transfected
with miR-7 mimics or miR-NC respectively to detect the effects on
various aspects of thyroid cancer biology. Our results showed that
the exogenous overexpression of miR-7 regulating by miR-7 mimics
inhibited proliferation and colony formation ability of thyroid
papillary cancer cells as measured by MTT and colony formation
assays. Moreover, cell migration and invasion ability was also
significantly inhibited by overexpression of miR-7. We also found
that miR-7 can distinctly induce G0/G1 cell cycle arrest. We tried
to explore the correlation between the expression levels of miR-7
and CKS2 in clinical thyroid papillary cancer specimens. As
expected, a negative correlation was observed between these two
variables. Luciferase reporter assay identified that miR-7 could
directly bind to the 3′-UTR of CKS2. In order to further explore
the molecular mechanism of the growth inhibition induced by miR-7,
we examined its effect on the expression of a panel of CKS2
downstream genes, namely cyclin B1, cyclin A and cdk1. Western blot
assays demonstrated that miR-7 was able to downregulate CKS2
protein, cyclin B1 protein and cdk1 protein. We concluded that
knockdown of endogenous CKS2 can mimic the result of miR-7
upregulation in thyroid papillary cancer cells. Collectively, the
experimental results showed that miR-7 inhibits the proliferation,
migration and invasion of thyroid papillary cancer cells via
targeting CKS2.
In recent years, several studies on miR-7 and cancer
were accomplished. It has been confirmed as a tumor-suppressor
factor in multiple cancers including those associated with poor
outcomes such as glioblastoma (34), non-small cell lung cancer (35) and gastric cancer (36) as well as some endocrine related
cancers such as breast, prostate and ovarian cancer (37). Glover et al found that miR-7
regulates adrenocortical carcinoma by targeting multiple cell
pathways and miR-7 replacement therapy is effective in reducing
tumor growth (38). Additionally,
they also demonstrated that miR-7 therapy in vivo could lead
to inhibition of cdk1, which is partially consistent with our
findings. In this experiment, we also tried to explore the
relationship between clinical data and miR-7 levels. The tumor size
and the number of metastatic lymph nodes was found to have a
negatively correlation with the relative miR-7 expression in our
cases, which suggested that miR-7 might be a reliable biological
marker in diagnosis of thyroid papillary carcinoma. Since this is a
pattern verified only in 20 clinical cases, the reliability is
relatively low and needs to be confirmed in a larger number of
samples.
In conclusion, our findings demonstrate that miR-7
is downregulated in papillary thyroid carcinoma tissues and cell
lines. This study also provides novel evidence that in a model of
papillary thyroid carcinoma, upregulation of miR-7 inhibits
cellular growth, suppresses cellular migration and invasion,
induces a G0/G1 arrest likely by targeting CKS2 and ultimately
regulating the expression of cyclinB1 and cdk, which could be
considered as a basis for the development of miRNA-targeted
therapies for papillary thyroid carcinoma.
Acknowledgements
This study was supported by grants from National
Natural Science Foundation of China (no. 82172240). We sincerely
thank all the teachers at the Central Laboratory of the Shanghai
Tenth People’s Hospital for their support.
References
|
1
|
Zhang J, Wang Y, Li D and Jing S: Notch
and TGF-β/Smad3 pathways are involved in the interaction between
cancer cells and cancer-associated fibroblasts in papillary thyroid
carcinoma. Tumour Biol. 35:379–385. 2014. View Article : Google Scholar
|
|
2
|
Lloyd RV, Buehler D and Khanafshar E:
Papillary thyroid carcinoma variants. Head Neck Pathol. 5:51–56.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Silver CE, Owen RP, Rodrigo JP, Rinaldo A,
Devaney KO and Ferlito A: Aggressive variants of papillary thyroid
carcinoma. Head Neck. 33:1052–1059. 2011. View Article : Google Scholar
|
|
4
|
Scheumann GF, Gimm O, Wegener G,
Hundeshagen H and Dralle H: Prognostic significance and surgical
management of locoregional lymph node metastases in papillary
thyroid cancer. World J Surg. 18:559–567; discussion 567–568. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mazzaferri EL and Kloos RT: Clinical
review 128: Current approaches to primary therapy for papillary and
follicular thyroid cancer. J Clin Endocrinol Metab. 86:1447–1463.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pellegriti G, Scollo C, Lumera G,
Regalbuto C, Vigneri R and Belfiore A: Clinical behavior and
outcome of papillary thyroid cancers smaller than 1.5 cm in
diameter: Study of 299 cases. J Clin Endocrinol Metab.
89:3713–3720. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Grant CS: Recurrence of papillary thyroid
cancer after optimized surgery. Gland Surg. 4:52–62.
2015.PubMed/NCBI
|
|
8
|
Lin X, Guan H, Li H, Liu L, Liu J, Wei G,
Huang Z, Liao Z and Li Y: miR-101 inhibits cell proliferation by
targeting Rac1 in papillary thyroid carcinoma. Biomed Rep.
2:122–126. 2014.PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen YT, Kitabayashi N, Zhou XK, Fahey TJ
III and Scognamiglio T: MicroRNA analysis as a potential diagnostic
tool for papillary thyroid carcinoma. Mod Pathol. 21:1139–1146.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kong X, Li G, Yuan Y, He Y, Wu X, Zhang W,
Wu Z, Chen T, Wu W, Lobie PE, et al: MicroRNA-7 inhibits
epithelial-to-mesenchymal transition and metastasis of breast
cancer cells via targeting FAK expression. PLoS One. 7:e415232012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li J, Zheng Y, Sun G and Xiong S:
Restoration of miR-7 expression suppresses the growth of Lewis lung
cancer cells by modulating epidermal growth factor receptor
signaling. Oncol Rep. 32:2511–2516. 2014.PubMed/NCBI
|
|
15
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fang Y, Xue JL, Shen Q, Chen J and Tian L:
MicroRNA-7 inhibits tumor growth and metastasis by targeting the
phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma.
Hepatology. 55:1852–1862. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Demetrick DJ, Zhang H and Beach DH:
Chromosomal mapping of the human genes CKS1 to 8q21 and CKS2 to
9q22. Cytogenet Cell Genet. 73:250–254. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Urbanowicz-Kachnowicz I, Baghdassarian N,
Nakache C, Gracia D, Mekki Y, Bryon PA and French M: ckshs
expression is linked to cell proliferation in normal and malignant
human lymphoid cells. Int J Cancer. 82:98–104. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lv M, Zhang X, Li M, Chen Q, Ye M, Liang
W, Ding L, Cai H, Fu D and Lv Z: miR-26a and its target CKS2
modulate cell growth and tumorigenesis of papillary thyroid
carcinoma. PLoS One. 8:e675912013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pallante P, Visone R, Ferracin M, Ferraro
A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M,
Negrini M, et al: MicroRNA deregulation in human thyroid papillary
carcinomas. Endocr Relat Cancer. 13:497–508. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ban Y, Yamamoto G, Takada M, Hayashi S,
Ban Y, Shimizu K, Akasu H, Igarashi T, Bando Y, Tachikawa T, et al:
Proteomic profiling of thyroid papillary carcinoma. J Thyroid Res.
2012:8150792012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aragon Han P, Weng CH, Khawaja HT,
Nagarajan N, Schneider EB, Umbricht CB, Witwer KW and Zeiger MA:
MicroRNA expression and association with clinicopathologic features
in papillary thyroid cancer: A systematic review. Thyroid.
25:1322–1329. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chou CK, Yang KD, Chou FF, Huang CC, Lan
YW, Lee YF, Kang HY and Liu RT: Prognostic implications of miR-146b
expression and its functional role in papillary thyroid carcinoma.
J Clin Endocrinol Metab. 98:E196–E205. 2013. View Article : Google Scholar
|
|
24
|
Ma Y, Qin H and Cui Y: miR-34a targets
GAS1 to promote cell proliferation and inhibit apoptosis in
papillary thyroid carcinoma via PI3K/Akt/Bad pathway. Biochem
Biophys Res Commun. 441:958–963. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pines J: Cell cycle: Reaching for a role
for the Cks proteins. Curr Biol. 6:1399–1402. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Spruck CH, de Miguel MP, Smith AP, Ryan A,
Stein P, Schultz RM, Lincoln AJ, Donovan PJ and Reed SI:
Requirement of Cks2 for the first metaphase/anaphase transition of
mammalian meiosis. Science. 300:647–650. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Frontini M, Kukalev A, Leo E, Ng YM,
Cervantes M, Cheng CW, Holic R, Dormann D, Tse E, Pommier Y, et al:
The CDK subunit CKS2 counteracts CKS1 to control cyclin A/CDK2
activity in maintaining replicative fidelity and neurodevelopment.
Dev Cell. 23:356–370. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Martinsson-Ahlzén HS, Liberal V,
Grünenfelder B, Chaves SR, Spruck CH and Reed SI: Cyclin-dependent
kinase-associated proteins Cks1 and Cks2 are essential during early
embryogenesis and for cell cycle progression in somatic cells. Mol
Cell Biol. 28:5698–5709. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen R, Feng C and Xu Y: Cyclin-dependent
kinase-associated protein Cks2 is associated with bladder cancer
progression. J Int Med Res. 39:533–540. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kita Y, Nishizono Y, Okumura H, Uchikado
Y, Sasaki K, Matsumoto M, Setoyama T, Tanoue K, Omoto I, Mori S, et
al: Clinical and biological impact of cyclin-dependent kinase
subunit 2 in esophageal squamous cell carcinoma. Oncol Rep.
31:1986–1992. 2014.PubMed/NCBI
|
|
31
|
Wang J, Xu L, Liu Y, Chen J, Jiang H, Yang
S and Tan H: Expression of cyclin kinase subunit 2 in human breast
cancer and its prognostic significance. Int J Clin Exp Pathol.
7:8593–8601. 2014.
|
|
32
|
Shen DY, Fang ZX, You P, Liu PG, Wang F,
Huang CL, Yao XB, Chen ZX and Zhang ZY: Clinical significance and
expression of cyclin kinase subunits 1 and 2 in hepatocellular
carcinoma. Liver Int. 30:119–125. 2010. View Article : Google Scholar
|
|
33
|
Dorée M and Hunt T: From Cdc2 to Cdk1:
When did the cell cycle kinase join its cyclin partner? J Cell Sci.
115:2461–2464. 2002.PubMed/NCBI
|
|
34
|
Kefas B, Godlewski J, Comeau L, Li Y,
Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, et
al: MicroRNA-7 inhibits the epidermal growth factor receptor and
the Akt pathway and is down-regulated in glioblastoma. Cancer Res.
68:3566–3572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xiong S, Zheng Y, Jiang P, Liu R, Liu X
and Chu Y: MicroRNA-7 inhibits the growth of human non-small cell
lung cancer A549 cells through targeting BCL-2. Int J Biol Sci.
7:805–814. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao X, Dou W, He L, Liang S, Tie J, Liu
C, Li T, Lu Y, Mo P, Shi Y, et al: MicroRNA-7 functions as an
anti-metastatic microRNA in gastric cancer by targeting
insulin-like growth factor-1 receptor. Oncogene. 32:1363–1372.
2013. View Article : Google Scholar
|
|
37
|
Kalinowski FC, Brown RAM, Ganda C, Giles
KM, Epis MR, Horsham J and Leedman PJ: MicroRNA-7: A tumor
suppressor miRNA with therapeutic potential. Int J Biochem Cell
Biol. 54:312–317. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Glover AR, Zhao JT, Gill AJ, Weiss J,
Mugridge N, Kim E, Feeney AL, Ip JC, Reid G, Clarke S, et al:
MicroRNA-7 as a tumor suppressor and novel therapeutic for
adrenocortical carcinoma. Oncotarget. 6:36675–36688.
2015.PubMed/NCBI
|