Introduction
Esophageal squamous cell carcinoma (ESCC) is one of
the most aggressive cancers and the major histological type of
esophageal cancer in Japan and East Asia (1–3).
ESCC cells frequently metastasize to the lymph nodes, liver, lungs
and bone (2–4). Despite the use of multimodality
therapies, the prognosis of patients with ESCC is still poor, with
an overall 5-year survival rate of approximately 20–30% (2,4).
Recently developed molecularly targeted therapeutics have not been
shown to have beneficial effects in patients with ESCC (2). Additionally, the molecular
pathogenesis of the aggressive phenotype in ESCC remains unclear.
Thus, in order to improve disease outcomes in patients with ESCC,
it is necessary to elucidate the molecular mechanisms of ESCC cell
aggressiveness using advanced genomic approaches.
The discovery of microRNAs (miRNAs) has resulted in
major advancements in cancer research (5,6).
miRNAs are small non-coding RNAs that function to fine tune the
expression of protein coding/non-coding RNAs by repressing
translation or cleaving RNA transcripts in a sequence-depending
manner (7). The unique
characteristic function of miRNAs is to regulate RNA transcripts in
human cells. Therefore, dysregulated expression of miRNAs can
disrupt tightly regulated RNA networks in cancer cells. Currently,
numerous studies have shown that miRNAs are aberrantly expressed in
several cancers, including ESCC (6,8).
Using miRNA expression signature analyses, we have sequentially
identified tumor-suppressive miRNAs and shown that these miRNAs
mediate novel cancer networks (9–13).
Our miRNA expression signatures revealed that
microRNA-375 (miR-375) is frequently downregulated in
several types of squamous cell carcinoma (10,13,14).
Moreover, our previous studies demonstrated that ectopic expression
of miR-375 suppressed cancer cell aggressiveness in several
types of cancer cells (15). In
ESCC cells, several studies have indicated that miR-375 has
antitumor roles through targeting oncogenic genes (16,17).
Moreover, miR-375-mediated cancer pathways are essential for
cancer cell initiation, development and aggressiveness.
Accordingly, in the present study, we aimed to
investigate the novel cancer networks regulated by miR-375
in ESCC cells. Our present data showed that matrix
metalloproteinase 13 (MMP13) was directly regulated by
miR-375 in ESCC cells. Overexpression of MMP13 was
observed in ESCC clinical tissues, and knockdown of MMP13
expression markedly inhibited ESCC cell migration and invasion,
indicating that MMP13 acted as a cancer-promoting gene in
ESCC cells. Moreover, the oncogenic genes CENPF,
KIF14 and TOP2 were found to function downstream of
MMP13. Taken together, these results showed that the
antitumor miR-375/oncogenic MMP13 axis had a pivotal
role in ESCC aggressiveness.
Materials and methods
Clinical ESCC specimens and ESCC cell
lines
Clinical specimens were collected from 25 patients
with ESCC. All patients underwent primary surgical treatment and
were pathologically proven to have ESCC at the Kagoshima University
Hospital from 2010 to 2014. The present study was approved by the
Bioethics Committee of Kagoshima University; written prior informed
consent and approval were obtained from all patients. The
clinicopathological characteristics of the patients are shown in
Table I.
 | Table IClinical features of patients with
ESCC. |
Table I
Clinical features of patients with
ESCC.
| No. | Age (years) | Gender |
Differentiation | T | N | M | Stage | ly | v | Recurrence |
|---|
| 1 | 68 | Male | Poor | 1b | 2 | 0 | IIIA | 1 | 3 | + |
| 2 | 72 | Male | Moderate | 1b | 0 | 0 | IA | 0 | 1 | − |
| 3 | 69 | Male | Moderate | 1b | 0 | 0 | IIIA | 0 | 0 | − |
| 4 | 62 | Male | Well | 3 | 2 | 0 | IIIB | 1 | 1 | + |
| 5 | 66 | Male | Moderate | 3 | 0 | 0 | IIA | 1 | 1 | − |
| 6 | 74 | Male | Moderate | 2 | 2 | 0 | IIIA | 3 | 1 | + |
| 7 | 56 | Male | Moderate | 2 | 0 | 0 | IB | 0 | 1 | − |
| 8 | 79 | Male | Moderate | 2 | 1 | 0 | IIB | 1 | 1 | − |
| 9 | 68 | Male | Moderate | 1b | 2 | 0 | IIIA | 1 | 1 | − |
| 10 | 52 | Male | Poor | 1b | 0 | 0 | IA | 1 | 1 | + |
| 11 | 67 | Male | Well | 3 | 2 | 0 | IIIB | 2 | 2 | + |
| 12 | 57 | Male | Poor | 3 | 3 | 0 | IIIC | 1 | 1 | + |
| 13 | 70 | Male | Moderate | 3 | 0 | 0 | IIA | 1 | 1 | + |
| 14 | 66 | Male | Moderate | 3 | 0 | 0 | IIA | 1 | 1 | − |
| 15 | 63 | Male | Well | 3 | 3 | 0 | IIIC | 2 | 1 | + |
| 16 | 55 | Male | Moderate | 3 | 2 | 0 | IIIB | 1 | 1 | + |
| 17 | 60 | Male | Well | 1b | 1 | 0 | IIB | 1 | 1 | − |
| 18 | 78 | Male | Well | 3 | 0 | 0 | IIA | 1 | 2 | − |
| 19 | 71 | Male | Well | 3 | 0 | 0 | IIA | 1 | 2 | − |
| 20 | 75 | Male | Moderate | 3 | 2 | 0 | IIIB | 1 | 1 | + |
| 21 | 60 | Male | Moderate | 2 | 1 | 0 | IIB | 1 | 2 | − |
| 22 | 62 | Male | Well | 1a | 1 | 0 | IIB | 0 | 0 | − |
| 23 | 71 | Male | Moderate | 1b | 1 | 0 | IIB | 0 | 0 | − |
| 24 | 69 | Male | Moderate | 1b | 0 | 0 | IA | 1 | 0 | − |
| 25 | 84 | Male | Well | 2 | 1 | 0 | IIB | 1 | 1 | − |
We used two ESCC cell lines: TE-8, which was
moderately differentiated; and TE-9, which was poorly
differentiated. Both of these cells lines were provided by Riken
BioResourse Center (Tsukuba, Japan).
Extraction of total RNA from clinical specimens and
cell lines was performed using ISOGEN (Nippon Gene, Tokyo, Japan)
according to the manufacturer's protocol. The quality of RNA was
checked using an Agilent 2100 Bioanalyzer (Agilent Technologies,
Santa Clara, CA, USA).
Quantitative real-time reverse
transcription polymerase chain reaction (qRT-PCR)
The procedure for PCR quantification was previously
described (13,18–20).
The expression levels of miR-375 (assay ID: 000564; Applied
Biosystems, Foster City, CA, USA) were analyzed by TaqMan qRT-PCR
assays (TaqMan MicroRNA assays; Applied Biosystems) and
RNU48 (assay ID: 001006) was used for normalization. TaqMan
probes and primers for MMP-13 (assay ID: Hs00233992_m1;
Applied Biosystems), CENPF (assay ID: Hs01118845_m1),
KIF14 (assay ID: Hs00978236_m1) and GUSB (the
internal control; assay ID: Hs00939627_ml; Applied Biosystems) were
used for gene expression analysis.
Transfection with mature miRNAs and small
interfering RNAs (siRNAs)
The following mature miRNA was used: Ambion Pre-miR
miRNA precursor for hsa-miR-375 (product ID: PM10327;
Applied Biosystems). The following siRNAs were used: Stealth Select
RNAi siRNA, si-MMP13 (cat nos. HSS106637 and HSS106638;
Invitrogen, Carlsbad, CA, USA), and negative control miRNA/siRNA
(P/N: AM17111; Applied Biosystems). RNAs were incubated with
Opti-MEM (Invitrogen) and Lipofectamine RNAiMax transfection
reagent (Invitrogen), as previously described (13,18–20).
Cell proliferation, migration and
invasion assays
TE-8 and TE-9 cells were transfected with 10 nM
miRNAs or siRNAs by reverse transfection. Cell proliferation,
migration and invasion assays were performed as previously
described (13,18–20).
Screening of miR-375 target genes using
in silico analysis and gene expression data
To identify miR-375 target genes, a
combination of genome-wide gene expression and in silico
analyses was conducted as previously described (13,18–20).
The microarray data were deposited into the GEO repository under
accession number GSE77790. Next, we selected putative miRNA target
genes using microRNA.org (August, 2010 release, http://www.microrna.org) databases. Our strategy for
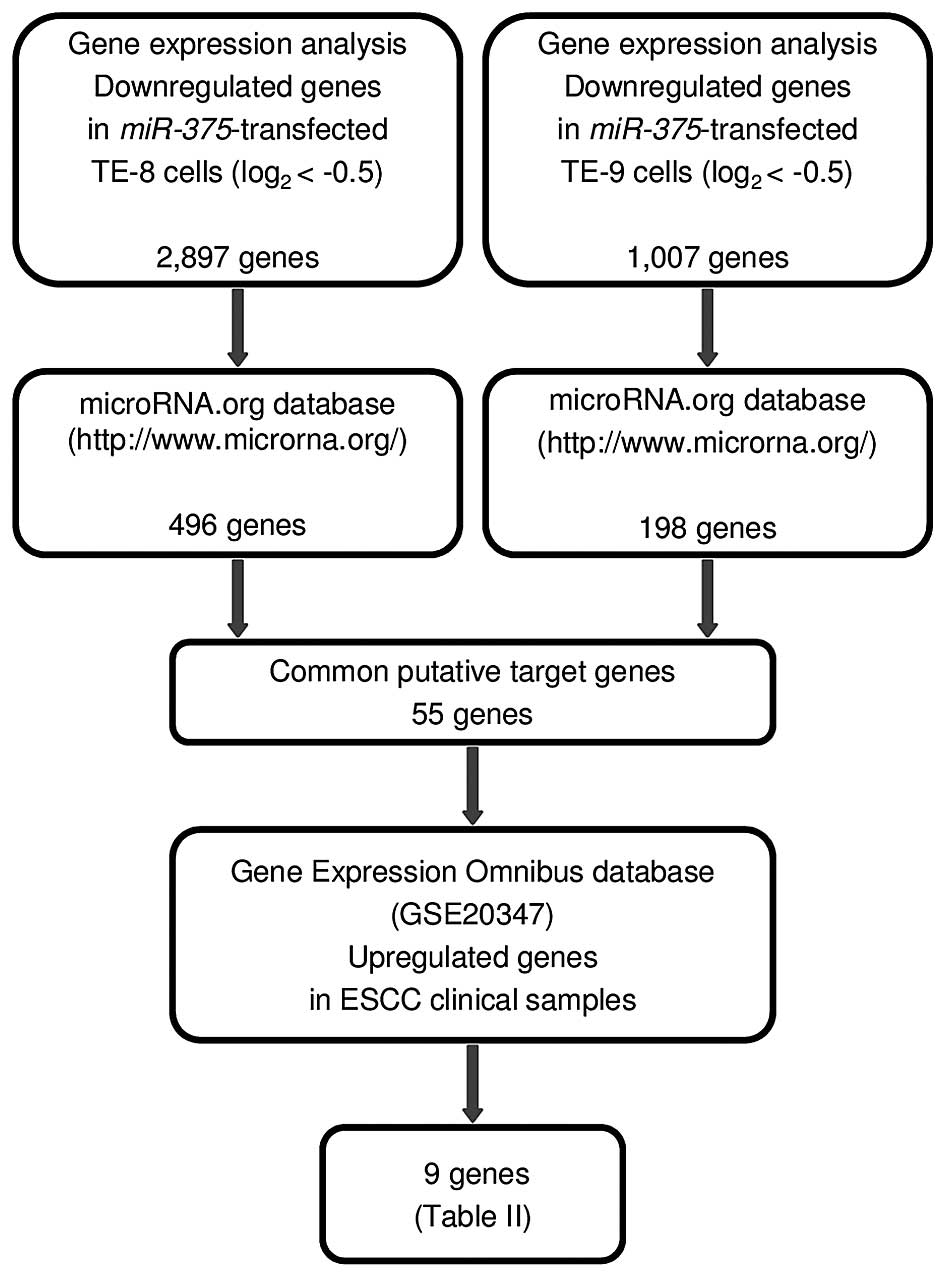
identification of miR-375 target genes is shown in Fig. 2.
Western blot analysis
Anti-human MMP-13 rabbit polyclonal IgG (1:1,000;
sc30073; Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used as
a primary antibody. Anti-human GAPDH mouse monoclonal IgG (1:5,000;
010–25521; Wako Pure Chemical Industries, Osaka, Japan) was used as
an internal loading control. The membrane was washed and incubated
with a horseradish peroxidase-conjugated secondary antibody. Bands
were visualized using Amersham ECL Prime Western Blotting detection
reagent (GE Healthcare Life Sciences, Uppsala, Sweden).
Immunohistochemistry
Tumor samples were fixed with 10% formaldehyde in
phosphate-buffered saline (PBS), embedded in paraffin and sectioned
into 4-μm-thick slices. The sections were incubated with rabbit
polyclonal anti-MMP-13 IgG (1:200; ab84594; Abcam, Cambridge, UK)
at 4°C overnight. The procedure for immunohistochemistry was
previously described (21).
Plasmid construction and Dual-luciferase
reporter assays
Partial wild-type sequences of the 3′ untranslated
region (UTR) of MMP13 containing the miR-375 target
site (positions 100–113 of the MMP13 3′ UTR) or sequences
with a deleted miR-375 target site were inserted between the
XhoI and PmeI restriction sites in the 3′ UTR of the
hRluc gene in the psiCHECK-2 vector (product ID: C8021;
Promega, Madison, WI, USA). TE-8 and TE-9 cells were transfected
with 50 ng of the vector and 10 nM miR-375 using
Lipofectamine 2000 (Thermo Fisher Scientific) in Opti-MEM (Thermo
Fisher Scientific). The activities of firefly and Renilla
luciferases were determined in lysates of transfected cells using a
Dual-luciferase reporter assay system according to the
manufacturer's recommendations (product ID: E1960; Promega). Data
were normalized to firefly luciferase activity (ratio of
Renilla/firefly luciferase activities).
Identification of downstream genes
mediated by MMP13 in ESCC cells
Gene expression analyses of
si-MMP13-transfected TE-8 and TE-9 cells revealed molecular
targets mediated by MMP13 in ESCC cells. This method is
described in more detail in previous studies (13,18–20).
Microarray results were deposited in the GEO database (accession
number GSE82108).
Statistical analysis
Relationships between two or three variables and
numerical values were analyzed using the Mann-Whitney U test or the
Bonferroni-adjusted Mann-Whitney test. Spearman's rank test was
used to evaluate the correlations between the expression levels of
miR-375 and MMP13. Expert StatView version 5.0 (SAS
Institute, Inc., Cary, NC, USA) was used in these analyses.
Results
Expression levels of miR-375 in ESCC
clinical specimens and cell lines
We evaluated the expression levels of miR-375
in ESCC tissues (n=25), normal esophageal specimens (n=13), and
ESCC cell lines (TE-8 and TE-9). The patient backgrounds and
clinicopathological characteristics are shown in Table I. The expression levels of
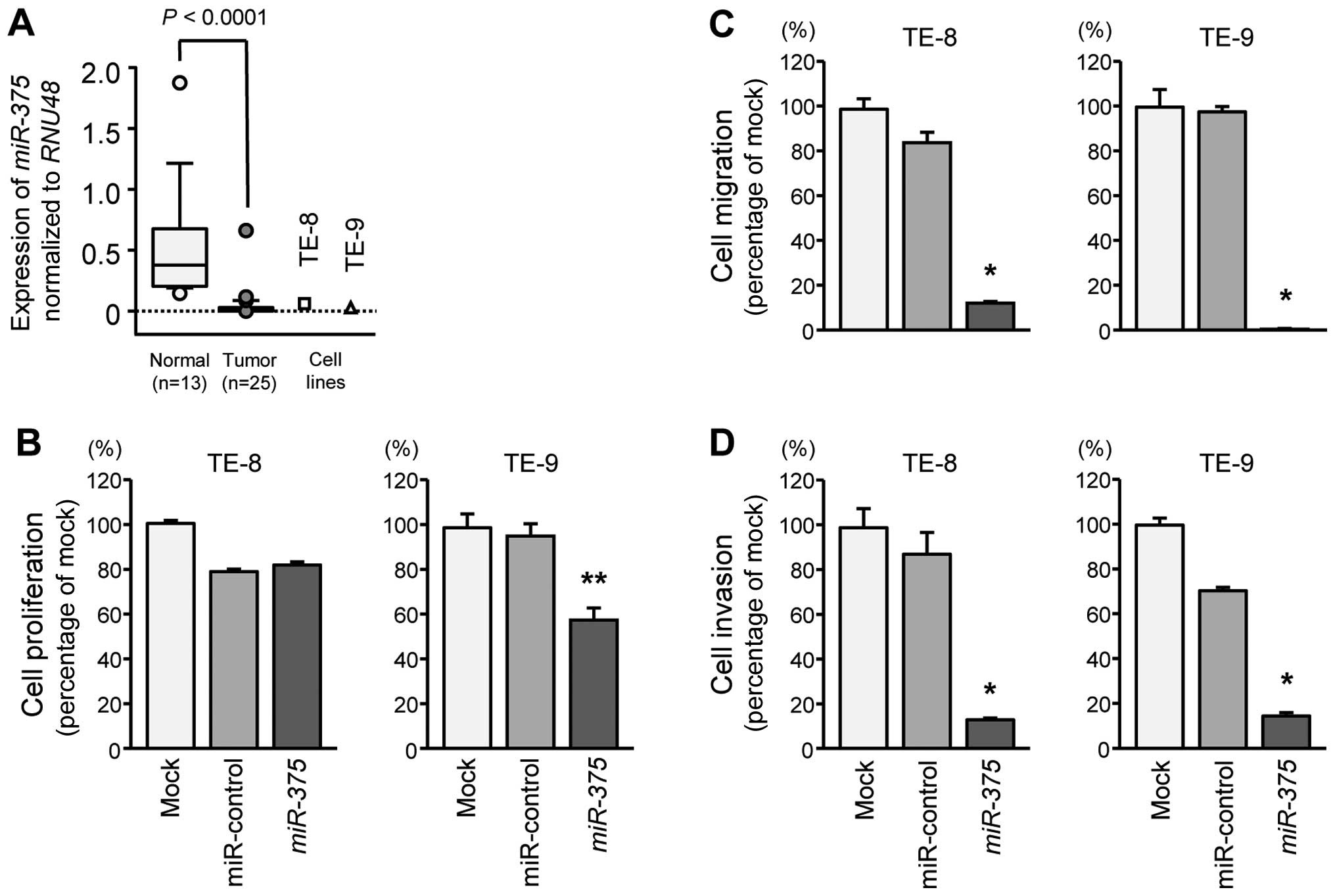
miR-375 were significantly downregulated in cancer tissues
and ESCC cell lines compared with those in normal tissues
(P<0.0001; Fig. 1A).
Additionally, there were no significant relationships between the
expression level of miR-375 and any of the
clinicopathological parameters examined in this study (recurrence,
T stage, N stage, vascular invasion, or survival rate).
Effects of miR-375 restoration on cell
proliferation, migration and invasion in ESCC cell lines
To investigate the antitumor functions of
miR-375, we performed gain-of-function studies using mature
miRNA transfection of TE-8 and TE-9 cells.
Cell proliferation was significantly suppressed by
miR-375 transfection in TE-9 cells in comparison with that
of mock or miR-control transfectants (Fig. 1B). However, no changes were
detected in TE-8 cells (Fig.
1B).
Migration assays showed that cell migration activity
was significantly inhibited by miR-375 transfection in TE-8
and TE-9 cells in comparison with that in mock or miR-control
transfectants (Fig. 1C).
Additionally, Matrigel invasion assays demonstrated that cell
invasion activity was significantly inhibited by miR-375
transfection in TE-8 and TE-9 cells in comparison with that in mock
or miR-control transfectants (Fig.
1D).
Identification of putative target genes
regulated by miR-375 in ESCC cells
To gain additional insights into the molecular
pathways regulated by antitumor miR-375 in ESCC cells, we
used a combination of in silico and gene expression
analyses. The strategy for identification of the
miR-375-regulated genes in ESCC cells is shown in Fig. 2.
In gene expression analyses, 2,897 and 1,007 genes
were downregulated (log2 ratio <-0.5) in TE-8 and
TE-9 miR-375 transfectants, respectively, in comparison with
that in control transfectants. Our present expression data were
deposited in the Gene Expression Omnibus (GEO accession number
GSE77790). Among these downregulated genes, we searched for genes
having putative miR-375 binding sites in their 3′ UTRs using
the microRNA.org database. A total of 55 genes were identified as
putative target genes of miR-375, and nine genes were
upregulated in ESCC clinical specimens, as determined using ESCC
expression data (GEO accession number: GSE20347; Table II).
 | Table IIHighly expressed genes putatively
regulated by miR-375 in ESCC. |
Table II
Highly expressed genes putatively
regulated by miR-375 in ESCC.
| Entrez Gene ID | Gene symbol | Description | miR-375
target sites | Expression in
miR-375 transfectants FC (Log2) | GEO data (GSE20347)
FC (Log2) |
|---|
|
|---|
| TE-8 | TE-9 |
|---|
| 4322 | MMP13 | Matrix
metalloproteinase 13 | 1 | −2.24 | −1.76 | 5.12 |
| 6004 | RGS16 | Regulator of
G-protein signaling 16 | 3 | −1.50 | −0.92 | 2.45 |
| 4920 | ROR2 | Receptor tyrosine
kinase-like orphan receptor 2 | 1 | −0.80 | −0.59 | 2.14 |
| 10202 | DHRS2 |
Dehydrogenase/reductase (SDR family)
member 2 | 3 | −3.07 | −0.83 | 2.02 |
| 1956 | EGFR | Epidermal growth
factor receptor | 1 | −0.93 | −0.78 | 1.58 |
| 655 | BMP7 | Bone morphogenetic
protein 7 | 1 | −0.85 | −0.74 | 1.54 |
| 23363 | OBSL1 | Obscurin-like
1 | 1 | −0.80 | −0.71 | 1.52 |
| 23035 | PHLPP2 | PH domain and
leucine rich repeat protein phosphatase 2 | 1 | −0.69 | −0.64 | 1.15 |
| 1896 | EDA | Ectodysplasin
A | 1 | −0.72 | −0.63 | 1.09 |
In this study, we focused on MMP13 because
its expression was most upregulated in ESCC clinical specimens and
most downregulated in miR-375 transfectants. Moreover,
previous studies have shown that the activation of MMPs is
associated with cancer cell aggressiveness (22).
Expression of MMP13 in ESCC clinical
specimens
Next, we validated the upregulation of MMP13
in the ESCC clinical specimens at both the mRNA and the protein
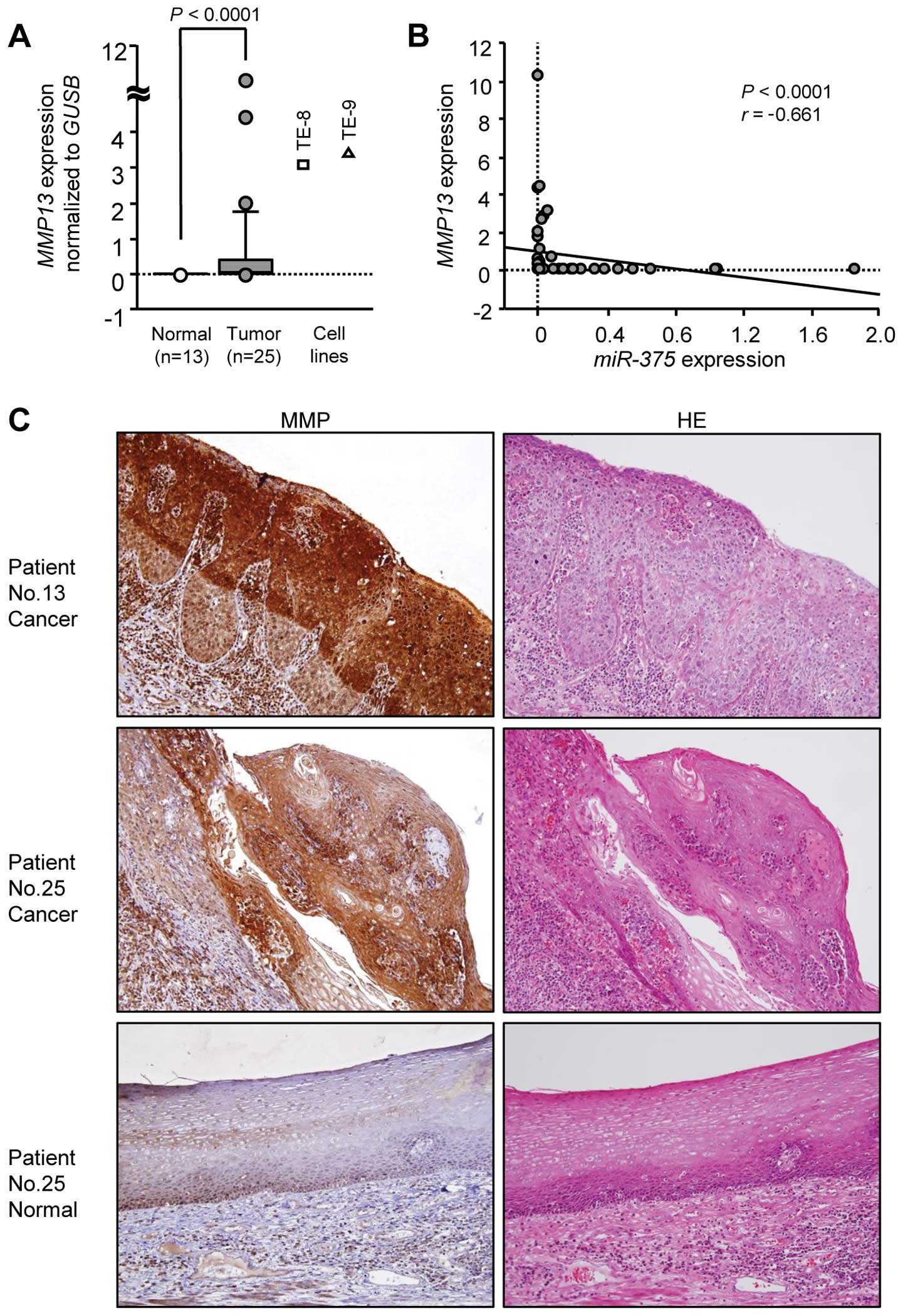
levels. The expression of MMP13 was significantly
upregulated in 25 ESCC specimens and ESCC cell lines compared with
that in 13 normal specimens (P<0.0001; Fig. 3A). The Spearman's rank tests showed
negative correlations between the expression of miR-375 and
that of MMP13 (r=−0.661, P<0.0001; Fig. 3B).
Immunohistochemistry showed that MMP13 tended to be
strongly expressed in ESCC lesions, whereas low expression was
observed in normal esophageal epithelium (Fig. 3C).
Direct regulation of MMP13 by miR-375 in
ESCC cells
We performed qRT-PCR to validate
miR-375-mediated repression of MMP13 expression in
ESCC cell lines. Our results showed that MMP13 mRNA was
significantly reduced in miR-375 transfectants in comparison
with that in mock or miR-control transfectants (P<0.0001;
Fig. 4A). MMP13 protein expression
was also repressed in miR-375 transfectants (Fig. 4B).
Next, we performed luciferase reporter assays using
TE-8 and TE-9 cells to determine whether MMP13 had an actual
target site for miR-375 binding. The microRNA.org database
predicted that there was one putative target site in the 3′ UTR of
MMP13 (Fig. 4C). Compared
with the miR-control, luminescence intensity was significantly
reduced by transfection with miR-375 at the miR-375
target site, positions 100–113, in the 3′ UTR of MMP13
(Fig. 4D).
Effects of silencing MMP13 on
proliferation, migration and invasion in ESCC cells
To investigate the functional roles of MMP13 in ESCC
cell lines, we performed loss-of-function assays by transfection of
si-MMP13 into TE-8 and TE-9 cells.
First, we evaluated the knockdown efficiency of
si-MMP13 transfection in ESCC cell lines. In the present
study, we used two siRNAs targeting MMP13 (si-MMP13-1
and si-MMP13-2). According to qRT-PCR and western blot
analyses, both siRNAs effectively downregulated MMP13
expression in both cell lines (Fig. 5A
and B).
Cell proliferation, migration and invasion assays
demonstrated that cell proliferation, migration, and invasion were
inhibited in si-MMP13-transfected cells compared with those
in mock- or siRNA-control-transfected cells (Fig. 5C–E).
Identification of downstream genes
regulated by MMP13 in ESCC cells
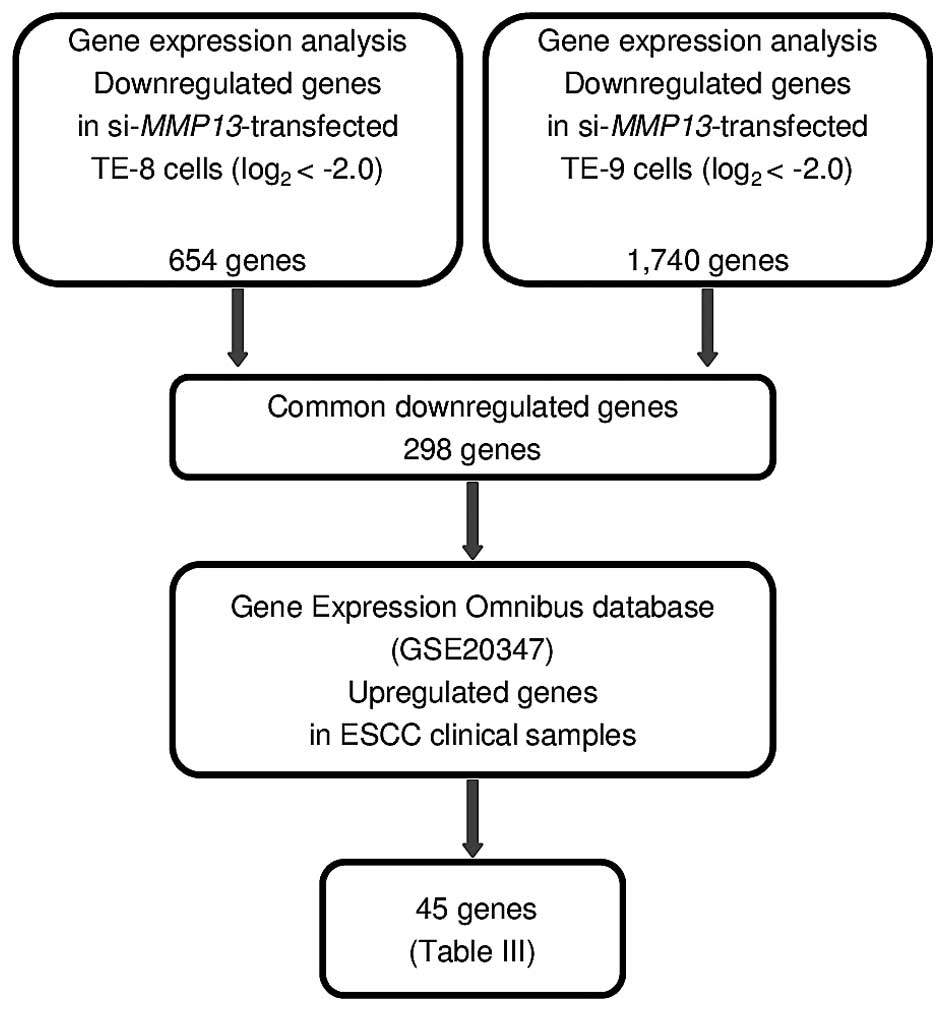
To determine which downstream genes were regulated
by MMP13, genome-wide gene expression and in silico analyses
were performed in TE-8 and TE-9 cells transfected with
si-MMP13.
Our expression analysis showed that a total of 298
genes were commonly downregulated (log2 ratio <-2.0)
in TE-8 and TE-9 cells following si-MMP13 transfection.
Among these genes, 52 were upregulated in ESCC clinical specimens,
as determined using ESCC expression data (GEO accession number:
GSE20347; Fig. 6 and Table III).
 | Table IIIDownregulated genes in
si-MMP13-transfected ESCC cell lines. |
Table III
Downregulated genes in
si-MMP13-transfected ESCC cell lines.
| Entrez gene ID | Gene symbol | Description | Expression in
si-MMP13 transfectants FC (log2) | GEO data (GSE20347)
FC (log2) |
|---|
|
|---|
| TE8 | TE9 |
|---|
| 4322 | MMP13 | Matrix
metallopeptidase 13 (collagenase 3) | −4.42 | −4.47 | 5.12 |
| 1063 | CENPF | Centromere protein
F, 350/400 kDa | −2.96 | −5.18 | 2.31 |
| 9928 | KIF14 | Kinesin family
member 14 | −2.28 | −4.66 | 2.14 |
| 2842 | GPR19 | G protein-coupled
receptor 19 | −2.67 | −3.74 | 2.12 |
| 983 | CDK1 | Cyclin-dependent
kinase 1 | −2.07 | −3.78 | 1.95 |
| 55165 | CEP55 | Centrosomal protein
55 kDa | −3.33 | −4.79 | 1.94 |
| 1033 | CDKN3 | Cyclin-dependent
kinase inhibitor 3 | −2.08 | −3.73 | 1.94 |
| 7153 | TOP2A | Topoisomerase (DNA)
II alpha 170 kDa | −3.36 | −5.01 | 1.91 |
| 10403 | NDC80 | NDC80 kinetochore
complex component | −2.19 | −3.69 | 1.76 |
| 9787 | DLGAP5 | Discs, large
(Drosophila) homolog-associated protein 5 | −2.27 | −3.32 | 1.72 |
| 55215 | FANCI | Fanconi anemia,
complementation group I | −2.27 | −3.97 | 1.70 |
| 23306 |
TMEM194A | Transmembrane
protein 194A | −2.31 | −2.79 | 1.68 |
| 4751 | NEK2 | NIMA-related kinase
2 | −2.70 | −3.84 | 1.66 |
| 2735 | GLI1 | GLI family zinc
finger 1 | −2.70 | −3.31 | 1.63 |
| 3161 | HMMR | Hyaluronan-mediated
motility receptor (RHAMM) | −4.06 | −5.29 | 1.60 |
| 259266 | ASPM | Asp (abnormal
spindle) homolog, microcephaly associated (Drosophila) | −2.17 | −3.81 | 1.56 |
| 4998 | ORC1 | Origin recognition
complex, subunit 1 | −2.23 | −3.08 | 1.53 |
| 57405 | SPC25 | SPC25, NDC80
kinetochore complex component | −2.16 | −4.12 | 1.48 |
| 28951 | TRIB2 | Tribbles
pseudokinase 2 | −2.28 | −2.35 | 1.44 |
| 9603 | NFE2L3 | Nuclear factor,
erythroid 2-like 3 | −2.00 | −2.51 | 1.42 |
| 9638 | FEZ1 | Fasciculation and
elongation protein zeta 1 (zygin I) | −2.27 | −2.97 | 1.42 |
| 9918 | NCAPD2 | Non-SMC condensin I
complex, subunit D2 | −2.12 | −2.79 | 1.38 |
| 7468 | WHSC1 | Wolf-Hirschhorn
syndrome candidate 1 | −2.43 | −3.36 | 1.33 |
| 100288413 |
ERVMER34-1 | Endogenous
retrovirus group MER34, member 1 | −2.76 | −3.78 | 1.32 |
| 1062 | CENPE | Centromere protein
E, 312 kDa | −2.60 | −3.91 | 1.29 |
| 55063 | ZCWPW1 | Zinc finger, CW
type with PWWP domain 1 | −3.19 | −3.44 | 1.25 |
| 81624 | DIAPH3 | Diaphanous-related
formin 3 | −2.22 | −3.54 | 1.25 |
| 6119 | RPA3 | Replication protein
A3, 14 kDa | −2.34 | −3.42 | 1.24 |
| 8318 | CDC45 | Cell division cycle
45 | −2.13 | −4.07 | 1.23 |
| 64151 | NCAPG | Non-SMC condensin I
complex, subunit G | −3.25 | −3.92 | 1.22 |
| 7083 | TK1 | Thymidine kinase 1,
soluble | −2.11 | −3.86 | 1.22 |
| 55732 |
C1orf112 | Chromosome 1 open
reading frame 112 | −2.06 | −2.62 | 1.22 |
| 1058 | CENPA | Centromere protein
A | −2.02 | −3.86 | 1.18 |
| 55635 | DEPDC1 | DEP domain
containing 1 | −2.33 | −3.44 | 1.18 |
| 3925 | STMN1 | Stathmin 1 | −2.66 | −4.51 | 1.17 |
| 3092 | HIP1 | Huntingtin
interacting protein 1 | −2.71 | −3.51 | 1.17 |
| 5427 | POLE2 | Polymerase (DNA
directed), epsilon 2, accessory subunit | −2.18 | −4.37 | 1.15 |
| 1719 | DHFR | Dihydrofolate
reductase | −2.46 | −3.63 | 1.14 |
| 54830 | NUP62CL | Nucleoporin 62 kDa
C-terminal like | −2.17 | −2.22 | 1.10 |
| 5062 | PAK2 | p21 protein
(Cdc42/Rac)-activated kinase 2 | −2.37 | −2.60 | 1.09 |
| 100129361 |
LOC100129361 | Chromosome X open
reading frame 69-like | −2.57 | −2.46 | 1.09 |
| 5933 | RBL1 | Retinoblastoma-like
1 | −3.24 | −4.43 | 1.08 |
| 4288 | MKI67 | Marker of
proliferation Ki-67 | −2.14 | −4.87 | 1.03 |
| 81691 |
LOC81691 | Exonuclease
NEF-sp | −2.62 | −3.61 | 1.03 |
| 675 | BRCA2 | Breast cancer 2,
early onset | −2.90 | −4.04 | 1.00 |
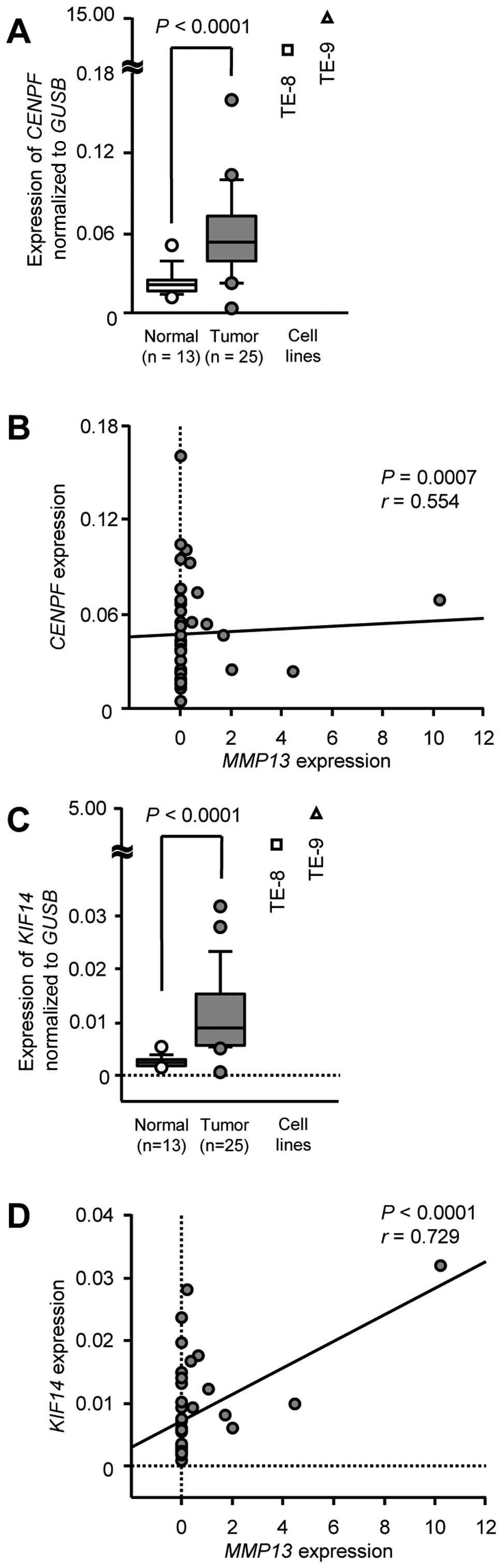
We then validated the upregulation of CENPF
and KIF14 mRNAs in ESCC clinical specimens. The expression
of CENPF and KIF14 mRNAs was significantly
upregulated in 25 ESCC specimens and ESCC cell lines compared with
that in 13 normal specimens (P<0.0001; Fig. 7A and C). The Spearman's rank tests
showed correlations between the expression of MMP13 and that
of CENPF or KIF14 (CENPF: r=0.554, P=0.0007, Fig. 7B; KIF14: r=0.729, P<0.0001,
Fig. 7D).
Discussion
Numerous studies of miRNA expression signatures in
ESCC have shown that miR-375 is frequently downregulated in
cancer tissues and functions as an antitumor miRNA (14,23).
In the present study, we confirmed that miR-375 was markedly
downregulated in cancer tissues and that ectopic expression of
miR-375 significantly suppressed cancer cell migration and
invasion. Thus, we found that loss of miR-375 expression
enhanced cancer cell aggressiveness in ESCC. Many previous studies
have shown that the expression of miR-375 is markedly
decreased in several types of cancers and that miR-375
functions as an antitumor miRNA (15,24).
In contrast to these antitumor activities, miR-375 is
upregulated in pediatric acute myeloid leukemia (AML) and prostate
cancer, suggesting that miR-375 acts as an oncogenic miRNA
in these diseases (25,26). The dual function of miR-375
is very unique; thus, it is important to identify
miR-375-regulated pathways in various cancer types.
It is also important to elucidate novel RNA networks
regulated by antitumor miR-375 in ESCC cells. Previous
studies have shown that insulin-like growth factor 1 receptor
(IGF1R), lactate dehydrogenase B (LDHB), and
astrocyte elevated gene-1/metadherin (AEG-1/MTDH) are
directly regulated by miR-375 in ESCC cells (16,17).
These target genes are upregulated in ESCC clinical specimens and
functioned as oncogenes in this disease. Another unique
characteristic of miRNAs is that a single miRNA can regulate a
large number of RNA transcripts in human cells (27,28).
Thus, the continuous identification of miR-375-regulated
oncogenes in ESCC cells is important for elucidation of the
molecular pathogenesis of ESCC.
In this study, we identified MMP13 as a
direct target of antitumor miR-375 in ESCC cells.
MMP13 (also known as collagenase 3) is a member of the
collagenase subfamily of MMPs and functions to degrade a wide range
of extracellular matrix components, including tenascin C,
fibronectin and type I–IV collagen (29). Thus, MMP13 has a wide range of
proteolytic functions, suggesting that MMP13 is involved in several
physiological and pathological processes (30). High expression of MMP13 has been
reported in rheumatoid arthritis, osteoarthritis and several types
of cancers (22). Previous studies
have also shown that high expression of MMP13 is associated with
vascular invasion and lymph node metastasis in ESCC (31). Our present data demonstrated that
knockdown of MMP13 markedly reduced cancer cell migration
and invasion in ESCC cells.
The MMP13 gene has also been reported to be
epigenetically regulated by several other miRNAs, including
miR-125b and miR-143, in cancer cells (32–34).
Notably, our miRNA signatures have shown that miR-125b and
miR-143 are down-regulated in ESCC and in oral and
hypopharyngeal squamous cell carcinoma (12–14).
Moreover, functional assays have indicated that these miRNAs act as
tumor suppressors in several cancers, including ESCC cells
(32–35). Loss of the expression of several
antitumor miRNAs and activation of MMP13 may enhance cancer
cell aggressiveness and metastasis. Thus, identification of
miR-375/MMP13-mediated cancer pathways may facilitate the
discovery of candidate therapeutic targets in ESCC.
Based on the above, we further investigated the
downstream genes mediated by MMP13 in ESCC cells using genome-wide
gene expression analysis. Our data showed that several
centromere-associated proteins were regulated by MMP13-mediated
genes, such as CENPF, CENPE, CENPA,
CEP55, NDC80 and SPC25. Moreover, cell
cycle-promoting genes, e.g., KIF14, CDK1,
TOP2A, CDC45 and PAK2, were also downregulated
by si-MMP13 in this study. Recent studies have reported that
several genes encoding mitotic apparatus components are upregulated
in cancer cells and contribute to cancer cell phenotypes (36,37).
Therefore, overexpression of genes encoding mitotic apparatus
components may represent a potential target for cancer drug
development (38). Several
compounds that inhibit centromere proteins and mitotic kinesins are
being tested as potential cancer therapies in clinical trials
(39).
Among these genes, we validated the overexpression
of CENPF and KIF14 in ESCC clinical specimens.
Previous studies have shown that CENPF is a master regulator
of prostate cancer malignancy and that high expression of
CEPNF is a prognostic indicator of poor survival and
metastasis in patients with ESCC (40). KIF14 is a member of the
kinesin superfamily of proteins and functions as a microtubule
motor protein involved in cytokinesis and chromosome segregation
(41). Overexpression of
KIF14 has been reported in several cancers, and its
expression is associated with cancer cell phenotypes (42,43).
An in-depth functional analysis of these genes in ESCC cells is
necessary to further characterize these pathways. Identification of
the downstream genes regulated by the miR-375/MMP13 axis may
lead to a better understanding of ESCC aggressiveness.
In conclusion, downregulation of miR-375 was
frequently observed in ESCC clinical specimens, and miR-375
was shown to function as an antitumor miRNA in ESCC cells. To the
best of our knowledge, this is the first report demonstrating that
MMP13 is directly regulated by antitumor miR-375 and
acts to regulate several cell cycle promoting genes. The
identification of novel molecular pathways and targets regulated by
the miR-375/MMP13 axis may lead to a better understanding of
ESCC molecular pathogenesis.
Acknowledgements
We wish to thank the Joint Research Laboratory,
Kagoshima University Graduate School of Medical and Dental
Sciences, for the use of their facilities. The present study was
supported by KAKENHI (C) grant 15K10801.
References
|
1
|
Hongo M, Nagasaki Y and Shoji T:
Epidemiology of esophageal cancer: Orient to Occident. Effects of
chronology, geography and ethnicity. J Gastroenterol Hepatol.
24:729–735. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ohashi S, Miyamoto S, Kikuchi O, Goto T,
Amanuma Y and Muto M: Recent advances from basic and clinical
studies of esophageal squamous cell carcinoma. Gastroenterology.
149:1700–1715. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Harada K, Baba Y, Ishimoto T, Shigaki H,
Kosumi K, Yoshida N, Watanabe M and Baba H: The role of microRNA in
esophageal squamous cell carcinoma. J Gastroenterol. 51:520–530.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kikkawa N, Hanazawa T, Fujimura L, Nohata
N, Suzuki H, Chazono H, Sakurai D, Horiguchi S, Okamoto Y and Seki
N: miR-489 is a tumour-suppressive miRNA target PTPN11 in
hypopharyngeal squamous cell carcinoma (HSCC). Br J Cancer.
103:877–884. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nohata N, Hanazawa T, Kikkawa N, Sakurai
D, Fujimura L, Chiyomaru T, Kawakami K, Yoshino H, Enokida H,
Nakagawa M, et al: Tumour suppressive microRNA-874 regulates novel
cancer networks in maxillary sinus squamous cell carcinoma. Br J
Cancer. 105:833–841. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nohata N, Hanazawa T, Kinoshita T, Inamine
A, Kikkawa N, Itesako T, Yoshino H, Enokida H, Nakagawa M, Okamoto
Y, et al: Tumour-suppressive microRNA-874 contributes to cell
proliferation through targeting of histone deacetylase 1 in head
and neck squamous cell carcinoma. Br J Cancer. 108:1648–1658. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fukumoto I, Kinoshita T, Hanazawa T,
Kikkawa N, Chiyomaru T, Enokida H, Yamamoto N, Goto Y, Nishikawa R,
Nakagawa M, et al: Identification of tumour suppressive
microRNA-451a in hypopharyngeal squamous cell carcinoma based on
microRNA expression signature. Br J Cancer. 111:386–394. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fukumoto I, Hanazawa T, Kinoshita T,
Kikkawa N, Koshizuka K, Goto Y, Nishikawa R, Chiyomaru T, Enokida
H, Nakagawa M, et al: MicroRNA expression signature of oral
squamous cell carcinoma: Functional role of microRNA-26a/b in the
modulation of novel cancer pathways. Br J Cancer. 112:891–900.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kano M, Seki N, Kikkawa N, Fujimura L,
Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M and
Matsubara H: miR-145, miR-133a and miR-133b: Tumor-suppressive
miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J
Cancer. 127:2804–2814. 2010. View Article : Google Scholar
|
|
15
|
Kinoshita T, Hanazawa T, Nohata N, Okamoto
Y and Seki N: The functional significance of microRNA-375 in human
squamous cell carcinoma: Aberrant expression and effects on cancer
pathways. J Hum Genet. 57:556–563. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Isozaki Y, Hoshino I, Nohata N, Kinoshita
T, Akutsu Y, Hanari N, Mori M, Yoneyama Y, Akanuma N, Takeshita N,
et al: Identification of novel molecular targets regulated by tumor
suppressive miR-375 induced by histone acetylation in esophageal
squamous cell carcinoma. Int J Oncol. 41:985–994. 2012.PubMed/NCBI
|
|
17
|
Kong KL, Kwong DL, Chan TH, Law SY, Chen
L, Li Y, Qin YR and Guan XY: MicroRNA-375 inhibits tumour growth
and metastasis in oesophageal squamous cell carcinoma through
repressing insulin-like growth factor 1 receptor. Gut. 61:33–42.
2012. View Article : Google Scholar
|
|
18
|
Matsushita R, Yoshino H, Enokida H, Goto
Y, Miyamoto K, Yonemori M, Inoguchi S, Nakagawa M and Seki N:
Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145
(miR-145-5p and miR-145-3p): Inhibition of bladder cancer cell
aggressiveness. Oncotarget. 7:28460–28487. 2016.PubMed/NCBI
|
|
19
|
Goto Y, Kojima S, Nishikawa R, Kurozumi A,
Kato M, Enokida H, Matsushita R, Yamazaki K, Ishida Y, Nakagawa M,
et al: MicroRNA expression signature of castration-resistant
prostate cancer: The microRNA-221/222 cluster functions as a tumour
suppressor and disease progression marker. Br J Cancer.
113:1055–1065. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Goto Y, Kojima S, Nishikawa R, Enokida H,
Chiyomaru T, Kinoshita T, Nakagawa M, Naya Y, Ichikawa T and Seki
N: The microRNA-23b/27b/24-1 cluster is a disease progression
marker and tumor suppressor in prostate cancer. Oncotarget.
5:7748–7759. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kita Y, Nishizono Y, Okumura H, Uchikado
Y, Sasaki K, Matsumoto M, Setoyama T, Tanoue K, Omoto I, Mori S, et
al: Clinical and biological impact of cyclin-dependent kinase
subunit 2 in esophageal squamous cell carcinoma. Oncol Rep.
31:1986–1992. 2014.PubMed/NCBI
|
|
22
|
Brinckerhoff CE, Rutter JL and Benbow U:
Interstitial collagenases as markers of tumor progression. Clin
Cancer Res. 6:4823–4830. 2000.
|
|
23
|
Yan JW, Lin JS and He XX: The emerging
role of miR-375 in cancer. Int J Cancer. 135:1011–1018. 2014.
View Article : Google Scholar
|
|
24
|
Hui AB, Bruce JP, Alajez NM, Shi W, Yue S,
Perez-Ordonez B, Xu W, O'Sullivan B, Waldron J, Cummings B, et al:
Significance of dysregulated metadherin and microRNA-375 in head
and neck cancer. Clin Cancer Res. 17:7539–7550. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Z, Hong Z, Gao F and Feng W:
Upregulation of microRNA-375 is associated with poor prognosis in
pediatric acute myeloid leukemia. Mol Cell Biochem. 383:59–65.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Szczyrba J, Nolte E, Wach S, Kremmer E,
Stöhr R, Hartmann A, Wieland W, Wullich B and Grässer FA:
Downregulation of Sec23A protein by miRNA-375 in prostate
carcinoma. Mol Cancer Res. 9:791–800. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kinoshita T, Yip KW, Spence T and Liu FF:
MicroRNAs in extracellular vesicles: Potential cancer biomarkers. J
Hum Genet. Jul 7–2016.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yonemori K, Kurahara H, Maemura K and
Natsugoe S: MicroRNA in pancreatic cancer. J Hum Genet. Jun
2–2016.(Epub ahead of print). View Article : Google Scholar
|
|
29
|
Knäuper V, López-Otin C, Smith B, Knight G
and Murphy G: Biochemical characterization of human collagenase-3.
J Biol Chem. 271:1544–1550. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vincenti MP and Brinckerhoff CE:
Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in
arthritis: Integration of complex signaling pathways for the
recruitment of gene-specific transcription factors. Arthritis Res.
4:157–164. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Etoh T, Inoue H, Yoshikawa Y, Barnard GF,
Kitano S and Mori M: Increased expression of collagenase-3 (MMP-13)
and MT1-MMP in oesophageal cancer is related to cancer
aggressiveness. Gut. 47:50–56. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu N, Zhang L, Meisgen F, Harada M,
Heilborn J, Homey B, Grandér D, Ståhle M, Sonkoly E and Pivarcsi A:
MicroRNA-125b down-regulates matrix metallopeptidase 13 and
inhibits cutaneous squamous cell carcinoma cell proliferation,
migration, and invasion. J Biol Chem. 287:29899–29908. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu D, Ding J, Wang L, Pan H, Zhou Z, Zhou
J and Qu P: microRNA-125b inhibits cell migration and invasion by
targeting matrix metallopeptidase 13 in bladder cancer. Oncol Lett.
5:829–834. 2013.PubMed/NCBI
|
|
34
|
Osaki M, Takeshita F, Sugimoto Y, Kosaka
N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T,
et al: MicroRNA-143 regulates human osteosarcoma metastasis by
regulating matrix metalloprotease-13 expression. Mol Ther.
19:1123–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu J, Mao Y, Zhang D, Hao S, Zhang Z, Li
Z and Li B: MiR-143 inhibits tumor cell proliferation and invasion
by targeting STAT3 in esophageal squamous cell carcinoma. Cancer
Lett. 373:97–108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yuen KW, Montpetit B and Hieter P: The
kinetochore and cancer: What's the connection? Curr Opin Cell Biol.
17:576–582. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sagona AP and Stenmark H: Cytokinesis and
cancer. FEBS Lett. 584:2652–2661. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rath O and Kozielski F: Kinesins and
cancer. Nat Rev Cancer. 12:527–539. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huszar D, Theoclitou ME, Skolnik J and
Herbst R: Kinesin motor proteins as targets for cancer therapy.
Cancer Metastasis Rev. 28:197–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Aytes A, Mitrofanova A, Lefebvre C,
Alvarez MJ, Castillo-Martin M, Zheng T, Eastham JA, Gopalan A,
Pienta KJ, Shen MM, et al: Cross-species regulatory network
analysis identifies a synergistic interaction between FOXM1 and
CENPF that drives prostate cancer malignancy. Cancer Cell.
25:638–651. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhu C, Zhao J, Bibikova M, Leverson JD,
Bossy-Wetzel E, Fan JB, Abraham RT and Jiang W: Functional analysis
of human microtubule-based motor proteins, the kinesins and
dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol
Cell. 16:3187–3199. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Corson TW, Zhu CQ, Lau SK, Shepherd FA,
Tsao MS and Gallie BL: KIF14 messenger RNA expression is
independently prognostic for outcome in lung cancer. Clin Cancer
Res. 13:3229–3234. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Thériault BL, Pajovic S, Bernardini MQ,
Shaw PA and Gallie BL: Kinesin family member 14: An independent
prognostic marker and potential therapeutic target for ovarian
cancer. Int J Cancer. 130:1844–1854. 2012. View Article : Google Scholar
|