Introduction
Tylosis is an extremely rare autosomal, dominantly
inherited disorder characterized by abnormal thickening of the
palmoplantar skin and a highly elevated risk of esophageal squamous
cell carcinoma (ESCC). Five families with high frequencies of
tylosis have been reported from the UK, USA, Germany, Spain and
Finland (1–7). Linkage and haplotype analyses in
these families have localized the tylosis esophageal cancer (TOC)
gene locus to a region of chromosome 17q25 (3–5,8,9). It
has also been reported that frequent loss of heterozygosity (LOH)
in 17q25.1 was observed in the sporadic form of ESCC (10,11).
These reports on inherited and sporadic ESCC indicated the presence
of a putative tumor suppressor gene for ESCC at this locus.
Although abnormalities of genes on 17q25.1, such as CYGB and
RHBDF2, have been demonstrated in families with tylosis,
abnormalities of these genes have not been clearly demonstrated in
sporadic forms of ESCC (12,13).
Therefore, additional genes on 17q25.1 other than CYGB and
RHBDF2 may also be involved in ESCC development.
Additionally, we recently reported the result of
whole-exome sequencing of paired DNA samples from 144 Japanese
patients with ESCC (14). The most
frequently mutated gene was TP53 (mutated in 93.1% of
patients), followed by NOTCH1, MLL2, NFE2L2,
ZNF750, FAT1 and PIK3CA (mutated in 10–20% of
patients), in line with the results of next-generation sequencing
analysis in other studies of sporadic ESCC (14–17).
Although 17,189 non-synonymous mutations in 10,552 genes were
identified in 144 ESCC samples in our previous study, recurrent
mutations were not observed in genes on chromosome 17q25.1,
including RHBDF2 (14).
Other studies did not uncover RHBDF2 mutations either in
sporadic ESCC (16,17).
In this study, we investigated the expression
patterns of genes in an ~1500 kb region on 17q25.1, including the
TOC locus, in tumor and corresponding normal tissues using RNA
sequence (RNA-seq) analysis data from patients with sporadic ESCC.
We demonstrated frequent downregulation of ST6
N-acetylgalactosaminide α-2,6-sialyltransferase 1
(ST6GALNAC1) on 17q25.1 in ESCC tissues compared to its
expression in corresponding normal tissues.
Patients and methods
Patients and sample collection
A total of 93 ESCC samples obtained by surgery were
used after obtaining written informed consent. All patients
underwent resection of the primary tumor at Iwate Medical
University, Kyushu University Beppu Hospital and affiliated
hospitals between 1992 and 2007. Resected cancer and corresponding
normal tissues were immediately cut and stored at −80°C until
DNA/RNA extraction. Total RNA and DNA were obtained using an RNeasy
mini kit and QIAamp DNA mini kit (Qiagen Inc., Valencia, CA, USA),
respectively.
Gene expression profiling on chromosome
17q25.1 using RNA-seq data of samples from three patients with
ESCC
The expression patterns of genes on chromosome
17q25.1 were analyzed using RNA-seq data for tumor and
corresponding normal tissues from three patients with ESCC. The
characteristics of the three patients were as follows:
well-differentiated SCC in the cervical esophagus (female, 70 years
old, T1N0M0, stage I), poorly differentiated SCC in the middle
thoracic esophagus (male, 73 years old, T3N0M0, stage II) and
moderately differentiated SCC in the lower thoracic esophagus
(male, 68 years old, T3N1M0, stage III). One microgram of extracted
RNA was used as a template to construct RNA-seq libraries. Detailed
protocols of RNA-seq analysis were described previously (18). Fold enrichment of the RNA-seq tags
in the samples was calculated for each mRNA using the assigned tag
counts and normalized to reads per kilobase (kb) mRNA.
Evaluation of ST6GALNAC1 and EVPL
expression in clinical samples
Quantitative real-time reverse transcription-PCR
(qRT-PCR) was performed to measure ST6GALNAC1, EVPL,
CYGB and glyceraldehyde 3-phosphate dehydrogenase
(GAPDH) mRNA expression using a LightCycler 480 Probes
Master kit (Roche Applied Science, Mannheim, Germany) according to
the manufacturer's protocol with specific primers and universal
probes that were designed at the Universal Probe Library's assay
design center (http://lifescience.roche.com/shop/CategoryDisplay?catalogId=10001&tab=Assay+Design+Center&identifier=Universal+Probe+Library&langId=-1).
Gene expression levels were normalized with respect to those of
GAPDH. Primer sequences and universal probe number for each
gene are listed in Table I.
 | Table IPrimer sequences and amplified
regions of ST6GALNAC1 gene in direct sequencing
analysis. |
Table I
Primer sequences and amplified
regions of ST6GALNAC1 gene in direct sequencing
analysis.
| Gene name | Forward primer | Reverse primer | Probe no. |
|---|
|
ST6GALNAC1 |
CGAAATAGGAGGCCTTCAGA |
AGAGAGTGAGGTTGGGCAGA | #50 |
| EVPL |
TACCGTGCCCTGTACGAGA |
GCGCAGACCTGCTTCTGT | #67 |
| CYGB |
CCGCTGCCTACAAGGAAGT |
GGGTGGAGTTAGGGGTCCT | #62 |
| GAPDH |
AGCCACATCGCTCAGACAC |
GCCCAATACGACCAAATCC | #60 |
Direct sequencing analysis of
ST6GALNAC1
In 46 cases of ESCC, coding exons of
ST6GALNAC1 were amplified using KOD FX (Toyobo, Tokyo,
Japan) according to the manufacturer's protocol and sequenced using
BigDye Terminator version 3.1 (Applied Biosystems, Foster City, CA,
USA) as previously described (19). Primer sequences are listed in
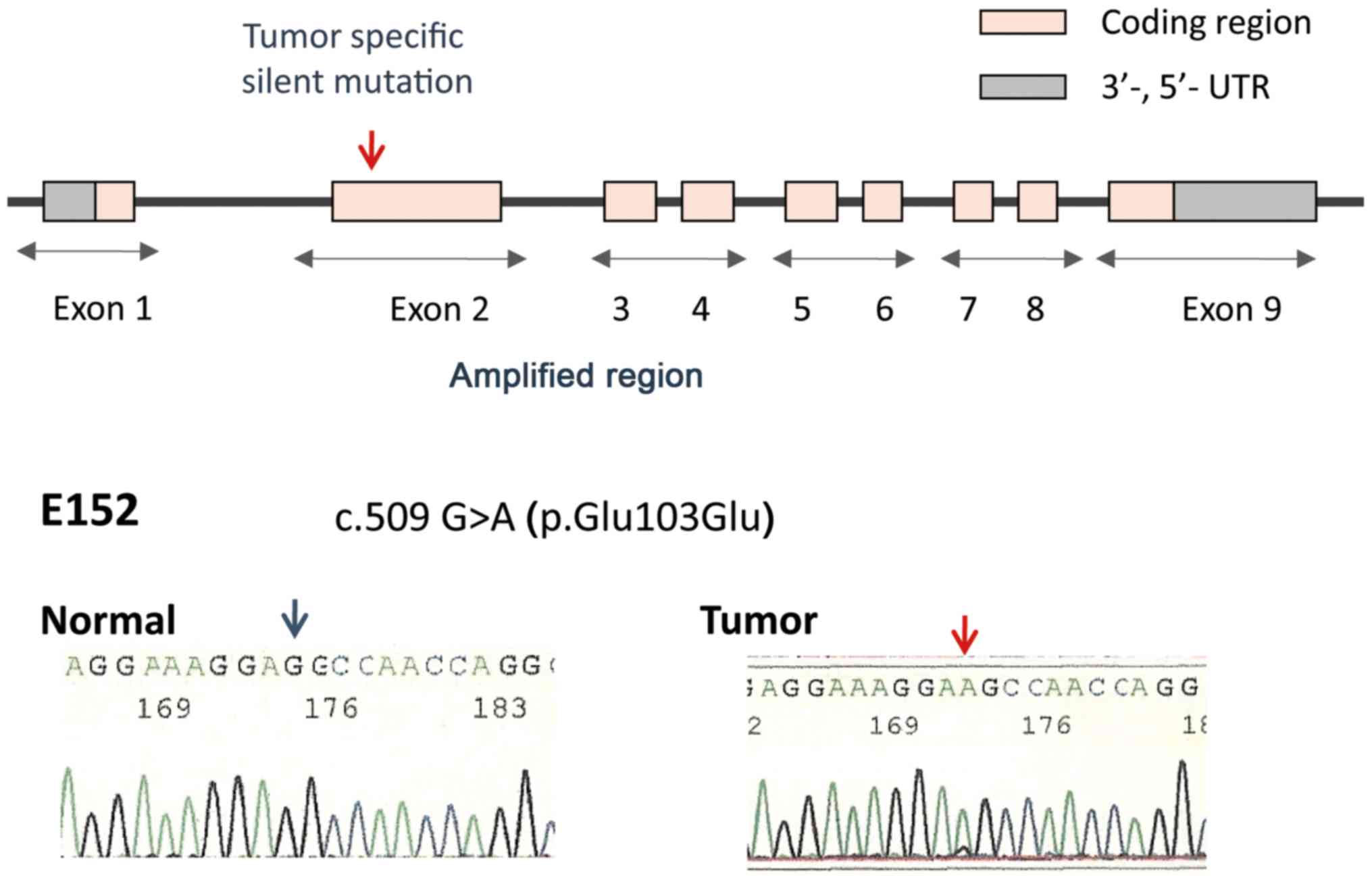
Table II and Fig. 1.
 | Table IIPrimer sequences and amplified
regions of ST6GALNAC1 gene in direct sequencing
analysis. |
Table II
Primer sequences and amplified
regions of ST6GALNAC1 gene in direct sequencing
analysis.
| Forward primer | Reverse primer | Amplified
region |
|---|
| Segment 1 |
CTTCCTTTAAGCCACGCCAGCTTAT |
TGAAATATGAGGAGTGGAAAGGACA | 3′UTR and Exon
1 |
| Segment 2 |
GCCTTCATCAAAGGTTATCTCTGTC |
AAAGTCTAGAAGCAGAGCCCAGGAG | Exon 2 |
| Segment 3 |
CCTTCCTGACTTCGTCCTCCTGTAT |
GAGCTTGGGTGGGGACAGCTTAC | Exons 3 and 4 |
| Segment 4 |
CCGTCTGATTGTGTCTTTCTGCAC |
CAACCCTTTAGAGCCACTCATGACA | Exons 5 and 6 |
| Segment 5 |
CCAGGTAAGGGAGCTGAGTCTGAAT |
CCAGGAGCTGTTTCTCCAGGTATTT | Exons 7 and 8 |
| Segment 6 |
CGTGATGTAGGTGAGTGCTTATGGC |
TTCACTGTAGAAAATTTATTTGCCT | Exon 9 and
5′UTR |
Microsatellite LOH analysis of the
ST6GALNAC1 locus
In samples from 34 patients with ESCC, PCR was
performed for three dinucleotide repeat microsatellite markers
(D17S2238, D17S2243 and D17S2245) within the ST6GALNAC1
locus using fluorescent primer pairs (Applied Biosystems). LOH was
analyzed using an ABI PRISM 3100 Genetic Analyzer and GeneScan
Analysis and Genotyper software version 3.7.1 (Applied
Biosystems).
Cell lines and cell culture
Ten human ESCC cell lines (KY150, KY270, KYSE410,
KYSE450, KYSE510, TE1, TE6, TE8, TE9 and TE10) were purchased from
the Japanese Collection of Research Bioresources Cell Bank and the
Riken Bioresource Center. Cells were maintained in RPMI-1640
containing 10% fetal bovine serum and cultured in a humidified 5%
CO2 incubator at 37°C.
Methylation levels and response to
5-aza-2′-deoxycytidine (5-Aza-dC) treatment in ESCC cell lines
ESCC cells were seeded at a density of
1×106 cells/10 cm dish and cultured for 24 h with an
inhibitor of DNA methyltransferase, 5-Aza-dC (Sigma-Aldrich, St.
Louis, MO, USA), at a final concentration of 2 µM. Control cells
were treated with the diluent phosphate-buffered saline (PBS)
alone. After 48 h of incubation, total RNA was extracted from
collected cells in each dish.
Statistical analysis
Data from RNA-seq analyses, qRT-PCR and methylation
assays were analyzed using JMP 12 software (SAS Institute, Inc.,
Cary, NC, USA) and GraphPad Prism 6 (GraphPad Software Inc., La
Jolla, CA, USA). Differences between the gene expression levels of
samples were estimated using the Wilcoxon rank test or paired
t-test. All differences were considered statistically significant
at the level of p<0.05.
Results
Differential gene expression profiling on
chromosome 17q25.1 in ESCC samples by RNA-seq analysis
In our RNA-seq analysis from three patients with
ESCC with differences in tumor stage, histological differentiation
and tumor location, 17,673 genes were detected with an RPKM value
of at least 2.0 in either normal or tumor tissues. The first 10
genes showed significant increase or decrease in their expression
in tumor tissues compared with their expression in normal tissues
and are listed in Table III.
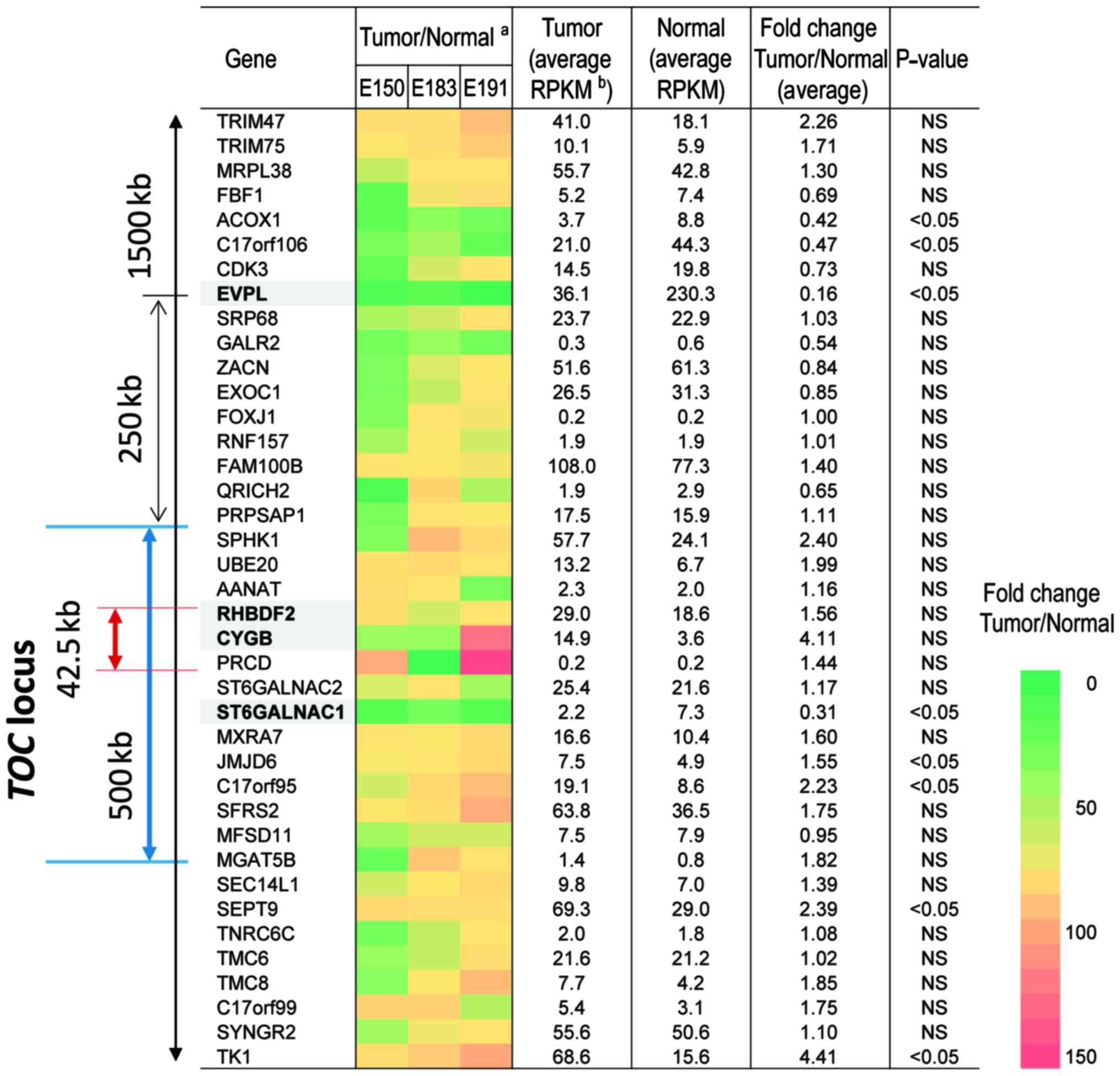
Among the expression data obtained by RNA-seq analysis, we focused
on the expression profile of genes in a 1500 kb region on 17q25.1
including the TOC locus, which has been mapped to the 500 kb region
in UK, USA and German pedigrees (8) and narrowed to a 42.5 kb region in the
UK pedigree (9). The differences
in expression levels between normal and tumor samples for 39 genes
in the region are shown in Fig. 2.
Among these genes, the expression levels of EVPL and
ST6GALNAC1 in tumor tissues were significantly decreased to
less than one-third of the levels in normal tissues (Fig. 2).
 | Table IIIUpregulated and downregulated genes
in ESCC tumor by RNA-seq analysis. |
Table III
Upregulated and downregulated genes
in ESCC tumor by RNA-seq analysis.
| Genes | Tumor (average
RPKM) | Normal (average
RPKM) | Fold-change
Tumor/normal |
|---|
| Upregulated |
| CST1 | 160.41 | 0.21 | 763.84 |
| GRP | 6.88 | 0.01 | 516.25 |
| OBP2A | 7.77 | 0.02 | 466.4 |
| IL-17C | 3.23 | 0.01 | 322.67 |
| MMP11 | 160.47 | 0.64 | 250.74 |
|
RNASE10 | 5.27 | 0.03 | 175.56 |
| MMP13 | 2.44 | 0.02 | 146.6 |
|
HIST1H3G | 3.61 | 0.03 | 135.25 |
| HOXD11 | 7.5 | 0.06 | 132.35 |
| MMP3 | 2.17 | 0.02 | 130 |
| Downregulated |
|
TMPRSS11B | 0.7 | 156.48 | 222.48 |
| SFTA2 | 0.22 | 42.3 | 192.26 |
| MUC21 | 3.24 | 494.75 | 152.54 |
| CRNN | 20.17 | 3015.85 | 149.5 |
| KRT4 | 75.48 | 11106.92 | 147.15 |
| MAL | 21.72 | 2735.73 | 125.97 |
| CWH43 | 0.21 | 15 | 70.3 |
| KRT78 | 9.69 | 617.84 | 63.76 |
| KRT13 | 610.37 | 37852.87 | 62.02 |
| CD207 | 0.06 | 3.79 | 59.89 |
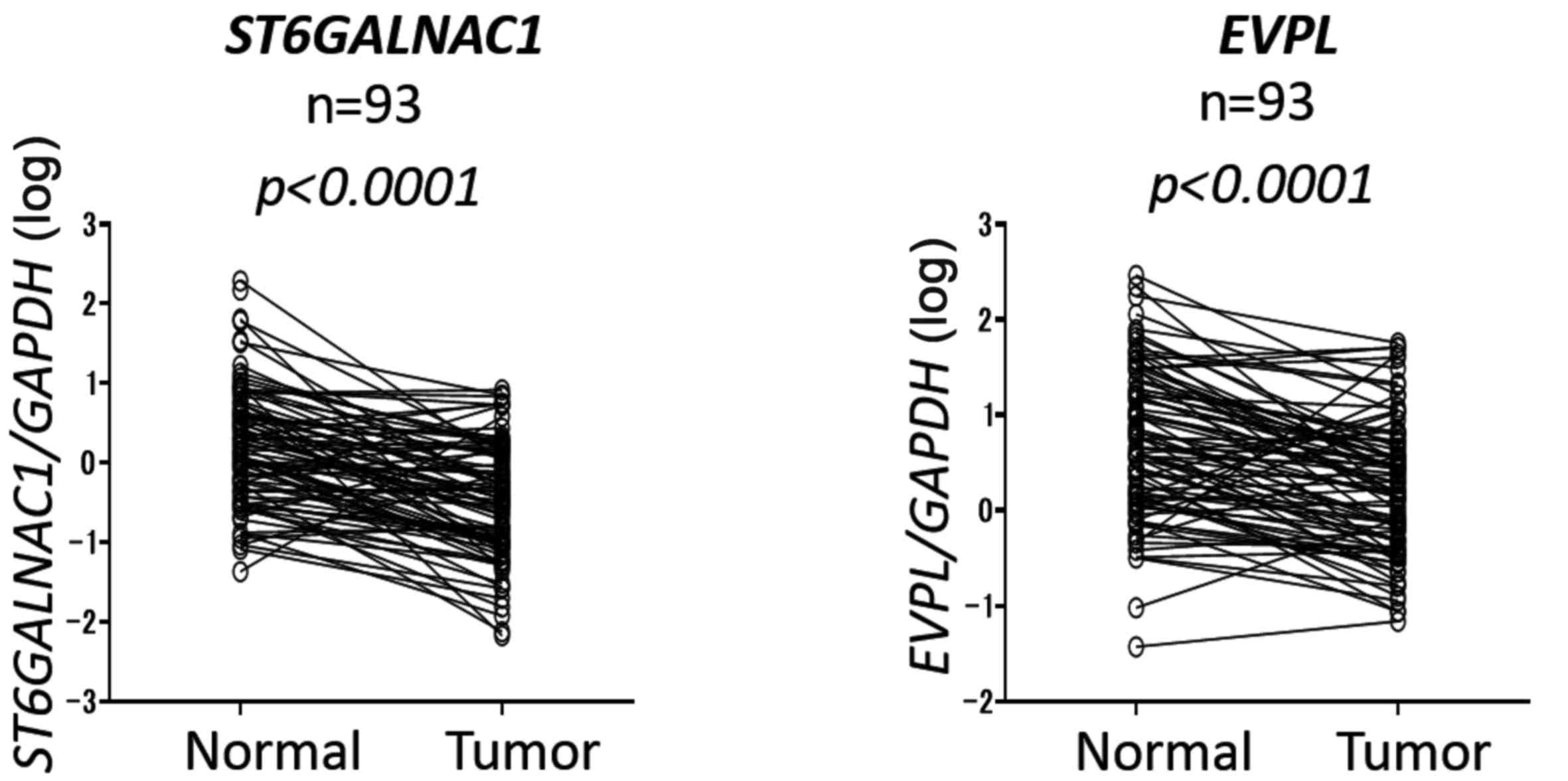
Downregulation of ST6GALNAC1 and EVPL in
ESCC tissues
We validated the expression levels of these two
genes using qRT-PCR in samples from 93 patients with ESCC. Both
EVPL and ST6GALNAC1 displayed significant
downregulation in tumor samples compared to their corresponding
normal tissue levels (p<0.0001) (Fig. 3). Although downregulation of
CYGB has been demonstrated in esophageal tissues in tylotic
patients compared with that in the normal esophagus (12), a significant difference in its
expression was not observed between sporadic ESCC and normal
samples by qRT-PCR analyses in our series (data not shown).
Nucleotide variants in ST6GALNAC1
Direct sequence analyses of samples from 46 patients
with ESCC revealed several nucleotide variants in ST6GALNAC1. Two
missense variants [c. 400 C>T (p.Pro67Leu) and c.724 C>T
(p.Thr175Met)] were observed in two patients. In four patients, 3
bp in-frame deletions [c.752_754delTGG (p.His184del)] were
observed. These variants were detected in both tumor and
corresponding normal tissues, and they have been registered in
dbSNP as rs143927446, rs138569950 and rs565363235. A one-base G
insertion in intron 3 registered as rs146144287 in dbSNP was also
detected in both tumor and normal tissues from six patients with
ESCC. Only one patient (1/46, 2.1%) displayed a tumor-specific
mutation in exon 2 of ST6GANAC1, although this mutation was
silent [c.509 G>A (p.Glu103Glu)] (Fig. 1). Therefore, no tumor-specific
non-synonymous mutations were observed in the coding region of
ST6GALNAC1.
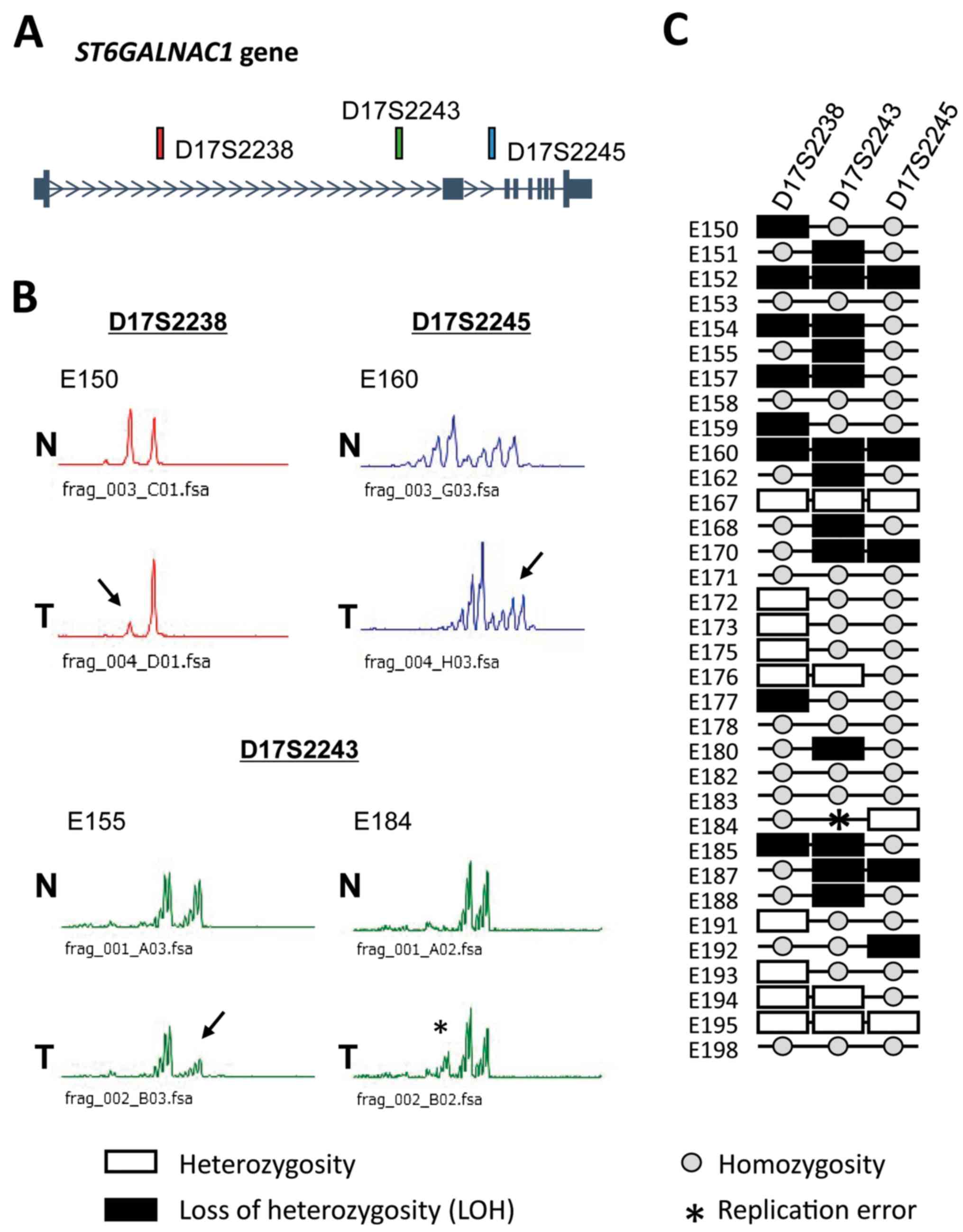
Frequent LOH was observed in the
ST6GALNAC1 locus in ESCC samples
Three microsatellite markers, D17S2238, D17S2243 and
D17S2245, were located in the introns of ST6GALNAC1
(Fig. 4A). PCR microsatellite
analysis of the ST6GALNAC1 locus in 34 patients using these
three markers demonstrated LOH in 17/27 (62.9%) informative cases
at one or more sites and a replication error in 1/27 (3.7%) cases
at D17S2243 (Fig. 4B and C).
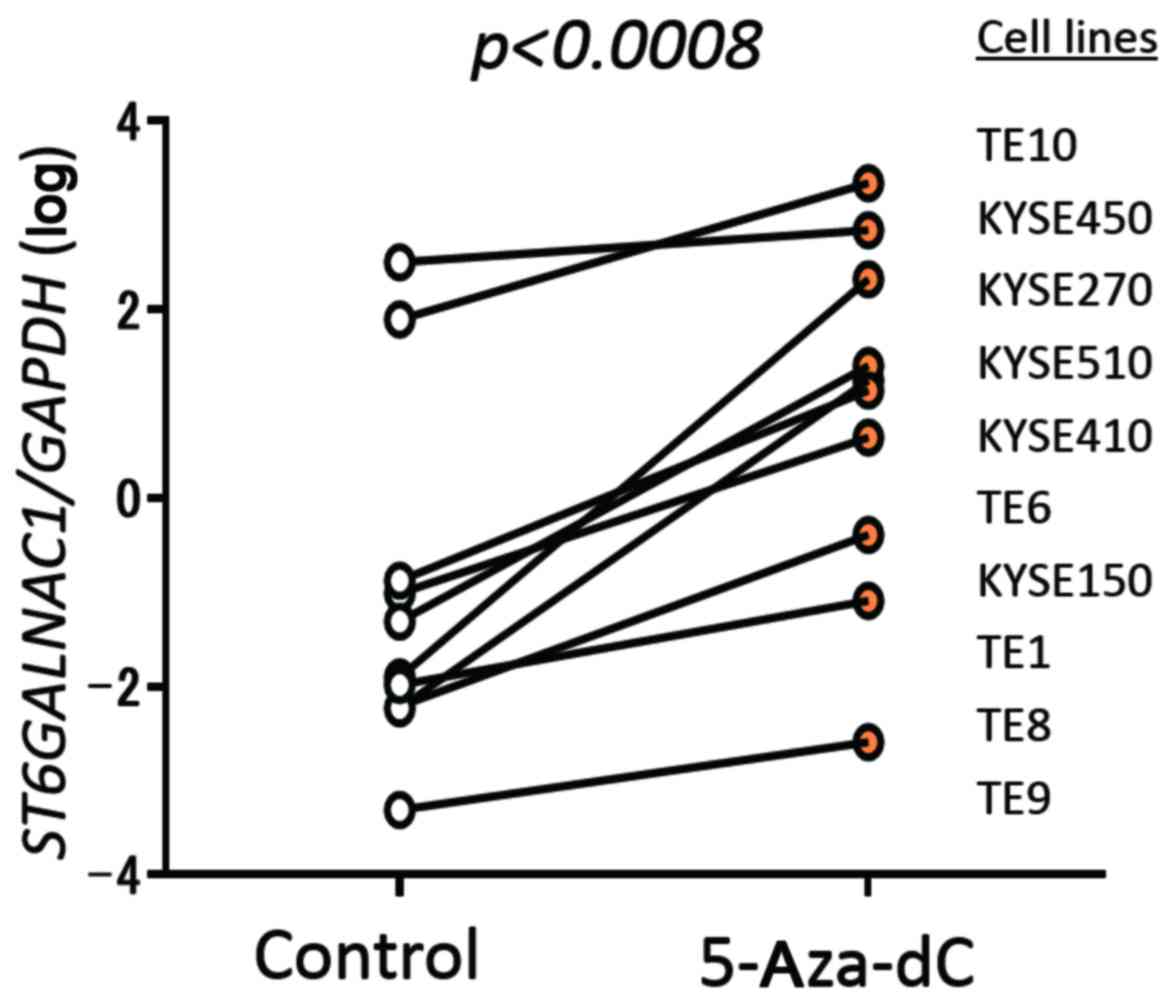
ST6GALNAC1 downregulation by methylation
in ESCC cell lines
Treatment with 5-Aza-dC significantly elevated
ST6GALNAC1 expression compared to the control level in all
10 ESCC cell lines assessed (Fig.
5).
Discussion
Because the region in chromosome 17q25.1 was
affected in both hereditary and sporadic forms of ESCC, it is
believed that a gene responsible for the oncogenesis or development
of ESCC existed in this region. Several genes in the TOC locus have
been studied in inherited and sporadic forms of ESCC. In our
present study, ST6GALNAC1 expression was significantly
decreased in ESCC tumor tissue compared with that in the
corresponding normal tissue, and the gene was located in the
vicinity of the minimal region of the TOC locus (Figs. 2 and 3). It was reported that ST6GALNAC1
was associated with biosynthesis of the sialyl-Tn (sTn) antigen in
cancer cells and that it was overexpressed in gastric, breast and
prostate cancer cell lines, inducing sTn expression (20). In contrast to these findings, our
results suggest that ST6GALNAC1 may have a tumor-suppressor
function in ESCC. In terms of the downregulation mechanisms of
ST6GALNAC1 expression, frequent LOH (62.9%) of the gene
locus was demonstrated in ESCC, although no tumor-specific mutation
was observed in the coding region (Figs. 1 and 4). Furthermore, demethylation using
5-Aza-dC recovered ST6GALNAC1 expression in all 10 ESCC cell
lines examined (Fig. 5). There are
no CpG islands in the promoter region of ST6GALNAC1.
However, it has been reported that ST6GALNAC1 was
downregulated by hyper-methylation of GC 2 bp upstream of the
transcription start site in estrogen receptor- and progesterone
receptor-positive breast cancers (21). These results suggest that
ST6GALNAC1 was inactivated by LOH and hyper-methylation of
the transcription start site.
Several candidate genes for sporadic and inherited
ESCC have been demonstrated. EVPL is a member of the
desmosomal plaque protein family that attaches to desmosomal
cadherin and keratin filaments. We previously reported infrequent
mutations and frequent LOH of this gene in sporadic ESCC (22). In this study, we demonstrated that
EVPL expression was significantly decreased in ESCC tissues
(Fig. 3). Downregulation of
EVPL may be involved in ESCC development, although the gene
is located 250 kb to the telomeric side of the minimal region of
the TOC locus (Fig. 2). It has
been demonstrated that CYGB was a tumor suppressor gene
inactivated by DNA hyper-methylation of its promoter in several
types of cancer, including ESCC (12,23).
However, downregulation of CYGB was not observed in our
series of tumor tissues from Japanese patients with ESCC compared
to its levels in the corresponding normal tissues (data not shown).
CYGB methylation has also been demonstrated in multiple
malignancies other than ESCC, such as leukaemia as well as breast,
bladder, lung and colon cancers (23). These findings indicated that
alterations of CYGB were limited to a subset of ESCCs and
that the tumor-suppressor role of CYGB may not be specific
in esophageal tissues but instead may be common among many types of
malignancies.
Recently, missense mutations of RHBDF2 in the
minimal region of the TOC locus were identified in patients with
tylosis from US/UK, German and Finnish families (7,13).
It may be clear that RHBDF2 is a responsible gene for
tylosis. Blaydon et al demonstrated that the altered
RHBDF2 represents a gain-of-function allele that results in
sustained EGFR signaling within the cells, and the signaling leads
to a hyper-proliferative phenotype. Furthermore, it was suggested
that RHBDF2 may also be dysregulated in a similar manner in
sporadic ESCC according to immunohistochemical data (13). In our previous study, however,
RHBDF2 mutation was not observed by whole-exome sequence
analysis using next-generation sequencing in 144 patients with
sporadic ESCC (14). This
sequencing analysis also demonstrated that recurrent mutations were
observed only in ZNF750, with the mutation rate of 16.7%, on
chromosome 17q. Frequent ZNF750 mutations were also
demonstrated in Chinese patients with sporadic ESCC by whole-exome
sequence analysis (15).
Furthermore, we found that the mutational APOBEC signature was
predominantly observed in the ESCC genome, and ZNF750
mutations were positively associated with the APOBEC signature
(14). ZNF750 was located
on 17q25.3 telomeric to the TOC locus, and its mutations were null
mutations accompanied by LOH (14). Therefore, ZNF750 may be a
strong candidate tumor suppressor gene for ESCC. It remains unclear
as to which gene dysregulation of in the chromosomal region is
essential for the development of hereditary and sporadic forms of
ESCC. Mutations in one allele of RHBDF2 gene induced
sustained EGFR signaling in the cells and led to a
hyperproliferative phenotype during wound repair in patients with
tylosis (13). Although
RHBDF2 mutation was not observed in sporadic ESCC, the EGFR
signaling were frequently dysregulated in sporadic ESCC cells
(14–17). Therefore, an abnormality in the
RHBDF2-EGFR pathway may lead to precancerous lesions in the
esophagus. In addition to the oncogenic change in RHBDF2-EGFR,
further inactivation of several tumor suppressor genes on 17q25,
such as EVPL, CYGB and ZNF750, by a two-hit
mechanism may induce ESCC.
In conclusion, ST6GALNAC1 was downregulated
in sporadic ESCC by hyper-methylation and LOH, and it may be a
candidate responsible gene for ESCC. Furthermore, our results on
sporadic ESCC and recent studies on tylotic families suggest that
multiple genes on chromosome 17q25 are involved in ESCC development
(Table IV).
 | Table IVSummary of altered genes on
chromosome 17q25 in sporadic or hereditary forms of esophageal
squamous cell carcinoma (ESCC). |
Table IV
Summary of altered genes on
chromosome 17q25 in sporadic or hereditary forms of esophageal
squamous cell carcinoma (ESCC).
| Genes | Locus | Position
(start-end) | Known function | Gene alterations in
sporadic ESCC or tylosis families |
|---|
| EVPL | 17q25.1 |
76004502–76027452 | Link the cornified
envelop to desmosomes and intermediated filaments | LOH and infrequent
mutation Sporadic ESCC |
| RHBDF2 | 17q25.1 |
76471069–76501790 | Activation of the
altered protein leading to constant EGF receptor signalling and
hyper-proliferation | Germ-line missense
mutations Tylosis families |
| CYGB | 17q25.1 |
76527356–76537905 | Collagen synthesis,
O2 sensing and transport or detoxification of reactive oxygen
species | LOH and
hyper-methylation Tylosis families |
|
ST6GALNAC1 | 17q25.1 |
76624763–76643838 | Synthesis of the
sialyl-Tn antigen in cancer cells | LOH and
hyper-methylation Sporadic ESCC |
| ZNF750 | 17q25.3 |
82828435–82840578 | Control terminal
epidermal differentiation via interactions with KDM1A, RCOR1 and
CTBP1/2 | LOH and frequent
mutation Sporadic ESCC |
Acknowledgments
This study was supported by JSPS KAKENHI (grant nos.
JP23591937 and JP26461994).
References
|
1
|
Ellis A, Field JK, Field EA, Friedmann PS,
Fryer A, Howard P, Leigh IM, Risk J, Shaw JM and Whittaker J:
Tylosis associated with carcinoma of the oesophagus and oral
leukoplakia in a large Liverpool family - a review of six
generations. Eur J Cancer B Oral Oncol. 30B:102–112. 1994.
View Article : Google Scholar
|
|
2
|
Stevens HP, Kelsell DP, Bryant SP, Bishop
DT, Spurr NK, Weissenbach J, Marger D, Marger RS and Leigh IM:
Linkage of an American pedigree with palmoplantar keratoderma and
malignancy (palmoplantar ectodermal dysplasia type III) to 17q24.
Literature survey and proposed updated classification of the
keratodermas. Arch Dermatol. 132:640–651. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hennies HC, Hagedorn M and Reis A:
Palmoplantar keratoderma in association with carcinoma of the
esophagus maps to chromosome 17q distal to the keratin gene
cluster. Genomics. 29:537–540. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kelsell DP, Risk JM, Leigh IM, Stevens HP,
Ellis A, Hennies HC, Reis A, Weissenbach J, Bishop DT, Spurr NK, et
al: Close mapping of the focal non-epidermolytic palmoplantar
keratoderma (PPK) locus associated with oesophageal cancer (TOC).
Hum Mol Genet. 5:857–860. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Risk JM, Field EA, Field JK, Whittaker J,
Fryer A, Ellis A, Shaw JM, Friedmann PS, Bishop DT, Bodmer J, et
al: Tylosis oesophageal cancer mapped. Nat Genet. 8:319–321. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Varela AB, Blanco Rodríguez MM, Boullosa
PE and Silva JG: Tylosis A with squamous cell carcinoma of the
oesophagus in a Spanish family. Eur J Gastroenterol Hepatol.
23:286–288. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saarinen S, Vahteristo P, Lehtonen R,
Aittomäki K, Launonen V, Kiviluoto T and Aaltonen LA: Analysis of a
Finnish family confirms RHBDF2 mutations as the underlying factor
in tylosis with esophageal cancer. Fam Cancer. 11:525–528. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Risk JM, Evans KE, Jones J, Langan JE,
Rowbottom L, McRonald FE, Mills HS, Ellis A, Shaw JM, Leigh IM, et
al: Characterization of a 500 kb region on 17q25 and the exclusion
of candidate genes as the familial tylosis oesophageal cancer (TOC)
locus. Oncogene. 21:6395–6402. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Langan JE, Cole CG, Huckle EJ, Byrne S,
McRonald FE, Rowbottom L, Ellis A, Shaw JM, Leigh IM, Kelsell DP,
et al: Novel microsatellite markers and single nucleotide
polymorphisms refine the tylosis with oesophageal cancer (TOC)
minimal region on 17q25 to 42.5 kb: Sequencing does not identify
the causative gene. Hum Genet. 114:534–540. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iwaya T, Maesawa C, Ogasawara S and Tamura
G: Tylosis esophageal cancer locus on chromosome 17q25.1 is
commonly deleted in sporadic human esophageal cancer.
Gastroenterology. 114:1206–1210. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
von Brevern M, Hollstein MC, Risk JM,
Garde J, Bennett WP, Harris CC, Muehlbauer KR and Field JK: Loss of
heterozygosity in sporadic oesophageal tumors in the tylosis
oesophageal cancer (TOC) gene region of chromosome 17q. Oncogene.
17:2101–2105. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
McRonald FE, Liloglou T, Xinarianos G,
Hill L, Rowbottom L, Langan JE, Ellis A, Shaw JM, Field JK and Risk
JM: Downregulation of the cytoglobin gene, located on 17q25, in
tylosis with oesophageal cancer (TOC): Evidence for trans-allele
repression. Hum Mol Genet. 15:1271–1277. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Blaydon DC, Etheridge SL, Risk JM, Hennies
HC, Gay LJ, Carroll R, Plagnol V, McRonald FE, Stevens HP, Spurr
NK, et al: RHBDF2 mutations are associated with tylosis, a familial
esophageal cancer syndrome. Am J Hum Genet. 90:340–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sawada G, Niida A, Uchi R, Hirata H,
Shimamura T, Suzuki Y, Shiraishi Y, Chiba K, Imoto S, Takahashi Y,
et al: Genomic landscape of esophageal squamous cell carcinoma in a
Japanese population. Gastroenterology. 150:1171–1182. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin DC, Hao JJ, Nagata Y, Xu L, Shang L,
Meng X, Sato Y, Okuno Y, Varela AM, Ding LW, et al: Genomic and
molecular characterization of esophageal squamous cell carcinoma.
Nat Genet. 46:467–473. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Song Y, Li L, Ou Y, Gao Z, Li E, Li X,
Zhang W, Wang J, Xu L, Zhou Y, et al: Identification of genomic
alterations in oesophageal squamous cell cancer. Nature. 509:91–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun
ZM, Zhang F, Zhao ZR, Li ZT, Liu ZY, et al: Genetic landscape of
esophageal squamous cell carcinoma. Nat Genet. 46:1097–1102. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Iwaya T, Fukagawa T, Suzuki Y, Takahashi
Y, Sawada G, Ishibashi M, Kurashige J, Sudo T, Tanaka F, Shibata K,
et al: Contrasting expression patterns of histone mRNA and microRNA
760 in patients with gastric cancer. Clin Cancer Res. 19:6438–6449.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yokobori T, Mimori K, Iwatsuki M, Ishii H,
Onoyama I, Fukagawa T, Kuwano H, Nakayama KI and Mori M:
p53-altered FBXW7 expression determines poor prognosis in gastric
cancer cases. Cancer Res. 69:3788–3794. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Munkley J: The role of Sialyl-Tn in
cancer. Int J Mol Sci. 17:2752016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li L, Lee KM, Han W, Choi JY, Lee JY, Kang
GH, Park SK, Noh DY, Yoo KY and Kang D: Estrogen and progesterone
receptor status affect genome-wide DNA methylation profile in
breast cancer. Hum Mol Genet. 19:4273–4277. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Iwaya T, Maesawa C, Kimura T, Ogasawara S,
Ikeda K, Kimura Y, Noda Y, Ishida K, Sato N, Saito K, et al:
Infrequent mutation of the human envoplakin gene is closely linked
to the tylosis oesophageal cancer locus in sporadic oesophageal
squamous cell carcinomas. Oncol Rep. 13:703–707. 2005.PubMed/NCBI
|
|
23
|
Shivapurkar N, Stastny V, Okumura N,
Girard L, Xie Y, Prinsen C, Thunnissen FB, Wistuba II, Czerniak B,
Frenkel E, et al: Cytoglobin, the newest member of the globin
family, functions as a tumor suppressor gene. Cancer Res.
68:7448–7456. 2008. View Article : Google Scholar : PubMed/NCBI
|