Introduction
Cancer continues to be a highly challenging
pathology that claims a prominent position as cause of annual
mortaliy worldwide due mainly to global aging, but also to more
numerous cases in developing countries (1). Current conventional chemotherapy has
various limitations (i.e., severe adverse effects and resistance to
treatment) (2); thus the
identification of novel effective and selective antitumor compounds
is mandatory. An excellent analysis of the current challenges and
future objectives in the treatment of cancer reported that the
continuously growing knowledge of molecular and cellular tumor
biology has revolutionized cancer treatment towards: i) the
technological improvement of tumor molecular profiling; and ii) the
discovery of predictive molecular targets (3).
The extensive biochemical research of multiple types
of cancer has revealed important enzymatic signaling pathways
responsible for tumor occurrence and progression, thus compelling
the need for the discovery of new means with which to block these
signaling cascades. Current approaches in the discovery of novel
anticancer therapies are oriented towards finding suitable
molecules that act as specific inhibitors of key enzymes which play
a significant role in carcinogenesis (4). The phosphoinositide 3-kinase/protein
kinase B (PI3K/AKT) pathway plays an important role in maintaining
relevant cellular functions, such as cell proliferation, survival
and cell translation (5,6) and its components are frequently
mutated, overexpressed or altered in common human cancers;
therefore, the targeting of the PI3K/AKT pathway by novel antitumor
drugs represents a promising alternative in cancer treatment
(7,8).
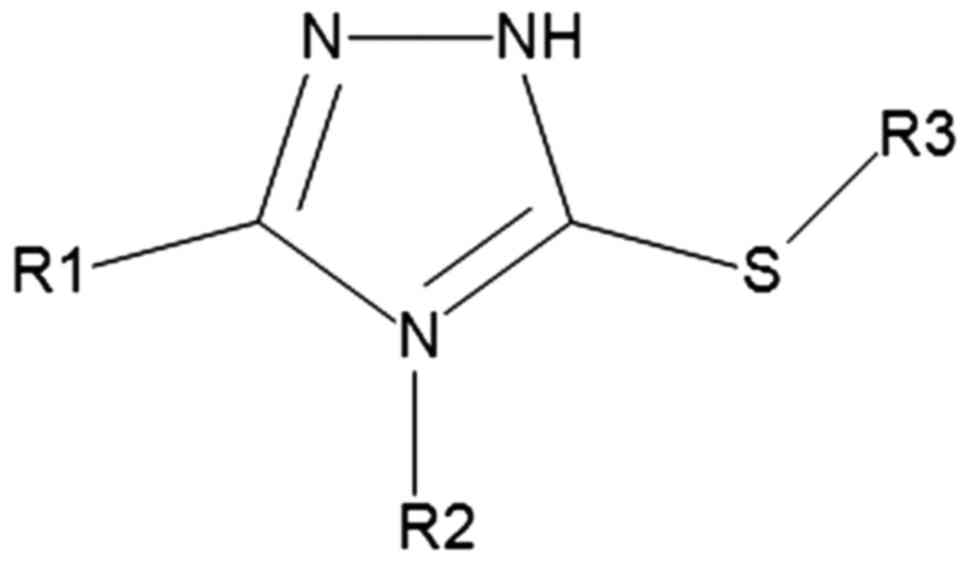
Triazoles are heterocyclic aromatic 5-membered ring
compounds (9) represented by two
isomers: 1,2,3-triazoles and 1,2,4-triazoles, each of them having
two tautomeric forms correlated with the nitrogen atom that has a
hydrogen atom bonded to it (Fig.
1).
Molecules bearing the 1,2,4-triazole moiety are
known to possess multiple biological activities, including
anticancer activity; the chemical modulation of the triazole ring
leads to the development of highly effective compounds with
improved selectivity (9). A search
carried out on the WOMBAT (10)
database has resulted in the identification of a large number of
biologically active molecules containing the 1,2,4-triazole ring.
Furthermore, recent studies have reported the significant
anticancer activity of newly synthesized 5-mercapto-1,2,4-triazole
derivatives against several tumor cell lines (11–13).
Molecular docking has aided in the design of 1,2,4-triazole
derivatives that act as methionine aminopeptidase type II (MetAp2)
inhibitors, thus exhibiting antiproliferative activity through the
induction of apoptosis (14). A
series of 25 compounds bearing the 1,2,4-triazole moiety have been
synthesized and assessed as antitumor agents through docking
simulation in target protein structures (RCSB PDB ID: 1YS1, 1FM6,
1BXL) and in in vitro antitumor screening, revealing
significant activity on leukemia, melanoma, lung and ovarian cancer
cell lines (15). In addition, a
significant number of synthesized 1,2,4-triazole derivatives has
been reported with significant antibacterial activity (16,17).
The aim of the current study was the design and
synthesis of 5-mercapto-1,2,4-triazole derivatives with eventually
predicted antiproliferative and antibacterian properties; the
antitumor activity is tentatively exerted through the inhibition of
PI3K protein, as established by means of molecular docking. The
biological activity of the identified triazole compounds was
evaluated in vitro on tumor cell lines, and on several
bacterial strains, respectively.
Materials and methods
Compound library building
In the current study, we aimed to create a compound
library for the purpose of their virtual screening against protein
targets which have been proven to be active in various types of
cancer, such as breast, lung and colon cancer. The created compound
library contains 5-mercapto-1,2,4-triazole derivatives (469
molecules) that were obtained by the substitution of various
radicals on the 1,2,4-triazole ring, in the fourth and fifth
position (R1 and R2) and on the thiol group from the third position
(Fig. 2).
The compound database was prepared using OMEGA
version 2.5.1.4 (OpenEye Scientific Software, Inc., Santa Fe, NM,
USA) (18) and filtered by means
of OMEGA's BlockBuster filter, using default parameters. After the
filtering process, 200 conformers were generated for each ligand.
Before the start of conformer generation, stereoisomers were
generated for compounds that possess asymmetric carbons in their
structure which were subsequently treated as independent
molecules.
Molecular docking
Molecular docking was carried out using OEDocking
HYBRYD version 3.0.1 (OpenEye Scientific Software, Inc.) (19), that uses the structure of a target
protein and the structure of the co-crystallized ligand to dock and
score molecules and also allows the selection of multiple protein
targets that can be used in the docking process. Docking results
interpretation was carried out using Discovery Studio 4.1 (Dassault
Systemes, BIOVIA Corp., San Diego, CA, USA).
Three-dimensional crystallographic structures of the
target proteins selected for this study [PI3Kα, AKT and mammalian
target of rapamycin (mTOR)] were obtained from the RCSB
ProteinDataBank (www.rcsb.org; accessed May, 2016)
(20). A set of multiple 3D
structures corresponding to each of the three protein targets, were
used for docking purposes, selected by the following criteria: i)
Protein structures with a co-crystallized ligand (as required by
the docking software); ii) protein structures that are non-mutant;
and iii) protein structures that have a Cruickshank DPI
(diffraction precision index) (21) under 0.5. Protein structures were
prepared as receptors, suitable for docking, using OEDocking's
MakeReceptor version 3.0.1 (OpenEye Scientific Software, Inc.)
(18).
The compound library was docked in each set of 3D
structures corresponding to each of the three protein targets.
Protein structures selected from the RCSB Protein Data Bank and
used in the docking process, were the following: i) For PI3Kα
protein, RCSB PDB ID's used: 4WAF, 4JPS, 4L2Y; ii) for AKT1
protein, RCSB PDB ID's used: 1H10, 1UNQ, 2UVM, 3CQU, 3CQW, 3MV5,
3MVH, 3O96, 3QKK, 3QKL, 3QKM, 4EJN, 4EKL, 4GV1; and iii) for mTOR
protein, RCSB PDB ID's used: 4DRH, 4DRI, 4DRJ, 3FAP, 4FAP, 2FAP,
1NSG, 1FAP. The mechanism of the docking process employed follows
four basic steps: i) A structure from the compound library is
screened against the set of receptors selected for docking; ii) the
molecule is docked in the protein structure which contains a
co-crystalized ligand that has the best shape, chemical and
topological similarities with the compound in question; iii) the
docked compound is scored by the docking software; and iv) all
compounds are docked and ranked according to the scoring function
score (Chemgauss4) (19).
Chemical synthesis
Thiosemicarbazide, 4-ethoxy-benzoic acid,
4-n-butoxibenzoic acid, cinnamic acid, and solvents (ethanol,
N,N-dimethylformamide, pyridine) were purchased [Acros Organics
(Geel, Belgium); Sigma-Aldrich, (St. Louis, MO, USA)] and used as
received. The acid chlorides (Fig. 3;
1a–c) were synthesized by treating the corresponding acids with
thionyl chloride at reflux and used without further
purification.
The synthesis of 1H-3-R-5-mercapto-1,2,4-triazoles
(Fig. 3, 3a–c), was carried out by acylation of
thiosemicarbazide with the corresponding acid chloride and pyridine
in N,N-dimethylformamide, followed by the cyclisation reaction of
the resulted 1-acyl-thiosemicarbazides (Fig. 3, 2a–c), in ethanolic NaOH, at reflux,
according to the previously published procedure (22). The synthesis route is shown in
Fig. 3.
Compound characterization
Melting points were determined on a Böetius PHMK
(Veb Analytik, Dresden, Germany) instrument, and thin-layer
chromatography was carried out on silica gel-coated plates 60 F254
Merck using hexane: ethyl acetate (various ratios) as eluents. IR
spectra were recorded in KBr pellet on a Jasco FT/IR-410
spectrophotometer. 1H-NMR and 13C-NMR spectra were recorded on a
Bruker Avance DRX 400 spectrometer in DMSO-d6, at room temperature.
The chemical shifts are reported as δ values (ppm) and were
referenced to the solvent residual peak (2.51 ppm for 1H
and 39.47 ppm for 13C). Elemental analysis was carried
out using a Vario EL cube (Elementar Analysensysteme GmbH, Hanau,
Germany). Mass spectra experiments were conducted on a 6120
Quadrupole LC/MS system from Agilent Technologies, Inc. (Santa
Clara, CA USA) equipped with a UV detector, ESI ionization source
and a Zorbax Rapid Resolution SB-C18 (1.8 µm; 50×2.1 mm)
column. LC-MS grade methanol was obtained from Merck Millipore
(Darmstadt, Germany) and used without further purification. All
samples were obtained by dissolving the solid samples in methanol.
All samples were analyzed using methanol as the isocratic mobile
phase, at a flow rate of 0.4 ml/min, temperature of 25°C and λ=250
nm. All mass spectra were recorded in the positive ion mode under
optimized ESI parameters using nitrogen nebulizer pressure at
35psi, nitrogen drying gas temperature at 250°C and flow rate at 12
l/min and capillary voltage of 3,000 V in the Scan mode.
Antibacterial assay
The three compounds were screened for their
antimicrobial activity against 6 bacterial strains:
Staphylococcus aureus ATCC 25923, Streptococcus
pyogenes ATCC 19615, Pseudomonas aeruginosa ATCC 27853,
Salmonella typhimurium ATCC 14028 and Proteus
vulgaris ATCC 13315 using the agar disk-diffusion method.
Culti-loops with ATCC strains were introduced into a tube with 1 ml
thioglycollate medium and these suspensions were then placed on
blood agar plates and incubated 24 h in a 37°C atmosphere. All the
bacterial strains were prepared from 24-h-old cultures. Bacterial
suspensions were adjusted to a McFarland's standard of 0.5 (a final
bacterial concentration of 1–2×108 CFU/ml). The
Mueller-Hinton agar plates were inoculated with bacterial
suspension using a sterile cotton swab. Within 15 min following
plate inoculation, sterile Whatman no. 1 filter paper disks (6 mm
in diameter) impregnated with the test compounds (3 µl
solution/disk) were distributed evenly on the plate surface leaving
at least 30 mm (center to center) between them. DMSO and gentamycin
were used as a negative and positive control, respectively. Plates
inoculated with the bacterial suspensions were incubated at 37°C
for 24 h. The inhibition zone diameters were measured in
millimeters using a ruler. For all bacterial strains duplicate
disk-diffusion tests were performed and the average reading was
taken into account.
Cell culture
The human tumor cell lines (A375 human melanoma
cells, Cat no. ATCC® CRL-1619™; A549 human lung
carcinoma cells, Cat no. ATCC® CCL-185™; and MDA-MB-231
human breast carcinoma cells, Cat no. ATCC® HTB-26™)
were purchased from the American Type Culture Collection (ATCC,
Manassas, VA, USA) via LGC Standards GmbH, Wesel Germany. The
B164A5 murine melanoma cell line and cell culture media, and
specific supplements were purchased from Sigma-Aldrich Co.
(Ayrshire, UK). Human keratinocytes (HaCaT) were a gift from the
University of Debrecen, Debrecen, Hungary. The cells were cultured
in Dulbecco's modified Eagle's Medium (DMEM) containing 4.5 g/l
glucose, L-glutamine and NaHCO3, and supplemented with
100 U/ml penicillin, 100 µg/ml streptomycin and 10% fetal
bovine serum (FBS). Cells were kept under standard conditions
(humidified atmosphere, 5% CO2, 37°C) and passaged every
second day. Cell number was determined by using the cell counting
chamber (Neubauer) in the presence of Trypan blue.
alamarBlue assay (viability assay)
Cell viability was measured by means of the
alamarBlue technique; cells (1×104/200 µl
medium/well) were seeded in a 96-well plate, allowed to attach and
then stimulated with various concentrations (10 and 50 µM)
of the test compounds for 24 h. At 24 h post-stimulation, a volume
of 20 µl/well of alamarBlue solution was added (10% of the
volume of cell culture medium present in each well-200 µl).
The plates were incubated at 37°C for 3 h and then
spectrophotometrically read at 570 and 600 nm, respectively, by
using a xMark™ Microplate Spectrophotometer (Bio-Rad, Berkeley, CA,
USA). Untreated cells and pure solvent were used as control. Cell
viability was calculated according to the previously published
formula (23).
Scratch assay (wound healing assay)
The migratory ability of various tumor cells (A375
human melanoma cells, A549 human lung cancer cells, MDA-MB-231
breast carcinoma cells and B164A5 murine melanoma cells) and
healthy cells (HaCaT human keratinocytes) was examined in
vitro by using a scratch assay technique. A number of
2×105 cells/well was cultivated in 12-well plates for 24
h prior to the experiment. A sterile pipette tip was used in order
to draw scratches in well-defined zones of the cells monolayer
(confluence of 80–90%). Cells that detached as a result of the
procedure were removed by washing with PBS before stimulation. The
cells were stimulated with the three test compounds: TZ53, TZ55 and
TZ3a following solubilization in DMSO (c1=10 µM
and c2=50 µM). Images of the cells in culture
were acquired at the starting point of the experiment, and after 3
and 24 h using an Optika Microscopes Optikam Pro Cool 5 and Optika
View software.
Statistical analysis
Measurements were carried out in triplicate for each
sample and the results are presented as the means ± standard error.
One-way ANOVA followed by Bonferroni's post-tests were used to
determine the statistical difference between the experimental and
control groups. A value of P<0.05 was considered to indicate a
statistically significant difference.
Results and Discussion
Molecular docking
The selection of the 5-mercapto-1,2,4-triazole
moiety for the building of the compound library was based on
previous studies that reported excellent antibacterial and
antitumor properties of structurally similar compounds. Nadeem
et al (24) reported the
synthesis of 4,5-disubstituted-1,2,4-triazoles-3-thiols that proved
active as antioxidants and exhibited antibacterial activity against
Gram-positive cocci, as well as significant cytotoxic activity
(24). In addition, in another
study, 1,2,4-triazole-3-thione derivatives bearing pyrazole
moieties were synthesized and tested against two human cancer cell
lines, MCF7 (breast carcinoma) and HeLa (cervix carcinoma) cells
using doxorubicin as a reference drug (25); all compounds revealed stronger
cytotoxic activity than doxorubicin accompanied by moderate
antibacterial activity.
Docking-based virtual screening (DBVS) aims to
compute the interactions between a potential ligand and the binding
site residues of a given protein-target. The binding affinity
towards the target is predicted using a scoring function (26). Virtual screening methods, such as
DBVS are extensively used in drug discovery to enhance
cost-efficiency (27).
Structure-based methods can be extensively applied due to numerous
3D crystallographic structures of key cancer targets. The docking
process in the current study used the following protein structures:
PI3Kα, AKT1 and mTOR.
PI3K is the enzyme that catalyzes the formation of
phosphatidylinositol 3,4,5-trisphosphate (PIP3) from
phosphatidylinositol 4,5-bisphosphate (PIP2) (28). PIP3 is responsible for the
activation of the enzyme, AKT, a serine/threonine-specific protein
kinase which phosphorylates downstream effectors within cell
survival, proliferation and metabolic pathways (29–31).
One of these effectors is mTOR, a serine/threonine protein kinase
from the phosphatidylinositol 3-kinase-related kinases (PIKK)
family which modulates protein synthesis via the regulation of the
phosphorylation state of translational proteins (Fig. 4), such as p70 ribosomal S6 kinase
(P70S6K) and eukaryotic translation initiation factor 4E-binding
protein 1 (4E-BP1) (32).
 | Figure 4Simplified scheme of the
PI3K/AKT/mTOR signaling pathway. PI3K, phosphoinositide
3-kinase/protein kinase B; PIP2, phosphatidylinositol
4,5-bisphosphate; PIP3, phosphatidylinositol 3,4,5-trisphosphate;
Akt, serine/threonine-specific protein kinase; mTOR, mammalian
target of tapamycin, a serine/threonine protein kinase; P70S6K, p70
ribosomal S6 kinase; 4E-BP1, eukaryotic translation initiation
factor 4E-binding protein 1. |
PI3K protein has been recognized as a major
signaling molecule involved in various cellular mechanisms, such as
cell proliferation and survival, as well as angiogenesis;
therefore, the abnormal signaling of the PI3K pathway has beeen
reported in almost 50% of human cancers (33). Mutations of the catalytic subunit
PI3KCA of PI3K protein occur in many types of cancer, including
colorectal cancer (34); in some
cases, PI3K mutations are associated with a poor clinical outcome
and prognosis.
Docked molecules were scored by the software's
scoring function (Chemgauss4). From all sets of 3D protein
structures corresponding to a target, the protein structure with
the highest number of docked molecules was selected for further
analysis. As the docking program has a ligand-based scoring
function, selecting the structure with the highest number of docked
compounds means that the respective protein binding site (or
binding site conformation) is the most adequate for later
receptor-target binding analysis. As such the following protein
structures were selected for further analysis: PI3Kα-PDB ID: 4L2Y,
AKT1-PDB ID: 3CQU and mTOR-PDB ID: 4DRH. In each case, the first 50
molecules ranked by the docking score were analyzed, taking into
account the molecular interactions with binding site residues and
the topological similarity with each cocrystalized ligand of the
protein. For mTOR, compounds were docked in the binding site of
rapamycin; given the fact that rapamycin is a very large molecule
compared to the structures from the compound library used for
docking, high scores were obtained for some of the structures.
Nevertheless we concluded that the molecular sizes of the triazole
derivatives were too small to accommodate well in the active
binding site of the mTOR protein. In the case of PI3K protein,
three small molecules TZ53, TZ55 and TZ3a, revealed good
co-planarity with the co-crystallized ligand of 4L2Y and
significant favorable interactions within the binding site.
Similarly to the co-crystallized ligand, compounds formed hydrogen
bonds (H-Bond) with the Asp810 and Tyr836 residues (Fig. 5).
Chemistry
As a result of docking analysis, compounds TZ53,
TZ55 and TZ3a were synthesized and further tested for their
toxicity and anticancer activity. Physicochemical analysis
confirmed the structure of the three triazole derivatives:
1H-3-(4-ethoxyphenyl)-5-mercapto-1,2,4-triazole (3a) (TZ53)
White powder; m.p.=225–227°C IR [KBr] (cm-1): 3093,
3022, 2996, 2979, 2938, 2916, 1615, 1596, 1570, 1522, 1457, 1394,
1300, 1259, 1229, 1181, 1118, 1045, 974, 838, 823, 731, 694, 549.
1H-NMR, 400.13 MHz, δ (ppm), DMSO-d6: 13.56 (s, br, H1, H6), 7.83
(d, J=8.4 Hz, 2H, H2′, H6′), 7.04 (d, J=8.4 Hz, 2H, H3′, H5′), 4.08
(q, J=6.9 Hz, 2H, H8′), 1.33 (t, J=7.0 Hz, 3H, H9′). 13C-NMR, 100.6
MHz, δ (ppm), DMSO-d6: 166.6 (C5), 160.4 (C4′), 150.2 (C3), 127.4
(C2′, C6′), 117.7 (C1′), 115.0 (C3′, C5′), 63.4 (C8′), 14.6 (C9′).
Elemental analysis, calculated for C10H11N3OS: C, 54.28; H, 5.01;
N, 18.99; found: C, 54.12; H, 5.43; N, 18.58. LC-MS Rt=0.431 min,
m/z = 222.0 [M+ H+]+, m/z =
244.0 [M+ Na+]+.
1H-3-(4-n-butoxyphenyl)-5-mercapto-1,2,4-triazoles (3b) (TZ55)
White powder, m.p.=240–250°C TLC: (Hexane:EA=1:1)
Rf=0,64 IR [KBr] (cm-1): 3065, 3026, 3000, 2957, 2936, 2909, 2871,
1616, 1568, 1522, 1484, 1456, 1295, 1252, 1228, 1189, 1070, 1025,
1008, 967, 831, 784, 735, 700, 553, 524. 1H-NMR, 400.13 MHz, δ
(ppm), DMSO-d6: 13.7, 13.6 (H1, H6), 7.83 (d, J=8.4 Hz, 2H,
H2′,6′), 7.04 (d, J=8.8 Hz, 2H, H3′,5′), 4.01 (t, J=6.4 Hz, 2H,
H8′), 1.69 (m, J=6.9 Hz, 2H, H9′), 1.42 (m, J=7.3 Hz, 2H, H10′),
0.92 (t, J=7.4 Hz, 3H, H11′). 13C-NMR, 100.6 MHz, δ (ppm), DMSO-d6:
166.6 (C5), 160.5 (C4′), 150.2 (C3), 127.4 (C2′, C6′), 117.7 (C1′),
115.0 (C3′, C5′), 67.5 (C8′), 30.6 (C9′), 18.8 (C10′), 13.7 (C11′).
Elemental analysis, calculated for C12H15N3OS: C, 57.81; H, 6.06;
N, 16.85; found: C, 58.01; H, 6.13; N, 16.37. LC-MS Rt = 0.426 min,
m/z = 272.1 [M+ Na+]+.
1H-3-styryl-5-mercapto-1,2,4-triazoles
(3c) (TZ3a)
White powder, m.p.=250–255°C (H2O); TLC:
(Hexan:EA=1:1) Rf=0,44 IR [KBr] (cm-1):3455, 3387, 3095, 3025,
2962, 2905, 2849, 1651, 1565, 1507, 1469, 1448, 1309, 1219, 1011,
958, 857, 758, 730, 683, 517, 499. 1H-NMR, 400.13 MHz, δ (ppm),
DMSO-d6: 13.59 (s, br, H1, H6), 7.59 (d, J=7.2 Hz, 2H, H4′, H8′),
7.47 (d, J=16.8 Hz, 1H, H2′), 7.43–7.34 (3H, H5′, H6′, H7′), 6.96
(d, J=16.8 Hz, 1H, H1′). 13C-NMR, 100.6 MHz, δ (ppm), DMSO-d6:
166.7 (C5), 150.1 (C3), 135.4 (C2′), 135.1 (C3′), 129.4 (C6′),
129.0 (C5′, C7′), 127.2 (C4′, C8′), 112.5 (C1′). Elemental
analysis, calculated for C10H9N3S: C, 59.09; H, 4.46; N, 20.67;
found: C, 59.07; H, 4.82; N, 20.41. LC-MS Rt = 0.433 min,
m/z = 226.0 [M+ Na+]+.
Antibacterial activity
The three compounds, TZ53, TZ55, TZ3a, were tested
as antibacterial agents by the disk-diffusion method; the results
are displayed in Table I.
 | Table IAntimicrobial activity of compounds
TZ53, TZ55, TZ3a using the disk-diffusion method.a |
Table I
Antimicrobial activity of compounds
TZ53, TZ55, TZ3a using the disk-diffusion method.a
| Test compound | Concentration
(µg/ml) | Streptococcus
pyogenes ATCC 19615 | Pseudomonas
aeruginosa ATCC 27853 | Staphylococcus
aureus ATCC 25923 | Salmonella
typhimurium ATCC 14028 | Proteus
vulgaris ATCC 13315 |
|---|
| TZ53 | 10 | 7 mm | 7 mm | 7 mm | 7 mm | 7 mm |
| 25 | 7 mm | 7 mm | 7 mm | 7 mm | 7 mm |
| 50 | 7 mm | 7 mm | 7 mm | 7 mm | 7 mm |
| 100 | 7 mm | 7 mm | 7 mm | 7 mm | 7 mm |
| TZ55 | 10 | 7 mm | 7 mm | 7 mm | 7 mm | 7 mm |
| 25 | 7 mm | 7 mm | 7 mm | 7 mm | 7 mm |
| 50 | 7 mm | 7 mm | 7 mm | 7 mm | 7 mm |
| 100 | 7 mm | 7 mm | 7 mm | 7 mm | 7 mm |
| TZ3a | 10 | 7 mm | 7 mm | 7 mm | 7 mm | 7 mm |
| 25 | 7 mm | 7 mm | 7 mm | 7 mm | 7 mm |
| 50 | 7 mm | 7mm | 7 mm | 7 mm | 7 mm |
| 100 | 7 mm | 7 mm | 7 mm | 7 mm | 7 mm |
The antimicrobial in vitro testing was
conducted on four Gram-positive (Streptococcus pyogenes,
Staphylococcus aureus) and Gram-negative (Pseudomonas
aeruginosa, Proteus vulgaris) bacterial strains and
Salmonella typhimurium. The triazole derivatives revealed a
similar moderate antifungal and antibacterial activity with no
visible variations between structures. Gupta and Jain (17) synthesized a set of Shiff bases,
also based on the 5-mercapto-1,2,4-triazole scaffold, showing good
antimicrobial activity; the moderate antimicrobial activity of our
three synthesized compounds can be explained by comparison, taking
into account the main structural similarities between the two sets
of compounds: i) The main scaffold represented by the
5-mercapto-1,2,4-triazole moiety; ii) the substituted phenyl
radical in the fifth position; and iii) a non-substituted thiol
group in the third position. Nevertheless there are two main
structural differences that consist in the absence of a radical in
the fourth position (substituted bezylideneamino radical) and of
halogen substitution of the phenyl radical from the fifth position
(17), differences that may
justify the weaker antibacterial activity of our triazole
compounds.
Antiproliferative activity
The antiproliferative activity of the three triazole
derivatives, TZ53, TZ55, TZ3a was tested on several tumor cell
lines: A375 (human melanoma cells), B164A5 (murine melanoma cells),
MDA-MB-231 (breast carcinoma cells) and A549 (lung carcinoma
cells), as well as on one healthy cell line, HaCaT keratinocytes by
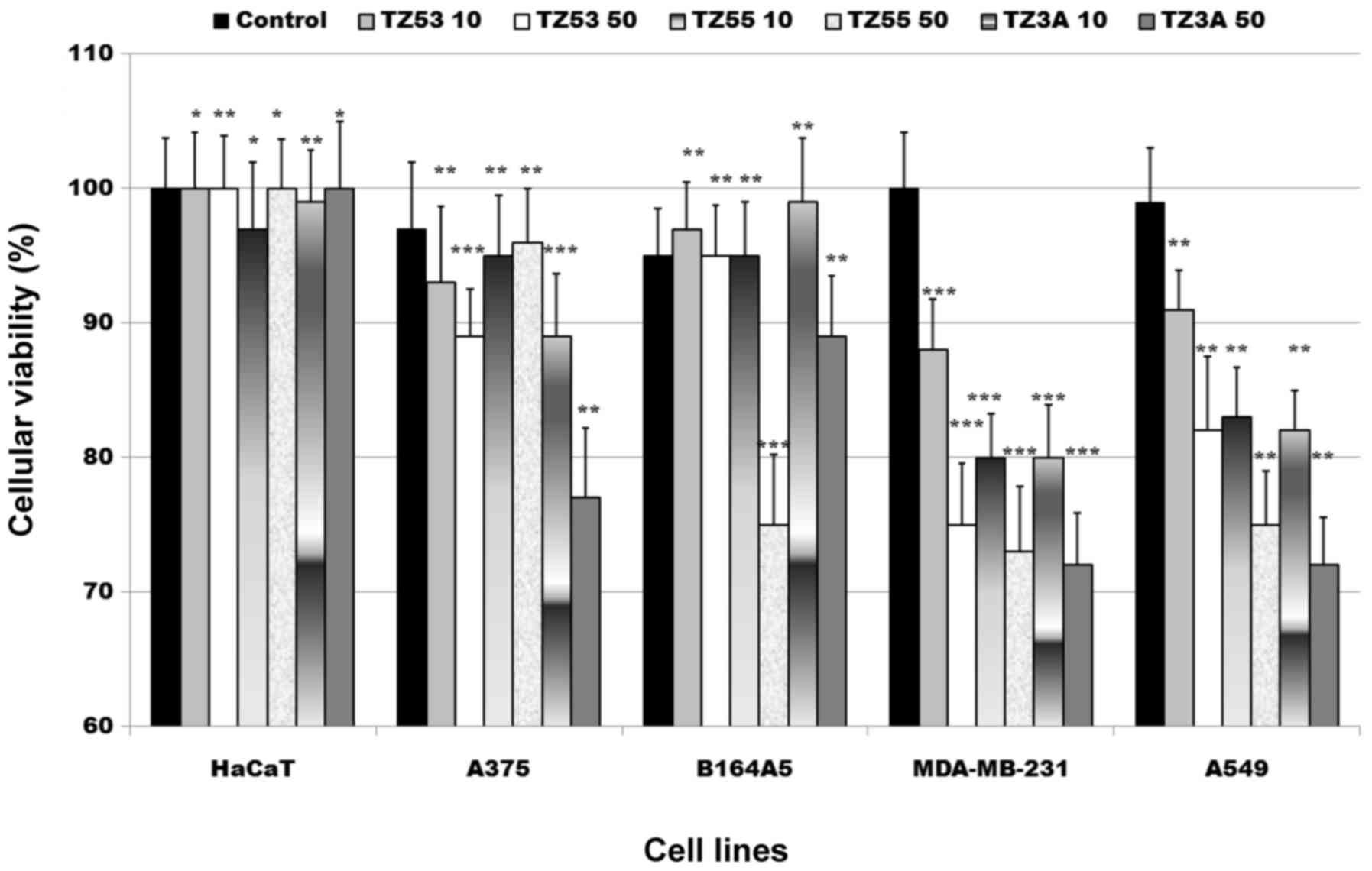
means of alamarBlue assay. The results are displayed in Fig. 6. The alamarBlue assay is a simple
test designed to quantitatively assess the number of viable cells;
it incorporates a cell growth indicator (alamarBlue) that reacts to
the cell metabolic activity by fluorescence and color change. More
specifically, as a result of cell growth, the non-fluorescent
oxidized blue form of the redox indicator turns into the
fluorescent reduced red form and the result is
spectrophotometrically evaluated (23). The graphical representation of the
alamarBlue assay results depicted in Fig. 6 shows a moderate antiproliferative
activity of the three compounds against the tumor cell lines used
in this study. The antiproliferative activity against the A375 and
B164A5 (melanoma cell lines, human and murine, respectively) was
relatively poor for all compounds. A stronger activity was noted
against the lung cancer cell line, A549, exerted in a
dose-dependent manner. NVP-BEZ235 is a PI3K-mTOR dual inhibitor
researched for its antiproliferative activity in various types of
cancer (35,36). Our result is consistent with that
of thye study by Gong et al (36), that assessed the antiproliferative
activity of multiple kinase inhibitors on various lung cancer cell
lines and revealed that NVP-BEZ235 inhibited the proliferation of
the A549 cell line. Therefore, we can presume that PI3K inhibition
may be accountable for the reported antiproliferative activity of
our compounds against A549 cancer cells.
The highest antiproliferative activity was recorded
against the breast cancer cell line, MDA-MB-231, an activity that
can be corroborated with the results of the molecular docking
analyses, which indicated the fitting of our triazole derivatives
in the active binding site of PI3K protein. MDA-MB-231 is a human
triple-negative breast cancer cell line characterized by the
absence of estrogen and progesterone receptors and also lacking
HER2 (receptor tyrosine-protein kinase erbB-2) amplification
(37); it exhibits a high
recurrence rate and poor prognosis. Thus far, there are only a few
studies regarding the discovery of therapeutic targets in the
treatment of triple-negative breast cancer. In 2014, Jia et
al (38) reported that
curcumin was cytotoxic to the MDA-MB-231 cell line, while another
breast cancer cell line (MCF7) proved to be resistant; the
antiproliferative activity of curcumin on MDA-MB-231 cells was
linked to its inhibitory activity on the PI3K/Akt pathway and,
moreover, the association of curcumin with a PI3K inhibitor,
wortmannin, further increased MDA-MB-231 sensitivity (38). Another study reported the
assessment of the cytotoxic activity of a selective PI3K inhibitor,
GDC-0941 (39); the MDA-MB-231
cancer cell line became sensitive to this inhibitor after the
siRNA-mediated silencing of the O-linked
N-acetylglucosamine (GlcNAc) transferase (OGT) gene, without
showing alterations of phosphorylated AKT levels. The combined
treatment of the MDA-MB-231 cell line with metformin and
carboplatin resulted in a synergistic inhibition of the AKT/mTOR
pathway, thus leading to decreased cell cycle and cell
proliferation (40). A highly
relevant study carried out by Liu et al (41) in 2011 may provide an explanation
regarding the major role the triazole ring plays in the selective
targeting of PI3K protein. According to their study, the selective
inhibition of PI3K protein is strongly required due to the high
toxicity associated with mTOR protein simultaneous inhibition which
disrupts cell metabolism; the authors reported the synthesis of an
excellent PI3K selective inhibitor containing the triazole ring.
This compound replaces the water molecule that forms a bridge
H-Bond, in the ATP binding site of the PI3Kα protein, between
Asp810 and Tyr836; the nitrogen in position 1 acts as an H-bond
acceptor thus interacting with Tyr836, while nitrogen in position 2
of the triazole ring serves as an H-bond donor thus interacting
with Asp810. The simultaneous presence of these interactions is
essential for a strong inhibition of the PI3Kα protein (41); our molecular docking studies
clearly revealed these two interactions (Fig. 5), thus reliably predicting the
biological results reported herein. Taking into account all these
experimental results, we can state that the inhibition of
MDA-MB-231 cell line is directly linked to the inhibition of PI3K
protein as indicated by molecular docking. It is presumable that
the triazole moiety plays an important role in the inhibition
mechanism through its favorable accommodation within the active
binding site of PI3K protein; since no signifi-cant differences
were reported between the three compounds in terms of
antiproliferative activity, the side chain does not seem to
influence the antiproliferative activity. An important aspect of
the in vitro testing is the toxicity assessment against
normal HaCaT cell line; the lack of cytotoxic activity indicates
low toxicity and selective antitumor activity.
Scratch assay
In light of the fact that concentrations of 10 and
50 µM of tested compounds induced a relatively moderate
activity against the viability of the four cancer cell lines, we
assumed that these concentrations might inflict an inhibitory
effect on tumor cells migration and proliferation. Therefore, the
two concentrations of the three test compounds, 10 and 50
µM, respectively, were used for the scratch assay method
which assesses cell migration and proliferation as a result of
stimulation.
The scratch assay results are consistent with the
antiproliferative results generated by the alamarBlue assay (data
not shown); the highest antimigratory activity was reported for
MDA-MB-231 cell line in both applied concentrations with no direct
correlation between concentration and biologic effect. Similar
results were recorded for A549 cell line where the inhibition of
migration was seen only for the higher concentration. As noticed
during the alamarBlue assay, A375 cell line exhibited the lowest
sensitivity towards all test compounds. In terms of toxicological
potential evaluated on HaCaT cells, all three compounds exhibited
no antimigratory effects.
In conclusion, in the present study, we described
the design and synthesis of three triazole derivatives with
antibacterial and antiproliferative activity. The three evaluated
structures emerged as a result of molecular docking which indicated
a highly favorable accommodation within the active binding site of
PI3K protein, thus acting as PI3K inhibitors and interfering with
the PI3K/AKT pathway, a signaling cascade present in numerous types
of cancer. The three compounds acted as antitumor agents against
triple negative breast cancer (MDA-MB-231 cell line) and exhibited
low toxicity and good selectivity thus offering promising
perspectives in anticancer therapy.
Acknowledgments
The authors are grateful to OpenEye Scientific
Software Inc. for providing academic license of their OEDocking
software. The authors wish to thank Dr Florin Borcan for his
assistance with the statistical analysis of thye experimental data.
The work of Sorin Avram was supported by Project 1.2 of the
Institute of Chemistry Timişoara of the Romanian Academy. This
study was financially supported by a grant from the Romanian
National Authority for Scientific Research and Innovation, CNCS –
UEFISCDI, project number: PN-II-RU-TE-2014-4-2842.
References
|
1
|
Chinthala Y, Thakur S, Tirunagari S,
Chinde S, Domatti AK, Arigari NK, K V N S S, Alam S, Jonnala KK,
Khan F, et al: Synthesis, docking and ADMET studies of novel
chalcone triazoles for anti-cancer and anti-diabetic activity. Eur
J Med Chem. 93:564–573. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Palumbo MO, Kavan P, Miller WH Jr, Panasci
L, Assouline S, Johnson N, Cohen V, Patenaude F, Pollak M, Jagoe
RT, et al: Systemic cancer therapy: achievements and challenges
that lie ahead. Front Pharmacol. 4:572013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zugazagoitia J, Guedes C, Ponce S, Ferrer
I, Molina-Pinelo S and Paz-Ares L: Current Challenges in Cancer
Treatment. Clin Ther. 38:1551–1566. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu P, Nielsen TE and Clausen MH:
FDA-approved small-molecule kinase inhibitors. Trends Pharmacol
Sci. 36:422–439. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Han F, Lin S, Liu P, Tao J, Yi C and Xu H:
Synthesis and structure-activity relationships of I3K//mTOR dual
inhibitors from a series of 2-amino-4-methylpyrido[2,3-d]pyrimidine
derivatives. Bioorg Med Chem Lett. 24:4538–4541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao HF, Wang J and Tony To SS: The
phosphatidylinositol 3-kinase/Akt and c-Jun N-terminal kinase
signaling in cancer: Alliance or contradiction? (Review). Int J
Oncol. 47:429–436. 2015.PubMed/NCBI
|
|
7
|
Hollande F, Pannequin J and Joubert D: The
long road to colorectal cancer therapy: Searching for the right
signals. Drug Resist Updat. 13:44–56. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Toren P and Zoubeidi A: Targeting the
I3K/Akt pathway in prostate cancer: Challenges and opportunities
(Review). Int J Oncol. 45:1793–1801. 2014.PubMed/NCBI
|
|
9
|
Sahu JK, Ganguly S and Kaushik A:
Triazoles: A valuable insight into recent developments and
biological activities. Chin J Nat Med. 11:456–465. 2013.PubMed/NCBI
|
|
10
|
Olah M, Rad R, Ostopovici L, Bora A,
Hadaruga N, Hadaruga D, Moldovan R, Fulias A, Mractc M and Oprea
TI: WOMBAT and WOMBAT-PK: bioactivity databases for lead and drug
discovery. Chemical biology: from small molecules to systems
biology and drug design. Schreiber SL, Kapoor TM and Wess G: 1–3.
Wiley-VCH Verlag GmbH; Weinheim: pp. 760–786. 2008, View Article : Google Scholar
|
|
11
|
Li X, Li XQ, Liu HM, Zhou XZ and Shao ZH:
Synthesis and evaluation of antitumor activities of novel chiral
1,2,4-triazole Schiff bases bearing γ-butenolide moiety. Org Med
Chem Lett. 2:262012. View Article : Google Scholar
|
|
12
|
Chowrasia D, Karthikeyan C, Choure L,
Sahabjada S, Gupta M, Arshad Md and Trivedi P: Synthesis,
characterization and anti cancer activity of some fluorinated
3,6-diaryl-[1,2,4] triazolo[3,4-b][1,3,4]thiadiazoles. Arab J Chem.
6–10. 2013.
|
|
13
|
Chand P, Chesney JA, Clem BF, Tapolsky GH,
Telang S and Trent JO: Small-Molecule Choline Kinase Inhibitors As
Anti-Cancer Therapeutics. Patent US 2011/0257211 A1
|
|
14
|
Hou YP, Sun J, Pang ZH, Lv PC, Li DD, Yan
L, Zhang HJ, Zheng EX, Zhao J and Zhu HL: Synthesis and antitumor
activity of 1,2,4-triazoles having 1,4-benzodioxan fragment as a
novel class of potent methionine aminopeptidase type II inhibitors.
Bioorg Med Chem. 19:5948–5954. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Georgiyants V, Perekhoda L, Saidov N and
Kadamov I: Docking studies and biological evaluation of anti-cancer
activity of new 1,2,4-triazole (4h) derivatives. Scr Sci Pharm.
1:46–53. 2014.
|
|
16
|
Holla BS, Sarojini BK, Rao BS, Akberali
PM, Kumari NS and Shetty V: Synthesis of some halogen-containing
1,2,4-triazolo-1,3,4-thiadiazines and their antibacterial and
anticancer screening studies - part I. Farmaco. 56:565–570. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gupta D and Jain DK: Synthesis, antifungal
and antibacterial activity of novel 1,2,4-triazole derivatives. J
Adv Pharm Technol Res. 6:141–146. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hawkins PCD, Skillman AG, Warren GL,
Ellingson BA and Stahl MT: Conformer generation with OMEGA:
Algorithm and validation using high quality structures from the
Protein Databank and Cambridge Structural Database. J Chem Inf
Model. 50:572–584. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McGann M: FRED and HYBRID docking
performance on standardized datasets. J Comput Aided Mol Des.
26:897–906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Berman HM, Westbrook J, Feng Z, Gilliland
G, Bhat TN, Weissig H, Shindyalov IN and Bourne PE: The protein
data bank. Nucleic Acids Res. 28:235–242. 2000. View Article : Google Scholar
|
|
21
|
Gurusaran M, Shankar M, Nagarajan R,
Helliwell JR and Sekar K: Do we see what we should see? Describing
non-covalent interactions in protein structures including
precision. IUCrJ. 1:74–81. 2013. View Article : Google Scholar
|
|
22
|
Bercean VN, Badea V, Sisu E, Bindila L and
Csunderlik C: A simplified method of obtaining
3-aryl-5-mercapto-1,2,4-triazoles. Rev Chim. 54:368–369. 2003.
|
|
23
|
Soica C, Oprean C, Borcan F, Danciu C,
Trandafirescu C, Coricovac D, Crăiniceanu Z, Dehelean CA and
Munteanu M: The synergistic biologic activity of oleanolic and
ursolic acids in complex with hydroxypropyl-γ-cyclodextrin.
Molecules. 19:4924–4940. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nadeem H, Mohsin M, Afzaal H, Riaz S,
Zahid A and Muhammad SA: Synthesis and in Vitro Biological
Activities of 4,5-Disubstituted 1,2,4-Triazole-3-Thiols. Adv
Microbiol. 3:366–375. 2013. View Article : Google Scholar
|
|
25
|
El-Sayed WA, Flefel EM and Morsy EMH:
Anticancer and antimicrobial activities of some synthesized
pyrazole and triazole derivatives. Pharmachem. 4:23–32. 2012.
|
|
26
|
Cheng T, Li Q, Zhou Z, Wang Y and Bryant
SH: Structure-based virtual screening for drug discovery: A
problem-centric review. AAPS J. 14:133–141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ferreira LG, dos Santos RN, Oliva G and
Andricopulo AD: Molecular Docking and Structure-Based Drug Design
Strategies. Molecules. 20:13384–13421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Oda A, Saijo K, Ishioka C, Narita K, Katoh
T, Watanabe Y, Fukuyoshi S and Takahashi O: Predicting the
structures of complexes between phosphoinositide 3-kinase (PI3K)
and romidepsin-related compounds for the drug design of
PI3K/histone deacetylase dual inhibitors using computational
docking and the ligand-based drug design approach. J Mol Graph
Model. 54:46–53. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Saini KS, Loi S, de Azambuja E,
Metzger-Filho O, Saini ML, Ignatiadis M, Dancey JE and
Piccart-Gebhart MJ: Targeting the PI3K/AKT/mTOR and Raf/MEK/ERK
pathways in the treatment of breast cancer. Cancer Treat Rev.
39:935–946. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nitulescu GM, Margina D, Juzenas P, Peng
Q, Olaru OT, Saloustros E, Fenga C, Spandidos DA, Libra M and
Tsatsakis AM: Akt inhibitors in cancer treatment: The long journey
from drug discovery to clinical use (Review). Int J Oncol.
48:869–885. 2016.
|
|
31
|
Häggblad Sahlberg S, Mortensen AC, Haglöf
J, Engskog MK, Arvidsson T, Pettersson C, Glimelius B, Stenerlöw B
and Nestor M: Different functions of AKT1 and AKT2 in molecular
pathways, cell migration and metabolism in colon cancer cells. Int
J Oncol. 50:5–14. 2017.
|
|
32
|
Vignot S, Faivre S, Aguirre D and Raymond
E: mTOR-targeted therapy of cancer with rapamycin derivatives. Ann
Oncol. 16:525–537. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fyffe C and Falasca M:
3-Phosphoinositide-dependent protein kinase-1 as an emerging target
in the management of breast cancer. Cancer Manag Res. 5:271–280.
2013.PubMed/NCBI
|
|
34
|
Cathomas G: PIK3CA in Colorectal Cancer.
Front Oncol. 4:352014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Moon G, Lee SE, Oh MM, Lee SC, Jeong SJ,
Hong SK, Yoon CY, Byun SS, Park HS and Cheon J: NVP-BEZ235, a dual
PI3K/mTOR inhibitor synergistically potentiates the antitumor
effects of cisplatin in bladder cancer cells. Int J Oncol.
45:1027–1035. 2014.
|
|
36
|
Gong HC, Wang S, Mayer G, Chen G, Leesman
G, Singh S and Beer DG: Signatures of drug sensitivity in nonsmall
cell lung cancer. Int J Proteomics. 2011:2154962011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yi YW, Hong W, Kang HJ, Kim HJ, Zhao W,
Wang A, Seong YS and Bae I: Inhibition of the PI3K/AKT pathway
potentiates cytotoxicity of EGFR kinase inhibitors in
triple-negative breast cancer cells. J Cell Mol Med. 17:648–656.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jia T, Zhang L, Duan Y, Zhang M, Wang G,
Zhang J and Zhao Z: The differential susceptibilities of MCF-7 and
MDA-MB-231 cells to the cytotoxic effects of curcumin are
associated with the PI3K/Akt-SKP2-Cip/Kips pathway. Cancer Cell
Int. 14:1262014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kwei KA, Baker JB and Pelham RJ:
Modulators of sensitivity and resistance to inhibition of PI3K
identified in a pharmacogenomic screen of the NCI-60 human tumor
cell line collection. PLoS One. 7:e465182012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu H, Scholz C, Zang C, Schefe JH, Habbel
P, Regierer AC, Schulz CO, Possinger K and Eucker J: Metformin and
the mTOR inhibitor everolimus (RAD001) sensitize breast cancer
cells to the cytotoxic effect of chemotherapeutic drugs in vitro.
Anticancer Res. 32:1627–1637. 2012.PubMed/NCBI
|
|
41
|
Liu KKC, Zhu J, Smith GL, Yin MJ, Bailey
S, Chen JH, Hu Q, Huang Q, Li C, Li QJ, et al: Highly Selective and
Potent Thiophenes as PI3K Inhibitors with Oral Antitumor Activity.
ACS Med Chem Lett. 2:809–813. 2011. View Article : Google Scholar : PubMed/NCBI
|