Introduction
Osteosarcoma is a malignant tumor originating from
mesenchymal tissues. It commonly arises in the distal femur and
proximal tibia; it is highly malignant and has poor prognosis. Its
clinical characteristics include early pulmonary metastasis, high
disability rate, and high recurrence rate. Osteosarcoma treatment
in 1970 was limited to amputation and radiotherapy, and most
patients died of lung metastasis within two years with a 5-year
survival rate of only 10–20% (1,2). In
the last 30 years, the wide application of combinatorial
chemotherapy and active surgical resection has greatly improved the
5-year survival rate for osteosarcoma. However, though chemotherapy
plays a pivotal role in osteosarcoma treatment, its overall
efficacy is still approximately 60%. Thus, osteosarcoma therapy is
now in a phase, where it is necessary to explore osteosarcoma
markers to determine its biological characteristics and prognosis,
and to determine novel clinical therapeutic targets for effective
molecular therapy.
Fibulin-4 (epidermal growth factor-containing
fibulin-like extracellular matrix protein 2, also known as EFEMP2,
MBP1, UPH1) is a member of the fibulin family of glycoproteins.
Fibulins, encoded by the FBLN genes, are extracellular matrix
proteins. Presently, the fibulin family contains seven members:
fibulin-1, -2, -3, -4, -5, -6, and -7 (3,4). It
is mainly involved in the formation and stability of membranes,
elastic fibers, and loose connective tissues. Fibulin-4 is widely
distributed in human tissues, and is closely associated with the
basement membrane and the extracellular matrix of elastic fibers.
Fibulin-4 plays an important role in the stabilization of
extracellular matrix structure (5,6).
Fibulin family members are involved in the processes
of cell morphology maintenance, growth, adhesion, and movement, and
are closely associated with the development of multiple tumors
(6). Fibulin-1 expression is
enhanced in ovarian cancer (7) and
breast cancer (8) and promotes
tumor development. However, in hepatocellular carcinoma (9), gastric cancer (10), and prostate cancer (11), fibulin-1 could suppress tumor
growth, induce apoptosis, and inhibit tumor angiogenesis (12). Fibulin-2 as a tumor suppressor
gene, could inhibit tumor cell growth and invasion in HCC and
breast cancer, and inhibited angiogenesis (13,14).
Fibulin-3 is reported to be highly expressed in pancreatic cancer
(15), cervical cancer (16,17),
and glioma (18), and promotes
tumor development. However, in nasopharyngeal carcinoma (19), breast cancer (20), and glioblastoma (21), fibulin-3 is downregulated, and can
inhibit tumor cell proliferation, invasion, and metastasis.
Fibulin-5 is widely considered a tumor suppressor gene that
inhibits tumor growth, invasion, and angiogenesis in the
development of most tumors (22–25).
At present, studies on the relationship between
tumors and fibulin-4 are still in the initial stage, and few
related studies are available. In our current study, we
investigated the function of fibulin-4 in human osteosarcoma
invasion and metastasis, and the the relationship between fibulin-4
and EMT.
Materials and methods
Cell culture
Osteosarcoma cell lines (HOS, MG63 and U-2OS) and
the normal osteoblastic cell line hFOB, were obtained from the
Shanghai Institute for Biological Sciences, Chinese Academy of
Sciences. All cell lines were cultured in complete growth media
containing DMEM/F12 (Gibco BRL, Rockville, MD, USA) supplemented
with 10% FBS (Gibco BRL) and 1% antibiotics, and were maintained at
37°C in an incubator with 5% CO2.
Isolation of MG63 cell subclones
MG63 cells at the logarithmic growth phase were
collected and diluted to approximately 10 cells/ml, and were then
seeded into a 96-well plate with 0.1 ml/well. Thus, as far as
possible, there was only one cell in each well. After 1 week at
37°C with 5% CO2, a single clone from one well was
selected and cultured as a subclone. Then the cell electrophoretic
mobility (EPM) of each clone was measured to study charge property
using microcapillary electrophoresis (microCE) chips according to
the method of Omasu et al (26). Moreover, the invasive and
proliferative abilities of the highly invasive and low invasive
subclones were analyzed by in vitro and in vivo
functional assays (27,28). All data are expressed as mean ±
standard error (SE).
Osteosarcoma tissue samples
With informed consent from patients, 290 specimens
were obtained from the Department of Pathology, Shandong Qilu
Hospital. None of these patients had undergone preoperative
radiation or chemotherapy. All patients received regular follow-up.
During the study period, contact with 15 patients was lost and 36
patients died. The follow-up period was from 2005 to 2014. This
study was approved by the Institutional Medical Ethics Committee of
Shandong University.
Immunohistochemistry (IHC) and
immunocytochemistry (ICC)
For IHC, paraffin-embedded sections were de-waxed in
xylene and rehydrated in ethanol. Heat-induced epitope retrieval
was then performed using a pressure cooker in 0.01 M citrate buffer
at pH 6.0. The sections were incubated for 3 min after the cooker
reached full pressure. For ICC, cells at 75–80% confluency were
seeded into a cell culture dish containing coverslips. After 24 h,
the coverslips were harvested, washed thrice with PBS, and fixed in
95% ethanol for 30 min. According to the procedure of the
streptavidin-peroxidase detection kit (ZSGB-BIO, Beijing, China),
the following steps were common for both IHC and ICC. All sections
and coverslips were treated with 3% hydrogen peroxide
(H2O2) and goat serum for 30 min
sequentially, to block endogenous peroxidase and the non-specific
binding sites, and then incubated with rabbit anti-human fibulin-4,
alkaline phosphatase (ALP), and cysteine-rich angiogenic inducer 61
(CYR61) antibodies (ab125073, ab108337, ab24448; Abcam) at working
dilutions of 1:200 overnight at 4°C. The sections and coverslips
were then incubated with the anti-rabbit biotin-conjugated
secondary antibody for 30 min at room temperature, stained for 1–5
min with the enzyme substrate 3′, 3-diaminobenzidine
tetrahydrochloride (DAB, Sigma-Aldrich, St. Louis, MO, USA), and
counterstained for 5 min with hematoxylin. Paraffin-embedded
sections of human ovarian cancer specimens (fibulin-4-positive)
were used as positive controls (29), and the negative control was
obtained by replacing the primary antibody with PBS. Brown granules
in the cytoplasm or stroma were considered as positive fibulin-4
expression.
Immunohistochemistry (IHC) and
immunocytochemistry (ICC) analysis
To assess fibulin-4 expression in IHC and ICC
experiments, the stained cell percentage and staining intensity
were measured. The percentage of positively stained cells was
scored from 0 to 4 (score 0, 0% cells stained; score 1, 1–25%;
score 2, 26–50%; score 3, 51–75%; or score 4, 76–100%), whereas the
staining intensity of fibulin-4 was scored as 0 (negative), 1
(weak), 2 (moderate), or 3 (strong) (30). Taken together, the intensity and
percentage scores made up of the final staining score (0–7), and
the scores of 0, 1–3, 4–5, and 6–7 were converted into sum indices
−, +, ++, and +++, respectively. For statistical analysis, low
fibulin-4 expression was defined as − or +, whereas high fibulin-4
expression was indicated by ++ or +++. Each tissue section was
independently analyzed by three pathologists. Immunohistochemical
expression was evaluated using Image-Pro Plus 6.0 (Media
Cybernetics, Inc., Rockville, MD, USA) to detect photodensity. In
brief, 5 positive fields within a section were selected at random
and read using Image-Pro Plus 6.0. The mean densities were
subsequently calculated. Using Pearson's product-moment correlation
coefficient, the associations between fibulin-4 vs. ALP and CYR61
were analyzed.
Lentivirus transfection
The pLVX-fibulin-4 vector and fibulin-4 shRNA as
well as a negative control were obtained from GeneChem Inc.
(Shanghai, China). According to the manufacturer's instructions,
prior to viral infection, target cells were plated at
0.5×105 cells per well in a 24-well plate and incubated
at 37°C in a CO2 incubator for 24 or 48 h until the
cells were 60–80% confluent. The cells were then infected by adding
the viral stock at a multiplicity of infection (MOI) of 100. In
addition, a transduction well with negative control viral
constructs was included. The cells were incubated overnight at 37°C
with 5% CO2; the transfection mixture was then replaced
with normal complete growth medium to avoid cell toxicity. At the
end of 48 h of incubation, the cells were assessed by fluorescence
microscopy. The transfection efficiency was confirmed by western
blotting, real-time quantitative RT-PCR, and immunocytochemistry
(ICC). The siRNA sequence for fibulin-4 was: sense
5′-CAGAUCCGUGCUGGAAACUCG-3′ and antisense
3′-AGUUUCCAGCACGGAUCUGAA-5′. The negative control (scrambled order)
was: sense 5′-AUCGACGAUCCCUAUUGGCGU-3′ and antisense
3′-GCCAAUAGGGAUCGUCGAUCU-5′.
Quantitative real-time-polymerase chain
reaction (qRT-PCR)
TRIzol® Reagent (Ambion™) was used to
isolate total RNA from cell and tissue samples. Reverse
Transcription was carried out with TaqMan® Reverse
Transcription Reagents (Applied Biosystems Inc.; Thermo Fisher
Scientific, Inc.). The procedure was based on the protocol provided
by Invitrogen. The real-time PCR mixture volume was 25 μl
including 12.5 μl SYBR green Mix (Power SYBR®
green PCR Master Mix, Applied Biosystems Inc.), 0.2 μl cDNA,
1 μl primer pair mix (5 pmol/μl each primer), and
11.3 μl DNAse/RNAse-free H2O. The experiment was
then set up with the following PCR program on ABI Prism SDS 7000
(Applied Biosystems Inc.; Thermo Fisher Scientific, Inc.): 50°C for
2 min, 1 cycle; 95°C for 10 min, 1 cycle; 40 cycles of 95°C for 15
sec → 60°C for 30 sec → 72°C for 30 sec; 72°C 10 min, 1 cycle.
Finally, the results were analyzed with SDS 7000 software. Specific
primers were designed by LightCycler® Probe Design
software (Roche Diagnostics, Basel, Switzerland) and were
synthesized by Takara Biotechnology Co., Ltd. The primer sequences
were as follows: fibulin-4: 5′-GCTGCTACTGTTGCTCTTGGG-3′,
5′-GGGATGGTCAGACACTCGTTG-3′; E-cadherin:
5′-GGATTGCAAATTCCTGCCATTC-3′, 5′-AACGTTGTCCCGGGTGTCA-3′;
N-cadherin: 5′-GTAGCTAATCTAACTGTGACCGATAAGG-3′,
5′-TTGGTTTGACCACGGTGACTAA-3′; vimentin:
5′-GCAGGAGGCAGAAGAATGGTA-3′, 5′-GGGACTCATTGGTTCCTTTAAGG-3′; Snail:
5′-TCGGAAGCCTAACTACAGCGA-3′, 5′-AGATGAGCATTGGCAGCGAG-3′; Slug:
5′-TGTGACAAGGAATATGTGAGCC-3′, 5′-TGAGCCCTCAGATTTGACCTG-3′; Twist:
5′-AGCAAGATTCAGACCCTCAAGCT-3′, 5′-CCTGGTAGAGGAAGTCGATGTACCT-3′;
β-actin: 5′-CCACGAAACTACCTTCAACTCCA-3′,
5′-GTGATCTCCTTCTGCATCCTGTC-3′.
Western blotting
Cells were lysed on ice in RIPA
(radio-immunoprecipitation assay) buffer with 1 mM PMSF
(phenylmethylsulfonyl fluoride). From the cell lysate, 40 μg
of total protein was loaded into the wells of the SDS-PAGE (sodium
dodecyl sulfate polyacrylamide gel elec trophoresis) gel, along
with a molecular weight marker. Electrophoresis was carried out for
1–2 h at 100 V, followed by transfer to PVDF (polyvinyl difluoride)
membranes, which were then blocked with 5% BSA (bovine serum
albumin). The membranes were then incubated overnight at 4°C with
primary antibodies (E-cadherin sc-8426, N-cadherin sc-7939,
vimentin sc-6260, Santa Cruz; fibulin-4 ab125073, Snail ab167609,
Slug ab27568, Twist ab50887, PI3K ab86714, p-PI3K ab182651, AKT
ab8805, P-AKT 38449, mTOR ab32028, p-mTOR ab109268; Abcam) at
working dilutions of 1:1000. After washing the membranes thrice
with TBST for 5 min each, the membranes were incubated with
conjugated secondary antibody diluted to 1:1000, at room
temperature for 1 h. Blots were developed using the enhanced
chemiluminescence method (Pierce™ ECL Western Blotting Substrate;
Thermo Fisher Scientific, Inc.).
Growth curves
Cells at the logarithmic phase were collected,
seeded into the wells of a 24-well plate (1×104
cells/well) and cultured at 37°C with 5% CO2. Three
wells were harvested every day and the cells were counted and
averaged. Growth curves were then plotted according to the average
cell counts of 7 consecutive days.
Soft agar colony formation assay
DMEM with 20% FBS (1.5 ml) mixed with 1.5 ml of 1.2%
agar was added to 3.5 cm dishes and solidified for the bottom
layer. Next, 1.5 ml of 0.7% agar was mixed with 1.5 ml DMEM (20%
FBS) and 200 μl of cell suspension (containing 600 cells),
and was immediately added to the above culture dishes. All the
dishes were incubated for 2 weeks at 37°C with 5% CO2.
The assay was repeated in triplicate. Under an inverted microscope
(Nikon Eclipse), the dish was divided into quadrants and in each
quadrant, colonies with diameters of more than 2 mm were counted
and the average was calculated. All the data are expressed as mean
± SE.
Cell invasion assay and migration
assay
The in vitro Matrigel invasion assay was
performed as previously described (31). The polyvinylpyrrolidone-free
polycarbonate (PVPF) membrane of Boyden chambers (BD Biosciences,
Bedford, MA, USA) were coated with 50 μl of Matrigel 1:3
diluted with serum-free media. Cell suspensions in volumes of 200
μl (2×105 cells) were seeded into the upper
chambers, and 600 μl of serum-free culture supernatant of
NIH3T3 cells was added to the lower chamber as a chemotactic
factor. The Boyden chambers were then incubated at 37°C for 24 h.
The non-invading cells on the upper surface of the membrane were
removed, and the cells on the lower surface were fixed with 4%
paraformaldehyde, stained with hematoxylin and eosin (H&E), and
counted in five random high-power fields (HPF) under an inverted
microscope. The cell migration assay was simultaneously performed
with the above steps, without the Matrigel coating on the membrane
and an incubation time of only 12 h. The cell invasion and
migration assays were both repeated in triplicate. All data are
expressed as mean ± SE.
Tumor xenografts in nude mice
BALB/C-nu/nu nude mice were purchased from the
National Resource Center for Rodent Laboratory Animal of China.
Each group included 5 nude mice, each of which was inoculated
subcutaneously with 5.0×106 cells. The mice were
maintained in a sterile animal facility and monitored daily for
tumor growth. Every week, the tumor volumes were measured using
vernier calipers, and calculated according to the formula, V =
length × width2 × 0.25. After 2 months, the mice were
sacrificed and the tumors were dissected and examined
histologically. All data are expressed as mean ± SE. The animal
experiment was approved by the Institutional Animal Care and Use
Committee of Shandong University and was in compliance with all
regulatory guidelines.
Statistical analysis
IHC data were analyzed using a χ2 test. A
two-tailed t-test was used to compare the means between two sets,
and a one-way analysis of variance was used to compare the means
among three groups. By the Kaplan-Meier method and the log-rank
test, survival curve analysis was performed to study the
relationship between fibulin-4 and the prognosis of patients with
osteosarcoma. The data were analyzed with SPSS software version
13.0 (SPSS Inc., Chicago, IL, USA). P<0.05 (two-sided) was
considered statistically significant.
Results
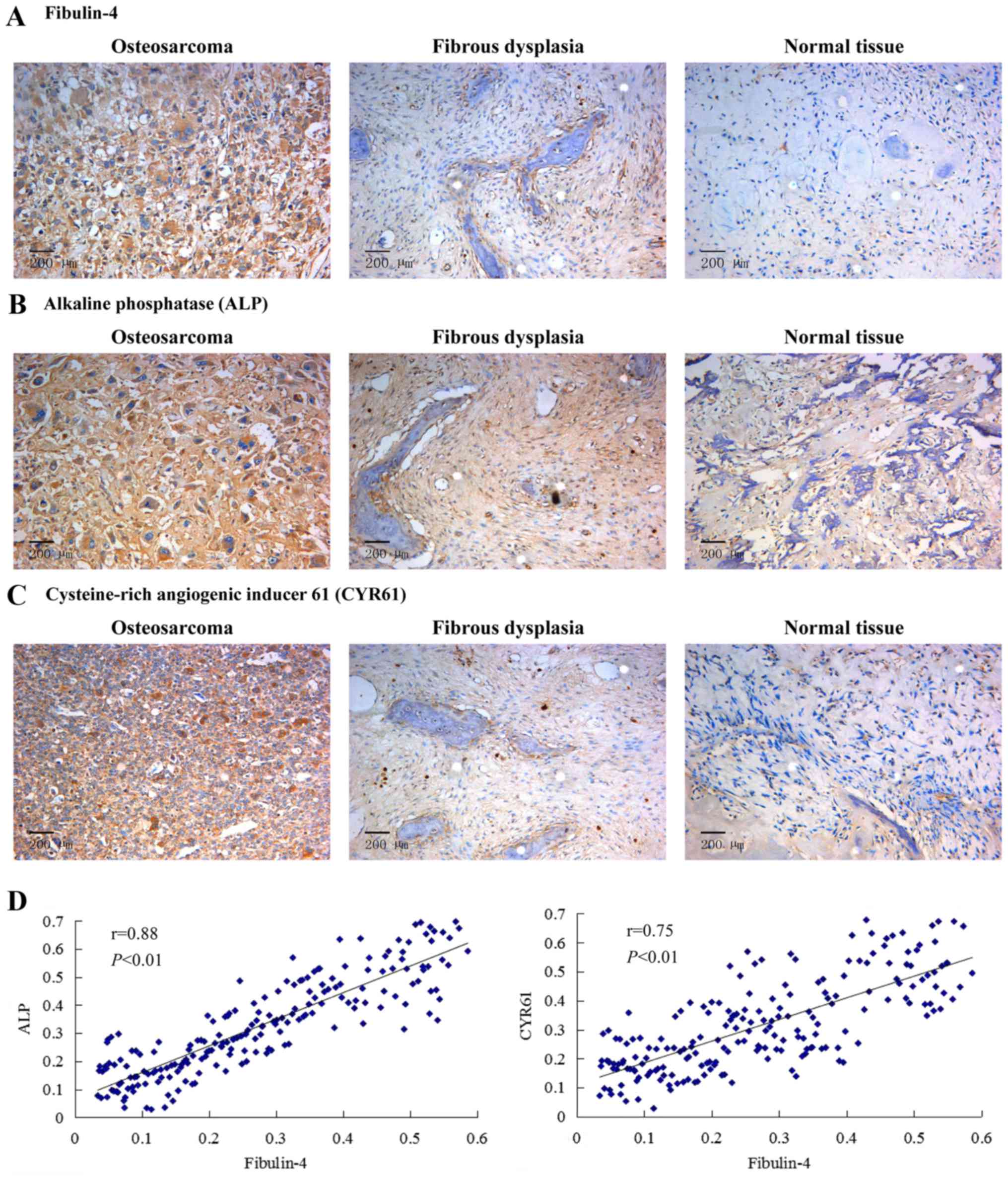
The expression of fibulin-4, ALP and
CYR61 in human osteosarcoma tissues
High fibulin-4 protein expression was detected in
osteosarcoma tissues, mainly in the stroma and in the osteosarcoma
cell cytoplasm. However, fibulin-4 immunoreactivity was very low in
most normal tissues (Fig. 1A).
Similar results were obtained in the detection of ALP and CYR61.
Compared to normal tissues, the expression levels of ALP and CYR61
in osteosarcoma tissues were significantly high (Fig. 1B and C). According to the Pearson's
product-moment correlation coefficient, the expression of CYR61 and
ALP vs. fibulin-4 exhibited strong positive correlations (Fig. 1D). The expression of fibulin-4 in
the extracellular matrix was found to be much less than that in the
cancer cell cytoplasm, which probably was due to that fibulin-4 has
an important role in development and integrity of extracellular
matrices (3), loss of fibulin-4 in
the extracellular matrix would reduce the stability of
extracellular matrix and promote cancer cell invasion and
metastasis. Moreover, high fibulin-4 expression was positively
associated with low differentiation and lymph node metastasis
(Table I). Similar results were
also observed in the qRT-PCR experiment. High fibulin-4 mRNA
expression was observed in osteosarcoma tissues and was correlated
with low tumor differentiation and positive nodal metastasis
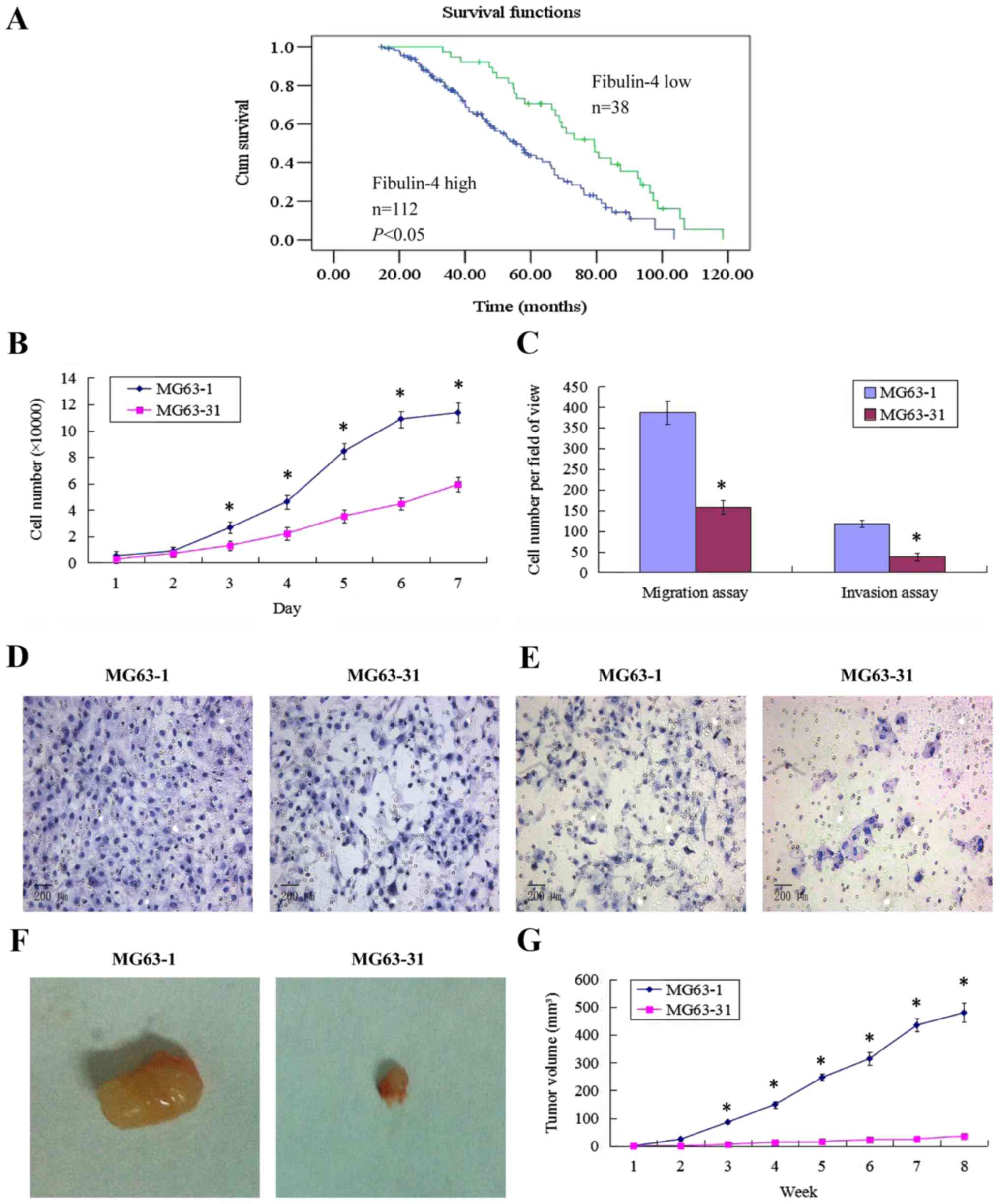
(Table II). Survival analysis was
performed by Kaplan-Meier analysis. This result showed that
patients with high fibulin-4 expression had poorer prognosis
compared to those with low fibulin-4 expression (log rank,
P<0.01; Fig. 2A).
 | Table IProtein expression of fibulin-4 in
human osteosarcoma tissues. |
Table I
Protein expression of fibulin-4 in
human osteosarcoma tissues.
| N | Fibulin-4 low (−/+)
| Fibulin-4 high
(++/+++)
| χ2 | P-value |
|---|
| n | % | n | % |
|---|
| Normal tissue | 60 | 55 | 91.7 | 5 | 8.3 | 101.5 | <0.01 |
| Fibrous
dysplasia | 80 | 62 | 77.5 | 18 | 22.5 | | |
| Osteosarcoma | 150 | 38 | 25.3 | 112 | 74.7 | | |
| Pathological
type | | | | | | 0.136 | >0.05 |
| Fibroblastic
osteosarcoma | 54 | 13 | 24.1 | 41 | 75.9 | | |
| Osteoblastic
osteosarcoma | 52 | 13 | 25 | 39 | 75 | | |
| Chondroblastic
osteosarcoma | 44 | 12 | 27.3 | 32 | 72.7 | | |
| Cell
differentiation | | | | | | 24.75 | <0.01 |
| High and
intermediate | 78 | 33 | 42.3 | 45 | 57.7 | | |
| Low | 72 | 5 | 6.9 | 67 | 93.1 | | |
| Nodal status | | | | | | 36.63 | <0.01 |
| Positive | 83 | 5 | 6 | 78 | 94 | | |
| Negative | 67 | 33 | 49.3 | 34 | 50.7 | | |
 | Table IImRNA expression of fibulin-4 in human
osteosarcoma tissues. |
Table II
mRNA expression of fibulin-4 in human
osteosarcoma tissues.
| N | Fibulin-4 mRNA
Normalized to β-actin reference | P-value |
|---|
| Normal tissue | 60 | 0.0114±0.0013 | |
| Fibrous
dysplasia | 80 | 0.0345±0.0032 | |
| Osteosarcoma | 150 | 0.0996±0.0094 | <0.05 |
| Pathology type | | | >0.05 |
| Fibroblastic
osteosarcoma | 54 | 0.0847±0.0075 | |
| Osteoblastic
osteosarcoma | 52 | 0.0913±0.0063 | |
| Chondroblastic
osteosarcoma | 44 | 0.0891±0.0082 | |
| Cell
differentiation | | | <0.05 |
| High and
medium | 78 | 0.0487±0.0037 | |
| Low | 72 | 0.0986±0.0091 | |
| Nodal status | | | <0.05 |
| Positive | 83 | 0.0958±0.0087 | |
| Negative | 67 | 0.0397±0.0053 | |
Establishment of highly invasive and low
invasive subclones
Using the single cell cloning technique, 31
subclones were obtained from MG63 cells. The subclone MG63-1, which
had the highest migration rate (19.59±0.56 μm/sec) showed
higher proliferative and invasive abilities, compared to the
subclone MG63-31, which showed the lowest migration rate (7.68±0.13
μm/sec). In vivo, the subcutaneous tumor formation
rate for the highly invasive subclone group was 100%, and was
accompanied by rapid tumor growth. However, the tumor formation
rate of the low invasive subclone group was only approximately 50%,
with very slow tumor growth. The tumor volume for MG63-1 was
479.82±34.31 mm3, much larger than that formed by
MG63-31 (35.91±3.73 mm3, P<0.01). These results are
shown in Fig. 2.
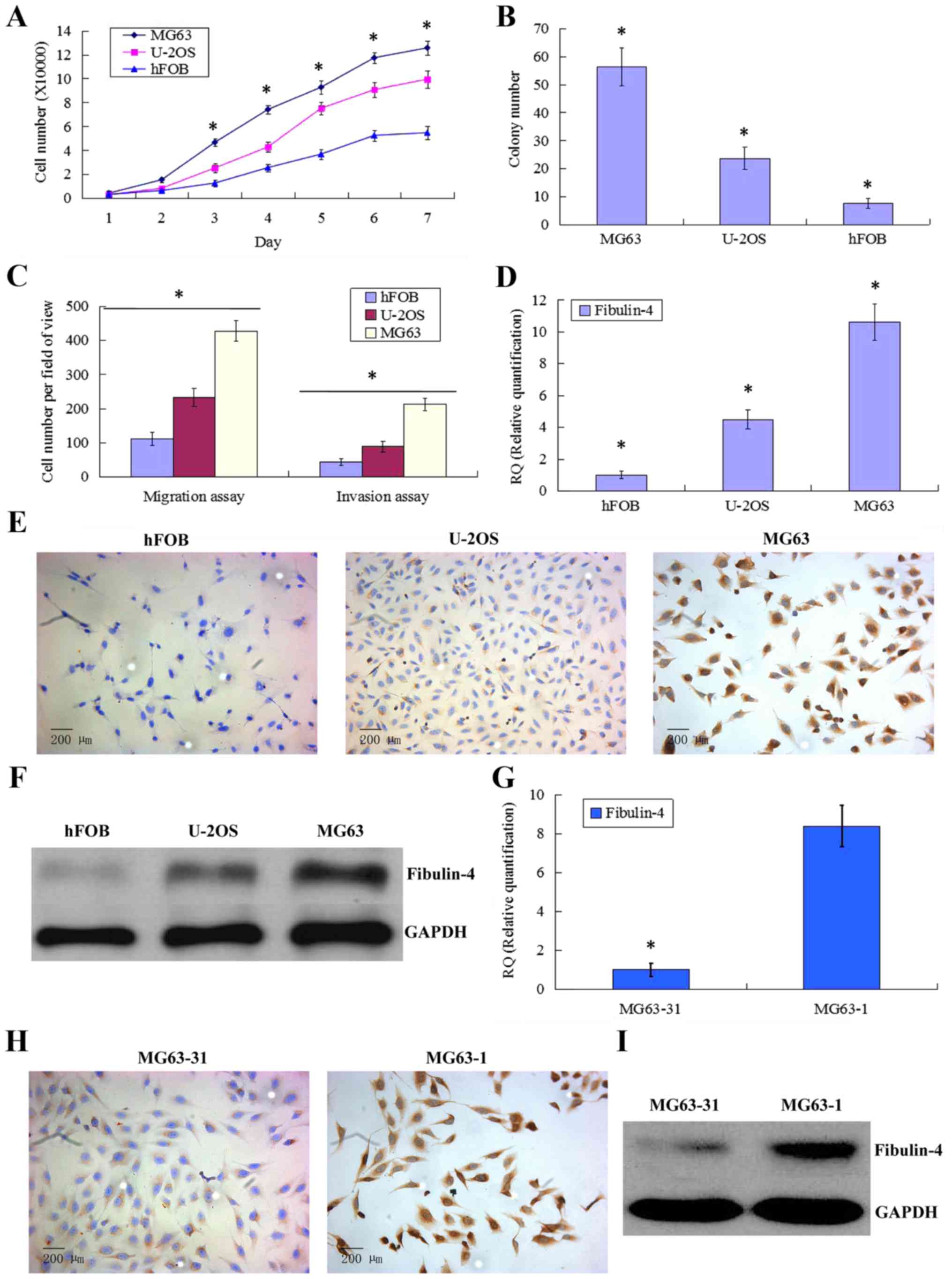
Different proliferation and invasion
abilities of human osteosarcoma cell lines and the normal
osteoblastic cell line
Compared to the normal osteoblastic cell line hFOB,
the human osteosarcoma cell lines U-2OS and MG63 showed stronger
proliferative abilities (Fig. 3A).
In the soft agar colony formation assay, the number of colonies
formed by MG63 and U-2OS was also significantly greater than that
formed by hFOB (Fig. 3B). In the
cell migration and Matrigel invasion assays, the average counts of
migrating and invading MG63 and U-2OS cells were both much higher
than those of hFOB (Fig. 3C). Upon
comparing the two osteosarcoma cell lines, we found that MG63 had
stronger proliferation and invasion abilities than those of
U-2OS.
Fibulin-4 expression in human
osteosarcoma cell lines and in differently invasive subclones
As shown in Fig.
3D–F, fibulin-4 was very weakly expressed in the normal
osteoblastic cell line hFOB compared to in the human osteosarcoma
cell lines MG63 and U-2OS. The strongest fibulin-4 expression was
detected in MG63, which showed the highest proliferation and
invasion abilities. Similar results were also observed upon
comparing subclones with differing invasive abilities (Fig. 3G–I). Compared with the low invasive
subclone MG63-31, high fibulin-4 expression was detected in the
highly invasive subclone MG63-1. These results indicate that high
fibulin-4 expression might be positively associated with the
proliferative and invasive abilities of osteosarcoma cells.
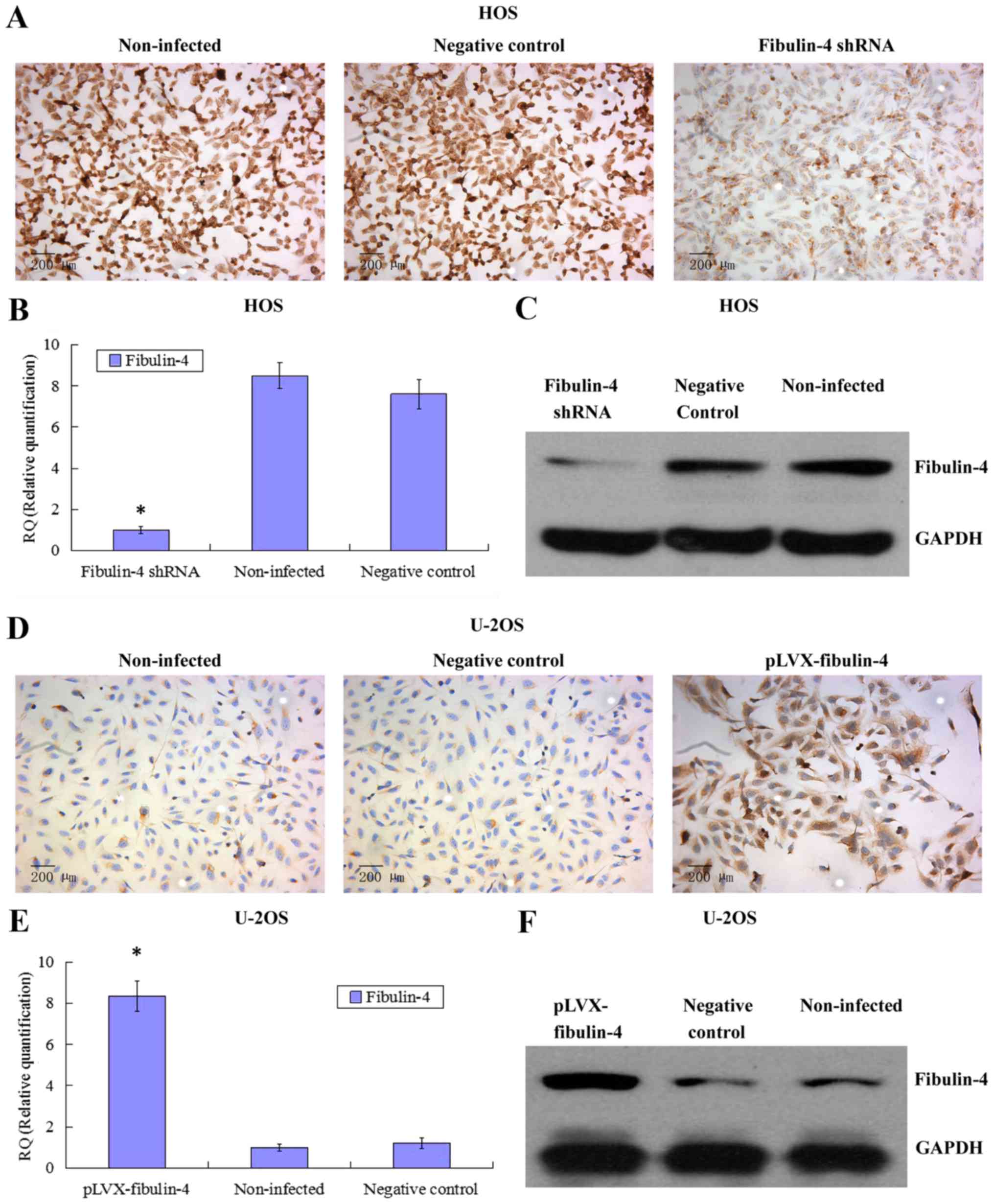
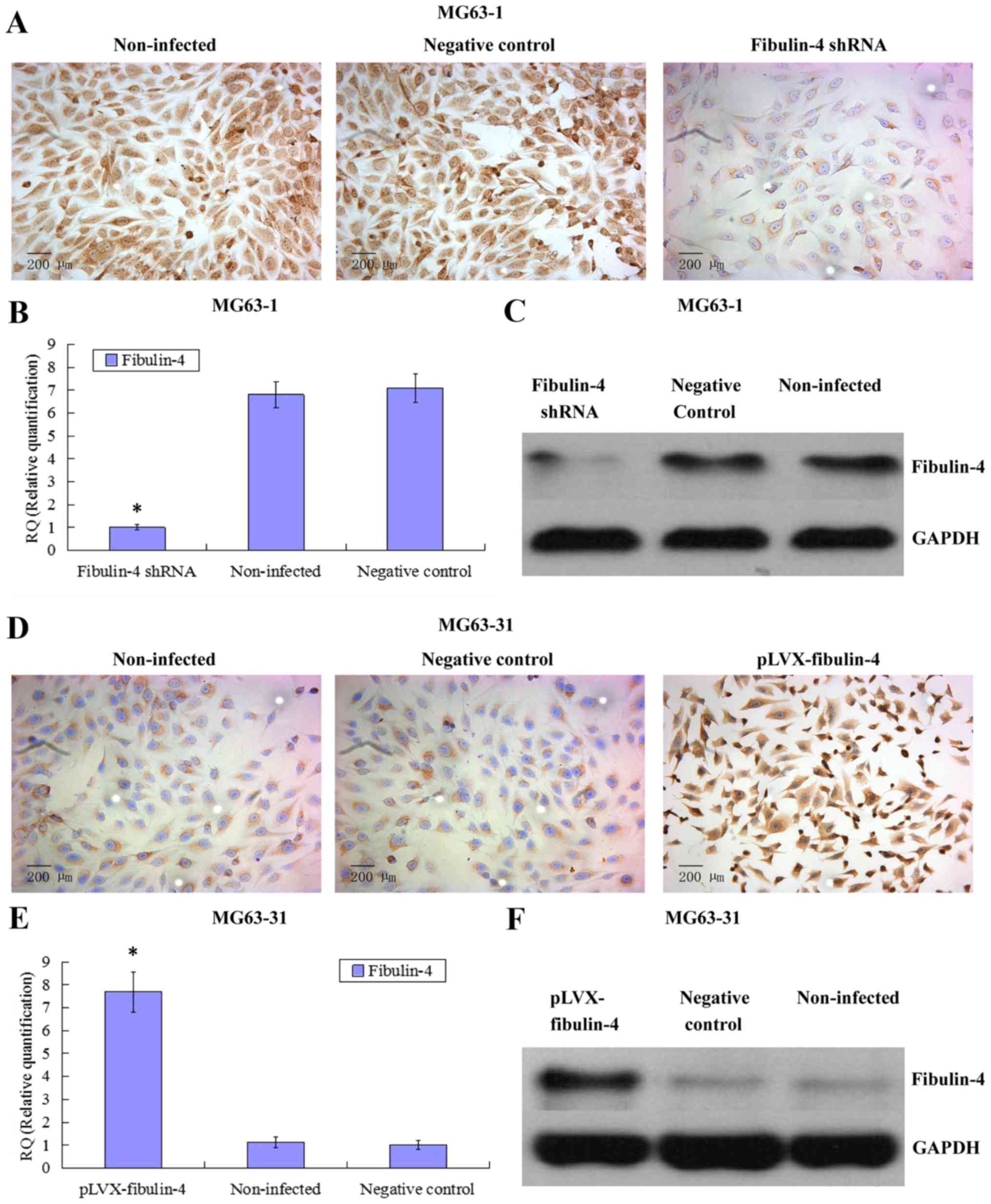
Identification of downregulated and
upregulated fibulin-4 expression in lentivirus transfection
systems
To further investigate the potential role of
fibulin-4 in osteosarcoma cell proliferation and invasion, we
decreased the expression of fibulin-4 in the highly invasive
osteosarcoma cell line HOS and subclone MG63-1, and increased
fibulin-4 expression in the low invasive osteosarcoma cell line
U-2OS and subclone MG63-31, by lentivirus transfection. After viral
infection, real-time q-RT-PCR, western blotting, and ICC were used
to confirm the altered expression of fibulin-4 at both mRNA and
protein levels, indicating the high efficiency of the lentivirus
transfections (Figs. 4 and
5).
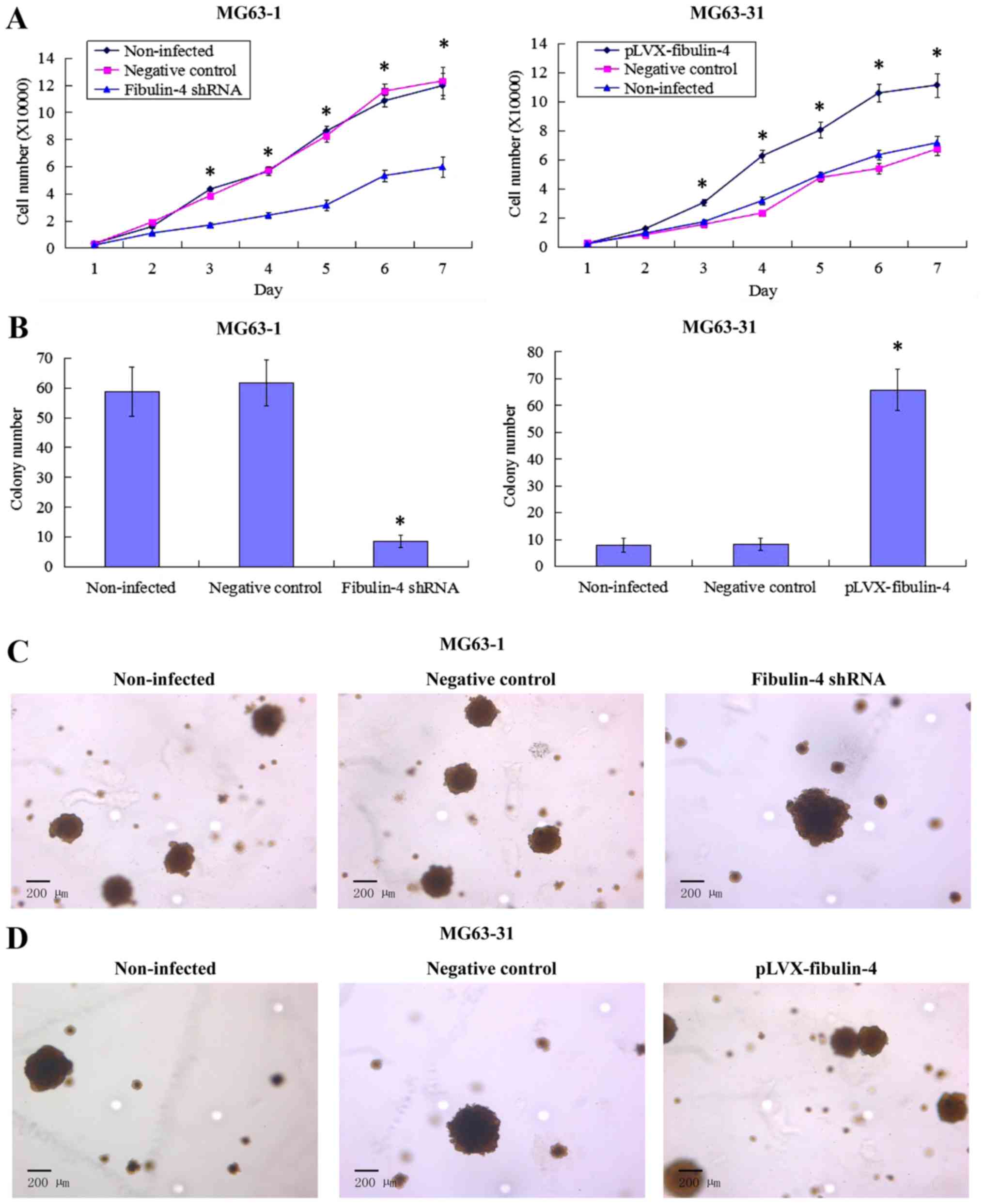
Effect of fibulin-4 knockdown and
overexpression on osteosarcoma cell proliferation
Downregulated fibulin-4 markedly inhibited cell
proliferation of the highly invasive subclone MG63-1, whereas
upregulated fibulin-4 significantly promoted cell proliferation of
the low invasive subclone MG63-31 (Fig. 6A). In the soft agar colony
formation assay, the colony forming efficiency of
fibulin-4-silenced cells was decreased, and conversely,
upregulation of fibulin-4 increased the colony forming efficiency
of the low invasive subclone (Fig. 6B
and C). No significant differences were observed in the
non-infected and negative control groups.
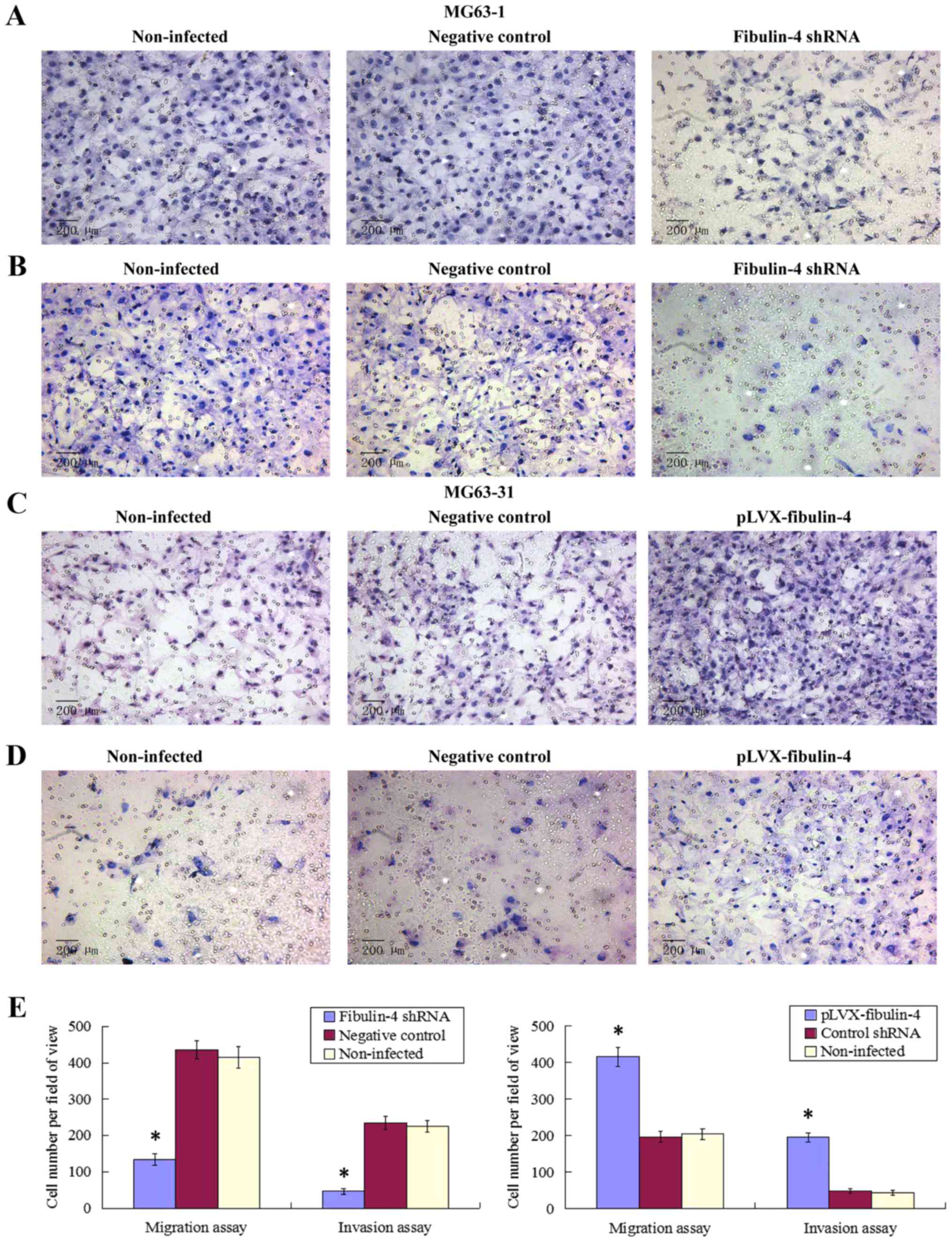
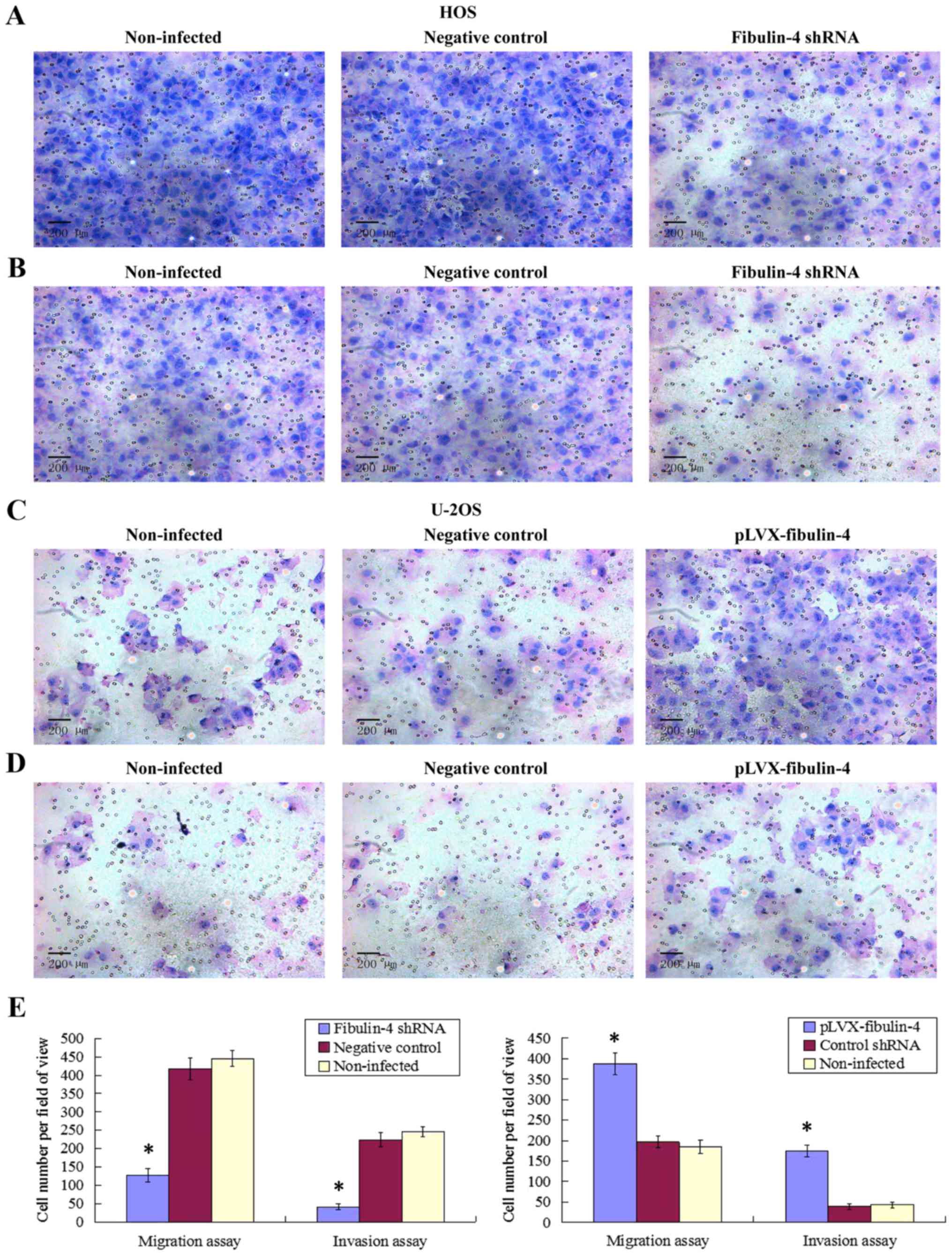
Effect of fibulin-4 knockdown and
overexpression on osteosarcoma cell migration and invasion
As shown in Figs. 7
and 8, fibulin-4 knockdown
inhibited osteosarcoma cell invasion and migration. The average
counts of migrating and invading fibulin-4 shRNA infected cells
were much lower than those of the negative controls and
non-infected groups (P<0.05), whereas, fibulin-4 overexpression
promoted the invasion and migration of osteosarcoma cells. The
average counts of migrating and invading pLVX-fibulin-4 infected
cells were much higher than those of the negative controls and the
non-infected groups (P<0.05). There were no significant
differences between the negative controls and non-infected
groups.
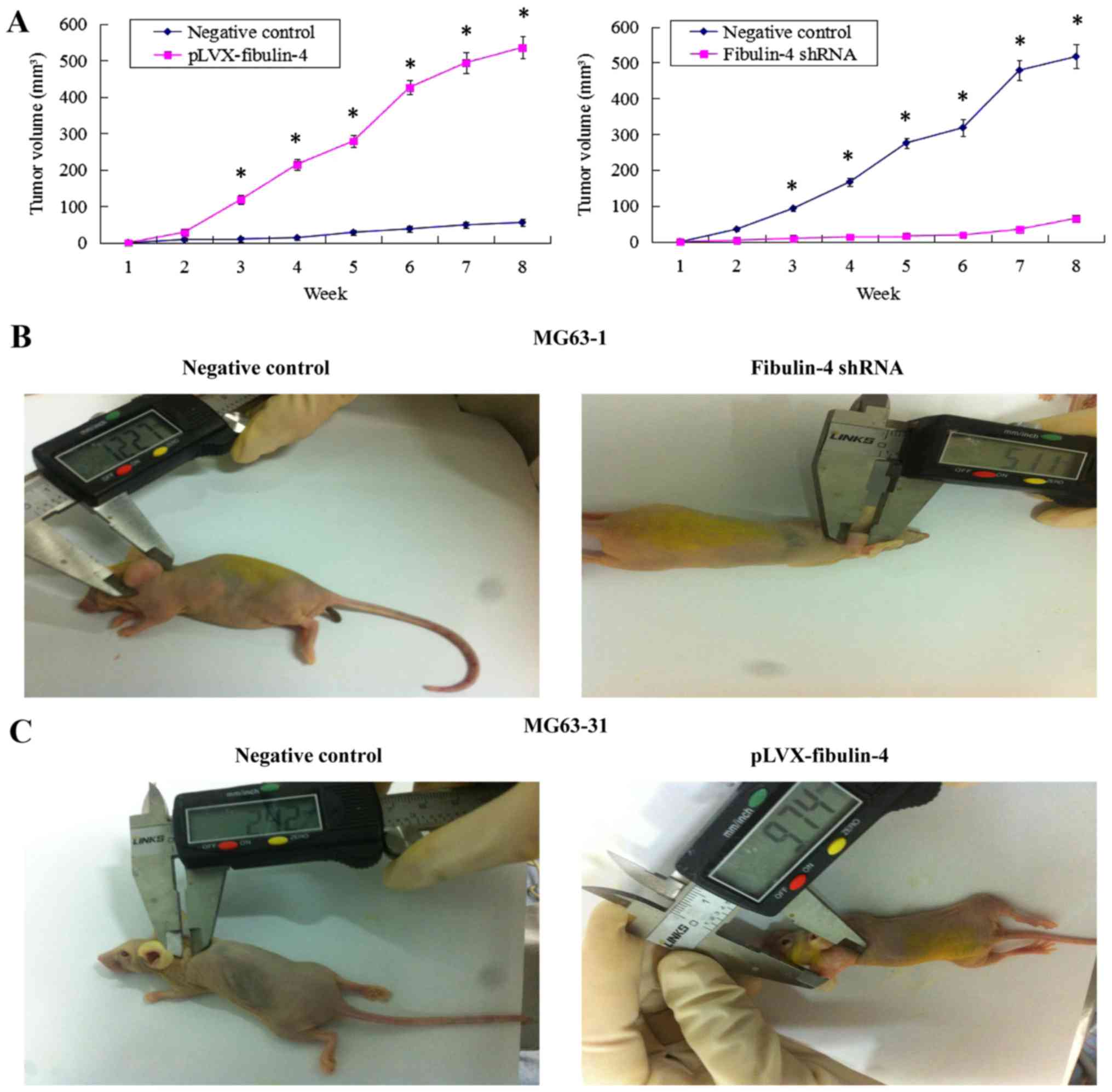
Effects of fibulin-4 knockdown and
overexpression on tumor growth in a xenograft model
The fibulin-4 shRNA infected cells, pLVX-fibulin-4
infected cells, negative control MG63-1, and negative control
MG63-31 were each inoculated subcutaneously in 5 nude mice,
respectively. The tumor formation rate of the negative control
MG63-1 was 100%, whereas the tumor formation rate in the fibulin-4
shRNA infected group was only 60%. Moreover, the average volumes of
the tumors formed in the fibulin-4 shRNA infected group were much
lower than those formed by the negative control MG63-1. Fibulin-4
knockdown inhibited tumor formation in nude mice. Simultaneously,
fibulin-4 overexpression promoted tumor growth in nude mice. The
tumor formation rate of pLVX-fibulin-4 infected cells was 100%,
whereas the tumor formation rate of the negative control MG63-31
was only 40%. Moreover, the average volumes of the tumors formed by
pLVX-fibulin-4 infected cells were much higher than those formed by
the negative control MG63-31 (Fig.
9).
Effects of fibulin-4 on key
epithelial-mesenchymal transition genes, CYR61 and ALP
CYR61 and ALP were significantly associated with
osteosarcoma development and progression. Therefore, we wondered if
fibulin-4 knockdown and upregulation affected these genes. As shown
in Fig. 10, using real-time
q-RT-PCR and western blotting, fibulin-4 knockdown significantly
inhibited the process of EMT, accompanied with increased E-cadherin
expression and decreased expression of N-cadherin, vimentin, Snail,
Slug, and Twist; in contrast, fibulin-4 upregulation could induce
EMT, with decreased E-cadherin expression, and increased expression
of N-cadherin, vimentin, Snail, Slug, and Twist. The expression
levels of CYR61 and ALP were also detected by western blotting in
fibulin-4 shRNA-infected and pLVX-fibulin-4-infected cells. CYR61
and ALP were downregulated following fibulin-4 shRNA infection, in
association with the knockdown of fibulin-4 expression, at the same
time, with fibulin-4 upregulation, CYR61 and ALP were increased
following pLVX-fibulin-4 infection. In conclusion, fibulin-4 could
promote the progression of human osteosarcomas.
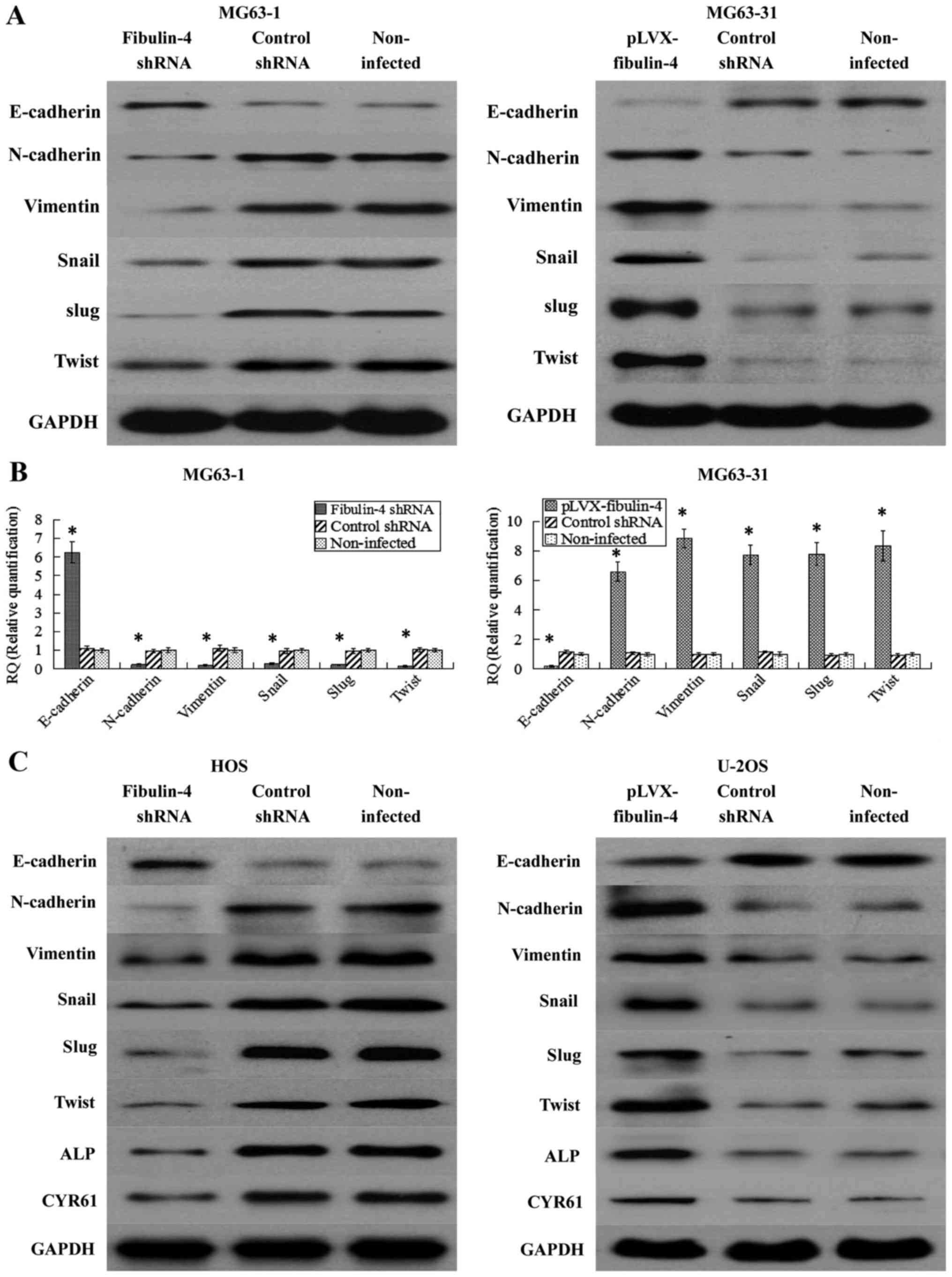 | Figure 10Effects of fibulin-4 knockdown and
overexpression on EMT genes, CYR61 and ALP, which were correlated
to tumor progression. After lentivirus transfections, EMT markers,
including E-cadherin, N-cadherin, vimentin, Snail, Slug and Twist
were measured by (A) western blotting and (B) real-time q-RT-PCR in
the lentivirus transfection systems of the differently invasive
osteosarcoma cell subclones. (C) EMT markers (E-cadherin,
N-cadherin, vimentin, Snail, Slug and Twist), CYR61 and ALP were
detected by western blotting in fibulin-4 shRNA-infected HOS cells
and pLVX-fibulin-4-infected U-2OS cells. Fibulin-4 knockdown
significantly increased the expression of E-cadherin, and decreased
the expression of N-cadherin, vimentin, Snail, Slug and Twist;
whereas fibulin-4 upregulation decreased the expression of
E-cadherin, and increased the expression of N-cadherin, vimentin,
Snail, Slug and Twist. CYR61 and ALP were downregulated following
fibulin-4 shRNA infection, in association with the knockdown of
fibulin-4 expression, moreover, with fibulin-4 upregulation, CYR61
and ALP were increased following pLVX-fibulin-4 infection.
*P<0.05. |
Effects of fibulin-4 on the PI3K/AKT
signaling pathway
PI3K-AKT-mTOR is one of the major signaling pathways
identified as important in cancer. We determined the expression of
the main signaling molecules PI3K, AKT, and mTOR, and the changes
in their phosphorylation levels. The results revealed that
fibulin-4 knockdown was able to reduce the PI3K, AKT, and mTOR
phosphorylation levels; in contrast, fibulin-4 up regulation could
increase their phosphorylation levels. Using the PI3K/AKT signaling
pathway inhibitor LY294002 (5, 10, and 20 μmol/l), we
treated the pLVX-fibulin-4-infected cells for 48 h. We found that
the inhibitor could significantly inhibit the PI3K/AKT pathway, and
EMT, both of which were activated by fibulin-4 up regulation.
Concentration-dependent increases were observed with increasing
doses (Fig. 11). In summary,
fibulin-4 could induce EMT to promote osteosarcoma invasion and
metastasis by the PI3K/AKT signaling pathway.
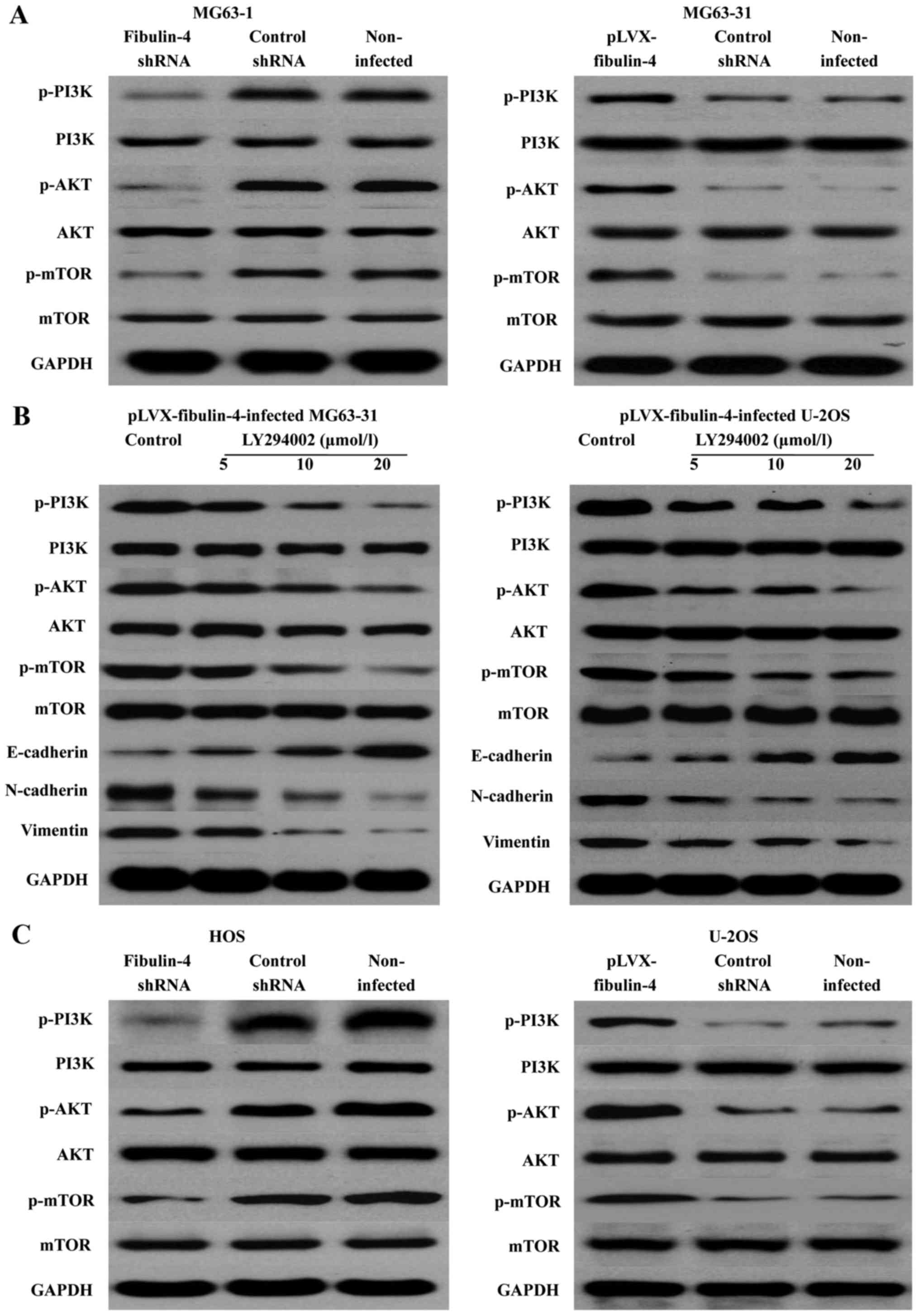 | Figure 11Effects of fibulin-4 on the PI3K/AKT
signaling pathway. (A) In fibulin-4 shRNA-infected MG63-1 cells,
fibulin-4 knockdown reduced the PI3K, AKT, and mTOR phosphorylation
levels; in contrast, in pLVX-fibulin-4-infected MG63-31 cells,
fibulin-4 upregulation increased their phosphorylation levels. (B)
The PI3K/AKT signaling pathway inhibitor LY294002 significantly
inhibited the PI3K/AKT pathway, and EMT, both of which were
activated by fibulin-4 upregulation. (C) In fibulin-4
shRNA-infected HOS cells, fibulin-4 knockdown could reduce the
PI3K, AKT, and mTOR phosphorylation levels; in contrast, in
pLVX-fibulin-4-infected U-2OS cells, fibulin-4 upregulation could
increase their phosphorylation levels. |
Discussion
In our study, we observed that high fibulin-4
expression was associated with poor prognosis of human
osteosarcoma, and that fibulin-4 was overexpressed in the highly
invasive osteosarcoma cell line and subclone. Fibulin-4 promoted
osteosarcoma cell invasion and metastasis by inducing EMT via the
PI3K/AKT/mTOR pathway.
From the protein and mRNA levels, we found that
fibulin-4 expression in osteosarcoma tissues and cell lines was
much higher than that in normal tissues and cell lines, especially
in the highly metastatic osteosarcoma cell line and subclone.
Moreover, high fibulin-4 expression was detected in low
differentiation and positive lymph node status. Similar results
have been reported by Li and Wang (32), wherein compared to normal specimens
and cell lines, upregulated fibulin-4 expression was found in
osteosarcoma clinical specimens and cell lines. In colon cancer,
fibulin-4 expression was enhanced, and it seemed to verify the
hypothesis of fibulin-4 as an oncogene (33). After further studies, fibulin-4 was
found to be a promising serum biomarker for the early detection of
colorectal cancer (34).
In a study of gynecologic tumors, fibulin-4
overexpression was associated with tumor progression and poor
prognosis in patients with cervical carcinoma and ovarian cancer
(29,35). In gliomas, fibulin-4 expression was
significantly increased in the tumor tissues compared to the
non-tumorous brain tissue (36).
However, fibulin-4 expression was found to be decreased in prostate
cancer (11). Therefore, we
suspect that like some members of the fibulin family, the role of
fibulin-4 in tumor development may exhibit tissue specificity,
wherein different tumor microenvironments determine different gene
functions (37). CYR61 is a
secreted, extracellular matrix (ECM)-associated signaling protein
of the CCN family. CYR61 is capable of regulating a broad range of
cellular activities, including cell adhesion, migration,
proliferation, differentiation, apoptosis, and senescence (38,39).
A previous study showed that Cyr61 expression is related to
osteosarcoma progression. In addition, Cyr61 could promote cell
migration and metastasis in osteosarcoma (40,41).
In the present study, by IHC, the expression levels of CYR61 and
ALP in osteosarcoma tissues were significantly high. According to
the Pearson's product-moment correlation coefficient, the
expression of CYR61 and fibulin-4 exhibited strong positive
correlations. In conclusion, we considered that fibulin-4 acted as
a facilitator for osteosarcoma to promote tumor growth and
dissemination.
Epithelial-mesenchymal transition (EMT) is a highly
conserved cellular program by which epithelial cells lose cell
polarity and cell-cell adhesion, and gain migratory and invasive
properties thus converting to motile mesenchymal cells. This
important process was initially recognized as a feature of
embryogenesis and promotes carcinoma invasion and metastasis
(42). Our results revealed that
fibulin-4 knockdown suppressed the process of EMT, with upregulated
expression of the epithelial marker E-cadherin, and decreased
expression of the mesenchymal markers N-cadherin and vimentin;
whereas fibulin-4 upregulation promoted EMT, accompanied by
decreased E-cadherin expression, and increased expression of
N-cadherin and vimentin. The transcription factors Snail (Snail-1),
Slug (Snail-2), and Twist which are involved in the process of EMT
were downregulated in fibulin-4 shRNA infected cells, and
upregulated in pLVX-fibulin-4-infected cells. These data
simultaneously indicate that fibulin-4 could promote the process of
EMT by affecting transcription factors, and thereby induce the
invasion and metastasis of osteosarcoma cells. The role of
fibulin-4 in the process of EMT has not yet been reported in other
cancers.
The PI3K/AKT/mTOR pathway is an intracellular
signaling pathway directly related to cellular quiescence,
proliferation, cancer, and longevity. In many cancers, this pathway
is overactive, thus reducing apoptosis, allowing proliferation, and
promoting invasion and metastasis (43,44).
In our study, overexpression levels of fibulin-4 could upregulate
the phospho-PI3K, phospho-AKT, and phospho-mTOR activities and
promote the process of EMT, whereas knockdown of its expression
could significantly downregulate phospho-PI3K, phospho-AKT, and
phospho-mTOR activities and inhibit EMT. Furthermore, we found that
the PI3K/AKT signaling pathway inhibitor LY294002 significantly
inhibited the PI3K/AKT pathway and EMT, both of which were
activated by fibulin-4 upregulation. This study demonstrated for
the first time that fibulin-4 could promote EMT through the
PI3K/AKT/mTOR signaling pathway. As the research on fibulin-4 is
still in its infancy, we could not find similar reports in other
cancers. However, EFEMP1, which shares high homology with fibulin-4
(45), affects the development of
a variety of tumors through the PI3/AKT pathway (19,46,47).
In conclusion, high fibulin-4 expression was
associated with poor prognosis in human osteosarcoma and the
malignant phenotype of osteosarcoma cells. Fibulin-4 has the
ability to promote proliferation, invasion, and metastasis of
osteosarcoma cells by inducing EMT through the PI3K/AKT/mTOR
signaling pathway. We believe that further research on fibulin-4
can contribute to more effective treatment for inhibiting invasion
and metastasis of osteosarcoma.
Acknowledgments
The study was supported by the fund of Suzhou
Science and Technology Project (XJ201536).
References
|
1
|
Geller DS and Gorlick R: Osteosarcoma: A
review of diagnosis, management, and treatment strategies. Clin Adv
Hematol Oncol. 8:705–718. 2010.
|
|
2
|
Messerschmitt PJ, Rettew AN, Brookover RE,
Garcia RM, Getty PJ and Greenfield EM: Specific tyrosine kinase
inhibitors regulate human osteosarcoma cells in vitro. Clin Orthop
Relat Res. 466:2168–2175. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Vega S, Iwamoto T and Yamada Y:
Fibulins: Multiple roles in matrix structures and tissue functions.
Cell Mol Life Sci. 66:1890–1902. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kobayashi N, Kostka G, Garbe JH, Keene DR,
Bächinger HP, Hanisch FG, Markova D, Tsuda T, Timpl R, Chu ML, et
al: A comparative analysis of the fibulin protein family.
Biochemical characterization, binding interactions, and tissue
localization. J Biol Chem. 282:11805–11816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Papke CL and Yanagisawa H: Fibulin-4 and
fibulin-5 in elastogenesis and beyond: Insights from mouse and
human studies. Matrix Biol. 37:142–149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Obaya AJ, Rua S, Moncada-Pazos A and Cal
S: The dual role of fibulins in tumorigenesis. Cancer Lett.
325:132–138. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moll F, Katsaros D, Lazennec G, Hellio N,
Roger P, Giacalone PL, Chalbos D, Maudelonde T, Rochefort H and
Pujol P: Estrogen induction and overexpression of fibulin-1C mRNA
in ovarian cancer cells. Oncogene. 21:1097–1107. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Greene LM, Twal WO, Duffy MJ, McDermott
EW, Hill AD, O'Higgins NJ, McCann AH, Dervan PA, Argraves WS and
Gallagher WM: Elevated expression and altered processing of
fibulin-1 protein in human breast cancer. Br J Cancer. 88:871–878.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kanda M, Nomoto S, Okamura Y, Hayashi M,
Hishida M, Fujii T, Nishikawa Y, Sugimoto H, Takeda S and Nakao A:
Promoter hypermethylation of fibulin 1 gene is associated with
tumor progression in hepatocellular carcinoma. Mol Carcinog.
50:571–579. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cheng YY, Jin H, Liu X, Siu JM, Wong YP,
Ng EK, Yu J, Leung WK, Sung JJ and Chan FK: Fibulin 1 is
downregulated through promoter hypermethylation in gastric cancer.
Br J Cancer. 99:2083–2087. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wlazlinski A, Engers R, Hoffmann MJ, Hader
C, Jung V, Müller M and Schulz WA: Downregulation of several
fibulin genes in prostate cancer. Prostate. 67:1770–1780. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie L, Palmsten K, MacDonald B, Kieran MW,
Potenta S, Vong S and Kalluri R: Basement membrane derived
fibulin-1 and fibulin-5 function as angiogenesis inhibitors and
suppress tumor growth. Exp Biol Med (Maywood). 233:155–162. 2008.
View Article : Google Scholar
|
|
13
|
Law EW, Cheung AK, Kashuba VI, Pavlova TV,
Zabarovsky ER, Lung HL, Cheng Y, Chua D, Lai-Wan Kwong D, Tsao SW,
et al: Anti-angiogenic and tumor-suppressive roles of candidate
tumor-suppressor gene, Fibulin-2, in nasopharyngeal carcinoma.
Oncogene. 31:728–738. 2012. View Article : Google Scholar
|
|
14
|
Yi CH, Smith DJ, West WW and Hollingsworth
MA: Loss of fibulin-2 expression is associated with breast cancer
progression. Am J Pathol. 170:1535–1545. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Seeliger H, Camaj P, Ischenko I, Kleespies
A, De Toni EN, Thieme SE, Blum H, Assmann G, Jauch KW and Bruns CJ:
EFEMP1 expression promotes in vivo tumor growth in human pancreatic
adenocarcinoma. Mol Cancer Res. 7:189–198. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Song EL, Hou YP, Yu SP, Chen SG, Huang JT,
Luo T, Kong LP, Xu J and Wang HQ: EFEMP1 expression promotes
angiogenesis and accelerates the growth of cervical cancer in vivo.
Gynecol Oncol. 121:174–180. 2011. View Article : Google Scholar
|
|
17
|
En-lin S, Sheng-Guo C and Hua-qiao W: The
expression of EFEMP1 in cervical carcinoma and its relationship
with prognosis. Gynecol Oncol. 117:417–422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu B, Thirtamara-Rajamani KK, Sim H and
Viapiano MS: Fibulin-3 is uniquely upregulated in malignant gliomas
and promotes tumor cell motility and invasion. Mol Cancer Res.
7:1756–1770. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hwang CF, Chien CY, Huang SC, Yin YF,
Huang CC, Fang FM, Tsai HT, Su LJ and Chen CH: Fibulin-3 is
associated with tumour progression and a poor prognosis in
nasopharyngeal carcinomas and inhibits cell migration and invasion
via suppressed AKT activity. J Pathol. 222:367–379. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sadr-Nabavi A, Ramser J, Volkmann J,
Naehrig J, Wiesmann F, Betz B, Hellebrand H, Engert S, Seitz S,
Kreutzfeld R, et al: Decreased expression of angiogenesis
antagonist EFEMP1 in sporadic breast cancer is caused by aberrant
promoter methylation and points to an impact of EFEMP1 as molecular
biomarker. Int J Cancer. 124:1727–1735. 2009. View Article : Google Scholar
|
|
21
|
Hu Y, Pioli PD, Siegel E, Zhang Q, Nelson
J, Chaturbedi A, Mathews MS, Ro DI, Alkafeef S, Hsu N, et al:
EFEMP1 suppresses malignant glioma growth and exerts its action
within the tumor extracellular compartment. Mol Cancer. 10:1232011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schluterman MK, Chapman SL, Korpanty G,
Ozumi K, Fukai T, Yanagisawa H and Brekken RA: Loss of fibulin-5
binding to beta1 integrins inhibits tumor growth by increasing the
level of ROS. Dis Model Mech. 3:333–342. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu Z, Ai Q, Xu H, Ma X, Li HZ, Shi TP,
Wang C, Gong DJ and Zhang X: Fibulin-5 is down-regulated in
urothelial carcinoma of bladder and inhibits growth and invasion of
human bladder cancer cell line 5637. Urol Oncol. 29:430–435. 2011.
View Article : Google Scholar
|
|
24
|
Yue W, Sun Q, Landreneau R, Wu C,
Siegfried JM, Yu J and Zhang L: Fibulin-5 suppresses lung cancer
invasion by inhibiting matrix metalloproteinase-7 expression.
Cancer Res. 69:6339–6346. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Albig AR and Schiemann WP: Fibulin-5
function during tumorigenesis. Future Oncol. 1:23–35. 2005.
View Article : Google Scholar
|
|
26
|
Omasu F, Nakano Y and Ichiki T:
Measurement of the electrophoretic mobility of sheep erythrocytes
using microcapillary chips. Electrophoresis. 26:1163–1167. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen J, Zhang J, Zhao Y, Li J and Fu M:
Integrin beta3 down-regulates invasive features of ovarian cancer
cells in SKOV3 cell subclones. J Cancer Res Clin Oncol.
135:909–917. 2009. View Article : Google Scholar
|
|
28
|
Chang XZ, Wang ZM, Yu JM, Tian FG, Jin W,
Zhang Y, Yu J, Li LF, Liu XF, Li ZW, et al: Isolation of a human
gallbladder cancer cell clone with high invasive phenotype in vitro
and metastatic potential in orthotopic model and inhibition of its
invasiveness by heparanase antisense oligodeoxynucleotides. Clin
Exp Metastasis. 24:25–38. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen J, Liu Z, Fang S, Fang R, Liu X, Zhao
Y, Li X, Huang L and Zhang J: Fibulin-4 is associated with tumor
progression and a poor prognosis in ovarian carcinomas. BMC Cancer.
15:912015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang X, Li D, Cheng S, Fan K, Sheng L,
Zhang J, Feng B and Xu Z: The correlation of bone morphogenetic
protein 2 with poor prognosis in glioma patients. Tumour Biol.
35:11091–11095. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Albini A, Iwamoto Y, Kleinman HK, Martin
GR, Aaronson SA, Kozlowski JM and McEwan RN: A rapid in vitro assay
for quantitating the invasive potential of tumor cells. Cancer Res.
47:3239–3245. 1987.PubMed/NCBI
|
|
32
|
Li R and Wang L: Fibulin-4 is a novel
Wnt/β-catenin pathway activator in human osteosarcoma. Biochem
Biophys Res Commun. 474:730–735. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gallagher WM, Greene LM, Ryan MP, Sierra
V, Berger A, Laurent-Puig P and Conseiller E: Human fibulin-4:
Analysis of its biosynthetic processing and mRNA expression in
normal and tumour tissues. FEBS Lett. 489:59–66. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yao L, Lao W, Zhang Y, Tang X, Hu X, He C,
Hu X and Xu LX: Identification of EFEMP2 as a serum biomarker for
the early detection of colorectal cancer with lectin affinity
capture assisted secretome analysis of cultured fresh tissues. J
Proteome Res. 11:3281–3294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen J, Zhang J, Liu X, Fang R, Zhao Y and
Ma D: Overexpression of fibulin-4 is associated with tumor
progression and poor prognosis in patients with cervical carcinoma.
Oncol Rep. 31:2601–2610. 2014.PubMed/NCBI
|
|
36
|
Wang L, Chen Q, Chen Z, Tian D, Xu H, Cai
Q, Liu B and Deng G: EFEMP2 is upregulated in gliomas and promotes
glioma cell proliferation and invasion. Int J Clin Exp Pathol.
8:10385–10393. 2015.PubMed/NCBI
|
|
37
|
Chen L, Sun B, Zhang S, Zhao X, He Y, Zhao
S, Lin T and Li X: Influence of microenvironments on
microcirculation patterns and tumor invasion-related protein
expression in melanoma. Oncol Rep. 21:917–923. 2009.PubMed/NCBI
|
|
38
|
Lau LF: CCN1/CYR61: The very model of a
modern matri-cellular protein. Cell Mol Life Sci. 68:3149–3163.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lau LF: Cell surface receptors for CCN
proteins. J Cell Commun Signal. 10:121–127. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hou CH, Lin FL, Hou SM and Liu JF: Cyr61
promotes epithelial-mesenchymal transition and tumor metastasis of
osteosarcoma by Raf-1/MEK/ERK/Elk-1/TWIST-1 signaling pathway. Mol
Cancer. 13:2362014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Habel N, Vilalta M, Bawa O, Opolon P,
Blanco J and Fromigué O: Cyr61 silencing reduces vascularization
and dissemination of osteosarcoma tumors. Oncogene. 34:3207–3213.
2015. View Article : Google Scholar
|
|
42
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li X, Wu C, Chen N, Gu H, Yen A, Cao L,
Wang E and Wang L: I3K/Akt/mTOR signaling pathway and targeted
therapy for glioblastoma. Oncotarget. 7:33440–33450.
2016.PubMed/NCBI
|
|
44
|
Gao Y, Yuan CY and Yuan W: Will targeting
PI3K/Akt/mTOR signaling work in hematopoietic malignancies? Stem
Cell Investig. 3:312016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang Y and Marmorstein LY: Focus on
molecules: Fibulin-3 (EFEMP1). Exp Eye Res. 90:374–375. 2010.
View Article : Google Scholar :
|
|
46
|
Kim IG, Kim SY, Choi SI, Lee JH, Kim KC
and Cho EW: Fibulin-3-mediated inhibition of
epithelial-to-mesenchymal transition and self-renewal of
ALDH+ lung cancer stem cells through IGF1R signaling.
Oncogene. 33:3908–3917. 2014. View Article : Google Scholar
|
|
47
|
Camaj P, Seeliger H, Ischenko I, Krebs S,
Blum H, De Toni EN, Faktorova D, Jauch KW and Bruns CJ: EFEMP1
binds the EGF receptor and activates MAPK and Akt pathways in
pancreatic carcinoma cells. Biol Chem. 390:1293–1302. 2009.
View Article : Google Scholar : PubMed/NCBI
|