Introduction
Epithelial ovarian cancer (EOC) is the leading cause
of death from gynaecologic malignancies (1–3). EOC
is highly lethal, primarily because it is diagnosed at an advanced
stage and is characterized by aggressive proliferation and
metastasis (2). Despite advances
in therapy, the high mortality and poor prognosis associated with
EOC remain unchanged (2).
Therefore, identifying new therapeutic targets for EOC and
understanding the molecular mechanisms underlying EOC
aggressiveness will yield novel strategies for overcoming this
malignancy.
Accumulating clinical and epidemiological evidence
has shown that oestrogen (E2) is responsible for promoting ovarian
cancer progression (4,5). In vitro and in vivo
experimental data have indicated that E2 enhances EOC cell
proliferation (6–11). The effects of E2 on EOC
proliferation are mainly attributable to E2-regulated target genes.
Previous studies have reported numerous E2-regulated proteins that
affect EOC cell proliferation and progression, such as cyclin D1,
c-myc and insulin-like growth factor-binding protein (IGFBP) family
members (5,12). Despite the identification of these
E2-regulated proteins, the exact effect of E2 on EOC progression is
not fully understood. Therefore, there is an urgent need to
identify novel E2-regulated target genes.
The discovery of long non-coding RNAs (lncRNAs,
>200 nucleotides) has opened a new avenue in cancer research.
Accumulating evidence highlights the importance of lncRNAs as a
novel class of oncogenes or tumour suppressor genes in the
development of many types of cancer (13–23),
including EOC (24–28). Further, some lncRNAs have recently
been reported to be involved in E2 signalling. For example, the
lncRNA-SRA1 was reported to function as a nuclear receptor
corepressor in E2 signalling (29), and HOTAIR is upregulated by E2 and
contributes to E2-supported breast cancer progression (30,31).
However, little is known about E2-regulated lncRNAs in ovarian
cancer.
We previously identified a novel E2-upregulated
lncRNA, TC0101441, based on microarray analysis and found that
TC0101441 contributes to E2-induced EOC cell migration (32). However, the detailed mechanism by
which E2 upregulates TC0101441 and the role of ElncRNA1 itself in
EOC progression have not been elucidated. In the present study, we
further evaluated TC0101441, designated as oestrogen-induced long
non-coding RNA-1 (ElncRNA1), and showed that E2 upregulated
ElncRNA1 at the transcriptional level through the oestrogen
receptor α (ERα)-oestrogen response element (ERE) pathway.
Clinically, ElncRNA1 levels were significantly higher in EOC
tissues than in normal ovarian surface epithelial tissues. In
vitro and in vivo assays revealed that ElncRNA1 promotes
EOC cell proliferation. This pro-proliferation effect of ElncRNA1
was partially mediated by the regulation of CDK4, CDK6 and cyclin
D1. These findings not only clarify the mechanism by which E2
upregulates ElncRNA1 but also underscore the important role of this
novel E2-upregulated lncRNA in EOC proliferation, thus providing a
connection between E2 and ovarian cancer from the perspective of
lncRNA.
Materials and methods
Cell lines and clinical tissue
samples
The human EOC cell lines (SKOV3, CAOV3, OVCAR3 and
HO8910) were a gift from University of Texas MD Anderson Cancer
Center (Houston, TX, USA). SKOV3, CAOV3 and HO8910 cells were
cultured in RPMI-1640 (Gibco, Gaithersburg, MD, USA). OVCAR3 cells
were maintained in McCoy's 5A supplemented with 10% foetal bovine
serum (Gibco) at 37°C in a 5% CO2 atmosphere. For E2
treatment, cells (plated at 20–30% confluence) were grown for 3
days in phenol red-free RPMI-1640 or McCoy's 5A containing 5%
charcoal-stripped foetal bovine serum (Serana, Bunbury, Australia).
Next, the cells were treated for 24 h with 10−8 M E2
(Sigma) or vehicle (DMSO, 0.01% of final volume) as a control.
All tissue samples were obtained during surgical
operations at the Obstetrics and Gynecology Hospital of Fudan
University between August 2005 and December 2008. All EOC tissues
were selected from patients who a) had not received preoperative
radiotherapy, chemotherapy, or hormonal therapy; and b) did not
have borderline ovarian tumours or two or more different
malignancies. All 40 EOC tissues analysed herein were serous
ovarian cancer tissues. In particular, according to the
International Federation of Gynecologists and Obstetricians (FIGO)
staging system, 6 cases were FIGO stage I, 6 cases were FIGO stage
II, 24 cases were FIGO stage III, and 4 cases were FIGO stage IV.
Additionally, according to histological grading guidelines, 6 cases
were grade I (G1), 12 cases were G2, and 22 cases were G3. The
normal ovarian epithelial tissues were obtained from participants
diagnosed with uterine fibroids who were scheduled to undergo a
hysterectomy and oophorectomy. Tissues were selected from
participants who did not have a history of ovarian cysts, ovarian
pathology, or ovarian surgery. All samples were pathologically
confirmed, immediately frozen in liquid nitrogen and stored at
−80°C. The study was approved by the Research Ethics Committee of
Fudan University, China. Informed consent was obtained from each
patient.
RNA extraction and quantitative real-time
polymerase chain reaction (qRT-PCR)
Total RNA was isolated with TRIzol reagent
(Invitrogen, Carlsbad, CA, USA) and then reverse transcribed into
cDNA using the ExScript RT-PCR kit (Takara, Otsu, Japan) following
the manu facturer's instructions. ElncRNA1 expression was measured
by qRT-PCR using the following primer sequences: forward,
5′-CAAGGCAGGTGAGAACGAGT-3′; and reverse,
5′-CTCGACTTAGGGAGCTGCAC-3′. Expression of the internal control,
glyceraldehyde 3-phosphate dehydrogenase (GAPDH), was measured with
the following primers: forward, 5′-TGACTTCAACAGCGACACCCA-3′; and
reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′. Amplification and detection
were conducted using a Prism 7900 system (Applied Biosystems,
Foster City, CA, USA) with the ExScript SYBR Green qRT-PCR kit
(Takara). All assays were repeated in triplicate, and statistical
analyses of the results were performed using the 2−ΔΔCt
relative quantification method.
Sequence analysis of ElncRNA1
The gene location was obtained from the University
of California Santa Cruz (UCSC) genome browser (hg18, http://genome.ucsc.edu/). The Basic Local Alignment
Search Tool (BLAST) was used for sequence alignment, and the
BLAST-Like Alignment Tool (BLAT) was used to map the cDNA to
chromosomes.
RNA stability assay
To measure RNA half-life, we added 10 μg/ml
actinomycin D (Sigma) to the cells to block transcription (nascent
RNA synthesis). The cells were pre-treated with actinomycin D prior
to E2 treatment. Total RNA was extracted from samples harvested at
the indicated time points. ElncRNA1 expression was quantified by
qRT-PCR. The half-life of ElncRNA1 RNA was calculated using data
from two independent experiments.
Search for EREs
A region 2,500 bp upstream and 500 bp downstream of
the transcriptional start site (TSS) was screened for transcription
factor binding sites (TFBSs) using the Patser program (http://ural.wustl.edu/src/patser-v3e.1.tar.gz) and
TRANSFAC 8.1 position weight matrices (PWMs) (http://www.gene-regulation.com/). A cut-off score
of 0.9 was employed, and both the forward and reverse genomic DNA
strands were investigated. EMBOSS/fuzznuc software (http://helixweb.nih.gov/emboss/html/fuzznuc.html)
was used to evaluate the putative EREs in the E2-regulated lncRNA.
The conserved ERE sequence is AGGTCAnnnTGACCT, and up to two
mismatches were allowed.
Plasmids and luciferase assays
A firefly luciferase (Luc) reporter driven by a
fragment of the 3′ untranslated regions (UTR) of ElncRNA1
containing the putative ERE [Luc-ElncRNA1-3′UTR wild-type (WT)] and
a Luc reporter driven by a corresponding mutant sequence
(Luc-ElncRNA1-3′UTR Mut) were synthesized. The primer sequences
were as follows: sense for Luc-ElncRNA1-3′UTR WT and
Luc-ElncRNA1-3′UTR Mut: GGGGTACCGTAGAGACAGGGTCTCACCACATT; antisense
for Luc-ElncRNA1-3′UTR WT: CGACGCGTGGAGCATCTGTCCCAGCTG; and
anti-sense for Luc-ElncRNA1-3′UTR Mut:
CGACGCGTGGAGCATCTGTCCCAGCTGGAGTCCCCTGTGGTGTTATCTGAGATTCAGCAGCTCTGCAAGAATATCATCTATGAC.
A plasmid expressing ERα was constructed by Hanyin Co. (Shanghai,
China). A plasmid expressing Renilla Luc was purchased from Promega
(Madison, WI, USA). SKOV3 cells were transfected with the above
plasmids or vectors for 24 h and then treated with E2; Luc assays
were performed after a 16 h E2 treatment using a Dual-Luciferase
Assay kit (Promega) according to the manufacturer's protocol.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed using a ChIP assay kit
(Millipore) according to the manufacturer's instruction. Specific
antibodies were used to immunoprecipitate either ERα or the
negative control IgG. Real-time PCR was performed using a SYBR
Green PCR kit (Qiagen). The primer sequences for amplifying the
ElncRNA1 promoter region flanking the ERα-ERE binding sites were as
follows: forward, 5′-GGAAGAACAGCTCCGTGAAG-3′; and reverse,
5′-CAGATTCAGGGCTCTTGAGG-3′.
Establishment of stable
ElncRNA1-knockdown cell lines
The two ElncRNA1-siRNA sequences were
5′-GCTCACATGAGAAAGCAAACT-3′ (siRNA1) and 5′-CUUGAGUUAUGAGGUAGCA-3′
(siRNA2). Lentiviral vectors encoding ElncRNA1-shRNA were designed
based on the two siRNA se quences (ElncRNA1-knockdown (KD)1 and
ElncRNA1-KD2) and were constructed by Hanyin Co. The recombinant
lentiviruses (KD1 and KD2) and the negative control (NC) lentivirus
(Hanyin Co.) were prepared at a titre of 109
transfection units (TU)/ml. To obtain stable cell lines, SKOV3 and
CAOV3 cells were seeded in 6-well plates and infected with virus
and polybrene the following day. Positive clones were selected with
puromycin for 14 days to establish the following new stable cell
lines: SKOV3-NC, SKOV3-KD1, SKOV3-KD2, CAOV3-NC, CAOV3-KD1 and
CAOV3-KD2.
MTT assay
Cell proliferation was assayed using a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
kit (Sigma-Aldrich, St. Louis, MO, USA) and a Synergy H4 Hybrid
Reader. Briefly, the culture medium was removed after 1, 2, 3, 4,
and 5 days; 0.5 mg/ml MTT in 200 μl of medium was added to
each well; and the plates were incubated for 4 h. Then, 150
μl of DMSO was added to the SKOV3 and CAOV3 cells for 10
min, and the optical density (OD) was measured at 490 nm. Each
experiment was repeated in triplicate.
Colony formation assay
Duplicate cultures of each cell type were maintained
at 37°C in a 5% CO2 atmosphere, and fresh medium was
added every 3 days. After 2 weeks, colonies in each well consisting
of >50 cells were counted. Each experiment was repeated in
triplicate.
Tumour formation in nude mice
The animal experiments were approved by the
Institutional Animal Care and Use Committee of Fudan University and
were performed according to the institutional guidelines and
protocols. SKOV3-KD1, SKOV3-KD2, and SKOV3-NC cells
(1×106) were subcutaneously injected into 5-week-old
BALB/c athymic nude mice (Department of Laboratory Animals, Fudan
University; n=7 for each cell line). The mice were sacrificed after
4 weeks and examined for the growth of subcutaneous tumours. The
tumour volume was calculated as previously described (33).
Western blotting
Western blot assays were performed using the
following primary antibodies: anti-cyclin E (Santa Cruz);
anti-cyclin D1 (Cell Signaling Technology); anti-CDK2 (Abways
Technology); anti-CDK4 (Abways Technology); anti-CDK6 (Abways
Technology); and β-actin (Proteintech). Briefly, stimulated cells
were lysed with RIPA buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl,
1% Triton X-100, 0.5% Na-deoxycholate] containing protease
inhibitors (Roche, Complete Mini); 20-30 μg samples of the
lysates were separated on 8-12% SDS-PAGE gels and transferred to
PVDF membranes. The membranes were incubated with primary
antibodies overnight at 4°C. The primary antibody incubation was
followed by incubation with an HRP-conjugated secondary antibody.
The bound antibodies were detected using an ECL kit (Pierce).
Statistical analysis
The data were processed using SPSS version 16.0
software (SPSS, Inc., Chicago, IL, USA). Continuous data were
compared between the two groups using independent t-tests. P-values
at <0.05 were considered statistically significant.
Results
A novel E2-induced lncRNA,
TC0101441/ElncRNA1
We previously identified a novel lncRNA, TC0101441,
which was significantly upregulated by E2 in SKOV3 ERα-positive
ovarian cancer cells (32). In
this study, we designated TC0101441 as E2-induced lncRNA 1
(ElncRNA1). Based on a bioinformatics analysis using the UCSC
genome browser (hg18, http://genome.ucsc.edu/), we noted that ElncRNA1 is
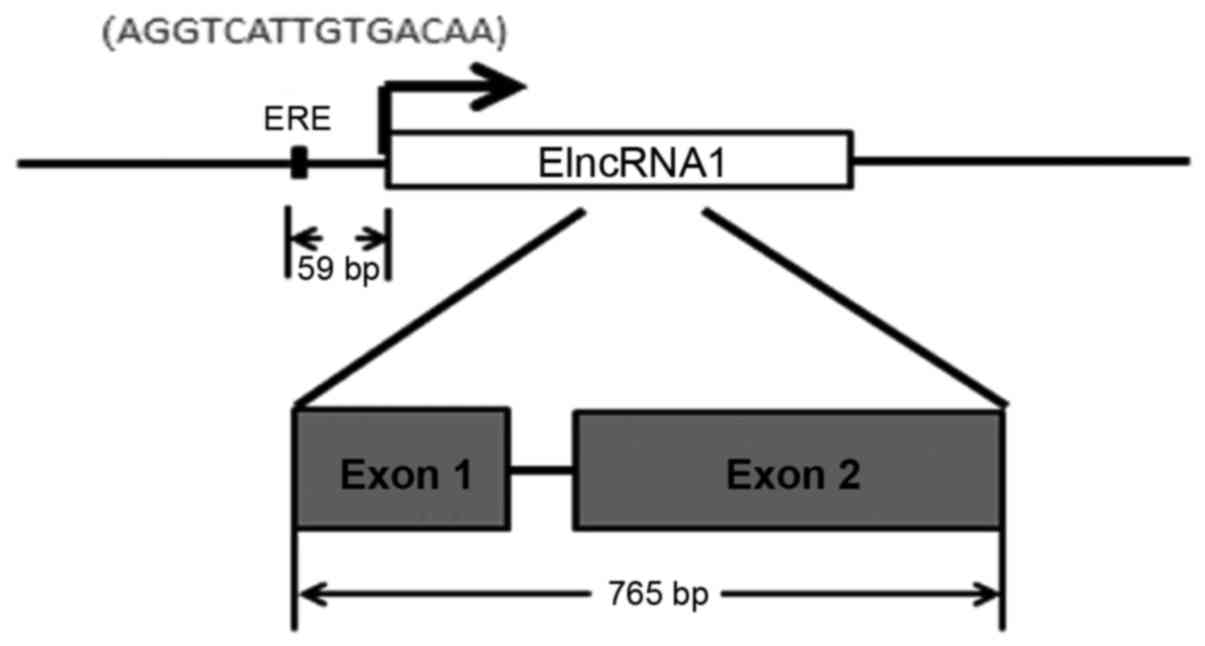
located on chromosome 1 (chr1: 202,377,159:202,378,011), contains
two exons and encodes a 765 bp lncRNA molecule (Fig. 1).
Effects of E2 on ElncRNA1 mRNA
stability
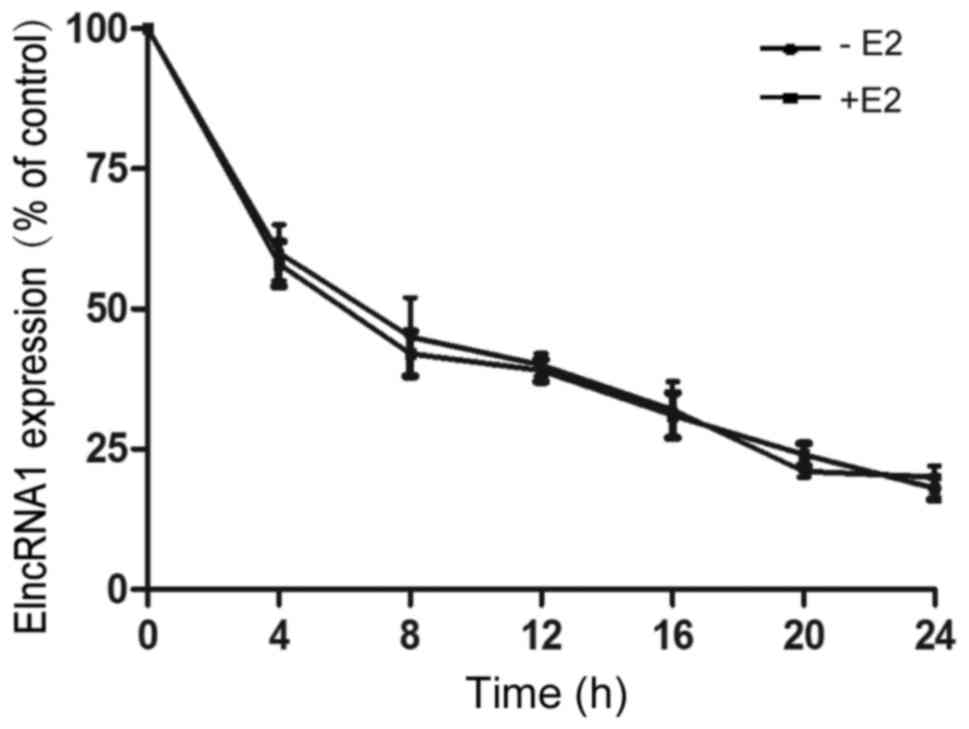
To determine whether E2-mediated changes in ElncRNA1
levels are related to transcription or mRNA stability, we treated
SKOV3 cells with actinomycin D to block nascent RNA synthesis prior
to E2 treatment and measured ElncRNA1 levels at various subsequent
time points. RNA stability was not significantly altered in SKOV3
cells in the absence or presence of E2 (Fig. 2). This result suggests that the
E2-mediated changes in ElncRNA1 levels are not due to altered mRNA
stability and implies that transcriptional regulation may be a
major mechanism by which ElncRNA1 is induced.
ERα-ERE binding is required for
transcriptional regulation of ElncRNA1 expression by E2
Given the results of our prior study, which
demonstrated that E2 induced ElncRNA1 expression in an
ERα-dependent manner (32), we
next screened the ElncRNA1 promoter for the presence of
ERα-responsive elements to confirm the transcriptional upregulation
of ElncRNA1 by E2. We identified a predicted ERE (−1725 to −1711)
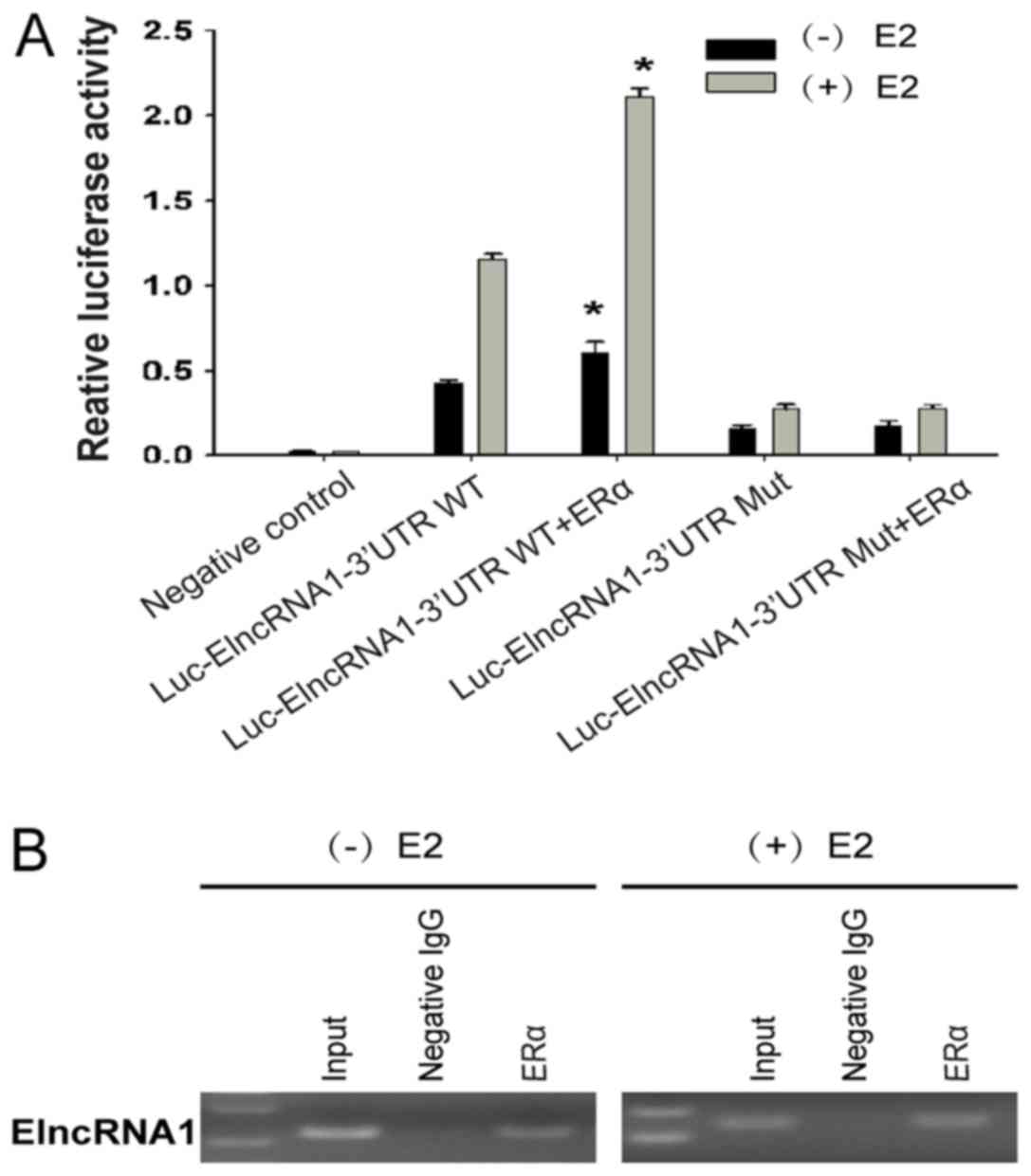
in a region 59 bp upstream of the TSS of ElncRNA1 (Fig. 1). We subsequently transfected SKOV3
cells with ERα and ElncRNA1 promoter reporters containing a mutated
or wild-type (WT) ERE and then measured Luc activity after E2
treatment. Cells expressing both WT ERE and ERα responded strongly
to E2 by inducing reporter activity (Fig. 3A), suggesting that both ERα and ERE
are required for E2 to upregulate ElncRNA1 expression.
We subsequently performed ChIP assays with
antibodies against ERα to confirm that ERα binds to the ERE region
of the ElncRNA1 promoter. As shown in Fig. 3B, enrichment of ERα-associated
promoter fragments was confirmed by RT-PCR, and this enrichment was
enhanced by E2. These results indicate that ERα can bind to the ERE
in the ElncRNA1 promoter in a manner that is enhanced by E2
treatment, thus inducing ElncRNA1 expression.
ElncRNA1 expression pattern in EOC
tissues
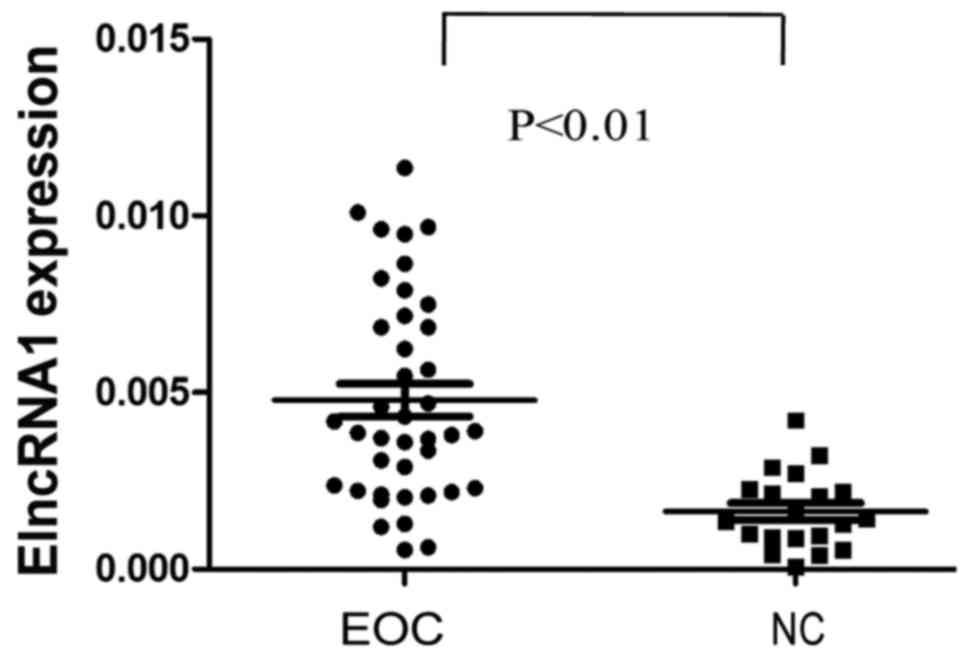
To investigate whether ElncRNA1 has clinical
implications, we used qRT-PCR to determine the ElncRNA1 expression
pattern in EOC. ElncRNA1 levels were significantly higher in 40 EOC
tissues than in 20 normal ovarian surface epithelial tissues
(P<0.01; Fig. 4). These results
suggest that ElncRNA1 over-expression may play a role in EOC
aggressiveness.
Silencing ElncRNA1 suppresses EOC cell
proliferation in vitro
To determine whether ElncRNA1 affects EOC cell
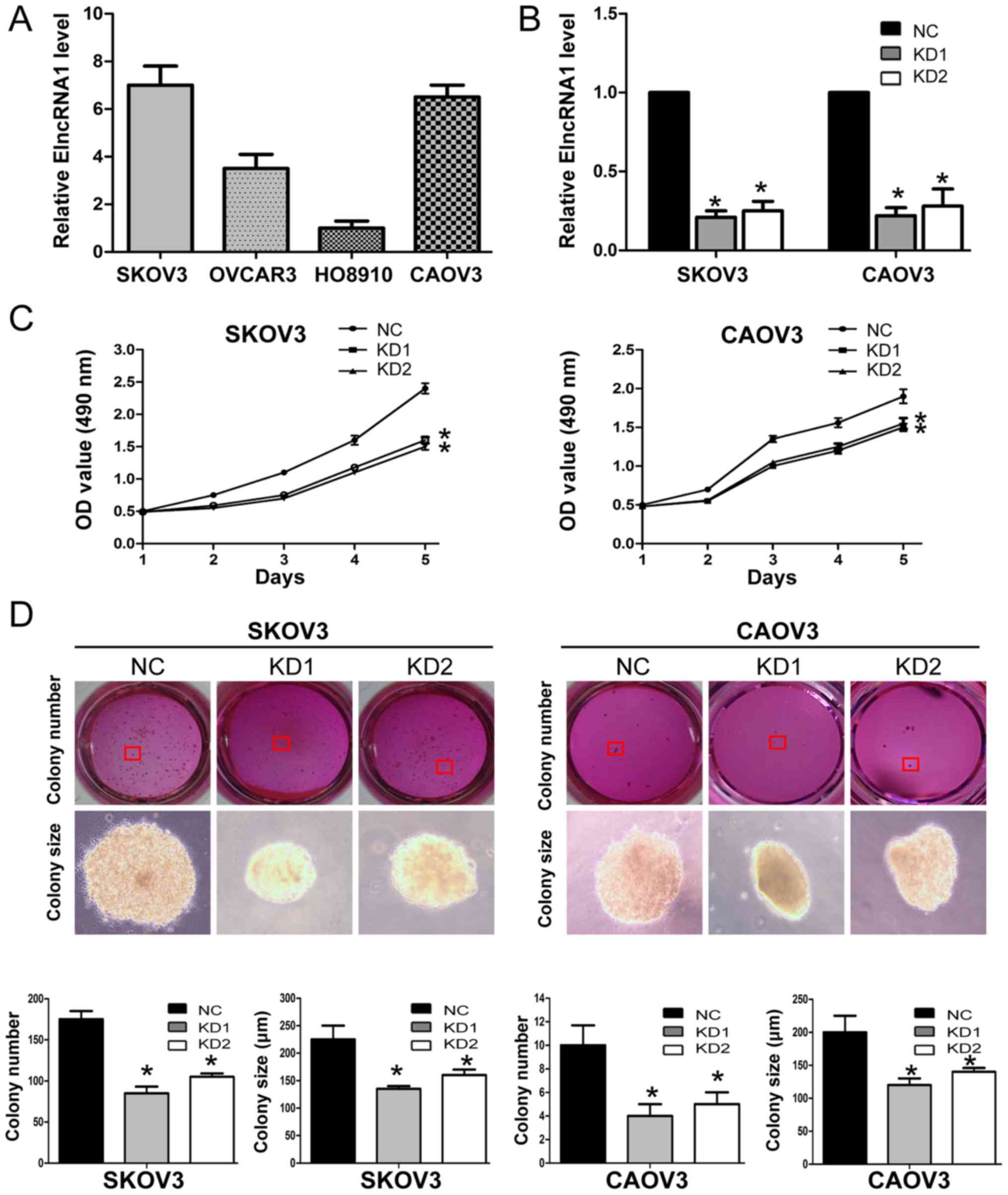
proliferation, we first examined the basal levels of ElncRNA1 in 4
EOC cell lines (SKOV3, CAOV3, OVCAR3 and HO8910) and found that
SKOV3 and CAOV3 cells expressed relatively higher ElncRNA1 levels
(Fig. 5A). Thus, we silenced
ElncRNA1 expression in these two cell lines to investigate the
effects of ElncRNA1 on cell proliferation. Since both siRNAs
efficiently silenced ElncRNA1 expression in SKOV3 and CAOV3 cells
(Fig. 5B), we constructed
lentiviral vectors to establish stable ElncRNA1-knockdown cell
lines (SKOV3-KD1, SKOV3-KD2, CAOV3-KD1, and CAOV3-KD2 cells) and
corresponding controls (SKOV3-NC and CAOV3-NC cells).
MTT assays showed that ElncRNA1 knockdown
significantly suppressed the proliferation of both SKOV3 and CAOV3
cells (Fig. 5C). Colony formation
assays demonstrated that ElncRNA1 knockdown reduced the number of
SKOV3 and CAOV3 colonies (Fig.
5D). These results indicate that silencing ElncRNA1 may inhibit
EOC cell proliferation in vitro.
Silencing ElncRNA1 inhibits EOC tumour
growth in vivo
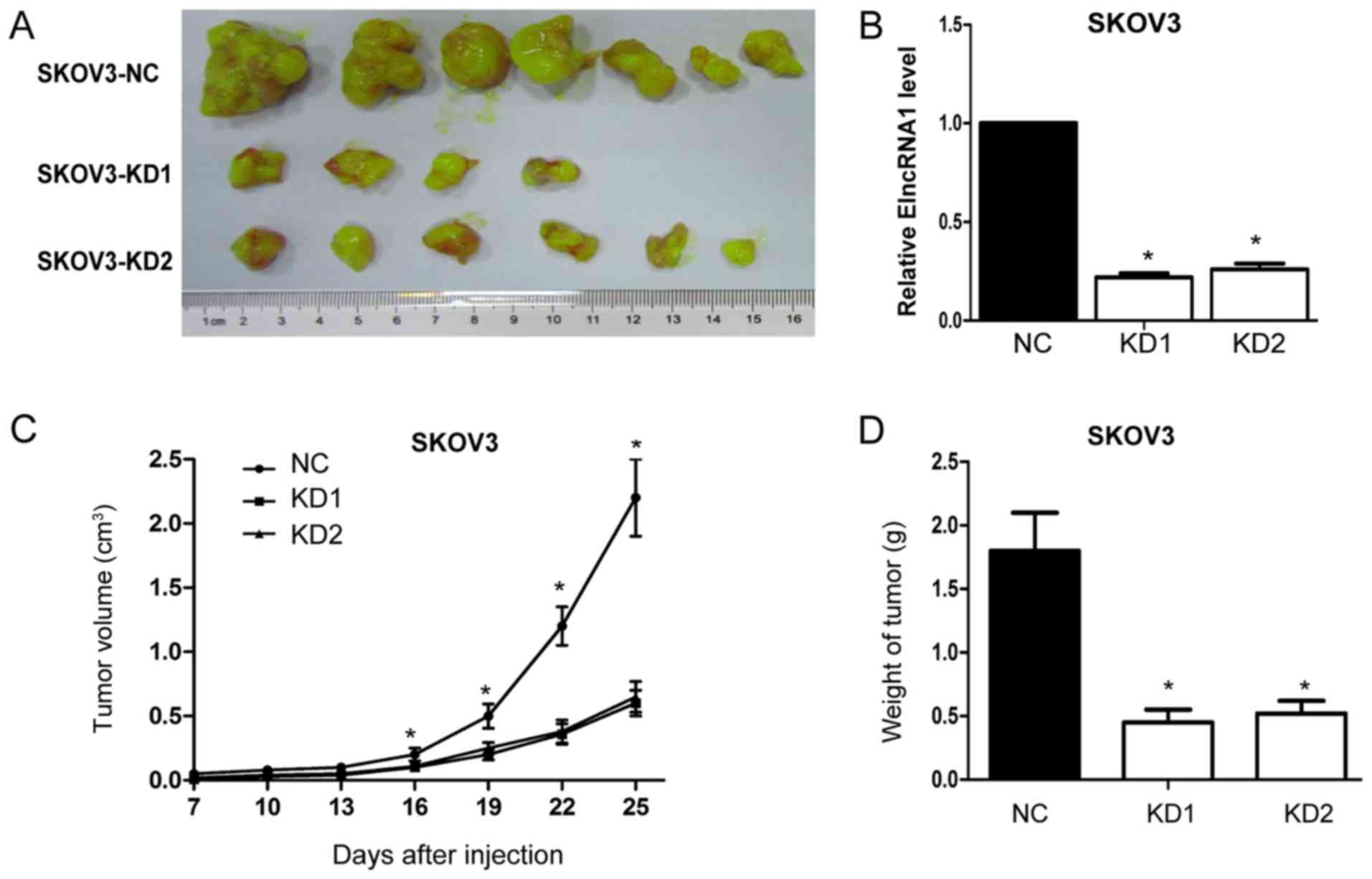
Next, we tested the effects of ElncRNA1 on EOC
tumour growth in vivo by injecting SKOV3-KD1, SKOV3-KD2 and
SKOV3-NC cells into nude mice. ElncRNA1 knockdown reduced the
frequency of tumour formation (SKOV3-NC, 7/7; SKOV3-KD1, 4/7; and
SKOV3-KD2, 6/7) (Fig. 6A).
Moreover, qRT-PCR analysis confirmed that ElncRNA1 expression
levels were significantly lower in tumour tissues from the
ElncRNA1-KD group relative to those from the control group
(Fig. 6B). Further, the volume and
average weight of tumours formed by SKOV3-KD1 and SKOV3-KD2 cells
were much lower compared to tumours formed by SKOV3-NC cells
(Fig. 6C). Together, these results
suggest that silencing ElncRNA1 inhibits EOC growth in vivo,
which corresponds with the in vitro results.
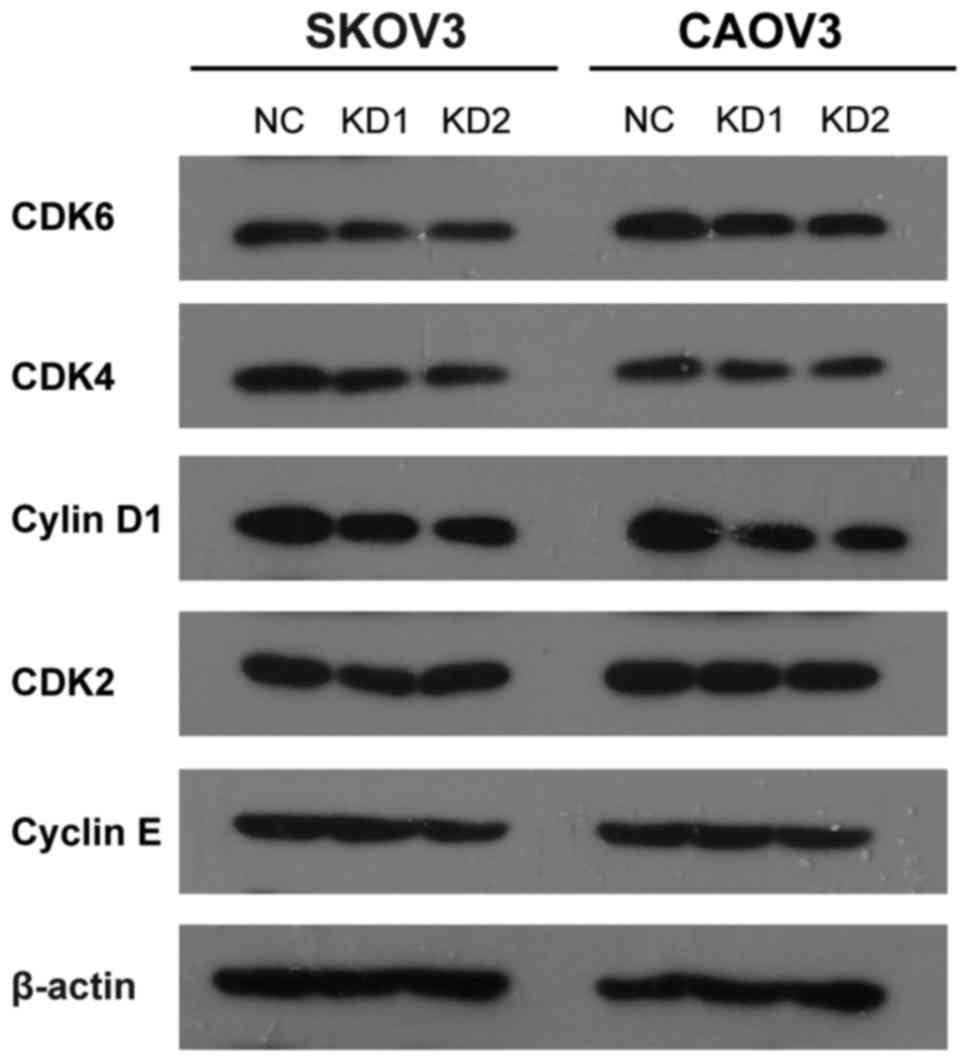
Certain proliferation-related proteins
are downstream mediators of ElncRNA1 activity affecting EOC cell
proliferation
To study possible mechanisms through which ElncRNA1
alters EOC cell proliferation further, we evaluated the expression
of certain key proliferation-related proteins, including cyclin D1,
cyclin E, CDK2, CDK4, CDK6, by western blot assay. The results
showed that ElncRNA1 knockdown in SKOV3 and CAOV3 cells resulted in
a significant decrease in CDK4, CDK6 and cyclin D1 protein levels
(Fig. 7), suggesting that CDK4,
CDK6 and cyclin D1 are the downstream mediators of ElncRNA1
activity affecting EOC cell proliferation. Together, these data
indicate that ElncRNA1 regulates EOC cell proliferation at least in
part by regulating CDK4, CDK6 and cyclin D1.
Discussion
Previously, we identified the novel E2-upregulated
lncRNA, ElncRNA1, based on microarray analysis. However, the
detailed mechanisms by which E2 upregulates ElncRNA1 and the role
of ElncRNA1 itself in EOC progression have not been determined
(32). In the present study, using
RNA stability assays, bioinformatics-based searches for ERE binding
sites, ChIP assays and dual luciferase reporter assays, we found
that E2 transcriptionally upregulated ElncRNA1 through the ERα-ERE
pathway. Clinically, ElncRNA1 levels were significantly higher in
EOC tissues than in normal ovarian surface epithelial tissues.
Further in vitro and in vivo assays revealed that
ElncRNA1 promoted EOC cell proliferation. This pro-proliferation
effect of ElncRNA1 was partially mediated by the regulation of
CDK4, CDK6 and cyclin D1. Together, these findings not only clarify
the mechanism by which E2 upregulates ElncRNA1 but also underscore
the important role of this novel E2-upregulated lncRNA in EOC
proliferation, thus providing a connection between E2 and ovarian
cancer from the perspective of lncRNA.
E2 plays an important role in regulating cancer
growth through various target genes, including protein coding genes
and non-coding RNAs. We previously identified a novel
E2-upregulated lncRNA (TC0101441, designated ElncRNA1) (32), but the molecular mechanisms by
which E2 upregulates ElncRNA1 have not been determined. So far,
several mechanisms have been proposed for E2-regulated gene
expression, including direct action of basal transcriptional
elements and the participation of cofactors (34-36).
Our prior study demonstrated that the E2-mediated induction of
ElncRNA1 expression is dependent on ERα (32), a ligand-activated transcription
factor in the nuclear receptor super-family that directly binds to
oestrogen-responsive elements (EREs) in the promoter regions of
target genes, thereby regulating their transcription (34,36,37).
Therefore, we hypothesized that ERα-ERE binding might contribute to
the transcriptional upregulation of ElncRNA1 expression by E2. To
address this hypothesis, we initially performed mRNA stability
experiments. The use of actinomycin D suggested that the E2-induced
changes in ElncRNA1 expression were not due to effects on mRNA
stability but were rather due to changes in E2-regulated
transcription. To confirm the transcriptional upregulation of
ElncRNA1 by E2, we screened the ElncRNA1 promoter and found an ERE
in a 59 bp region upstream of the TSS of ElncRNA1. Subsequent ChIP
assays indicated that ERα bound to the ERE in the ElncRNA1
promoter. Moreover, dual luciferase reporter assays revealed that
cells expressing both WT ERE and ERα responded strongly to E2 by
inducing reporter activity, suggesting that both ERα and ERE are
required for E2 to upregulate ElncRNA1 expression. Together, these
findings indicate that E2 acts through its nuclear receptor ERα,
which binds directly to ERE in the promoter region of ElncRNA1,
thereby inducing transcriptional upregulation of ElncRNA1
expression. Additionally, we can conclude that E2 transcriptionally
upregulates ElncRNA1 through the ERα-ERE pathway.
However, the role of this novel E2-upregulated
lncRNA, ElncRNA1, remained to be determined. Accumulating evidence
indicates that aberrantly expressed lncRNAs play oncogenic or
tumour suppressor roles in human cancer. For example, HOTAIR
promotes tumour growth in cervical cancer (38), and MEG3 inhibits cell proliferation
in prostate cancer (39). As
lncRNAs are emerging as key components of cancer progression, it
was reasonable to hypothesize that ElncRNA1 may contribute to EOC
progression. Our previous work showed that ElncRNA1 contributes to
E2-induced EOC metastasis. However, the role of ElncRNA1 itself in
EOC has not been determined, and an association between ElncRNA1
and aspects of EOC progression other than metastasis has not been
demonstrated. In the present study, ElncRNA1 levels were clearly
higher in EOC tissues than in noncancerous tissues, suggesting that
ElncRNA1 may play a role in the aggressiveness of EOC. In
vitro, ElncRNA1 knockdown suppressed cell proliferation and
reduced the number of SKOV3 and CAOV3 colonies. Additionally, in
vivo experiments confirmed that ElncRNA1 depletion inhibited
tumour growth in nude mice. Taken together, these data suggest that
ElncRNA1 plays an important role in promoting EOC growth and can
function as an oncogene, thereby contributing to the proliferative
effects of E2.
Previous study demonstrated that the key
proliferation-related genes 'cyclin D1-CDK4/6', associated with
E2/ER signaling pathway, promoted breast cancer proliferation
(40). In the present study, we
found that ElncRNA1 knockdown in SKOV3 and CAOV3 cells resulted in
a significant decrease in CDK4, CDK6 and cyclin D1 protein levels
instead of cyclin E and CDK2 levels, suggesting that ElncRNA1
regulates EOC cell proliferation at least in part by regulating
'cyclin D1-CDK4/6'. One limitation of this study is that we did not
study the exact mechanism by which ElncRNA1 regulates cyclin
D1-CDK4/6 in EOC growth. Further studies are required to explore
the exact molecular mechanism of E2/ERα- ElncRNA1-'cyclin
D1-CDK4/6' pathway.
In conclusion, our study provides the first evidence
that E2 transcriptionally upregulates ElncRNA1 through the ERα-ERE
pathway and that this novel E2-upregulated lncRNA has an oncogenic
role in EOC growth. The addition of ElncRNA1 to the E2-ERα-ERE
signalling pathway may provide greater insight into the oestrogenic
effects on EOC progression from the perspective of lncRNA.
Acknowledgments
This study was supported by funding from the
National Natural Science Foundation of China (81370689 and
81571404; to K.-Q.H.), the National Natural Science Foundation for
Young Scholars of China (81502240; to J.-J.Q.), and the Shanghai
Science and Technology Development Funds for the Talents
(15YF1401400; to J.-J.Q.).
References
|
1
|
Hollis RL and Gourley C: Genetic and
molecular changes in ovarian cancer. Cancer Biol Med. 13:236–247.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Doubeni CA, Doubeni AR and Myers AE:
Diagnosis and management of ovarian cancer. Am Fam Physician.
93:937–944. 2016.PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsilidis KK, Allen NE, Key TJ, Dossus L,
Kaaks R, Bakken K, Lund E, Fournier A, Dahm CC, Overvad K, et al:
Menopausal hormone therapy and risk of ovarian cancer in the
European prospective investigation into cancer and nutrition.
Cancer Causes Control. 22:1075–1084. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cunat S, Hoffmann P and Pujol P: Estrogens
and epithelial ovarian cancer. Gynecol Oncol. 94:25–32. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Park SH, Cheung LW, Wong AS and Leung PC:
Estrogen regulates Snail and Slug in the down-regulation of
E-cadherin and induces metastatic potential of ovarian cancer cells
through estrogen receptor alpha. Mol Endocrinol. 22:2085–2098.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hua K, Din J, Cao Q, Feng W, Zhang Y, Yao
L, Huang Y, Zhao Y and Feng Y: Estrogen and progestin regulate
HIF-1α expression in ovarian cancer cell lines via the activation
of Akt signaling transduction pathway. Oncol Rep. 21:893–898.
2009.PubMed/NCBI
|
|
8
|
Hua K, Feng W, Cao Q, Zhou X, Lu X and
Feng Y: Estrogen and progestin regulate metastasis through the
PI3K/AKT pathway in human ovarian cancer. Int J Oncol. 33:959–967.
2008.PubMed/NCBI
|
|
9
|
Ding JX, Feng YJ, Yao LQ, Yu M, Jin HY and
Yin LH: The reinforcement of invasion in epithelial ovarian cancer
cells by 17 beta-Estradiol is associated with up-regulation of
Snail. Gynecol Oncol. 103:623–630. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Spillman MA, Manning NG, Dye WW, Sartorius
CA, Post MD, Harrell JC, Jacobsen BM and Horwitz KB:
Tissue-specific pathways for estrogen regulation of ovarian cancer
growth and metastasis. Cancer Res. 70:8927–8936. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Laviolette LA, Garson K, Macdonald EA,
Senterman MK, Courville K, Crane CA and Vanderhyden BC:
17beta-estradiol accelerates tumor onset and decreases survival in
a transgenic mouse model of ovarian cancer. Endocrinology.
151:929–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
O'Donnell AJ, Macleod KG, Burns DJ, Smyth
JF and Langdon SP: Estrogen receptor-alpha mediates gene expression
changes and growth response in ovarian cancer cells exposed to
estrogen. Endocr Relat Cancer. 12:851–866. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar
|
|
14
|
Du Y, Kong G, You X, Zhang S, Zhang T, Gao
Y, Ye L and Zhang X: Elevation of highly up-regulated in liver
cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell
proliferation via down-regulating p18. J Biol Chem.
287:26302–26311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Matouk IJ, DeGroot N, Mezan S, Ayesh S,
Abu-lail R, Hochberg A and Galun E: The H19 non-coding RNA is
essential for human tumor growth. PLoS One. 2:e8452007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou Y, Zhang X and Klibanski A: MEG3
noncoding RNA: A tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qiao HP, Gao WS, Huo JX and Yang ZS: Long
non-coding RNA GAS5 functions as a tumor suppressor in renal cell
carcinoma. Asian Pac J Cancer Prev. 14:1077–1082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fang J, Sun CC and Gong C: Long noncoding
RNA XIST acts as an oncogene in non-small cell lung cancer by
epigenetically repressing KLF2 expression. Biochem Biophys Res
Commun. 478:811–817. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li J, Yu H, Xi M and Lu X: Long noncoding
RNA C17orf91 is a potential prognostic marker and functions as an
oncogene in ovarian cancer. J Ovarian Res. 9:492016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liao T, Qu N, Shi RL, Guo K, Ma B, Cao YM,
Xiang J, Lu ZW, Zhu YX, Li DS, et al: BRAF-activated LncRNA
functions as a tumor suppressor in papillary thyroid cancer.
Oncotarget. 8:238–247. 2017.
|
|
23
|
Sang Y, Zhou F, Wang D, Bi X, Liu X, Hao
Z, Li Q and Zhang W: Up-regulation of long non-coding HOTTIP
functions as an oncogene by regulating HOXA13 in non-small cell
lung cancer. Am J Transl Res. 8:2022–2032. 2016.PubMed/NCBI
|
|
24
|
Tanos V, Prus D, Ayesh S, Weinstein D,
Tykocinski ML, De-Groot N, Hochberg A and Ariel I: Expression of
the imprinted H19 oncofetal RNA in epithelial ovarian cancer. Eur J
Obstet Gynecol Reprod Biol. 85:7–11. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rangel LB, Sherman-Baust CA, Wernyj RP,
Schwartz DR, Cho KR and Morin PJ: Characterization of novel human
ovarian cancer-specific transcripts (HOSTs) identified by serial
analysis of gene expression. Oncogene. 22:7225–7232. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Silva JM, Boczek NJ, Berres MW, Ma X and
Smith DI: LSINCT5 is over expressed in breast and ovarian cancer
and affects cellular proliferation. RNA Biol. 8:496–505. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qiu JJ, Wang Y, Liu YL, Zhang Y, Ding JX
and Hua KQ: The long non-coding RNA ANRIL promotes proliferation
and cell cycle progression and inhibits apoptosis and senescence in
epithelial ovarian cancer. Oncotarget. 7:32478–32492.
2016.PubMed/NCBI
|
|
28
|
Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW,
Jin HY, Zhang Y, Li Q and Hua KQ: Overexpression of long non-coding
RNA HOTAIR predicts poor patient prognosis and promotes tumor
metastasis in epithelial ovarian cancer. Gynecol Oncol.
134:121–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chooniedass-Kothari S, Vincett D, Yan Y,
Cooper C, Hamedani MK, Myal Y and Leygue E: The protein encoded by
the functional steroid receptor RNA activator is a new modulator of
ER alpha transcriptional activity. FEBS Lett. 584:1174–1180. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bhan A and Mandal SS: Estradiol-induced
transcriptional regulation of Long non-coding RNA, HOTAIR. Methods
Mol Biol. 1366:395–412. 2016. View Article : Google Scholar
|
|
31
|
Xue X, Yang YA, Zhang A, Fong KW, Kim J,
Song B, Li S, Zhao JC and Yu J: LncRNA HOTAIR enhances ER signaling
and confers tamoxifen resistance in breast cancer. Oncogene.
35:2746–2755. 2016. View Article : Google Scholar :
|
|
32
|
Qiu J, Ye L, Ding J, Feng W, Zhang Y, Lv
T, Wang J and Hua K: Effects of oestrogen on long noncoding RNA
expression in oestrogen receptor alpha-positive ovarian cancer
cells. J Steroid Biochem Mol Biol. 141:60–70. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Y, Wang Z, Qi Z, Yin S, Zhang N, Liu
Y, Liu M, Meng J, Zang R, Zhang Z, et al: The negative interplay
between Aurora A/B and BRCA1/2 controls cancer cell growth and
tumorigenesis via distinct regulation of cell cycle progression,
cytokinesis, and tetraploidy. Mol Cancer. 13:942014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin CY, Vega VB, Thomsen JS, Zhang T, Kong
SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, et al:
Whole-genome cartography of estrogen receptor alpha binding sites.
PLoS Genet. 3:e872007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kushner PJ, Agard DA, Greene GL, Scanlan
TS, Shiau AK, Uht RM and Webb P: Estrogen receptor pathways to
AP-1. J Steroid Biochem Mol Biol. 74:311–317. 2000. View Article : Google Scholar
|
|
36
|
Carroll JS, Meyer CA, Song J, Li W,
Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC,
Hall GF, et al: Genome-wide analysis of estrogen receptor binding
sites. Nat Genet. 38:1289–1297. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang Y, Li X and Muyan M: Estrogen
receptors similarly mediate the effects of 17β-estradiol on
cellular responses but differ in their potencies. Endocrine.
39:48–61. 2011. View Article : Google Scholar
|
|
38
|
Lee M, Kim HJ, Kim SW, Park SA, Chun KH,
Cho NH, Song YS and Kim YT: The long non-coding RNA HOTAIR
increases tumour growth and invasion in cervical cancer by
targeting the Notch pathway. Oncotarget. 7:44558–44571.
2016.PubMed/NCBI
|
|
39
|
Luo G, Wang M, Wu X, Tao D, Xiao X, Wang
L, Min F, Zeng F and Jiang G: Long non-coding RNA MEG3 inhibits
cell proliferation and induces apoptosis in prostate cancer. Cell
Physiol Biochem. 37:2209–2220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Herrera-Abreu MT, Palafox M, Asghar U,
Rivas MA, Cutts RJ, Garcia-Murillas I, Pearson A, Guzman M,
Rodriguez O, Grueso J, et al: Early adaptation and acquired
resistance to CDK4/6 inhibition in estrogen receptor-positive
breast cancer. Cancer Res. 76:2301–2313. 2016. View Article : Google Scholar : PubMed/NCBI
|