Introduction
Distant metastasis is a major clinical determinant
of the survival of individuals with gastrointestinal cancer. Liver
metastasis occurs in 70% of patients with pancreatic cancer and
confers a very poor prognosis. Occult liver metastases may already
be present at the time of surgery (1) and recur, resulting in early mortality
after surgical resection (2).
Gal-9, a member of the β-galactoside-binding animal
lectin family, is a promising agent for the treatment of
immune-related and neoplastic diseases. Gal-9 is a
tandem-repeat-type galectin family member consisting of two
carbohydrate recognition domains connected by a linker peptide
(3) and was first described as an
eosinophil chemoattractant (4).
This protein has been reported to contribute to various biological
processes, including cell aggregation, adhesion, chemoattraction,
and apoptosis (5). Furthermore,
Gal-9 has been tested as a potential therapeutic agent for various
autoimmune diseases and allergic diseases (6,7). The
functions of Gal-9 have been reported to include the induction of
apoptosis in T-cells, particularly CD4+ Th1 and Th17
cells, and the stimulation of regulatory T-cell activity (8-10).
Recently, it has been reported that Gal-9 suppresses tumor growth
in various types of human cancer (10,11),
such as melanoma (12) and chronic
myelogenous leukemia (13).
Regarding the mechanism underlying the Gal-9-induced inhibition of
cell growth in various cancers, Gal-9 has been shown to induce
cancer cell death via an apoptotic signaling pathway (12,13).
This apoptotic signaling in multiple myeloma is caspase-dependent
and is induced by activation of the JNK and p38 MAP kinases
(14). Alternatively, Gal-9
induces apoptosis in chronic myelogenous leukemia by increasing
Noxa expression (13). Recently,
it was revealed that Gal-9 shows potent cytotoxic activity toward
KRAS mutant colon cancer via the induction of frustrated autophagy
(15). Furthermore, loss of Gal-9
expression is associated with metastatic progression in various
epithelial cancers, including breast cancer and hepatocellular
carcinoma (16,17).
We have previously shown that Gal-9 suppresses cell
proliferation and tumor growth in various types of gastrointestinal
tumors, such as hepatocellular carcinoma (18), cholangiocarcinoma (19), gallbladder carcinoma (20), and gastric cancer (21) by inducing apoptosis, and we have
identified several microRNAs (miRNAs) that are associated with the
antitumor effect of Gal-9.
miRNAs, which are small, endogenous, non-coding RNAs
of 21–30 nucleotides in length, modulate the expression of various
target genes at the post-transcriptional and translational levels
(22,23). A total of 1,881 human miRNAs are
registered at miRbase as of release 21 (http://microrna.sanger.ac.uk/). However, little is
known about the association of certain miRNAs with the antitumor
effects of Gal-9 on liver metastasis.
Therefore, the purpose of the present study was to
determine whether Gal-9 suppresses the tumor growth of liver
metastases and to identify the mechanism of the antitumor
effect.
Materials and methods
Chemicals
A mutant form of Gal-9 lacking the entire linker
region that is stable against proteolysis was recombinantly
produced as previously described (24).
Cell lines and culture
Three human liver metastasis cell lines were derived
from pancreatic cancer (KMP2, KMP7 and KMP8) and obtained from the
Japanese Collection of Research bioresources (Osaka, Japan). The
cells were cultured in RPMI-1640 (Gibco, Tokyo, Japan) and Ham's
F12 medium (Sigma-Aldrich, St. Louis, MO, USA) (1 to 1 mix) with 5%
heat-inactivated fetal bovine serum (Wako Pure Chemical Industries,
Osaka, Japan) and 100 mg/l penicillin-streptomycin (Invitrogen,
Tokyo, Japan), and the cells were incubated in a humidified
atmosphere containing 5% CO2 at 37°C.
Cell proliferation assay
Cell proliferation assays were conducted using a
cell counting kit-8 (CCK-8) kit (Dojindo Laboratories, Kumamoto,
Japan) according to the manufacturer's instructions. Cells
(5.0×103) from each cell line were seeded into the wells
of a 96-well plate and cultured in 100 μl of the
corresponding medium. After 24 h, the seeded cells were treated
with 0, 0.1 or 0.3 μM Gal-9 diluted in the culture medium.
At the indicated time-points, the medium was changed to 100
μl of medium containing the CCK-8 reagent, and the cells
were incubated for 3 h. The absorbance at a wavelength of 450 nm
was measured in each well using an automated microplate reader.
Enzyme-linked immunosorbent assay (ELISA)
for quantification of apoptosis
KMP8 was chosen as a model cell line because it was
most sensitive to Gal-9 in vitro. Caspase-cleaved
cytokeratin 18 (cCK18) was evaluated using the M30 Apoptosense
ELISA kit obtained from PEVIVA AB (Bromma, Sweden) (25). KMP8 cells (5×103) were
seeded in a 96-well plate and cultured for 6, 24, or 48 h following
the addition of 0.3 μM Gal-9. The cells were then lysed in
polyoxyethylene octylphenyl ether (NP-40) (Wako Pure Chemical
Industries). The subsequent ELISAs were performed according to the
manufacturer's instructions. The absorbance at a wavelength of 450
nm was measured in each well using an automated microplate reader.
The abundance of the antigen in the control and unknown samples was
calculated via interpolation from a standard curve.
Analysis of early apoptosis
Analysis of early apoptosis was performed using an
Annexin V-FITC Early Apoptosis Detection kit (#6592; Cell Signaling
Technology, Boston, MA, USA), which can distinguish early apoptotic
cells within a cell population. KMP8 cells (1.0×106
cells in a 60-mm-diameter dish) were treated with or without 0.3
μM Gal-9 for 6 h. because of the high sensitivity of the
cells to Gal-9, they were not suitable for analysis at the 12- and
24-h time-points. Apoptotic and necrotic cell death was analyzed
through double staining with FITC-conjugated Annexin V and
propidium iodide (PI); this staining method is based on the binding
of Annexin V to apoptotic cells with exposed phosphatidyl-serines
and the PI labeling of late apoptotic/necrotic cells with membrane
damage. Staining was performed according to the manufacturer's
instructions. Flow cytometry was conducted using a Cytomics FC 500
flow cytometer (Beckman Coulter, Indianapolis, IN, USA). Cell
percentages were analyzed using Kaluza software (Beckman Coulter).
The experiment was repeated five times, and a test for significant
difference was carried out.
Analysis of apoptosis-related protein
profiles using an antibody array
Cells were seeded in 100-mm culture dishes and were
then treated with 0.3 μM Gal-9 for 6 h, followed by lysis in
PRO-PREP (iNtRON Biotechnology, Sungnam, Korea). A human apoptosis
antibody array kit (R&D Systems, Minneapolis, MN, USA) was
subsequently used to measure apoptosis-related proteins according
to the manufacturer's instructions. Briefly, proteins were captured
by antibodies spotted on a nitrocellulose membrane. Next, the
levels of apoptosis-related proteins were assessed using an
HRP-conjugated antibody, followed by detection via
chemiluminescence. Finally, each array membrane was exposed to
X-ray film using a chemiluminescence detection system (Perkin-Elmer
Co. Waltham, MA, USA).
Gel electrophoresis and western blot
analysis
KMP8 cells (1.0×106/dish) were seeded in
100-mm culture dishes and cultured for 6 or 12 h, after which 0 or
0.3 μM Gal-9 was added. The cells were subsequently lysed
using a protease inhibitor cocktail (PRO-PREP complete protease
inhibitor mixture; iNtRON biotechnology). Furthermore, to
investigate cytoplasmic proteins, a Cell Fractionation kit (Cell
Signaling Technology) was used according to the manufacturer's
instructions. Briefly, the cells (cultured for 6 h after the
addition of 0 or 0.3 μM Gal-9) were separated into three
distinct fractions: cytoplasmic, membrane/organelle, and
nuclear/cytoskeletal, using the appropriate isolation buffer for
each process. Next, the samples were subjected to sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12%
agarose gels, and the proteins were transferred to nitrocellulose
membranes. After blocking, the membranes were first incubated with
primary antibodies, followed by secondary antibodies. The
immunoreactive proteins were visualized on X-ray film using an
enhanced chemiluminescence detection system (Perkin-Elmer Co.).
Primary antibodies against caspase-3 (8G10) (#9665),
cleaved caspase-3 (D175) (#1661), caspase-7 (D2Q3L) (#12827),
caspase-9 (C9) (#9508), cleaved caspase-9 (Asp330) (#7237), PARP
(#9542), cleaved-PARP (D64E10) (#5625), cytochrome c (D18C7)
(#11940), Smac/Diablo (#2954), HtrA2/Omi (D20A5) (#9745), and GAPDH
(D16H11) XP (#5174) were purchased from Cell Signaling Technology.
Additionally, an anti-β-actin monoclonal antibody (A5441;
Sigma-Aldrich) and antibodies against cyclin D1 (RB-9041; Thermo
Fisher Scientific, Waltham, MA, USA), cyclin E (HE12) (MA5-14336;
Thermo Fisher Scientific), Cdk6 (sc-177; Santa Cruz Biotechnology,
Santa Cruz, CA), Cdk4 (sc-749; Santa Cruz Biotechnology), and Cdk2
(sc-163; Santa Cruz Biotechnology) were used. The secondary
antibodies included horseradish peroxidase (HRP)-linked anti-mouse
and anti-rabbit IgG (GE Healthcare, UK).
Analysis of miRNA arrays
Five culture dishes of KMP8 cells were treated with
0.3 μM Gal-9 for 6 h; another five dishes of KMP8 cells were
used as a control without Gal-9 treatment. Those cells were stored
in the RNAprotect reagent (Qiagen, Venlo, The Netherlands). All
samples were processed for total RNA extraction using miRNeasy Mini
kit (Qiagen) according to the manufacturer's instructions. After
the assessment of RNA quantity and quality using the RNA 6000 Nano
kit (Agilent Technologies, Santa Clara, CA, USA), the samples were
labeled using the miRCURY Hy3 Power Labeling kit (Exiqon, Vedbaek,
Denmark) and then hybridized to a human miRNA Oligo chip (v.21;
Toray Industries, Tokyo, Japan). Scanning was conducted using a
3D-Gene Scanner 3000 (Toray Industries). 3D-Gene Extraction version
1.2 software (Toray Industries) was employed to calculate the raw
intensity of the images. To determine the difference in miRNA
expression between the Gal-9-treated and control samples, the raw
data were analyzed using GeneSpring GX 10.0 software (Agilent
Technologies). For the raw data that were above the background
level, quantile normalization was performed. Differentially
expressed miRNAs were determined with the Mann-Whitney U test.
Unsupervised hierarchical clustering was performed on the
normalized data, which consisted of ten observations of the five
treated and five control experiments, using the farthest neighbor
method employing the absolute uncentered Pearson's correlation
coefficient as a metric. A heat map was produced using the relative
expression intensity for each miRNA, in which the base-2 logarithm
of the intensity was median centered for each row.
Statistical analysis
All of the analyses were conducted using GraphPad
Prism software version 6.0 (GraphPad Software, San Diego, CA, USA).
Unpaired t-tests were conducted for comparisons between groups. A
P-value of 0.05 was considered to indicate a significant difference
between the groups.
Results
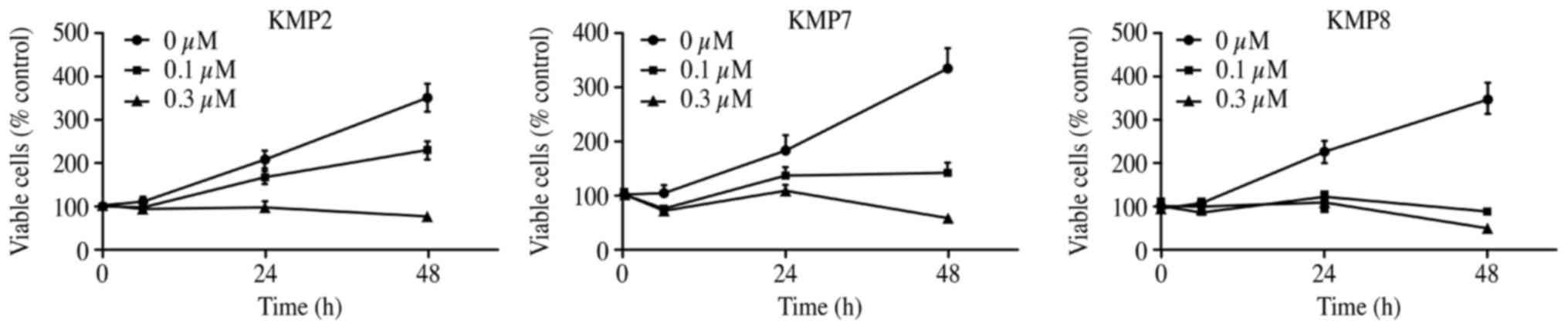
Gal-9 suppresses the proliferation of
human liver metastasis cells (KMP2, KMP7 and KMP8 cells)
To evaluate the effect of Gal-9 on the growth
activity of human liver metastasis cells in vitro, we
examined the effect of Gal-9 on cell proliferation in three liver
metastasis cell lines (KMP2, KMP7 and KMP8 cells) cultured for 48 h
with 0, 0.1 or 0.3 μM Gal-9 in the corresponding medium. We
found that Gal-9 suppressed cell proliferation in the three liver
metastasis cell lines in a dose- and time-dependent manner
(Fig. 1).
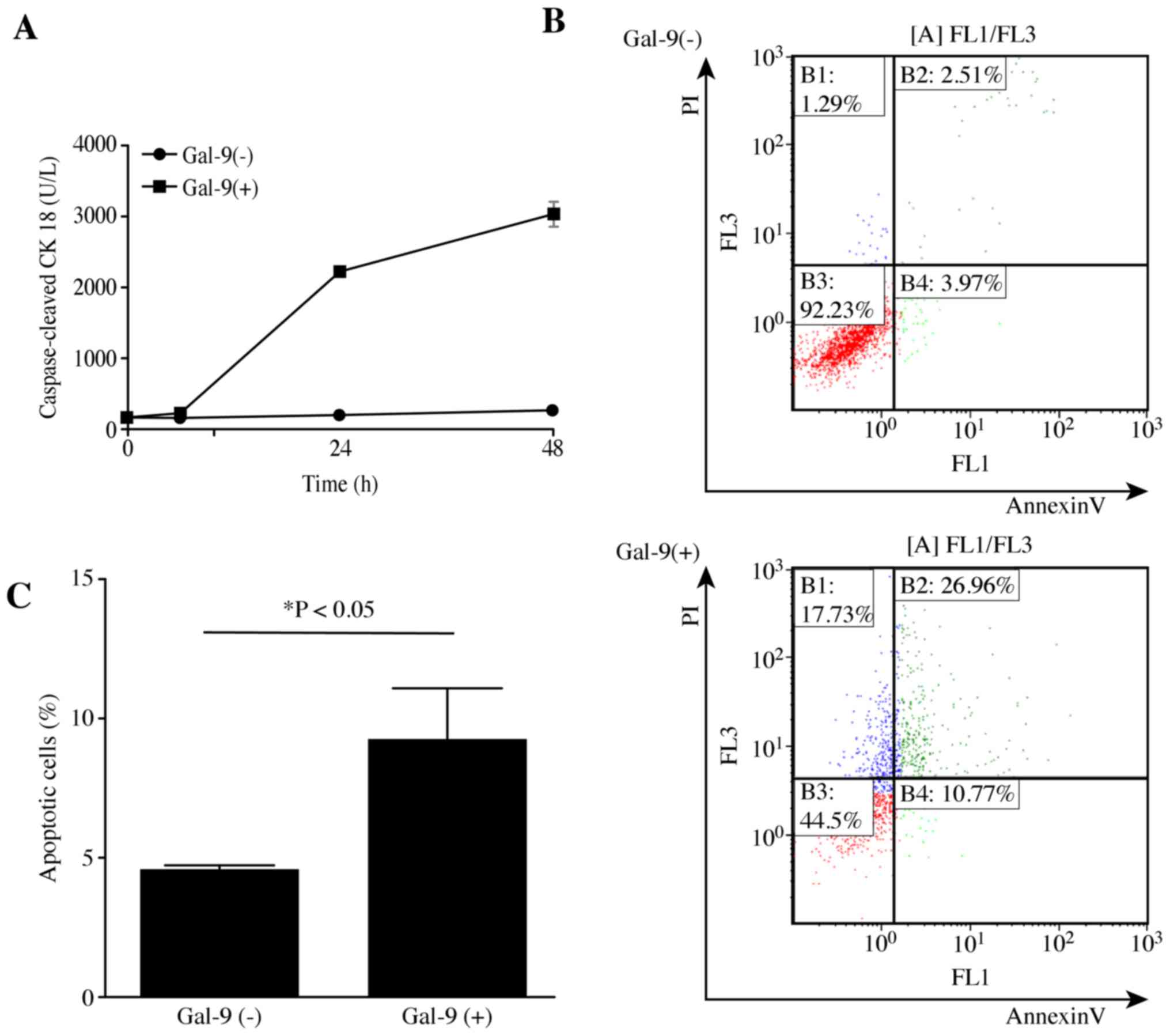
Gal-9 induces apoptosis of liver
metastasis cells
To determine whether Gal-9 induced apoptosis, KMP8
cells were treated with or without 0.3 μM Gal-9, and the
levels of cCK18 following treatment were determined using the M30
ELISA kit. We found that Gal-9 significantly increased the levels
of cCK18 in KMP8 cells in a time-dependent manner (Fig. 2A). The experiments were repeated
three times, and the same results were obtained. Additionally,
Gal-9 induced early apoptosis of KMP8 cells, as determined by
Annexin V-FITC/PI staining and flow cytometry. The different
quadrants in Fig. 2B represent
living cells (lower left quadrant), early apoptotic cells (lower
right quadrant), and late apoptotic cells (upper right quadrant).
As demonstrated by the increased numbers of early-phase apoptotic
cells, the antitumor effects of Gal-9 begin at an early stage, with
a significant difference observed (Fig. 2C).
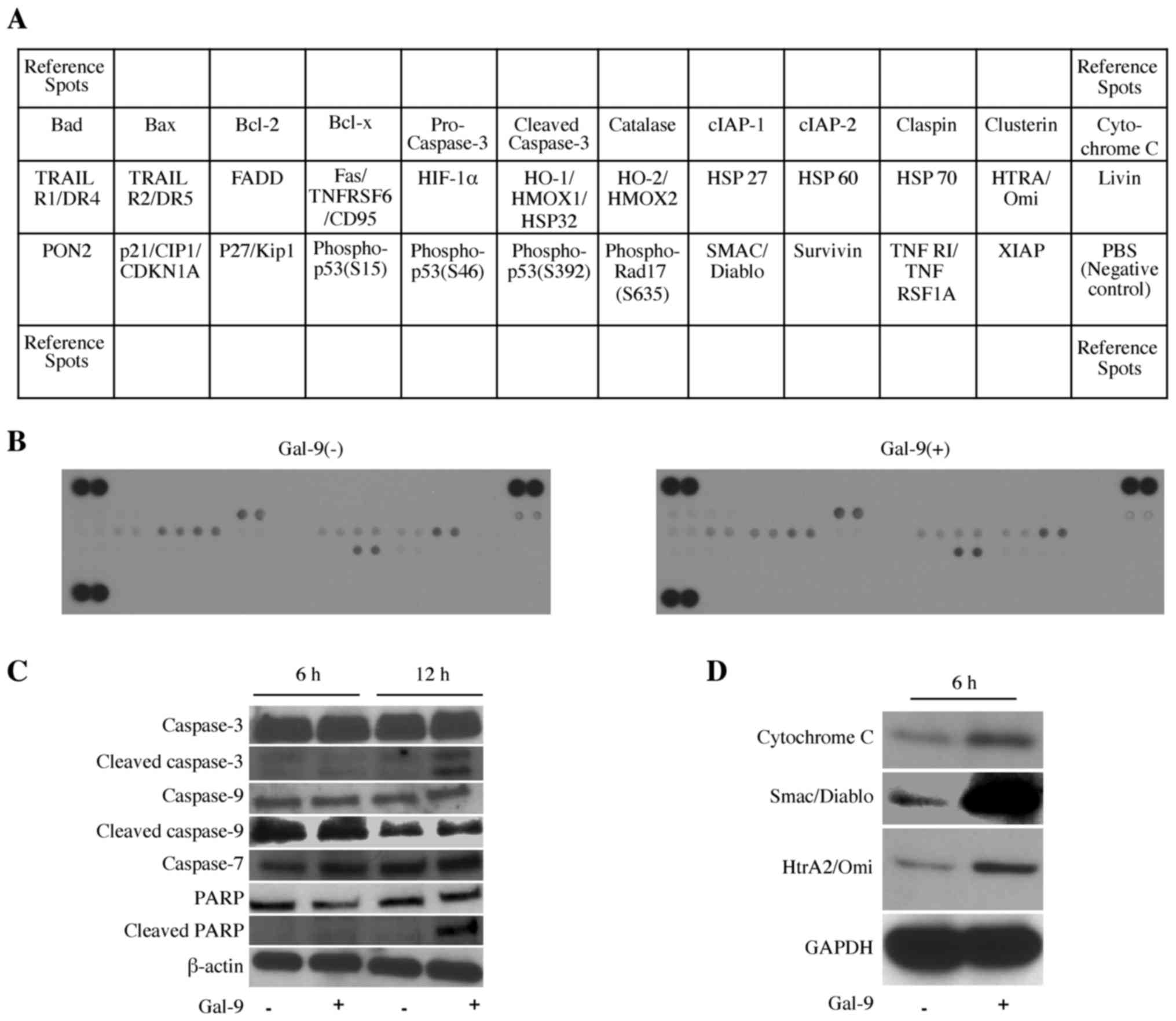
Effects of Gal-9 on the levels of
apoptosis-associated proteins
Next, we used an apoptosis array system to identify
which apoptosis-associated proteins are involved in the antitumor
effects of Gal-9. Using an antibody array enabled screening of the
expression of 35 apoptosis-associated proteins in KMP8 cells in the
presence or absence of Gal-9, but there was no change in this array
system (Fig. 3A and B).
Furthermore, we investigated apoptosis-associated
proteins including caspase-3, -7, -9, PARP, cleaved caspase-3 and
cleaved PARP; Gal-9 activated caspase-7 slightly at 6 h, and
cleaved caspase-3 and cleaved PARP were activated at 12 h after
Gal-9 treatment (Fig. 3C). Next,
we examined the intracellular distribution of the apoptosis-related
molecules. In cytoplasmic fractions, the levels of cytochrome
c, Smac/Diablo and HtrA2 were increased in the Gal-9-treated
group (Fig. 3D). These results
demonstrated potential apoptotic mechanisms, including involvement
of the mitochondrial apoptotic pathway. Thus, Gal-9 suppressed the
proliferation of KMP8 cells by inducing apoptosis via the intrinsic
apoptosis pathway.
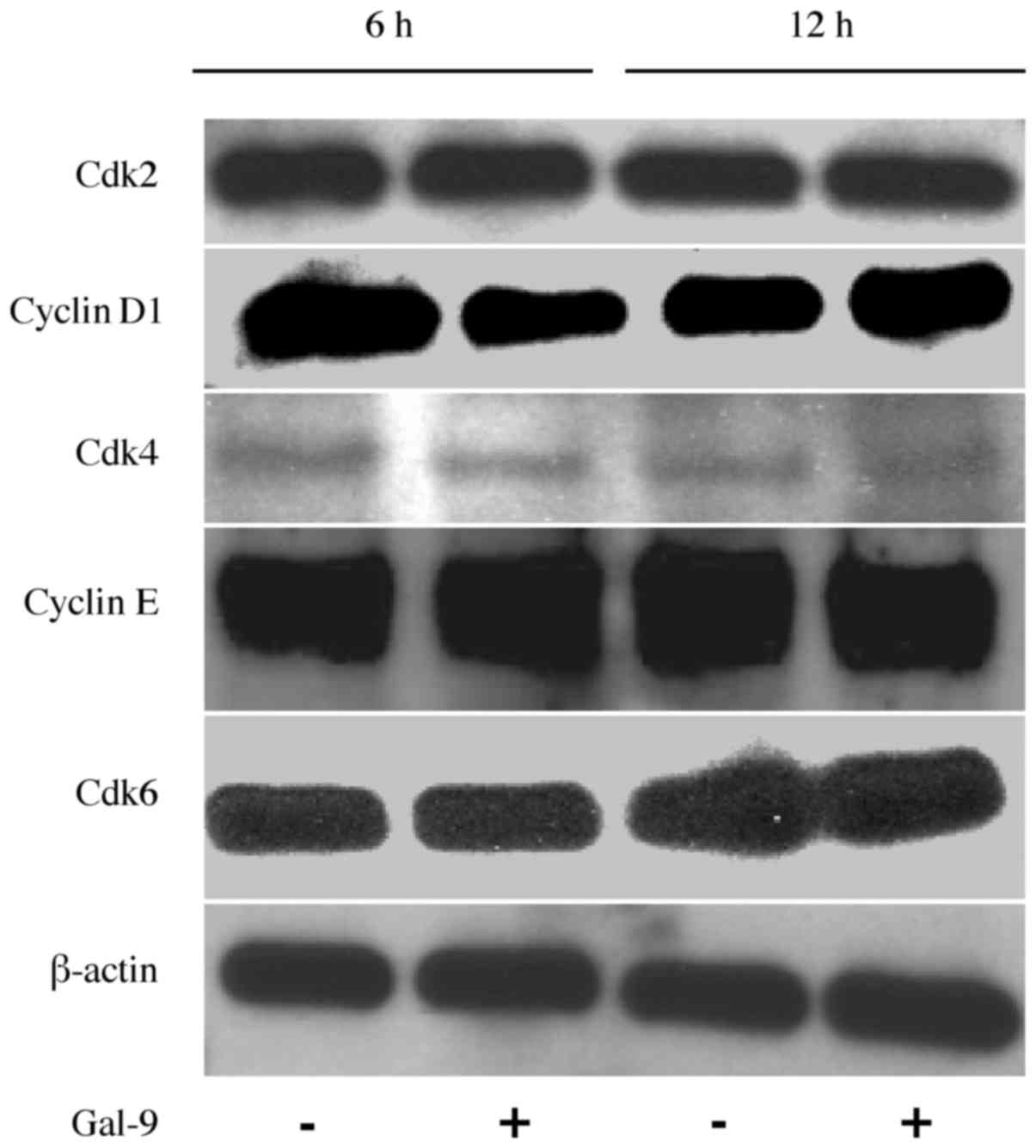
No specific effects of Gal-9 were
observed on cell cycle regulatory proteins in liver metastasis
cells
As Gal-9 suppressed cell proliferation very
significantly, we considered whether there was a route other than
apoptosis induction and investigated the effects of Gal-9 on the
cell cycle. The effects of Gal-9 on the expression of various cell
cycle-related proteins in KMP8 cells were evaluated through western
blotting. Cells were treated with 0 or 0.3 μM Gal-9 for 6-12
h, and we then examined the levels of various cell cycle-related
molecules, such as cyclin D1, Cdk4, Cdk6, cyclin E, and Cdk2.
However, no significant differences in the expression of these cell
cycle-related proteins were detected (Fig. 4). These results suggested that
Gal-9 suppresses the proliferation of KMP8 cells predominantly by
inducing apoptosis, and not by promoting cell cycle arrest.
Effects of Gal-9 on miRNA expression in
KMP8 cells
To further examine the antitumor effect of Gal-9, we
screened the expression levels of miRNAs in KMP8 cells and compared
the miRNA profiles obtained with or without Gal-9 treatment. KMP8
cells were treated with 0 or 0.3 μM Gal-9 for 6 h.
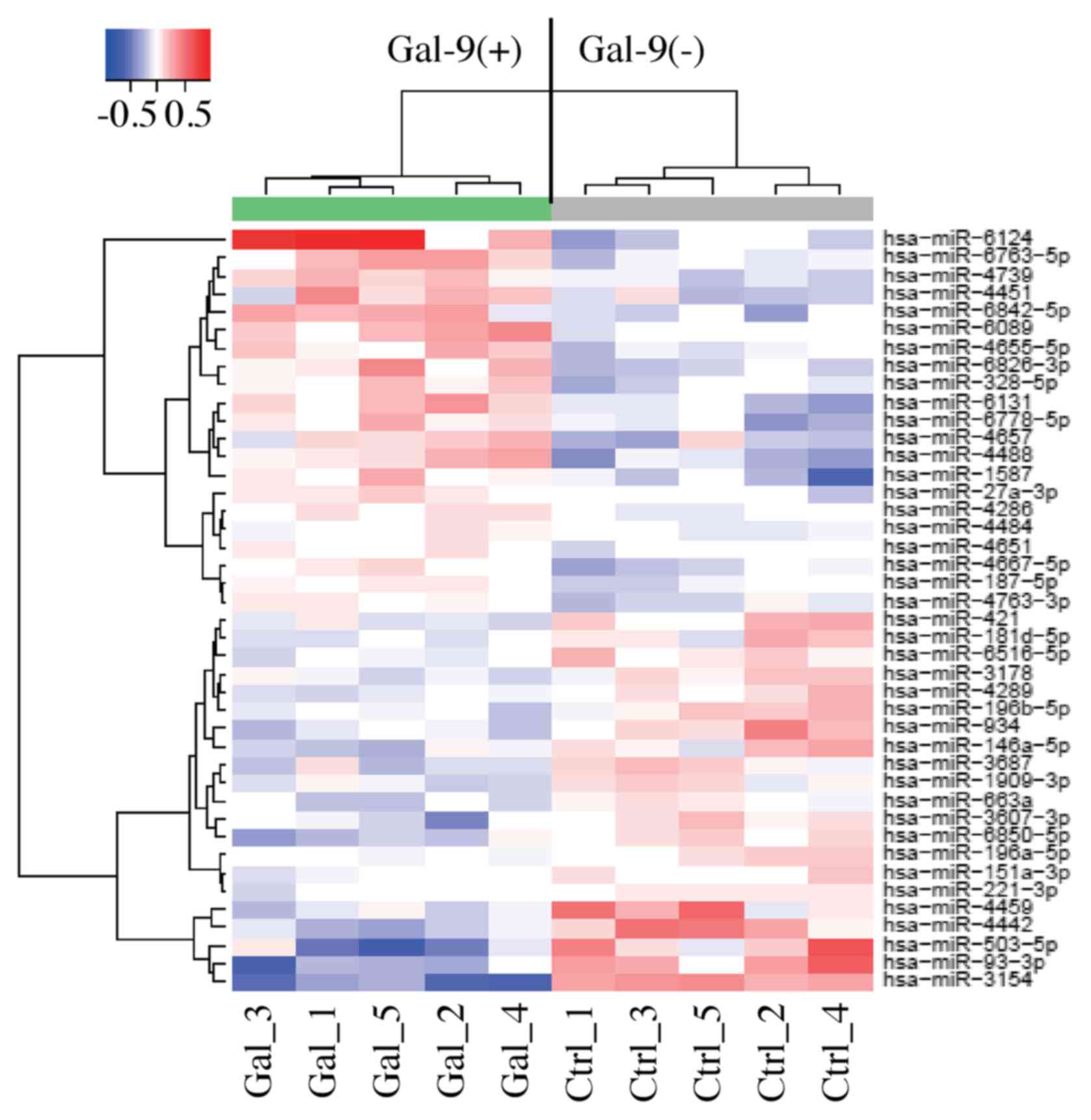
Unsupervised hierarchical clustering analysis showed that the
treated group clustered separately from the control group (Fig. 5). We identified 42 miRNAs that were
differentially expressed between the two groups of KMP8 cells among
the 2,555 miRNAs (21 upregulated miRNAs and 21 down-regulated
miRNAs) (Table I).
 | Table 1Statistical results for miRNAs in
KMP8 cells treated with Gal-9 compared with untreated KMP8
cells. |
Table 1
Statistical results for miRNAs in
KMP8 cells treated with Gal-9 compared with untreated KMP8
cells.
| Upregulated
microRNAs | Fold-change (Gal-9
treated/non-treated) | P-value | Chromosomal
localization |
|---|
| hsa-miR-6124 | 1.70 | 0.0079 | 11 |
| hsa-miR-4488 | 1.30 | 0.0079 | 11 |
|
hsa-miR-6842-5p | 1.25 | 0.0317 | 8 |
| hsa-miR-6131 | 1.25 | 0.0119 | 5 |
|
hsa-miR-6826-3p | 1.23 | 0.0159 | 3 |
| hsa-miR-4451 | 1.22 | 0.0465 | 4 |
| hsa-miR-1587 | 1.21 | 0.0278 | Xp11.4 |
| hsa-miR-4739 | 1.21 | 0.0117 | 17 |
|
hsa-miR-6763-5p | 1.20 | 0.0208 | 12 |
|
hsa-miR-6778-5p | 1.20 | 0.0079 | 17 |
| hsa-miR-4657 | 1.19 | 0.0465 | 7 |
| hsa-miR-6089 | 1.18 | 0.0259 | Xp22.3 |
|
hsa-miR-4655-5p | 1.17 | 0.0079 | 7 |
| hsa-miR-328-5p | 1.16 | 0.0119 | 16q22.1 |
|
hsa-miR-4667-5p | 1.12 | 0.0345 | 9 |
|
hsa-miR-4763-3p | 1.10 | 0.0361 | 22 |
| hsa-miR-187-5p | 1.08 | 0.0269 | 18q12.2 |
| hsa-miR-27a-3p | 1.08 | 0.0238 | 19p13.13 |
| hsa-miR-4286 | 1.07 | 0.0413 | 8 |
| hsa-miR-4651 | 1.05 | 0.0196 | 7 |
| hsa-miR-4484 | 1.05 | 0.0445 | 10 |
|
| Downregulated
microRNAs | Fold-change (Gal-9
treated/non-treated) | P-value | Chromosomal
localization |
|
| hsa-miR-3154 | 0.59 | 0.0119 | 9 |
| hsa-miR-93-3p | 0.67 | 0.0079 | 7q22.1 |
| hsa-miR-503-5p | 0.67 | 0.0317 | Xq26.3 |
| hsa-miR-4442 | 0.72 | 0.0079 | 3 |
| hsa-miR-4459 | 0.77 | 0.0465 | 5 |
| hsa-miR-934 | 0.81 | 0.0079 | Xq26.3 |
|
hsa-miR-6850-5p | 0.84 | 0.0317 | 8 |
|
hsa-miR-146a-5p | 0.85 | 0.0465 | 5q33.3 |
|
hsa-miR-3607-3p | 0.87 | 0.0356 | |
| hsa-miR-3687 | 0.87 | 0.0317 | 21 |
| hsa-miR-421 | 0.87 | 0.0317 | Xq13.2 |
|
hsa-miR-196b-5p | 0.88 | 0.0465 | 7p15.2 |
|
hsa-miR-6516-5p | 0.89 | 0.0119 | 17 |
|
hsa-miR-181d-5p | 0.89 | 0.0459 | 19p13.13 |
| hsa-miR-3178 | 0.90 | 0.0426 | 16 |
|
hsa-miR-1909-3p | 0.90 | 0.0317 | 19p13.3 |
| hsa-miR-4289 | 0.90 | 0.0273 | 9 |
| hsa-miR-663a | 0.91 | 0.0465 | 20p11.1 |
|
hsa-miR-196a-5p | 0.93 | 0.0189 | |
| hsa-miR-221-3p | 0.94 | 0.0118 | Xp11.3 |
|
hsa-miR-151a-3p | 0.94 | 0.0196 | 8q24.3 |
Discussion
The present study revealed that Gal-9 led to dose-
and time-dependent inhibition of cell proliferation in liver
metastasis cell lines. In this study, Gal-9 increased the levels of
cCK18 in a time-dependent manner. A neo-epitope in cytokeratin 18
becomes available upon an early caspase cleavage event during
apoptosis, and the monoclonal antibody M30, which is specific for
this site, can be utilized to specifically detect apoptotic cells,
and not necrotic cells (26). This
assay measures the accumulation of cCK18 in cells; thus, we
investigated the initiation of apoptosis using Annexin V-FITC/PI
staining and flow cytometry. Annexin V-FITC is an early apoptosis
marker, and PI indicates necrotic cells. Staining of cells with
both PI and Annexin V-FITC demonstrates later-stage apoptosis and
early necrosis. Gal-9 induced early apoptosis of KMP8 cells, as
determined by Annexin V-FITC/PI staining and flow cytometry. Next,
we investigated the changes in apoptotic molecules with or without
Gal-9. Cleaved caspase-3, cleaved PARP, and caspase-7, which are
apoptosis-related proteins (27),
were activated by Gal-9. Subsequently, we investigated the
intracellular distribution of apoptosis-related molecules and Gal-9
activated cytochrome c, Smac/Diablo and HtrA2 in cytoplasmic
fractions. Cytochrome c released from damaged mitochondria
can initiate the activation cascade of caspases once it is released
into the cytosol (28). This is a
very early event in the intrinsic apoptosis pathway and contributes
to caspase-9 activation. Furthermore, mitochondria release multiple
pro-apoptotic molecules, such as Smac/Diablo and HtrA2, in addition
to cytochrome c. Smac/Diablo and HtrA2/Omi, which are
mitochondrial proteins that are released together with cytochrome
c, suppress the inhibitor of apoptosis protein (IAP) and
increase caspase activity (29–31).
In contrast, Gal-9 treatment for 6 or 12 h did not affect the G0–G1
transition or the expression levels of cell cycle-related proteins.
These data suggest that Gal-9 suppresses cell proliferation in
liver metastasis cell lines by inducing apoptosis, and not by
promoting cell cycle arrest.
The miRNAs associated with the antitumor effects of
Gal-9 were analyzed using miRNA expression arrays. It has become
apparent that miRNA expression is associated with various cancers.
Our previous studies revealed that miRNAs lead to apoptosis as a
consequence of the antitumor effect of Gal-9 in various
gastrointestinal cancers (18–21).
Hierarchical cluster analyses were performed to clarify the
alteration of the expression of miRNAs due to Gal-9 treatment. We
identified 42 differentially expressed miRNAs (21 upregulated and
21 downregulated) in KMP8 cells that were either subjected to Gal-9
treatment or not. Among the miRNAs downregulated by Gal-9, miR-196a
overexpression has been observed in several types of cancers, and
high expression of miR-196a has been associated with a poor
prognosis in pancreatic cancer (32). miR-221 targets p27 and DNA
damage-inducible transcript 4 (DDIT4), and overexpression of
miR-221 stimulates liver tumorigenesis (33). In addition, miR-93 inhibits the
expression of Programmed cell death 4 (PDCD4), a tumor suppressor
gene, and consequently inhibits apoptosis in gastric cancer cells
(34). These findings suggest that
Gal-9 induces apoptosis by altering the expression of several
miRNAs.
In conclusion, Gal-9 suppresses the cell
proliferation and tumor growth of human liver metastases in
vitro. The antitumor effect of Gal-9 appears to depend on
several pathways, such as the induction of apoptosis in cancer
cells via multiple pro-apoptotic molecules, including Smac/Diablo
and HtrA2, and cytochrome c release from mitochondria.
However, Gal-9 was found to have little or no effect on the cell
cycle in this study and in our previous studies (18–21).
The statistical superiority or non-inferiority of certain
chemotherapy regimens has been demonstrated by phase III trials of
unresectable pancreatic cancer (including distant metastasis
cases), including gemcitabine (35), S-1 (36), oxaliplatin plus irinotecan,
leucovorin, fluorouracil (FOLFIRINOX) (37), and nab-paclitaxel plus gemcitabine
(38). Paclitaxel promotes the
accelerated assembly of excessively stable microtubules and induces
apoptosis, which are the main mechanisms of its antitumor effect
(39). Furthermore, many antitumor
drugs, such as gemcitabine and fluorouracil, have an effect on the
cell cycle (40). These results
suggest that a combination of the apoptosis inducer Gal-9 with
other antitumor drugs that induce cell cycle arrest would be more
effective for cancer therapy.
Thus, Gal-9 may represent a novel therapeutic agent
as an adjunct to conventional chemotherapy for the treatment of
liver metastasis.
Glossary
Abbreviations
Abbreviations:
|
Gal-9
|
Galectin-9
|
|
CCK-8
|
cell counting kit-8
|
|
cCK18
|
caspase-cleaved cytokeratin 18
|
|
FITC
|
fluorescein isothiocyanate
|
|
PI
|
propidium iodide
|
|
miRNA
|
microRNA
|
Acknowledgments
We thank Ms. Kayo Hirose, Ms. Keiko Fujikawa, Ms.
Megumi Okamura, Ms. Fuyuko Kokado and Ms. Miwako Watanabe for
providing technical assistance.
References
|
1
|
Qian Y, Sang Y, Wang FX, Hong B, Wang Q,
Zhou X, Weng T, Wu Z, Zheng M, Zhang H, et al: Prognostic
significance of B7-H4 expression in matched primary pancreatic
cancer and liver metastases. Oncotarget. 7:72242–72249.
2016.PubMed/NCBI
|
|
2
|
Takamori H, Hiraoka T, Kanemitsu K, Tsuji
T, Hamada C and Baba H: Identification of prognostic factors
associated with early mortality after surgical resection for
pancreatic cancer - under-analysis of cumulative survival curve.
World J Surg. 30:213–218. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miyanishi N, Nishi N, Abe H, Kashio Y,
Shinonaga R, Nakakita S, Sumiyoshi W, Yamauchi A, Nakamura T,
Hirashima M, et al: Carbohydrate-recognition domains of galectin-9
are involved in intermolecular interaction with galectin-9 itself
and other members of the galectin family. Glycobiology. 17:423–432.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Matsumoto R, Matsumoto H, Seki M, Hata M,
Asano Y, Kanegasaki S, Stevens RL and Hirashima M: Human ecalectin,
a variant of human galectin-9, is a novel eosinophil
chemoattractant produced by T lymphocytes. J Biol Chem.
273:16976–16984. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hirashima M, Kashio Y, Nishi N, Yamauchi
A, Imaizumi TA, Kageshita T, Saita N and Nakamura T: Galectin-9 in
physiological and pathological conditions. Glycoconj J. 19:593–600.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Seki M, Oomizu S, Sakata KM, Sakata A,
Arikawa T, Watanabe K, Ito K, Takeshita K, Niki T, Saita N, et al:
Galectin-9 suppresses the generation of Th17, promotes the
induction of regulatory T cells, and regulates experimental
autoimmune arthritis. Clin Immunol. 127:78–88. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Niki T, Tsutsui S, Hirose S, Aradono S,
Sugimoto Y, Takeshita K, Nishi N and Hirashima M: Galectin-9 is a
high affinity IgE-binding lectin with anti-allergic effect by
blocking IgE-antigen complex formation. J Biol Chem.
284:32344–32352. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu C, Anderson AC, Schubart A, Xiong H,
Imitola J, Khoury SJ, Zheng XX, Strom TB and Kuchroo VK: The Tim-3
ligand galectin-9 negatively regulates T helper type 1 immunity.
Nat Immunol. 6:1245–1252. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Oomizu S, Arikawa T, Niki T, Kadowaki T,
Ueno M, Nishi N, Yamauchi A and Hirashima M: Galectin-9 suppresses
Th17 cell development in an IL-2-dependent but Tim-3-independent
manner. Clin Immunol. 143:51–58. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wiersma VR, de Bruyn M, Helfrich W and
Bremer E: Therapeutic potential of Galectin-9 in human disease. Med
Res Rev. 33(Suppl 1): E102–E126. 2013. View Article : Google Scholar
|
|
11
|
Fujihara S, Mori H, Kobara H, Rafiq K,
Niki T, Hirashima M and Masaki T: Galectin-9 in cancer therapy.
Recent Pat Endocr Metab Immune Drug Discov. 7:130–137. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kageshita T, Kashio Y, Yamauchi A, Seki M,
Abedin MJ, Nishi N, Shoji H, Nakamura T, Ono T and Hirashima M:
Possible role of galectin-9 in cell aggregation and apoptosis of
human melanoma cell lines and its clinical significance. Int J
Cancer. 99:809–816. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kuroda J, Yamamoto M, Nagoshi H, Kobayashi
T, Sasaki N, Shimura Y, Horiike S, Kimura S, Yamauchi A, Hirashima
M, et al: Targeting activating transcription factor 3 by Galectin-9
induces apoptosis and overcomes various types of treatment
resistance in chronic myelogenous leukemia. Mol Cancer Res.
8:994–1001. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kobayashi T, Kuroda J, Ashihara E, Oomizu
S, Terui Y, Taniyama A, Adachi S, Takagi T, Yamamoto M, Sasaki N,
et al: Galectin-9 exhibits anti-myeloma activity through JNK and
p38 MAP kinase pathways. Leukemia. 24:843–850. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wiersma VR, de Bruyn M, Wei Y, van Ginkel
RJ, Hirashima M, Niki T, Nishi N, Zhou J, Pouwels SD, Samplonius
DF, et al: The epithelial polarity regulator LGALS9/galectin-9
induces fatal frustrated autophagy in KRAS mutant colon carcinoma
that depends on elevated basal autophagic flux. Autophagy.
11:1373–1388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Irie A, Yamauchi A, Kontani K, Kihara M,
Liu D, Shirato Y, Seki M, Nishi N, Nakamura T, Yokomise H, et al:
Galectin-9 as a prognostic factor with antimetastatic potential in
breast cancer. Clin Cancer Res. 11:2962–2968. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang ZY, Dong JH, Chen YW, Wang XQ, Li
CH, Wang J, Wang GQ, Li HL and Wang XD: Galectin-9 acts as a
prognostic factor with antimetastatic potential in hepatocellular
carcinoma. Asian Pac J Cancer Prev. 13:2503–2509. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fujita K, Iwama H, Sakamoto T, Okura R,
Kobayashi K, Takano J, Katsura A, Tatsuta M, Maeda E, Mimura S, et
al: Galectin-9 suppresses the growth of hepatocellular carcinoma
via apoptosis in vitro and in vivo. Int J Oncol. 46:2419–2430.
2015.PubMed/NCBI
|
|
19
|
Kobayashi K, Morishita A, Iwama H, Fujita
K, Okura R, Fujihara S, Yamashita T, Fujimori T, Kato K, Kamada H,
et al: Galectin-9 suppresses cholangiocarcinoma cell proliferation
by inducing apoptosis but not cell cycle arrest. Oncol Rep.
34:1761–1770. 2015.PubMed/NCBI
|
|
20
|
Tadokoro T, Morishita A, Fujihara S, Iwama
H, Niki T, Fujita K, Akashi E, Mimura S, Oura K, Sakamoto T, et al:
Galectin-9: An anticancer molecule for gallbladder carcinoma. Int J
Oncol. 48:1165–1174. 2016.PubMed/NCBI
|
|
21
|
Takano J, Morishita A, Fujihara S, Iwama
H, Kokado F, Fujikawa K, Fujita K, Chiyo T, Tadokoro T, Sakamoto T,
et al: Galectin-9 suppresses the proliferation of gastric cancer
cells in vitro. Oncol Rep. 35:851–860. 2016.PubMed/NCBI
|
|
22
|
Zamore PD and Haley B: Ribo-gnome: The big
world of small RNAs. Science. 309:1519–1524. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Morishita A and Masaki T: miRNA in
hepatocellular carcinoma. Hepatol Res. 45:128–141. 2015. View Article : Google Scholar
|
|
24
|
Nishi N, Itoh A, Fujiyama A, Yoshida N,
Araya S, Hirashima M, Shoji H and Nakamura T: Development of highly
stable galectins: Truncation of the linker peptide confers
protease-resistance on tandem-repeat type galectins. FEBS Lett.
579:2058–2064. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schutte B, Henfling M, Kölgen W, Bouman M,
Meex S, Leers MP, Nap M, Björklund V, Björklund P, Björklund B, et
al: Keratin 8/18 breakdown and reorganization during apoptosis. Exp
Cell Res. 297:11–26. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Leers MP, Kölgen W, Björklund V, Bergman
T, Tribbick G, Persson B, Björklund P, Ramaekers FC, Björklund B,
Nap M, et al: Immunocytochemical detection and mapping of a
cytokeratin 18 neo-epitope exposed during early apoptosis. J
Pathol. 187:567–572. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cohen GM: Caspases: The executioners of
apoptosis. Biochem J. 326:1–16. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cai J, Yang J and Jones DP: Mitochondrial
control of apoptosis: The role of cytochrome c. Biochim Biophys
Acta. 1366:139–149. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Du C, Fang M, Li Y, Li L and Wang X: Smac,
a mitochondrial protein that promotes cytochrome c-dependent
caspase activation by eliminating IAP inhibition. Cell. 102:33–42.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Verhagen AM, Ekert PG, Pakusch M, Silke J,
Connolly LM, Reid GE, Moritz RL, Simpson RJ and Vaux DL:
Identification of DIABLO, a mammalian protein that promotes
apoptosis by binding to and antagonizing IAP proteins. Cell.
102:43–53. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Suzuki Y, Imai Y, Nakayama H, Takahashi K,
Takio K and Takahashi R: A serine protease, HtrA2, is released from
the mitochondria and interacts with XIAP, inducing cell death. Mol
Cell. 8:613–621. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang J, Chen J, Chang P, Leblanc A, Li D,
Abbruzzesse JL, Frazier ML, Killary AM and Sen S: MicroRNAs in
plasma of pancreatic ductal adenocarcinoma patients as novel
blood-based biomarkers of disease. Cancer Prev Res (Phila).
2:807–813. 2009. View Article : Google Scholar
|
|
33
|
Pineau P, Volinia S, McJunkin K, Marchio
A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM and
Dejean A: miR-221 overexpression contributes to liver
tumorigenesis. Proc Natl Acad Sci USA. 107:264–269. 2010.
View Article : Google Scholar :
|
|
34
|
Liang H, Wang F, Chu D, Zhang W, Liao Z,
Fu Z, Yan X, Zhu H, Guo W, Zhang Y, et al: miR-93 functions as an
oncomiR for the downregulation of PDCD4 in gastric carcinoma. Sci
Rep. 6:237722016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Burris HA III, Moore MJ, Andersen J, Green
MR, Rothenberg ML, Modiano MR, Cripps MC, Portenoy RK, Storniolo
AM, Tarassoff P, et al: Improvements in survival and clinical
benefit with gemcitabine as first-line therapy for patients with
advanced pancreas cancer: A randomized trial. J Clin Oncol.
15:2403–2413. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ueno H, Ioka T, Ikeda M, Ohkawa S,
Yanagimoto H, Boku N, Fukutomi A, Sugimori K, Baba H, Yamao K, et
al: Randomized phase III study of gemcitabine plus S-1, S-1 alone,
or gemcitabine alone in patients with locally advanced and
metastatic pancreatic cancer in Japan and Taiwan: GEST study. J
Clin Oncol. 31:1640–1648. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Conroy T, Desseigne F, Ychou M, Bouché O,
Guimbaud R, Bécouarn Y, Adenis A, Raoul JL, Gourgou-Bourgade S, de
la Fouchardière C, et al Groupe Tumeurs Digestives of Unicancer;
PRODIGE Intergroup: FOLFIRINOX versus gemcitabine for metastatic
pancreatic cancer. N Engl J Med. 364:1817–1825. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Von Hoff DD, Ervin T, Arena FP, Chiorean
EG, Infante J, Moore M, Seay T, Tjulandin SA, Ma WW, Saleh MN, et
al: Increased survival in pancreatic cancer with nab-paclitaxel
plus gemcitabine. N Engl J Med. 369:1691–1703. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Milross CG, Mason KA, Hunter NR, Chung WK,
Peters LJ and Milas L: Relationship of mitotic arrest and apoptosis
to antitumor effect of paclitaxel. J Natl Cancer Inst.
88:1308–1314. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Toyota Y, Iwama H, Kato K, Tani J, Katsura
A, Miyata M, Fujiwara S, Fujita K, Sakamoto T, Fujimori T, et al:
Mechanism of gemcitabine-induced suppression of human
cholangiocellular carcinoma cell growth. Int J Oncol. 47:1293–1302.
2015.PubMed/NCBI
|