Introduction
Head and neck squamous cell carcinoma (HNSCC) is the
sixth most common group of malignancies worldwide, and is generally
classified into four independent types of cancer (1). Hypopharyngeal squamous cell carcinoma
(HSCC), which accounts for ~3–5% of all HNSCCs (2), has high rates of recurrence and poor
survival rates, and is regarded as the most malignant form of HNSCC
(3,4). At the time of diagnosis, ~80% of HSCC
patients are at an advanced stage of disease. However, there is
only limited understanding of the underlying molecular mechanisms
that lead to a highly malignant phenotype and ultimately result in
the unfavorable prognosis in HSCC patients. Thus, it is urgent to
investigate the pathogenesis of HSCC, to identify new biomarkers
and explore innovative treatment strategies.
Tumor progression is a multistep process that
involves multiple genes, including the inactivation of tumor
suppressor genes and the activation of oncogenes (5). Two important features that are
directly related to the severity of tumors, including HSCC, are
unlimited cellular proliferation and tumor metastasis, which can
occur due to the aberrant expression of key regulators controlling
cell proliferation, survival and motility (6). Forkhead box M1 (FoxM1), belongs to
the Fox transcription factor family, which are characterized by the
presence of a 'Forkhead box' or 'winged helix' DNA-binding domain.
FoxM1 acts as a key regulator of the cell cycle by influencing the
phase transitions from G1 to S and G2 to M (7,8). In
addition, a previous study showed that FoxM1 acts as a regulator of
a wide range of other biological processes, including apoptosis,
migration and angiogenesis (9). It
has been reported that a variety of tumors, such as gastric
(10), non-small cell lung cancer
(NSCLC) (11) and nasopharyngeal
carcinoma (NPC) (12), exhibit
increased expression of FoxM1. In addition, the elevated expression
of FoxM1 is reported to be closely associated with poor prognosis
in patients with certain types of malignant tumors, and is regarded
as an independent predictor of poor survival in various solid
tumors (13–15). Additionally, it has been shown that
decreased expression of FoxM1 leads to a reduction in the
proliferation of breast tumor (16) and leukemia cells (17), and increased apoptosis of laryngeal
squamous cell carcinoma (LSCC) cells (18). Crosstalk between the FoxM1/Cav-1
axis and the epithelial-mesenchymal transition (EMT) was
demonstrated to be a critical molecular mechanism in regulating the
metastasis of pancreatic cancer (19). Furthermore, Xue et al
(20) have highlighted the
critical interaction of FoxM1 and SMAD3 for controlling TGF-β
signaling during breast cancer metastasis. Collectively, these
findings suggest that FoxM1 may act as a promising therapeutic
target in numerous types of human malignancies. To the best of our
knowledge, there have so far been no reports on the expression and
functional role of FoxM1 in HSCC.
The aim of the present study was to clarify the
expression of FoxM1 in HSCC tissues, to determine the clinical
significance of FoxM1 in primary HSCC, and to evaluate the
relationship between the FoxM1 expression and the prognosis of HSCC
patients. In addition, to assess the function of FoxM1 in HSCC cell
lines, we examined the effects of siRNA-mediated FoxM1 suppression
on the proliferation, apoptosis, migration and EMT in the human
HSCC cell line Fadu in vitro.
Materials and methods
Patients and tissue samples
A total of 7 fresh primary HSCC tumor tissues and 2
adjacent normal tissue samples were resected during surgery at the
Affiliated Hospital of Nantong University (Nantong, China) between
March 2015 and May 2016. None of the patients had received
treatment prior to surgery. In addition, we retrospectively
collected biopsy samples from 63 HSCC patients with complete
clinical and pathological data and who had received primary
treatment in our hospital between August 2009 and August 2016,
during the same period, 20 adjacent normal tissue specimens were
also collected. The detailed clinical characteristics of the 63
HSCC patients are summarized in Table
I. Paraffin-embedded tissue blocks were obtained and sectioned
in the Department of Pathology. The follow-up time ranged from 3 to
71 months, with a median time of 22 months. The overall survival
(OS) time was calculated from the date of surgery to the date of
death or last follow-up. The disease-free survival (DFS) time was
calculated from the date of surgery to the date of recurrence or
last follow-up. The last follow-up of these patients was in
November 2016. This study was approved by the ethics committee of
the Affiliated Hospital of Nantong University and all patients
provided written informed consent. The pathological samples were
obtained from the surgically resected tissue specimens to avoid
disadvantaging the health or prognosis of the patients, and the
privacy of the patients' personal information was maintained.
 | Table IThe relationship between FoxM1
expression and clinicopathological factors in HSCC. |
Table I
The relationship between FoxM1
expression and clinicopathological factors in HSCC.
| Clinicopathological
features | No. of cases | FoxM1
expression | P-value |
|---|
| Age (years) | | | 0.851 |
| <60 | 23 | 7.39±2.78 | |
| ≥60 | 40 | 7.54±3.21 | |
| Sex | | | 0.321 |
| Male | 3 | 5.00±3.46 | |
| Female | 60 | 7.61±2.99 | |
|
Differentiation | | | 0.004a |
| High
differentiation | 30 | 6.37±2.67 | |
| Poor
differentiation | 33 | 8.51±3.02 | |
| Tumor size
(cm) | | | 0.002a |
| ≤2 | 25 | 6.08±2.70 | |
| >2 | 38 | 8.41±2.92 | |
| Clinical stage | | | 0.001a |
| I–II | 20 | 5.65±2.73 | |
| III–IV | 43 | 8.34±2.81 | |
| Lymph node
metastasis | | | 0.002a |
| Negative | 30 | 6.30±2.84 | |
| Positive | 33 | 8.57±2.84 | |
| Treatment | | | 0.045b |
| Surgery only | 29 | 6.64±2.91 | |
| Surgery and
radiation | 19 | 7.74±2.71 | |
| Surgery,
radiation, chemotherapy | 12 | 8.18±3.42 | |
| Other
treatment | 3 | 11.33±1.15 | |
| Ki-67
expression | | | <0.001a |
| Low
expression | 26 | 5.35±2.01 | |
| High
expression | 37 | 8.99±2.73 | |
Immunohistochemistry (IHC)
Formalin-fixed and paraffin-embedded sections with a
thickness of 4 µm were dried at 60°C for 8 h, followed by
dewaxing in xylene, dehydration in a graded alcohol series, and
washing in double-distilled water. After pretreatment in sodium
citrate buffer (pH 6.0, 30 min, at 100°C) to facilitate antigen
retrieval, followed by natural cooling, endogenous peroxidase
activity was blocked with 0.3% H2O2
(ZSGB-Bio, Beijing, China) for 15 min at room temperature away from
light. Subsequently, the slides were incubated with 10% normal goat
serum for 40 min. Primary antibodies against FoxM1 (1:100; Bioworld
Technology, Inc., Nanjing, China) and Ki-67 (1:100; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) were applied overnight in a
moist chamber at 4°C. After 20 h, the slides were washed three
times with phosphate-buffered saline (PBS), then were incubated
with a 2-step Plus Poly-HRP Anti-Mouse/Rabbit IgG detection system
(ZSGB-Bio) at room temperature for 30 min. 3,3′-diaminobenzidine
(DAB) (ZSGB-Bio) was used for the visualization of
immunoreactivity, which appeared as a pale brown color. Nuclei were
stained blue with hematoxylin.
Immunostained tissue sections were reviewed and
scored independently by two pathologists without knowledge of the
patient characteristics. The percentage of staining and the
staining intensity were recorded as follows: 0 (0–4%), 1 (5–25%), 2
(26–50%), 3 (51–75%) and 4 (76–100%); and 0 (negative), 1 (weak), 2
(moderate) and 3 (strong). The products of these two scores were
used as the final staining scores (ranging from 0 to 12). According
to the final score, the expression levels of FoxM1 or Ki-67 were
categorized as low (<6) or high (≥6).
Cell culture and siRNA transfection
The HSCC cell line Fadu was purchased from the ATCC
(Shanghai, China) and cultured in Dulbecco's modified Eagle's
medium (DMEM; GE Healthcare Life Sciences-HyClone Laboratories,
Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS;
Gibco-Thermo Fisher Scientific, Cambridge, MA, USA), in a
humidified incubator with 5% CO2 at 37°C.
Small interfering RNAs (siRNAs) targeting FoxM1 and
a control siRNA were obtained from Biomics Biotechnologies, Co.,
Ltd. (Nantong, China). Fadu cells were placed in 96- or 6-well
plates in complete DMEM overnight. When the cells reached ~50%
confluence, they were transfected with the siRNAs using
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer's instructions. After 6 h of transfection, the medium
containing siRNAs and Lipofectamine 2000 was replaced with 100
µl (96-well plates) or 2 ml (6-well plates) of complete
DMEM. RNAs and proteins were obtained at 48 and 72 h after
transfection, respectively. The sequences of FoxM1 siRNAs
(si-FoxM1) were as follows: #1, 5′-GGAAAUGCUUGUGAUUCAAdTdT-3′
(sense) and 5′-UUGAAUCACAAGCAUUUCCdTdT-3′ (antisense); #2,
5′-GGAUGUGAAUCUUCCUAGAdTdT-3′ (sense) and
5′-UCUAGGAAGAUUCACAUCCdTdT-3′ (antisense); #3,
5′-CCAACAGGAGUCUAAUCAdTdT-3′ (sense) and
5′-UUGAUUAGACUCCUGUUGGdTdT-3′ (antisense); and #4,
5′-GGAUUUCAGCCCAGUACAAdTdT-3′ (sense) and
5′-UUGUACUGGGUGAAAUCCdTdT-3′ (antisense). The sequences of the
negative control siRNA (si-NC) were 5′-UUCUCCGAACGUGUCACGUdTdT-3′
(sense) and 5′-ACGUGACACGUUCGGAGAAdTdT-3′ (antisense).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cultured cells using
TRIzol® reagent (Vazyme, Nanjing, China) and reverse
transcribed into complementary DNA (cDNA) using a cDNA synthesis
kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the
manufacturer's instructions. qPCR was performed using the
AceQ® qPCR SYBR Green-Master Mix kit (Vazyme). The
primer sequences were as follows: Human GAPDH,
5′-GAAGGTGAAGGTCGGAGTC-3′ (forward) and 5′-GAAGATGGTGATGGGATTTC-3′
(reverse); and FoxM1, 5′-CAGACTATCAAGGAGGAAG-3′ (forward) and
5′-CCAGGAGTGAGATGATTC-3′ (reverse). The qPCR conditions were 95°C
for 10 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 30
sec and 72°C for 30 sec. The levels of FoxM1 mRNA were quantified
using the 2−ΔΔCT method and normalized against the
levels of GAPDH.
Protein extraction and western blot
analysis
Tissue and cell samples were lysed in lysis buffer
(PMSF: RIPA, 1:100; Beyotime Institute of Biotechnology, Haimen,
China). Protein concentrations were quantified using the
bicinchoninic acid (BCA) protein assay method, and 20 µg
protein samples were separated by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
electroblotted onto PVDF membranes (Millipore, Bedford, MA, USA).
After blocking in 5% skimmed milk in Tris-buffered saline Tween-20
(TBST) for 2 h, the membranes were incubated with the primary
antibodies overnight at 4°C [anti-FoxM1 (1:1,000; Bioworld
Technology), anti-GAPDH (1:8,000; Abways Technology, Wynne, AR,
USA), anti-proliferating cell nuclear antigen (PCNA) (#ab2426,
1:2,000; Abcam), anti-cyclin A1 (#sc-751; 1:300; Santa Cruz
Biotechnology), anti-E-cadherin (#RLT1453, 1:300; Suzhou Ruiying
Biological Technology, Co., Ltd., Jiaozuo, China) and anti-vimentin
(#RLT4879, 1:300; Suzhou Ruiying Biological Technology)], followed
by incubation for 1 h with the corresponding secondary antibody
(1:1,000) at room temperature. An ECL Plus kit (ZSbio, Beijing,
China) was used to detect the immunoreactive bands.
Cell Counting kit-8 (CCK-8) cell
proliferation assay
Fadu cells were seeded into a 96-well plate at a
density of 5×103 cells/well and cultured in complete
medium overnight. Subsequently, the cells were transfected with
FoxM1-siRNAs or control siRNA. After 6 h of transfection, the
medium was replaced with 100 µl of complete DMEM containing
10 µl CCK-8 solution (CCK-8 kit; Beyotime Institute of
Biotechnology) and the cells were incubated for 1.5 h. The end of
this incubation was defined as 0 h, and other subsequent
time-points (6, 12, 24, 48 and 72 h) were also analyzed. The
optical density (OD) value of each well was measured at 450 nm
using a microplate reader (Hitachi, Ltd., Tokyo, Japan).
Flow cytometric analyses of the cell
cycle and cell apoptosis
Cells were serum-deprived for 72 h, followed by
incubation with complete medium for 6, 12, 24 and 36 h; cells were
collected at every time-point. The cells were then seeded into a
6-well plate and cultured overnight, then transfected with si-FoxM1
or si-NC. After 48 h, the cells were collected and fixed in 70%
ice-cold ethylalcohol (precooled at 4°C) for at least 24 h at
−20°C. Following centrifugation, the cells were washed with
ice-cold PBS three times to remove the fixation fluid.
Subsequently, with the addition of 1 ng/ml RNaseA, the cells were
incubated in the dark for 20 min at 4°C. Propidium iodide (PI;
Becton-Dickinson, San Jose, CA, USA) was then added for staining.
Finally, cell cycle distribution was analyzed with a FACSCalibur
flow cytometer (Becton-Dickinson) and CellQuest acquisition and
analysis programs (Becton-Dickinson). Each experiment was performed
in triplicate.
For the analysis of apoptosis, cells were
double-stained with an Annexin V-FITC/PI apoptosis detection kit
(BBI) according to the manufacturer's protocol and analyzed using
the FACSCalibur flow cytometer.
Wound healing assay
Fadu cells were seeded into a 6-well plate and
transfected with si-FoxM1 or si-NC. After 24 h, cell confluence
reached 90% and wounds were created by scraping the cells with a
100 µl pipette tip. Subsequently, 1X PBS was used to remove
the free-floating cells and serum-free medium was added to the
6-well plate. After 48 h, a microscope (Olympus, Tokyo, Japan) was
used to observe the migrated distance of the cells. Duplicate wells
of the same treatment groups were examined.
Immunofluorescence analysis
The expression of FoxM1, EMT markers and β-tubulin
were observed by immunostaining and imaging. After transfection
with si-FoxM1 or si-NC for 48 h, cells were seeded on coverslips,
which were pre-placed in a 24-well plate and fixed with 4%
paraformaldehyde after 24 h adherent growth. After 40 min, cells
were washed with 1X PBS and blocked in Immunol Staining Blocking
Buffer (#P0102; Beyotime Institute of Biotechnology) for 2 h at
room temperature. Cells were incubated with primary antibodies
against FoxM1 (1:100; Bioworld Technology), cyclin A1 (#sc-751,
1:100; Santa Cruz Biotechnology), PCNA (#ab2426, 1:100; Abcam),
E-cadherin (#RLT1453, 1:100; Suzhou Ruiying Biological Technology)
vimentin (#RLT4879, 1:100; Suzhou Ruiying Biological Technologyl)
and β-tubulin (#RLM3030, 1:100; Suzhou Ruiying Biological
Technology) for 20 h, and then incubated with Alexa
Fluor-conjugated secondary antibodies (1:1000; Invitrogen/Thermo
Fisher Scientific) and Hoechst stain (Sigma-Aldrich, St. Louis, MO,
USA) for 2 h at room temperature in the dark. The images were
viewed and recorded with a fluorescence microscope (Leica
Microsystems, Wetzlar, Germany).
Immunocytochemical staining
Cells were seeded on coverslips that were pre-placed
in a 24-well plate and treated with si-FoxM1or si-NC for 72 h. The
cells were then fixed with 4% paraformaldehyde for 40 min and
incubated with 4% BSA for 2 h, followed by incubation with primary
antibodies against FoxM1 (1:100; Bioworld Technology), E-cadherin
(#RLT1453, 1:100; Ruiying Biological) and vimentin (#RLT4879,
1:100; Ruiying Biological) overnight at 4°C. Cells were washed with
PBS for 15 min (3×5 min) and then incubated with the 2-step Plus
Poly-HRP Anti-Mouse/Rabbit IgG detection system. Subsequently,
after rinsing with PBS, the cells were stained with DAB and
hematoxylin solution was used to stain the nuclei. The cells were
imaged and analyzed under a microscope (Leica Microsystems).
Statistical analysis
All statistical analyses were performed using the
SPSS software, version 22.0 (IBM Corp., Armonk, NY, USA). Student's
t-test or ANOVA were performed to analyze the associations between
FoxM1 expression and the clinicopathological features of the HSCC
patients, as well as the results of the in vitro
experiments, while a Pearson's correlation analysis was performed
to evaluate the correlation between FoxM1 expression and Ki-67
expression in HSCC patients. Kaplan-Meier analysis was used to
estimate overall or recurrence-free survival times; the differences
between the survival curves were analyzed using the log-rank test.
Univariate and multivariate analyses were performed using the Cox
proportional hazards model. In all analyses, P<0.05 was
considered to indicate a statistically significant result.
Continuous data are presented as the mean ± standard deviation.
Results
FoxM1 is highly expressed in HSCC and is
associated with tumor progression
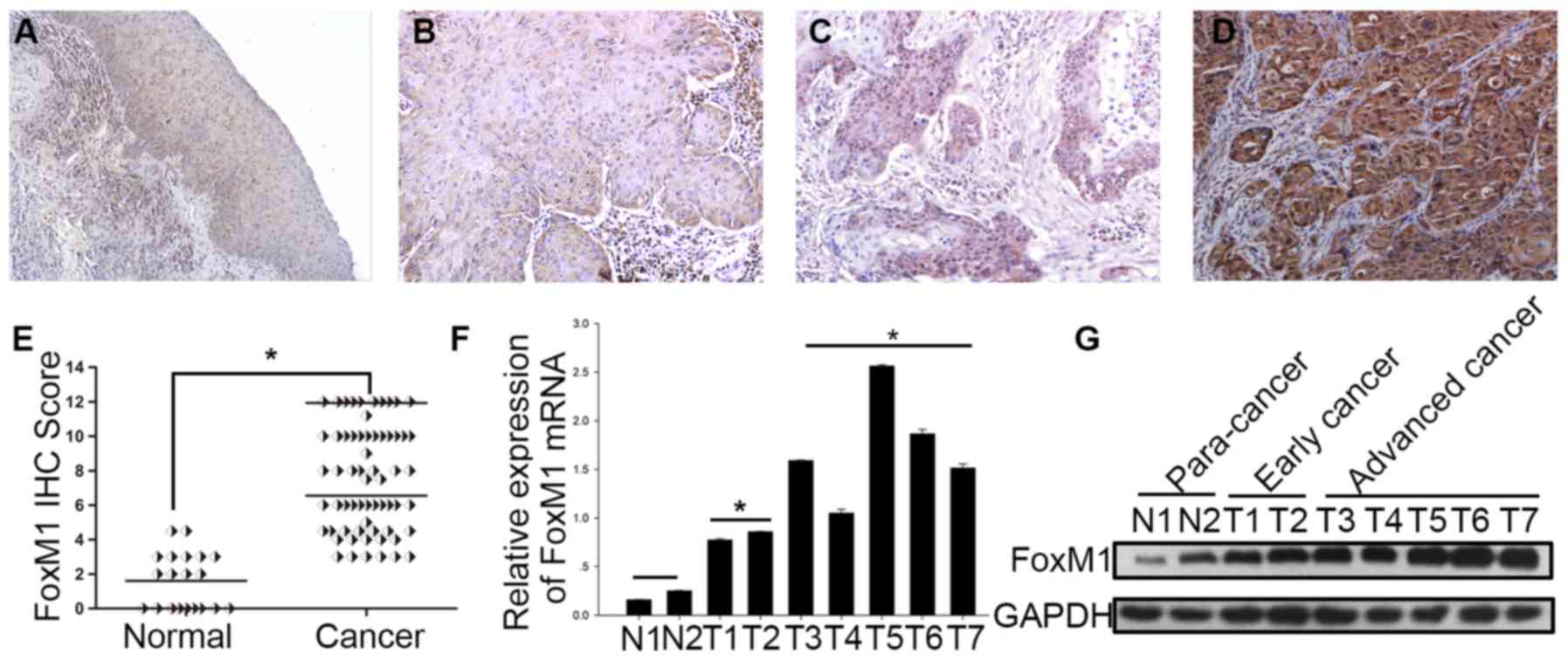
To evaluate the expression of FoxM1 in HSCC, IHC was
used to examine FoxM1 expression in 63 HSCC specimens at different
levels of malignancy and 20 adjacent normal tissue specimens. FoxM1
was expressed predominantly in the nuclei, and mixed nuclear and
cytoplasmic expression was also observed in some tumor cells.
Notably, all HSCC tissues showed positive FoxM1 staining, by
contrast, all of the adjacent tissues showed negative or weak
staining of FoxM1 (P<0.001; Fig.
1A–E). In addition, fresh tissue samples from 7 HSCC patients
[consisting of 2 early-stage (designated T1-2), 5 advanced-stage
(designated T3-7) and 2 para-cancer samples (designated N1-2)] were
collected to examine the protein and mRNA expression levels of
FoxM1. The results showed that the advanced-stage tumors exhibited
higher expression levels of FoxM1 compared with the early-stage
tumors and para-cancer tissues (Fig.
1F and G).
To investigate potential associations between the
expression level of FoxM1 and the clinicopathological
characteristics of the HSCC patients, we used a Student's t-test or
an ANOVA to compare the mean expression levels of FoxM1 between
patients grouped according to their clinicopathological
characteristics. The mean FoxM1 expression levels (IHC final
scores) were significantly increased in poorly differentiated vs.
well-differentiated tumors (P=0.004), in tumor size >2 vs. ≤2 cm
(P=0.002), in tumors of clinical stage III–IV vs. I–II (P=0.001),
in cases with vs. without lymph node metastasis (P=0.002), and
according to the treatment method (surgery vs. surgery + radiation;
surgery, radiation + chemoradiotherapy; and other treatment,
P=0.045), whereas there were no significant differences associated
with patient's age or sex (Table
I). All of these results indicate that FoxM1 is highly
expressed in HSCC tissues, and that the overexpression of FoxM1 is
involved in the degree of tumor malignancy in HSCC.
Elevated FoxM1 expression in HSCC
patients is associated with poor prognosis
Ki-67, a nuclear protein, is widely utilized as a
proliferation marker in tumor specimens, and the Ki-67 index has
been reported to be prognostic in various malignancies (21). Our results showed that there was a
positive correlation between FoxM1 and Ki-67 expression (Pearson's
correlation coefficient: r=0.637, P<0.001; Fig. 2A–C). Furthermore, we divided 63
patients with hypopharyngeal cancer into two groups, the results of
the Kaplan-Meier analysis and log-rank test showed that patients
with high FoxM1 expression (N=33, FoxM1 IHC score >6) had
shorter overall survival (P=0.0004) and recurrence-free survival
(P=0.001) times compared with those patients with low FoxM1
expression (N=30, FoxM1 IHC score ≤6) (Fig. 2D–F).
In addition, a Cox proportional hazards model was
applied to estimate the effect of FoxM1 expression on HSCC patient
survival. Univariate Cox regression analysis identified treatment
method and the expression of FoxM1 and Ki-67 as significant
prognostic factors. Using multivariate analysis, FoxM1 (HR, 5.051;
95% CI, 1.079–23.262; P=0.038), Ki-67 and tumor size were found to
be independent prognostic factors for overall survival (Table II). Collectively, the results
indicate that FoxM1 expression may act as an independent predictor
for poor patient prognosis.
 | Table IIUnivariate and multivariate analysis
of overall survival. |
Table II
Univariate and multivariate analysis
of overall survival.
|
Characteristics | Univariate analysis
| Multivariate
analysis
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (years) | | | | | | |
| <60/≥60 | 2.016 | 0.768–5.295 | 0.155 | – | – | – |
| Sex | | | | | | |
| Male/female | 21.895 |
0.001–890135.642 | 0.569 | – | – | – |
|
Differentiation | | | | | | |
| High/poor | 1.642 | 0.629–4.285 | 0.311 | 1.066 | 0.355–3.201 | 0.909 |
| Tumor size
(cm) | | | | | | |
| ≤2/>2 | 1.343 | 0.515–3.504 | 0.547 | 0.146 | 0.027–0.796 | 0.026a |
| Clinical stage | | | | | | |
| I–II/III–IV | 3.442 | 0.795–14.910 | 0.098 | 2.825 | 0.435–18.342 | 0.277 |
| Lymph node
metastasis | | | | | | |
|
Negative/positive | 2.171 | 0.785–6.001 | 0.135 | 2.402 | 0.516–11.193 | 0.264 |
| Treatment | | | | | | |
| Surgery only | 1.883 | 1.182–3.000 | 0.008b | 1.093 | 0.598–1.998 | 0.771 |
| Surgery and
radiation | | | | | | |
| Surgery,
radiation, chemotherapy | | | | | | |
| Other
treatment | | | | | | |
| Ki-67
expression | | | | | | |
| Low/high | 6.707 | 1.880–23.928 | 0.003b | 6.487 | 1.196–35.171 | 0.030a |
| FoxM1
expression | | | | | | |
| Low/high | 8.788 | 2.031–38.021 | 0.004b | 5.051 | 1.097–23.262 | 0.038a |
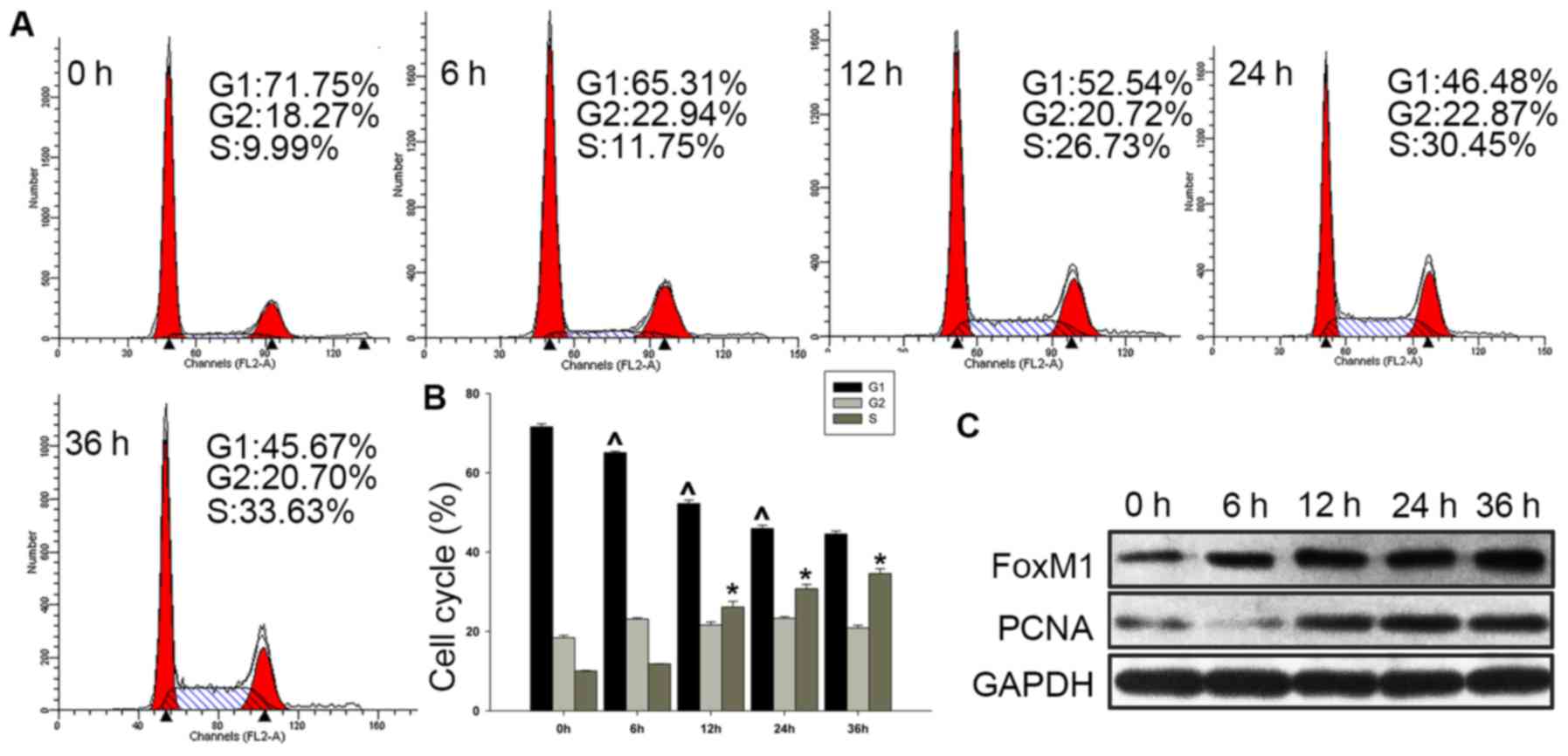
FoxM1 is highly expressed in
proliferating Fadu cells
As the clinical data supported the hypothesis that
high FoxM1 expression is associated with the proliferation of HSCC,
further studies in the HSCC cell line Fadu were performed to verify
this. We detected the expression of FoxM1 during cell cycle
progression. Fadu cells were serum-deprived for 72 h, and serum was
then added to the cultures for 6, 12, 24 and 36 h. It was observed
that the proportion of cells in G1 phase decreased (from 71.58±0.76
to 44.56±0.77%), and those in S phase cells gradually increased
(from 10.03±0.14 to 34.57±1.24%) with the addition of serum
(Fig. 3A and B). In addition, the
expression of FoxM1 was found to be increased concomitantly with
the upregulation of the cell proliferation marker PCNA (Fig. 3C).
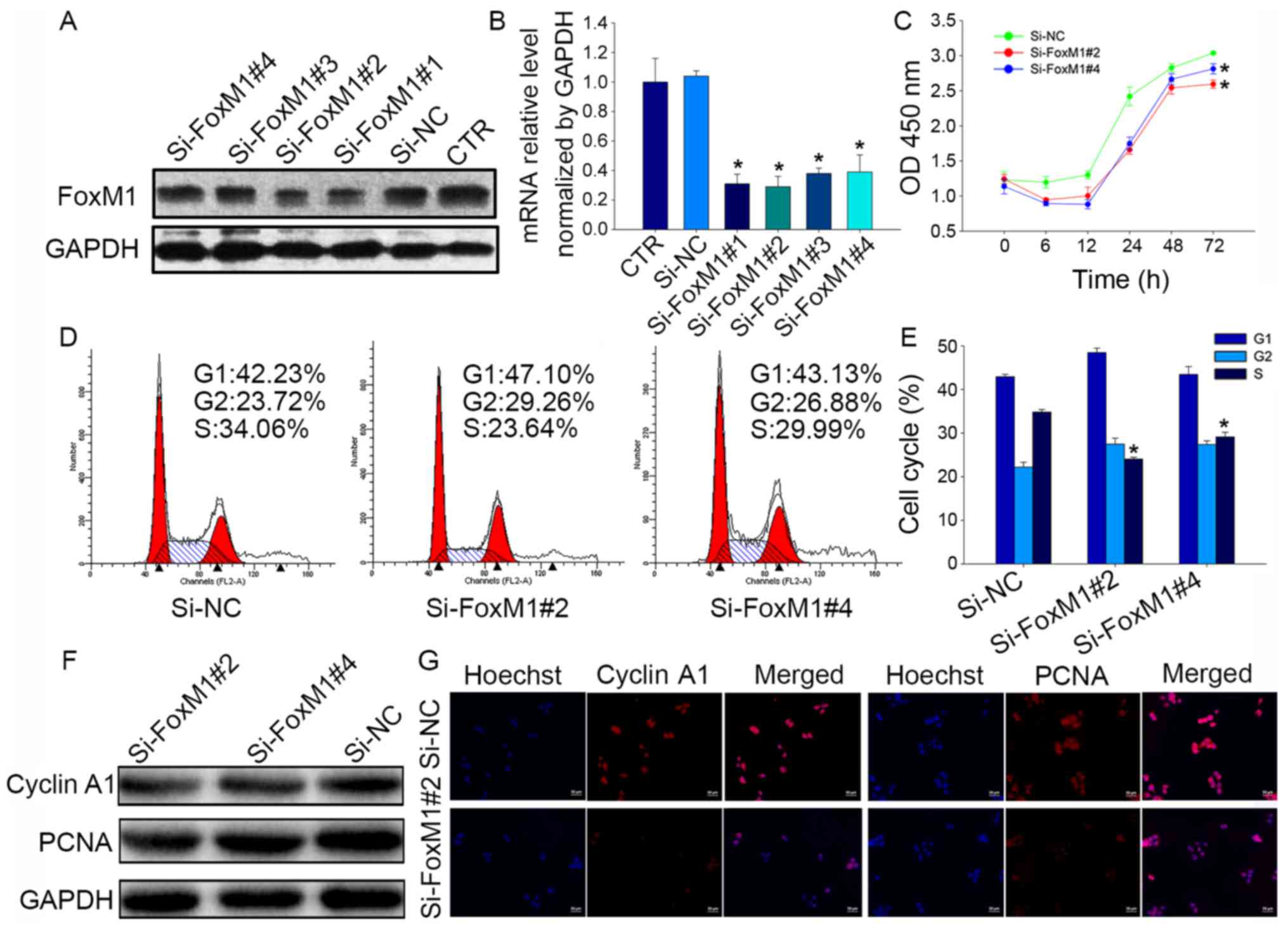
Knockdown of FoxM1 with siRNA blocks cell
proliferation and induces cell cycle arrest in Fadu cells
To further identify the effect of FoxM1 on the
biological functions of Fadu cells, cells were transfected with a
negative control siRNA (si-NC) or one of four siRNAs targeting
FoxM1, which were used to knockdown FoxM1 expression (Fig. 4A and B). The results showed that
si-FoxM1#2 was the most effective at decreasing FoxM1 mRNA
expression (reduction of 74±13%), and that si-FoxM1#4 was
moderately effective (reduction of 65±11%); therefore, these two
siRNAs were selected for use in subsequent experiments.
CCK-8 assays showed that FoxM1 knockdown resulted in
a significant decrease in Fadu cell proliferation compared with the
proliferation of si-NC-transfected cells (P<0.05; Fig. 4C). Flow cytometry was used to
analyze the effects of FoxM1 on cell cycle distribution, which
revealed a reduced proportion of cells in S phase following
treatment with si-FoxM1#2 (24.07±0.39%; P<0.05) or si-FoxM1#4
(29.15±1.03%; P<0.05) compared with si-NC treatment
(34.84±0.58%; Fig. 4D and E). In
addition, the protein expression levels of cyclin A1 and PCNA,
which are closely related to cell proliferation, were also
downregulated after treatment with si-FoxM1 (Fig. 4F and G) compared with si-NC
treatment. Taken together, these data confirm that FoxM1 may
regulate cell proliferation by influencing cell cycle
progression.
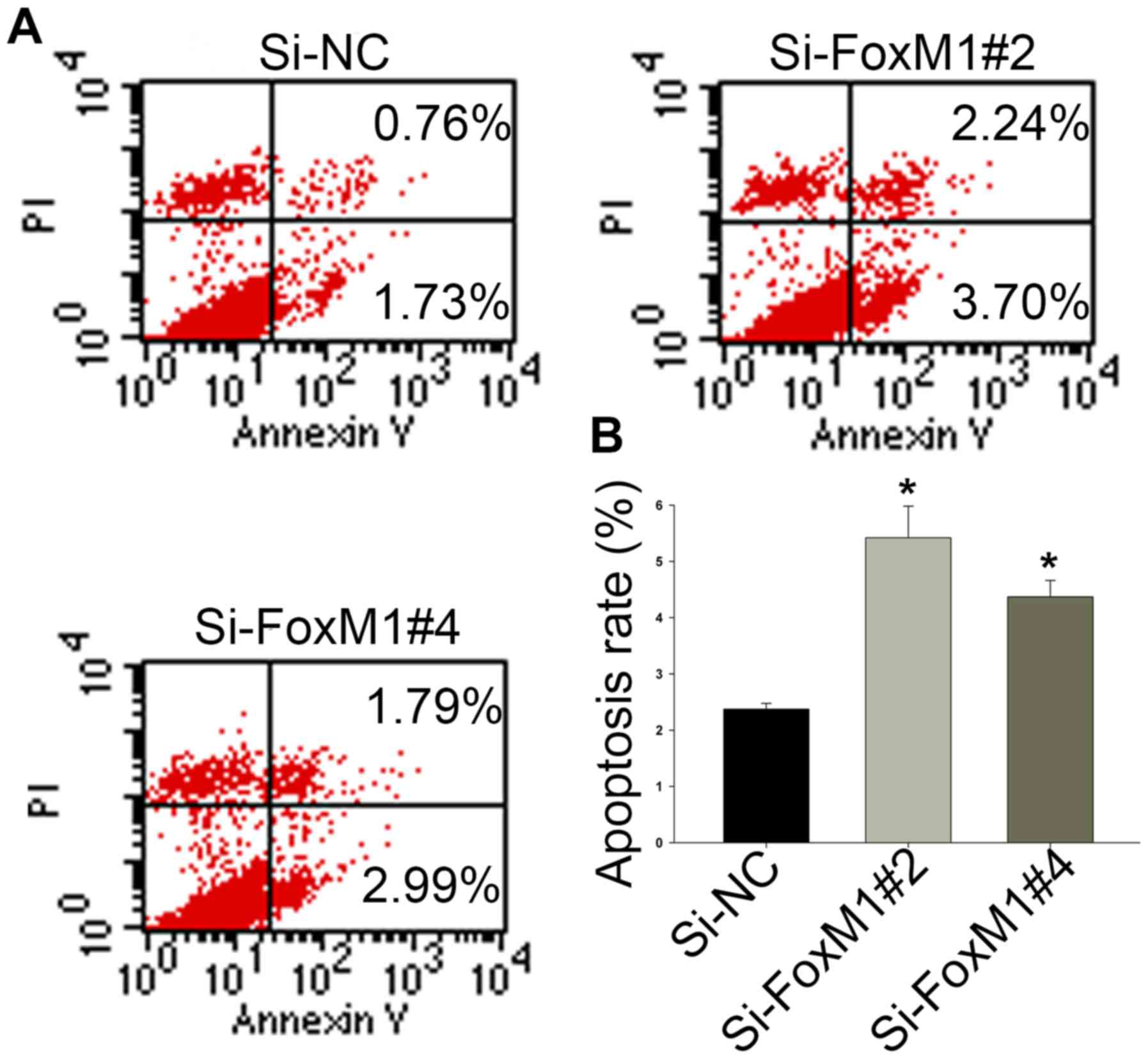
Knockdown of FoxM1 with siRNA promotes
apoptosis in Fadu cells
Annexin V/PI dual staining was used to confirm that
FoxM1 knockdown could induce apoptosis in HSCC cells. As shown in
Fig. 5A and B, after transfection
with si-FoxM1 for 48 h, the proportion of cells undergoing
apoptosis was increased from 2.37±0.10 (si-NC) to 4.37±0.29
(si-FoxM1#4) or 5.42±0.56 (si-FoxM1#2). These results indicate that
higher levels of FoxM1 decrease the rate of cell apoptosis in
HSCC.
FoxM1 enhances the migration of Fadu
cells by promoting EMT
The migratory capacities of Fadu cells transfected
with si-NC or si-FoxM1 were detected with a wound-healing assay.
Following culture in serum-free medium for 48 h, the migratory
activities of cells in the si-FoxM1#2 group were notably impaired
compared with those of the si-NC group (Fig. 6A and B). These results indicated
that FoxM1 may stimulate cell migration in HSCC.
EMT, as an essential cell biological process related
to embryonic development, also contributes to cancer metastasis and
tumor progression (22). During
EMT, proliferating epithelial cells acquire the features of a more
invasive mesenchymal phenotype, including increased migratory
activity and motility (23). In
order to investigate whether FoxM1 contributes to EMT progression
in HSCC cells, we evaluated the protein expression levels of FoxM1
and epithelial (E-cadherin) and mesenchymal (vimentin) markers in
Fadu cells. Western blot analysis revealed that cells treated with
si-FoxM1 exhibited increased E-cadherin expression and decreased
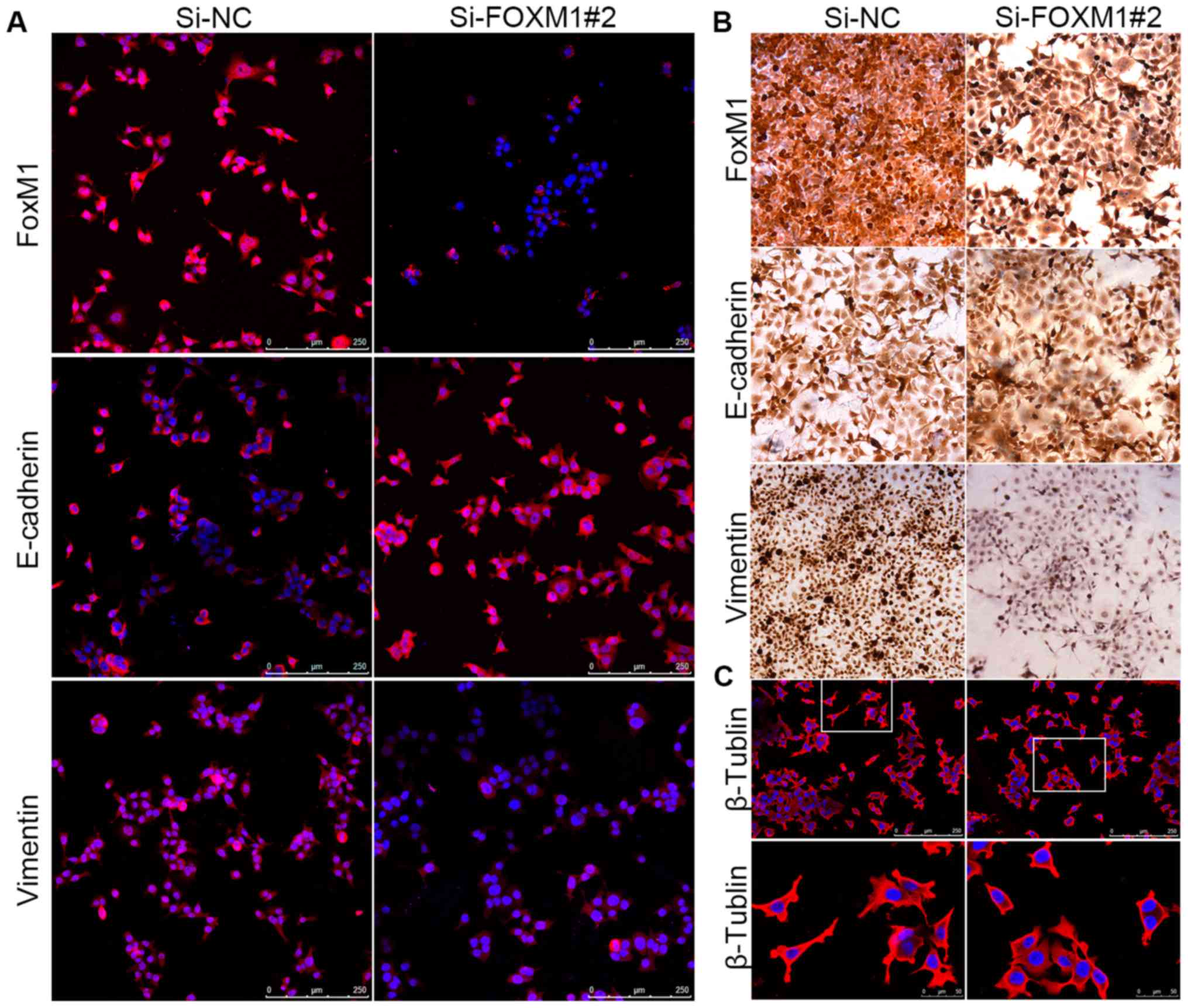
vimentin expression (Fig. 6C).
Consistent with the results of the western blot analysis, the data
from the immunofluorescence assays showed a similar molecular
expression pattern, indicating phenotypic changes between Fadu
cells transfected with si-NC and si-FoxM1 (Fig. 7A). The results were also confirmed
by immunocytochemical staining; following FoxM1 knockdown, the
staining intensity of vimentin was decreased and the staining
intensity of E-cadherin was increased (Fig. 7B). Collectively, these results
indicated that FoxM1 overexpression could partially induce the
transition to a mesenchymal phenotype, which may be a potential
mechanism regulating migration in HSCC cells. Furthermore, using
immunofluorescence microscopy, we detected the expression of
β-tubulin, which plays an essential role in maintaining cell
morphology and promoting the migration of Fadu cells. Cells treated
with si-FoxM1 showed fewer fibers and pseudopodia compared with the
si-NC group (Fig. 7C), indicating
that FoxM1 may be involved in the formation and growth of
invadopodia and, thus, the initiation of cell migration (24).
Discussion
The progression of carcinomas is affected by complex
gene regulatory networks. At present, little has been established
regarding the molecular mechanisms involved in HSCC. A previous
study by Mochizuki et al (25) demonstrated that CD271 was a marker
for tumor initiation and was associated with a poor prognosis in
human HSCC. Additionally, STK33 was found to be a potential
oncogene and a promising diagnostic marker for HSCC (26,27).
However, these two molecules alone do not explain the pathogenesis
of HSCC.
Previous evidence has shown that aberrant expression
of FoxM1 is involved in tumorigenesis and tumor progression
(9,28). Overexpression of FoxM1 has been
shown to be associated with larger tumor size, lymph node
metastasis, advanced tumor stage and poor disease-free and overall
survival times in patients with estrogen receptor-positive breast
cancer (29). In addition,
patients with FoxM1-overexpressing angiosarcoma had significantly
shorter disease-specific and event-free survival times compared
with patients with low FoxM1 expression (30). Thus, a high level of FoxM1 in tumor
patients appears to consistently indicate a high degree of
malignancy and predict a poor prognosis. In the present study,
FoxM1 was expressed to a significantly higher level in HSCC
compared with para-tumor tissues in paraffin-embedded specimens. In
addition, patients with advanced-stage HSCC had higher FoxM1
expression at the protein and mRNA levels compared with those
patients with early-stage disease. Furthermore, analysis of the
associations between FoxM1 expression and the clinicopathological
features of 63 patients with HSCC revealed that patients defined as
having poorly differentiated tumors, large tumor size,
advanced-stage disease, positive lymph node metastasis and high
Ki-67 expression exhibited higher FoxM1 expression. The results of
Kaplan-Meier analysis and univariate and multivariate Cox
regression analyses suggested that patients with high FoxM1
expression had a higher rate of mortality and tendency for
recurrence. Collectively, these results indicate that high
expression of FoxM1 may be a biological marker for malignant
transformation and an indicator of poor prognosis in HSCC.
Abnormal proliferation has been demonstrated to be a
key feature for cellular malignant transformation, and to play a
vital role in tumorigenesis and development. FoxM1 is a typical
proliferation-associated transcription factor involved in promoting
the entry of cells into S phase and M phase, thereby maintaining
the proper execution of mitosis (7,31).
Previous studies have reported that tumor cells with elevated FoxM1
expression showed high proliferative ability, whereas inhibition of
FoxM1 expression led to the suppression of proliferative activity.
It was shown that knockdown of FoxM1 expression significantly
diminished NPC cell proliferation in vitro and inhibited the
growth of NPC tumors in vivo (32). Furthermore, by using shRNA to
diminish FoxM1 expression, Yang et al (33) found that reduced FoxM1 expression
blocked the anchorage-independent growth and proliferation of
MCF-7, MDA-MB-231 and ZR-75-30 breast cancer cells. In addition,
FoxM1 was revealed to serve a pivotal role in tumor cell cycle
progression. Previous studies also found that downregulation of
FoxM1 inhibited cell proliferation and induced cell cycle arrest,
with reduced the expression levels of cyclin B1, cyclin D1, and
cyclin-dependent kinase 2 in clear cell renal cell carcinoma
(34). Moreover, Jiang et
al (18) found that
thiostrepton-mediated downregulation of FoxM1 induced cell cycle
arrest at S phase and inhibited DNA synthesis in LSCC cells in a
dose- and time-dependent manner. Consistent with these prior
studies, the present results demonstrated that Fadu cells with
inhibited FoxM1 expression showed a decreased rate of proliferation
and a reduced proportion of cells in S phase, along with decreased
levels of PCNA and cyclin A1 expression, thus, confirming the role
of FoxM1 in the regulation of tumor cell proliferation via its
influence on cell cycle progression.
The survival and growth of cancer cells does not
depend solely on proliferation, but also on resistance to cell
death signals. FoxM1 was previously reported to act as an oncogene
by regulating cell apoptosis (35). In the present study, we found that
knockdown of FoxM1 expression markedly reduced the rate of Fadu
cell apoptosis, validating the stimulatory effect of FoxM1 on HSCC
aggressiveness.
Metastasis, which is regarded as a significant event
during the malignant progression of a tumor, has been widely
researched; however, its detailed mechanisms are not fully
understood. Various evidence supports the hypothesis that FoxM1
plays an active role in tumor cell migration and metastasis in
malignancies such as liver (36)
and colorectal cancer (37). In
the present study, we observed that FoxM1 overexpression was
related to the metastasis of HSCC. Accumulating data have revealed
that EMT leads to increased cell migration in several types of
cancer (38). In colorectal
cancer, EMT was found to be a key process in tumor metastasis
(37,39). In gastric cancer, it was
demonstrated that Grhl2 reduced cell migration by inhibiting
TGF-β-induced EMT in vitro and in vivo (40). In the present study, altered cell
migratory capacity together with changes in the expression of EMT
markers (E-cadherin and vimentin) in response to FoxM1 knockdown
indicated that FoxM1 had a notable effect on the process of EMT in
HSCC cells.
As one of the primary components and functional
units of the cytoskeleton, β-tubulin is expressed in a wide variety
of eukaryotic cells and is involved in invadopodium formation,
which ultimately influences cell movement (41). We observed that Fadu cells
transfected with si-NC had long erpseudopodia compared with those
treated with si-FoxM1. Therefore, FoxM1-mediated EMT may be a
pivotal step in HSCC metastasis.
There exist some limitations in this study. Firstly,
due to low rates of HSCC, the number of samples is relatively
small, with available follow-up data. Secondly, all of our samples
were obtained from the same hospital. And then, the intricate
mechanisms of FoxM1 in promoting HSCC progression need further
studies to confirm. Furthermore, studies are also needed to enhance
our results in vivo.
In conclusion, the present results demonstrated that
FoxM1 was highly expressed in HSCC tissues, and that upregulated
FoxM1 was significantly associated with the malignant phenotype and
poor prognosis of HSCC patients, indicating a key role of FoxM1 in
the regulation of HSCC progression. We also demonstrated that FoxM1
may serve as a promoter of HSCC cell cycle progression, thereby
inducing cell proliferation. In addition, FoxM1 appeared to protect
Fadu cells from apoptosis, and facilitate HSCC cell migration,
possibly by promoting EMT. Collectively, these findings provide
evidence for a novel molecular mechanism underlying the development
of HSCC, and may lead to improvements in molecular targeted
therapies for this type of tumor.
Acknowledgments
The present study was supported by the Scientific
and Innovative Research Project of Nantong (MS32016015).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cooper JS, Porter K, Mallin K, Hoffman HT,
Weber RS, Ang KK, Gay EG and Langer CJ: National Cancer Database
report on cancer of the head and neck: 10-year update. Head Neck.
31:748–758. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Day D, Hansen AR and Siu LL:
Hypopharyngeal cancer: looking back, moving forward. Curr Oncol.
23:221–222. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hall SF, Groome PA, Irish J and O'Sullivan
B: The natural history of patients with squamous cell carcinoma of
the hypopharynx. Laryngoscope. 118:1362–1371. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martinez-Outschoorn UE, Peiris-Pages M,
Pestell RG, Sotgia F and Lisanti MP: Cancer metabolism: A
therapeutic perspective. Nat Rev Clin Oncol. 14:11–31. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wierstra I: The transcription factor FOXM1
(Forkhead box M1): Proliferation-specific expression, transcription
factor function, target genes, mouse models, and normal biological
roles. Adv Cancer Res. 118:97–398. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Petrovic V: FoxM1 regulates cell cycle
progression through multiple mechanisms. University of Illinois at
Chicago; Dissertations & Theses - Gradworks. 2008
|
|
9
|
Halasi M and Gartel AL: FOX(M1) news: it
is cancer. Mol Cancer Ther. 12:245–254. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Miao L, Xiong X, Lin Y, Cheng Y, Lu J,
Zhang J and Cheng N: Down-regulation of FoxM1 leads to the
inhibition of the epithelial-mesenchymal transition in gastric
cancer cells. Cancer Genet. 207:75–82. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kong FF, Qu ZQ, Yuan HH, Wang JY, Zhao M,
Guo YH, Shi J, Gong XD, Zhu YL, Liu F, et al: Overexpression of
FOXM1 is associated with EMT and is a predictor of poor prognosis
in non-small cell lung cancer. Oncol Rep. 31:2660–2668. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang L, Wang P and Chen H: Overexpression
of FOXM1 is associated with metastases of nasopharyngeal carcinoma.
Ups J Med Sci. 119:324–332. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun HC, Li M, Lu JL, Yan DW, Zhou CZ, Fan
JW, Qin XB, Tang HM and Peng ZH: Overexpression of Forkhead box M1
protein associates with aggressive tumor features and poor
prognosis of hepatocellular carcinoma. Oncol Rep. 25:1533–1539.
2011.PubMed/NCBI
|
|
14
|
Wu XR, Chen YH, Liu DM, Sha JJ, Xuan HQ,
Bo JJ and Huang YR: Increased expression of forkhead box M1 protein
is associated with poor prognosis in clear cell renal cell
carcinoma. Med Oncol. 30:3462013. View Article : Google Scholar
|
|
15
|
Xia JT, Wang H, Liang LJ, Peng BG, Wu ZF,
Chen LZ, Xue L, Li Z and Li W: Overexpression of FOXM1 is
associated with poor prognosis and clinicopathologic stage of
pancreatic ductal adenocarcinoma. Pancreas. 41:629–635. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ahmad A, Wang Z, Kong D, Ali S, Li Y,
Banerjee S, Ali R and Sarkar FH: FoxM1 down-regulation leads to
inhibition of proliferation, migration and invasion of breast
cancer cells through the modulation of extra-cellular matrix
degrading factors. Breast Cancer Res Treat. 122:337–346. 2010.
View Article : Google Scholar
|
|
17
|
Nakamura S, Hirano I, Okinaka K, Takemura
T, Yokota D, Ono T, Shigeno K, Shibata K, Fujisawa S and Ohnishi K:
The FOXM1 transcriptional factor promotes the proliferation of
leukemia cells through modulation of cell cycle progression in
acute myeloid leukemia. Carcinogenesis. 31:2012–2021. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang L, Wu X, Wang P, Wen T, Yu C, Wei L
and Chen H: Targeting FoxM1 by thiostrepton inhibits growth and
induces apoptosis of laryngeal squamous cell carcinoma. J Cancer
Res Clin Oncol. 141:971–981. 2015. View Article : Google Scholar
|
|
19
|
Huang C, Qiu Z, Wang L, Peng Z, Jia Z,
Logsdon CD, Le X, Wei D, Huang S and Xie K: A novel FoxM1-caveolin
signaling pathway promotes pancreatic cancer invasion and
metastasis. Cancer Res. 72:655–665. 2012. View Article : Google Scholar :
|
|
20
|
Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu
M, Feng XH, Sawaya R, Medema RH, Hung MC and Huang S: Sustained
activation of SMAD3/SMAD4 by FOXM1 promotes TGF-β-dependent cancer
metastasis. J Clin Invest. 124:564–579. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ghanim B, Klikovits T, Hoda MA, Lang G,
Szirtes I, Setinek U, Rozsas A, Renyi-Vamos F, Laszlo V, Grusch M,
et al: Ki67 index is an independent prognostic factor in
epithelioid but not in non-epithelioid malignant pleural
mesothelioma: A multicenter study. Br J Cancer. 112:783–792. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mego M, Reuben J and Mani S:
Epithelial-mesenchymal transition (EMT) and cancer stem cells
(CSCs): The Traveling Metastasis. Liquid Biopsies in Solid Tumors.
Cancer Drug Discovery and Development. Cristofanilli M: Humana
Press; Totowa, NJ: pp. 67–80. 2017, View Article : Google Scholar
|
|
23
|
Chen T, You Y, Jiang H and Wang ZZ:
Epithelial-mesenchymal transition (EMT): A biological process in
the development, stem cell differentiation, and tumorigenesis. J
Cell Physiol. Jan 12–2017, (Epub ahead of print). https://doi.org/10.1002/jcp.25797.
View Article : Google Scholar
|
|
24
|
Chodniewicz D and Klemke RL: Guiding cell
migration through directed extension and stabilization of
pseudopodia. Exp Cell Res. 301:31–37. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mochizuki M, Tamai K, Imai T, Sugawara S,
Ogama N, Nakamura M, Matsuura K, Yamaguchi K, Satoh K, Sato I, et
al: CD271 regulates the proliferation and motility of
hypopharyngeal cancer cells. Sci Rep. 6:307072016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang L, Chen C, Zhang G, Ju Y, Zhang J,
Wang H and Li J: STK33 overexpression in hypopharyngeal squamous
cell carcinoma: Possible role in tumorigenesis. BMC Cancer.
15:132015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen C, Huang L, Zhang G, Li Y, Li L, Bai
X, Liu W, Wang H and Li J: STK33 potentiates the malignancy of
hypopharyngeal squamous carcinoma: Possible relation to calcium.
Cancer Biol Ther. 17:976–984. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wierstra I: FOXM1 (Forkhead box M1) in
tumorigenesis: Overexpression in human cancer, implication in
tumorigenesis, oncogenic functions, tumor-suppressive properties,
and target of anticancer therapy. Adv Cancer Res. 119:191–419.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ahn H, Sim J, Abdul R, Chung MS, Paik SS,
Oh YH, Park CK and Jang K: Increased expression of forkhead box M1
is associated with aggressive phenotype and poor prognosis in
estrogen receptor-positive breast cancer. J Korean Med Sci.
30:390–397. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ito T, Kohashi K, Yamada Y, Iwasaki T,
Maekawa A, Kuda M, Hoshina D, Abe R, Furue M and Oda Y: Prognostic
significance of forkhead Box M1 (FOXM1) expression and antitumor
effect of FOXM1 inhibition in angiosarcoma. J Cancer. 7:823–830.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wierstra I and Alves J: FOXM1, a typical
proliferation-associated transcription factor. Biol Chem.
388:1257–1274. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen H, Yang C, Yu L, Xie L, Hu J, Zeng L
and Tan Y: Adenovirus-mediated RNA interference targeting FOXM1
transcription factor suppresses cell proliferation and tumor growth
of nasopharyngeal carcinoma. J Gene Med. 14:231–240. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang C, Chen H, Yu L, Shan L, Xie L, Hu J,
Chen T and Tan Y: Inhibition of FOXM1 transcription factor
suppresses cell proliferation and tumor growth of breast cancer.
Cancer Gene Ther. 20:117–124. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xue Y, Zhang G, Xu R, Zou X, Zhong X, Wu
G, Wang X, Long D, Wu Y, Xu H, et al: Overexpression of FoxM1 is
associated with tumor progression. J Urol. 189:e7832013. View Article : Google Scholar
|
|
35
|
Bhat UG, Halasi M and Gartel AL: Thiazole
antibiotics target FoxM1 and induce apoptosis in human cancer
cells. PLoS One. 4:e5592. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Park HJ, Gusarova G, Wang Z, Carr JR, Li
J, Kim KH, Qiu J, Park YD, Williamson PR, Hay N, et al:
Deregulation of FoxM1b leads to tumour metastasis. EMBO Mol Med.
3:21–34. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang C, Wang Y, Feng Y, Zhang Y, Ji B,
Wang S and Sun Y, Zhu C, Zhang D and Sun Y: Gli1 promotes
colorectal cancer metastasis in a Foxm1-dependent manner by
activating EMT and PI3K-AKT signaling. Oncotarget. 7:86134–86147.
2016.PubMed/NCBI
|
|
38
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen B, Zeng X, He Y, Wang X, Liang Z, Liu
J, Zhang P, Zhu H, Xu N and Liang S: STC2 promotes the
epithelial-mesenchymal transition of colorectal cancer cells
through AKT-ERK signaling pathways. Oncotarget. 7:71400–71416.
2016.PubMed/NCBI
|
|
40
|
Xiang J, Fu X, Ran W and Wang Z: Grhl2
reduces invasion and migration through inhibition of TGFβ-induced
EMT in gastric cancer. Oncogenesis. 6:e2842017. View Article : Google Scholar
|
|
41
|
Ganguly A and Cabral F: The arresting
action of microtubules in cell motility. Cell Cycle. 10:2614–2615.
2011. View Article : Google Scholar : PubMed/NCBI
|