Introduction
Pancreatic cancer is one of the most lethal human
cancers, and it is usually discovered and diagnosed at its advanced
stage (1). Pancreatic ductal
adenocarcinoma (PDAC) is the most common subtype of pancreatic
cancer, with a low 5-year survival rate of 7.7% (2). Despite considerable advances in
neoadjuvant chemotherapy, surgical techniques and perioperative
care, the prognosis for PDAC has not improved significantly
(3). Hence, prognosis markers are
very important for PDAC patients, they provide valuable prognostic
and treatment information.
Recently, a growing number of prognostic markers
have been discovered, including mRNAs, non-coding RNA, circulating
tumor DNA and tumor-derived exosomes (3–7).
These prognostic biomarkers associate with tumor clinical stage,
overall survival (OS) time, and disease-free survival (DFS) time by
regulating tumor biological behavior. For example, integrin β1
modulates tumor resistance to gemcitabine and serves as an
independent prognostic factor in PDAC (8). Regenerating family member proteins
promote acinar-to-ductal metaplasia and act as novel diagnostic and
prognostic markers in PDAC (9).
Vav guanine nucleotide exchange factor 3 and DIX domain containing
1 are linked to poor prognosis of pancreatic cancers and promote
the proliferation, motility and invasiveness of pancreatic cancer
cells (10,11). However, there are no prognostic
markers for use in the clinic. Thus, there is still a growing need
for prognostic markers and therapeutic targets to improve the
outcomes of PDAC patients.
In the present study, we found that stress
associated endoplasmic reticulum protein 1 (SERP1) was
significantly upregulated in PDAC tissues. SERP1, also known as
ribosome-associated membrane protein 4 (RAMP4), is a
Sec61-associated polypeptide that is induced by ER stress (12). High level of SERP1 was correlated
with advanced stage and poor prognosis of PDAC. SERP1
downregulation promoted cell apoptosis via upregulating
apoptosis-related protein SRP receptor β subunit (SRPRB) expression
and inhibiting nuclear factor-κB (NF-κB) activation. Previous
studies have verified that NF-κB is a known regulator of
anti-apoptotic molecules (13,14),
is constitutively activated by phosphorylation modification in
various tumors including PDAC (15–17).
NF-κB phosphorylation exist in PDAC cells such as PANC-1, BxPC-3
and AsPC-1, and contributing to their resistance to apoptosis and
high metastatic potential (14).
The inhibition of NF-κB phosphorylation has been shown to sensitize
cells to apoptosis in pancreatic cancer cells (14). Our results are the first to show
SERP1 is involved in regulating NF-κB activation and apoptosis,
helping to elucidate the possible mechanism of SERP1 in predicting
prognosis of PDAC.
Materials and methods
Database analysis
The mRNA expression data of 14 pairs of PDAC and
adjacent tissues were obtained from GEO profiles database
(http://www.ncbi.nlm.nih.gov/geo/,
GSE41368, GSE16515). Mutation information and clinical data were
downloaded from cBioPortal database (www.cbioportal.org). The TCGA data on mRNA (RNASeq V2)
levels in PDAC patients were obtained from https://cancergenome.nih.gov/. SERP1, SRPRB and NF-κB
mRNA level were used in the present study. The IHC-based protein
expression data including high-resolution images were viewed and
downloaded from the Human Protein Atlas web portal (www.proteinatlas.org). The sum IOD of IHC images were
measured by Image-Pro Plus software (version 6.0; Media
Cybernetics).
Gene set enrichment analysis (GSEA)
The association between SERP1 and biological
processes/signaling pathway gene set was analyzed using Gene set
enrichment analysis (GSEA v2.2, http://www.broad.mit.edu/gsea/). GSEA calculates a
gene set enrichment score (ES) that estimates genes from
pre-defined gene set. Thresholds for significance were determined
by permutation analysis (1,000 permutations). A gene set is
considered significantly enriched when the P-value is <0.05.
Cell culture and siRNA transfection
Human PDAC cell line PANC-1 was purchased from the
Cell Resource Center of Beijing Xiehe (Beijing, China) and
cultivated in an incubator at 37°C with 5% CO2. PANC-1
cells were maintained in high-glucose Dulbecco's modified Eagle's
medium (DMEM; Cell Resource Center of Beijing Xiehe, Beijing,
China) supplemented with 10% fetal bovine serum (FBS; HyClone
Laboratories, Inc., Logan, UT, USA) as well as penicillin (100
U/ml; Thermo Fisher Scientific, San Jose, CA, USA). The small
interfering RNA targeting SERP1 and its negative control (NC)
sequences were designed and synthesized by GenePharma Biotech, Co.,
Ltd. (Shanghai, China). The expression plasmid pcDNA3.0-SRPRB and
corresponding control plasmid were preserved in our laboratory.
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used for
siRNA and plasmid transfection according to the manufacturer's
protocol. After 48 h of transfection, cells were used for
subsequent experiments.
Apoptosis assay
Annexin V-FITC apoptosis detection kit I (BD
Biosciences, San Jose, CA, USA) was used to detect cell apoptosis.
PANC-1 cells were washed twice with cold phosphate-buffered saline,
and then cells were resuspend in Annexin V binding buffer at a
concentration of 0.5×107 cells/ml. Transfer of cell
suspension (100 μl) was made to a 1 ml test tube, and 5
μl FITC Annexin V and 10 μl propidium iodide solution
was added to the test tube. The cells were gently vortexed and
incubated for 15 min at room temperature (25°C) in the dark. Then
400 μl of Annexin V binding buffer was added to each tube,
and analyzed by flow cytometry (Accuri C6; BD Biosciences, Franklin
Lakes, NJ, USA) within 1 h.
Western blot analysis
Cells were collected and lysed in
radioimmunoprecipitation buffer (Beijing CoWin Biotech Co., Ltd.,
Beijing, China) with protease inhibitors for 30 min to extract
total proteins from cells with SRPRB overexpression or SERP1
knockdown. Protein levels were quantified by bicinchoninic acid
assays (Beijing CoWin Biotech Co., Ltd.). Thirty micrograms of
protein from each sample was resolved by 12% sodium dodecyl
sulphate-polyacrylamide gel electrophoresis (Beijing CoWin Biotech
Co., Ltd.). Proteins were transferred to nitrocellulose membranes
(Sigma-Aldrich, St. Louis, MO, USA), which were blocked for 1 h in
bovine serum albumin blocking buffer (Invitrogen). Subsequently,
the membranes were incubated overnight at 4°C with primary
antibodies targeting SERP1 (1:1,000 dilution, cat. no. ab130974;
Abcam, Cambridge, MA, USA), SRPRB (1:300 dilution, cat. no.
D223153; Sangon Biotech), phospho-NF-κB p65 (Ser536)(1:1,000
dilution, cat. no. 3033, Cell Signaling Technology, Beverly, MA,
USA), NF-κB p65 (1:1,000 dilution, cat. no. 8242, Cell Signaling
Technology), or GAPDH (1:1,000 dilution, cat. no. 70699; Abcam),
were followed by incubation with an HRP-conjugated goat anti-rabbit
secondary antibody (1:10,000 dilution, cat. no. CW0103; Beijing
CoWin Biotech Co., Ltd.) for 1 h at room temperature.
Immunocomplexes were detected using an enhanced chemiluminescence
kit (Thermo Fischer Scientific), and images were analyzed using
ImageJ software (version 1.62; National Institute of Health,
Bethesda, MD, USA).
Protein-protein interaction (PPI) network
construction
Search Tool for the Retrieval of Interacting
genes/Proteins (STRING; Search Tool for the Retrieval of
Interacting Genes, http://string-db.org/) is a database of known and
predicted protein interactions that may aid in the comprehensive
description of cellular mechanisms and functions (18). The PPI network interacting with
SERP1 in pancreatic cancer was constructed using the STRING
database.
Gene ontology (GO) analysis
To explore the functional annotation enrichment of
genes interacted with SERP1, GO analysis to determine clusters of
these mRNAs with enriched molecular functions were performed. We
used the database for annotation, visualization and integrated
discovery (DAVID) v6.7 online tool (http://david.abcc.ncifcrf.gov) to functionally
annotate input genes, classify gene functions, and identify gene
conversions, and to perform the GO analyses. A P-value of <0.05
was considered significant (19).
Statistical analysis
The statistical analysis was carried out using the
SPSS 21.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 5
(GraphPad Software Inc., San Diego, CA, USA). mRNA expression data,
IHC and apoptosis rate were analyzed using independent sample
t-tests. Survival curves between different groups were obtained
from Kaplan-Meier method and log-rank test. After the univariate
analysis, Cox proportional hazards model was used to identify the
independent prognostic factors for DFS and OS. P-values <0.05
were considered statistically significant.
Results
Expression level of SERP1 is upregulated
in PDAC tissues
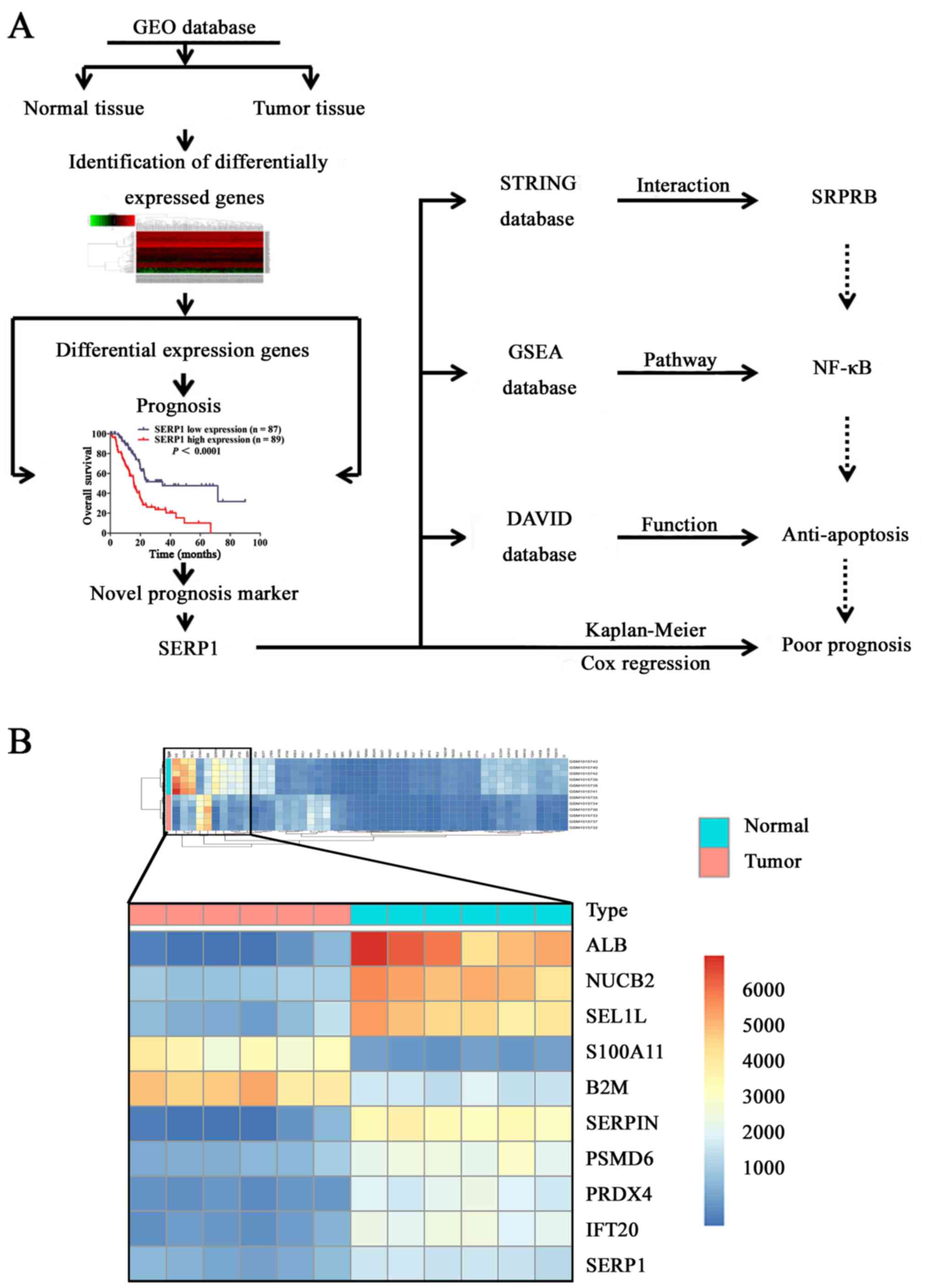
As shown in flow chart of derivation and functional
analysis of SERP1 (Fig. 1A), to
identify the novel differentially expressed mRNAs related with PDAC
occurrence and development, six pairs of PDAC tissues and adjacent
normal pancreas tissues from GEO database were compared, top 10
significantly differently expressed mRNAs (P<0.01; FC >1 and
FC <−1) are shown in Fig. 1B.
To evaluate the correlation of differentially expressed mRNAs with
prognosis, we performed survival analysis of top 10 mRNAs. Among
the 10 dysregulated mRNAs, SERP1, S100A11 and SEL1L were strongly
correlated with survival time. However, SERP1 was one of rarely
reported upregulated mRNAs in PDAC (Fig. 2A), and attracted our interest.
Furthermore, SERP1 was again substantially upregulated in PDAC
tissues by GEO (n=8) (Fig. 2B). In
Human Protein Atlas web portal database, we equally observed that
SERP1 was significantly upregulated in PDAC tissues (n=11) compared
to normal ductal epithelial cells of pancreas (n=3), and it was
localized mainly in the cytoplasm of tumor cells (Fig. 2C and D). Analysis of molecular
genetic alterations revealed that amplification of SERP1 exist in
numerous malignant tumors, including PDAC, and we presume
amplification may be a potential cause of high expression of SERP1
in PDAC (Fig. 2E).
Upregulated SERP1 is associated with
advanced stage and poor prognosis of PDAC
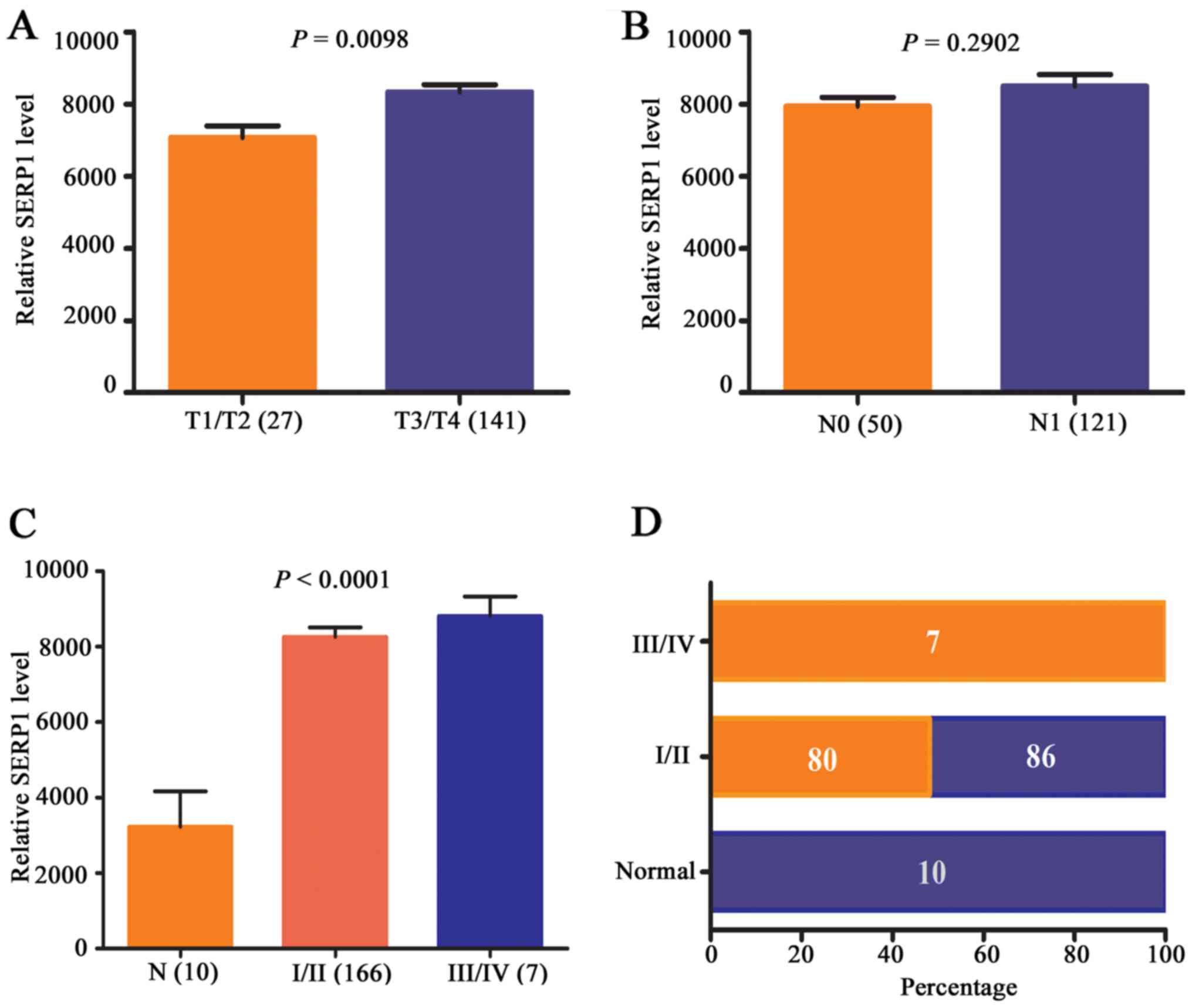
To further elucidate the role of SERP1 in pancreatic
cancer progression, the expression level of SERP1 in different
stages of pancreatic cancer patients was analyzed by TCGA database.
The results revealed that with the increase of T stage (P=0.0098)
and clinical stage (P<0.0001) in PDAC, but not N stage
(P<0.2902), SERP1 expression level increased accordingly
(Fig. 3A–C). Next, constituent
ratio analysis was performed in an expanded set of 173 primary PDAC
tissues and 10 normal samples, revealing that none of the cases
exhibited high expression level for SERP1 in normal pancreas
tissues. However, in I/II and III/IV stage of pancreatic cancer
tissues, 80/166 (56%) of cases and 7/7 (100%) of cases showed
higher expression level for SERP1 comparing to normal tissues
(P<0.01) (Fig. 3D). The above
results suggested that SERP1 was upregulated in PDAC and associated
with advanced stage, which may have prognostic significance for
PDAC patients.
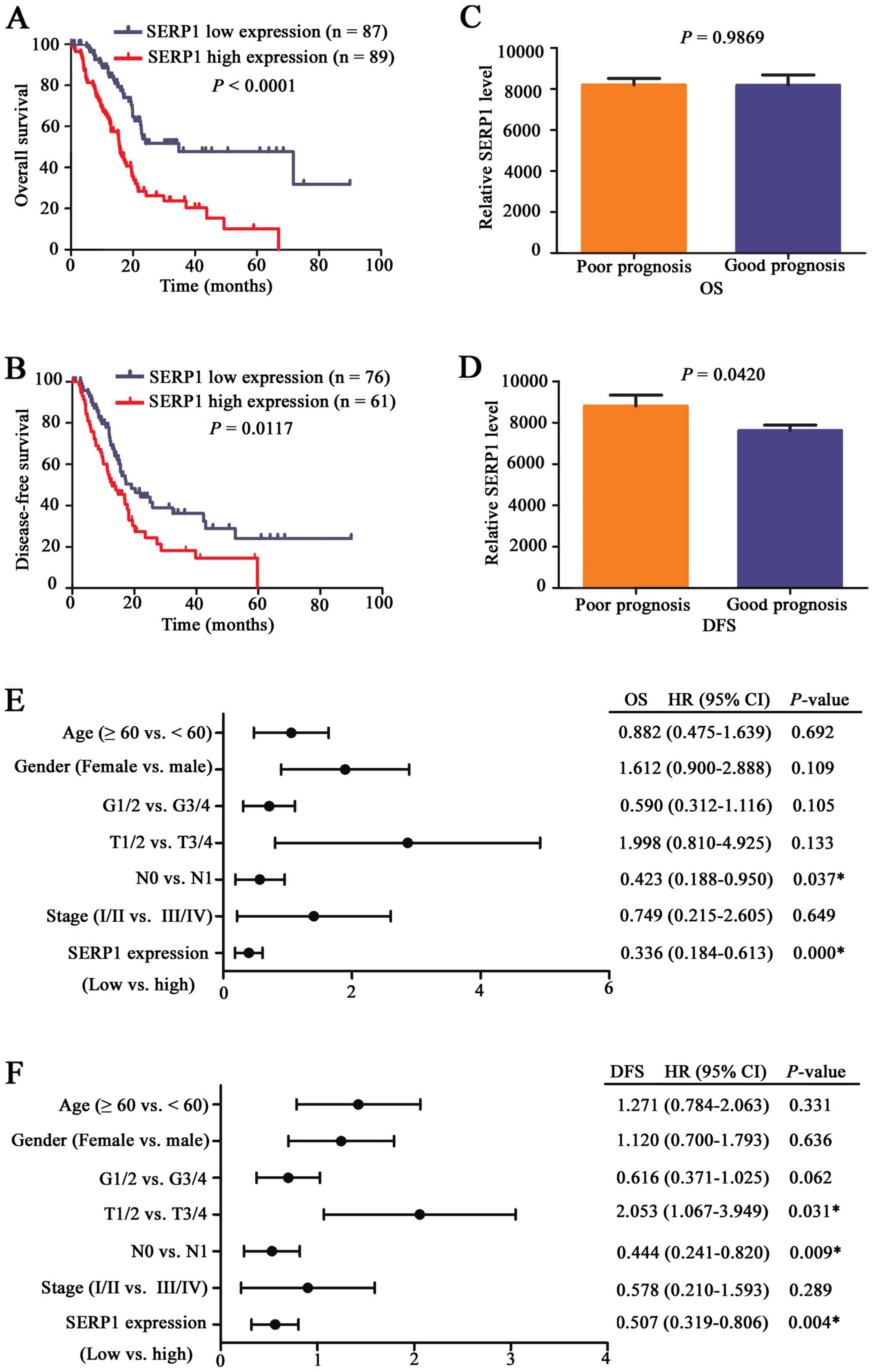
Next, patients were divided into SERP1 high
expression and SERP1 low expression groups, prognostic role of
SERP1 was further investigated. Kaplan-Meier curve showed that
SERP1 high expression group had shorter OS and DFS compared with
SERP1 low expression group in terms of survival duration
(P<0.05) (Fig. 4A and B),
particularly in OS. Next, patients were divided into good and poor
survival groups, and results revealed that the expression level of
SERP1 was higher in patients with shorter DFS (P=0.0420) (Fig. 4C and D). Moreover, multivariate Cox
regression analysis further revealed SERP1 was an independent
prognostic marker for PDAC patients (Fig. 4E and F).
Downregulated SERP1 promotes PDAC cell
apoptosis
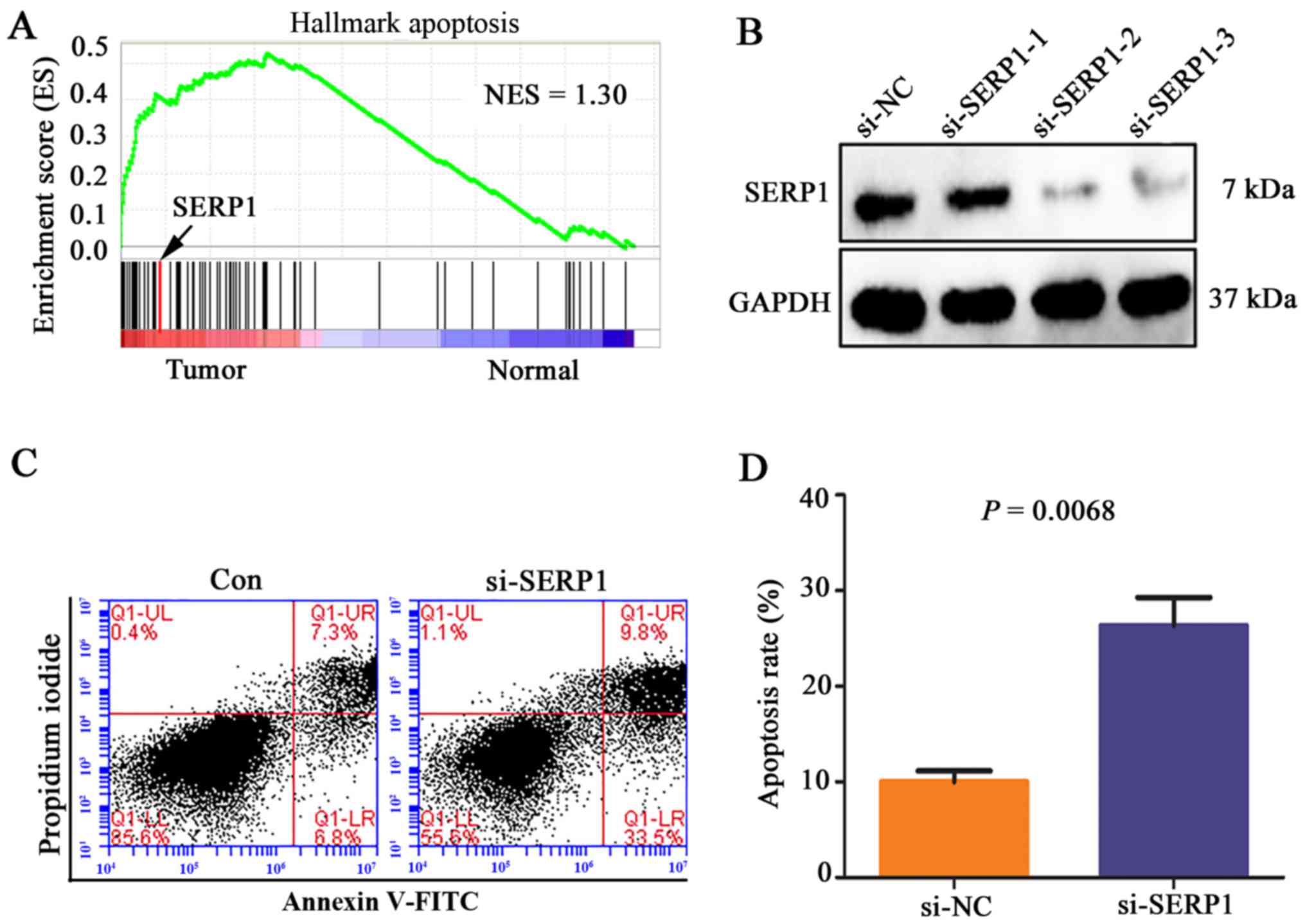
In order to gain a better understanding of the
potential mechanisms of SERP1 influencing prognosis of PDAC, GSEA
was performed to elucidate the functions and signaling pathways
involved in these differentially expressed genes in PDAC. GSEA
results showed that the gene sets related to apoptosis were
enriched in PDAC tissues, including SERP1 (Fig. 5A). In vitro, si-SERP1 and
corresponding negative control were transfected into PANC-1 cells.
The expression level of SERP1 was significantly decreased in PANC-1
cells transfected with SERP1 siRNA (Fig. 5B), which promoted cell apoptosis
compared with negative control group (P=0.0068) (Fig. 5C and D).
Downregulated SERP1 increases apoptosis
related protein SRPRB expression
Owing to protein-protein interaction playing an
important role in regulating tumor biological characteristics,
STRING was used to find the interaction genes with SERP1. As
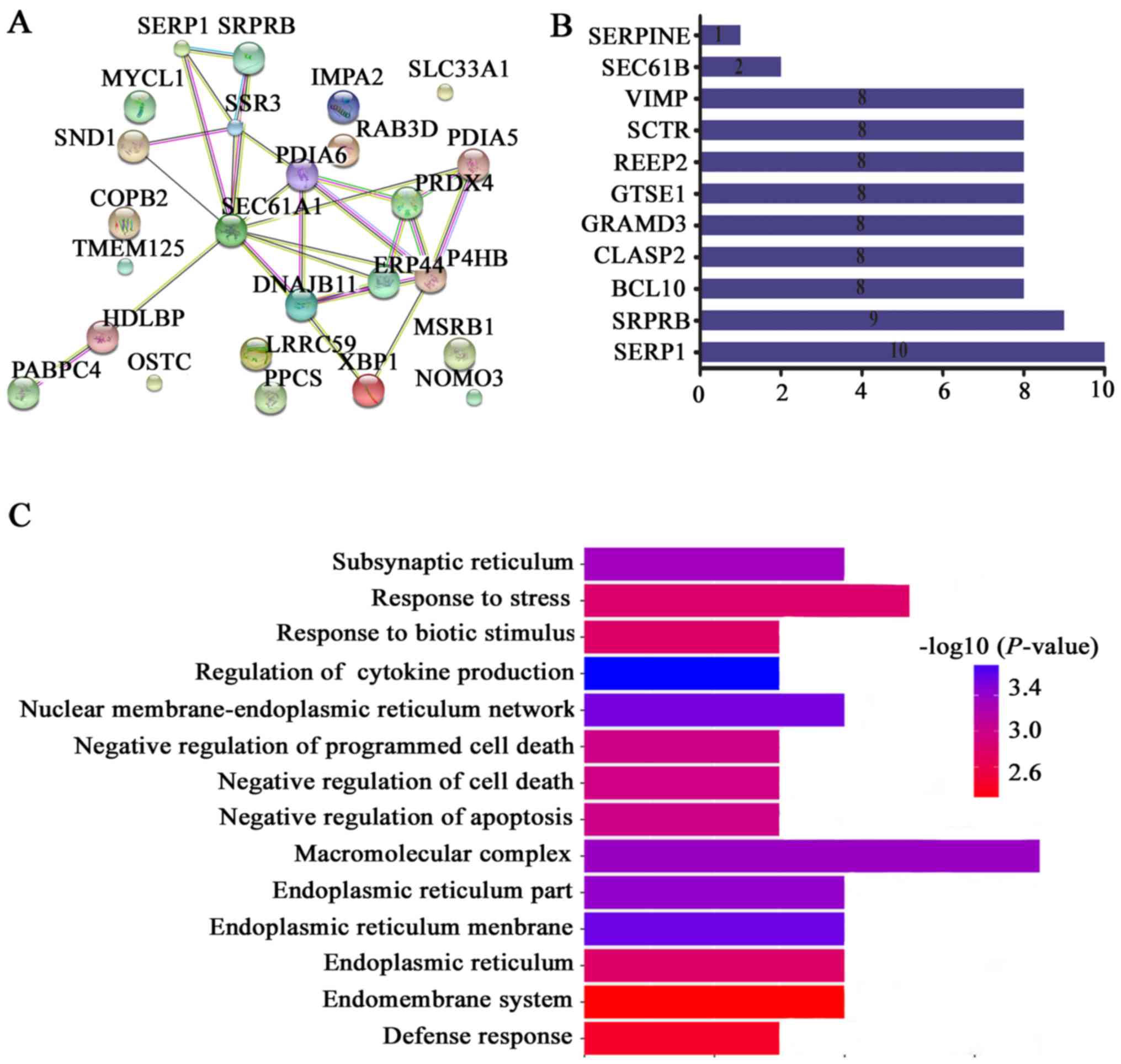
Fig. 6A and B showed, these genes
include SRPRB, BCL10, CLASP2, GRAMD3, GTSE1, REEP2, SCTR, VIMP,
SEC61B and SERPINE. We further performed GO analysis for these
interaction genes. Consistent with GSEA analysis, we noted that
these genes were especially enriched in functions of regulating
cell death regulation and apoptosis (Fig. 6C). Especially, apoptosis related
protein SRP receptor β subunit (SRPRB) was the highest ranked
interaction genes with SERP1 (Fig.
6B), and was one of significantly co-expressed genes with SERP1
(Pearson =0.63) (Table I).
 | Table ICo-expression genes with SERP1 from
cBioPortal database. |
Table I
Co-expression genes with SERP1 from
cBioPortal database.
| Gene symbol | Cytoband | Pearson score | Spearman score |
|---|
| SSR3 | 3q25.31 | 0.83 | 0.61 |
| XBP1 | 22q12.1|22q12 | 0.71 | 0.31 |
| SEC61A1 | 3q21.3 | 0.71 | 0.46 |
| HDLBP | 2q37.3 | 0.7 | 0.49 |
| IMPA2 | 18p11.2 | 0.68 | 0.36 |
| SLC33A1 | 3q25.31 | 0.68 | 0.44 |
| PABPC4 | 1p34.2 | 0.67 | 0.34 |
| RAB3D | 19p13.2 | 0.67 | 0.29 |
| MYCL | 1p34.2 | 0.66 | 0.21 |
| OSTC | 4q25 | 0.66 | 0.52 |
| P4HB | 17q25 | 0.65 | 0.36 |
| TMEM125 | 1p34.2 | 0.65 | 0.22 |
| COPB2 | 3q23 | 0.63 | 0.5 |
| MSRB1 | 16p13.3 | 0.63 | 0.43 |
| LRRC59 | 17q21.33 | 0.63 | 0.43 |
| SRPRB | 3q22.1 | 0.63 | 0.55 |
| PPCS | 1p34.2 | 0.62 | 0.33 |
| ERP44 | 9q31.1 | 0.62 | 0.37 |
| SND1 | 7q31.3 | 0.62 | 0.37 |
| PRDX4 | Xp22.11 | 0.61 | 0.39 |
| PDIA5 | 3q21.1 | 0.61 | 0.35 |
| PDIA6 | 2p25.1 | 0.6 | 0.44 |
| NOMO3 | 16p13 | 0.6 | 0.28 |
| DNAJB11 | 3q27.3 | 0.6 | 0.55 |
To explore the relationship between SERP1 and SRPRB,
the Human Protein Atlas Database was applied. In clinical level,
previous results found that SERP1 upregulated in PDAC tissues. On
the contrary, SRPRB protein expression levels were significantly
downregulated (P=0.0199) in PDAC tissue compared with normal
pancreas tissues, it was also localized mainly in the cytoplasm of
tumor cells. Interestingly, SRPRB was obviously upregulated in
stromal fibroblasts of tumor tissues, but almost no expression was
found of the SRPRB gene in stromal fibroblasts of normal pancreas
(P=0.0090) (Fig. 7A and B). In
vitro, si-SERP1 was transfected into PANC-1 cells and the
expression level of SRPRB was increased with SERP1 downregulation
in PANC-1 cells (Fig. 7C). Next,
in order to identify the effect of upregulated SRPRB on PDAC cell
apoptosis, pcDNA3.0-SRPRB and corresponding negative control were
transfected into PANC-1 cells, respectively. The SRPRB expression
level of PANC-1 cells transfected with SRPRB overexpression plasmid
was upregulated (Fig. 7D), and the
apoptosis rate of PANC-1 was obviously increased (P=0.0016)
(Fig. 7E and F). The above results
showed that downregulated SERP1 promoted cell apoptosis probably
via upregulating SRPRB expression.
SRPRB promotes cell apoptosis through
NF-κB activation
NF-κB activation plays an important role in
controlling the survival of tumor cells (20–22),
the abnormal upregulation were thought to promote tumor cell
survival (23). In the present
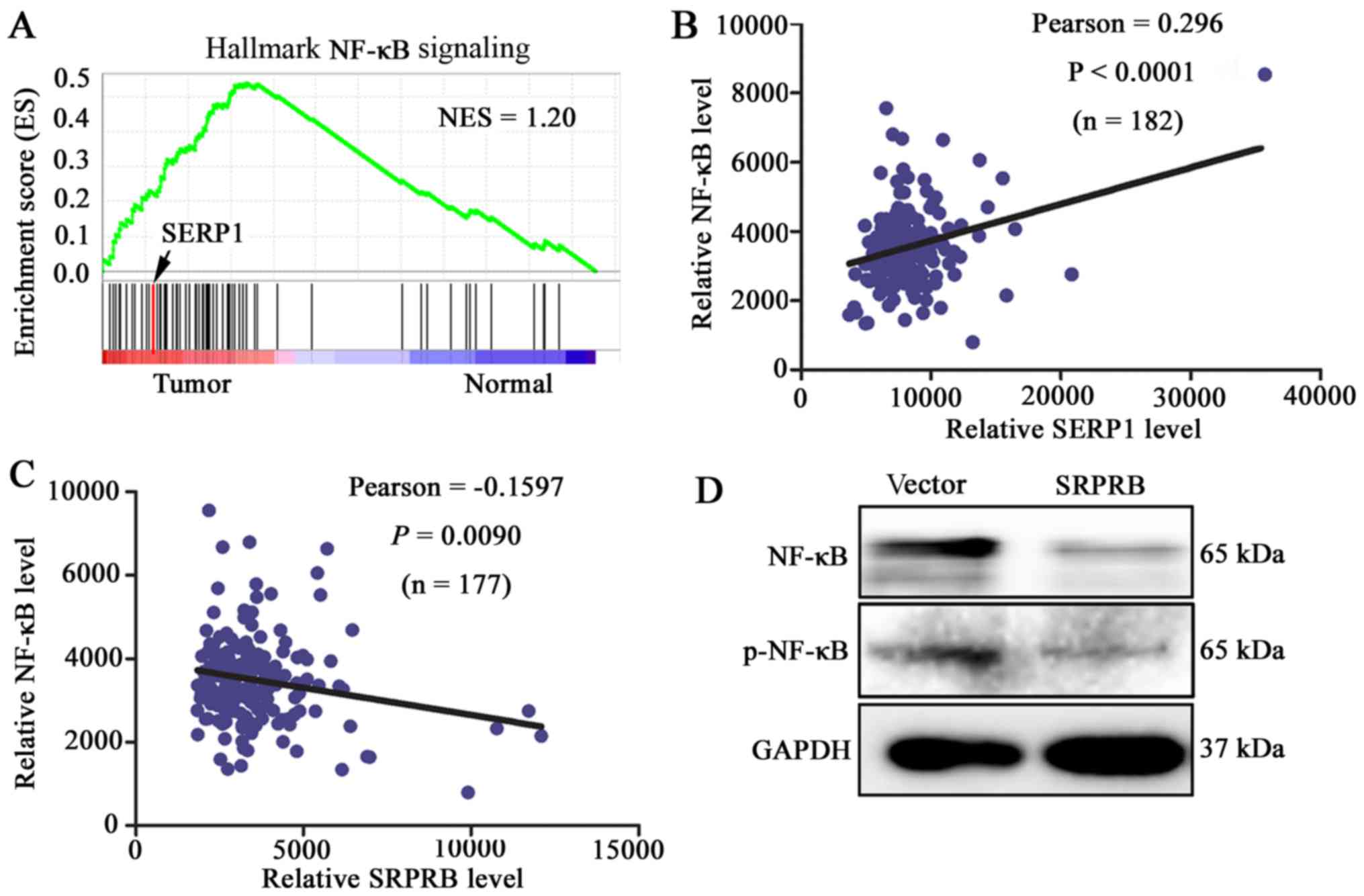
study, we found that gene sets related to NF-κB signaling pathway
were enriched in PDAC tissues by GSEA (Fig. 8A). Moreover, the expression level
of NF-κB was positively correlated with the SERP1 expression
(P<0.0001) (Fig. 8B) and
negatively correlated with SRPRB in 178 PDAC patients from TCGA
database (P=0.0090) (Fig. 8C). To
validate that upregulated SRPRB can suppress the expression of
NF-κB, pcDNA3.0-SRPRB was transfected into PANC-1 cells and the
expression levels of p-NF-κB and NF-κB were detected. The results
demonstrated that the expression levels of the phosphorylation
NF-κB and NF-κB were reduced with SRPRB upregulation in PANC-1
cells (Fig. 8D). The above data
suggested that downregulated SERP1 prompted apoptosis possibly via
upregulating SRPRB expression and inhibiting the expression of
p-NF-κB and NF-κB in PDAC cells.
Discussion
In the present study, the prognostic value of SERP1
in PDAC is reported. We demonstrated that SERP1 expression level
was significantly upregulated in human PDAC tissues compared to
adjacent noncancerous tissues. High expression of SERP1 was
positively associated with advanced clinical stage and shorter
survival time (OS and DFS), indicating poor prognosis in PDAC.
In vitro, downregulated SERP1 expression significantly
promoted PANC-1 cell apoptosis which was mainly carried out by
upregulating SRPRB expression and inhibiting the activation of
NF-κB. Therefore, we speculated SERP1 may be a novel prognosis
marker of PDAC patients via anti-apoptosis, which may be associated
with the expression of SRPRB.
A growing number of new biomarkers have been found,
such as noncoding RNA, microRNAs and liquid biopsies. Among these
biomarkers, C-reactive protein, mutational status of P53, KRAS or
BRCA2 are the most useful biomarkers in clinical practice (24). However, prognostic markers for
pancreatic cancer are still limited in clinical practice. Previous
studies confirmed a significant correlation between high SERP1
levels and poor patient outcome in glioblastoma patients (25), no studies were found in other
tumors. In present study, we first found that SERP1 was strongly
correlated with prognosis of PDAC patients. A previous study found
that SERP1 expression level was enhanced in vitro by hypoxia
and/or reoxygenation or other forms of stress, and associated with
unfolded proteins in endoplasmic reticulum (ER) stress (26). Many different physiological
processes, highly secretory cells such as pancreatic β-cells,
plasma B lymphocytes, and pathological conditions such as hypoxia,
ER Ca2+ depletion, and cancer can cause an imbalance
between ER protein folding load and capacity, leading to
accumulation of unfolded proteins in the ER lumen, a condition
known as 'ER stress' (27). In
addition, SERP1 reintroduction could reverse the hypoxic cell death
and plays an important role in promoting tumor cell survival
(25). The status of ER stress
control early biogenesis of membrane proteins (28), which is very important in cell
death, for example, inhibition of ER stress leads to rescue of
triptolide-mediated pancreatic cancer MIAPaCa-2 cell death
(27). In this study, PPI and GO
analysis suggested that SERP1 and its interactional proteins are
mainly involved in regulating cell death and apoptosis, and we
observed that downregulated SERP1 expression actually promoted cell
apoptosis compared with negative control group in vitro,
which may be a potential cause of influencing prognosis of PDAC
patients.
We found that SRPRB was an important interactional
protein in the whole PPI network, and hypothesized that SRPRB may
have an important role in regulating cell apoptosis. SRPRB is a
novel human gene whose transcripts were upregulated in apoptotic
MCF-7 cells (29), and generally
localized in the cytoplasm. Tissue-specific expression of SRPRB was
found in various human tumors. For example, positive staining of
SRPRB was found in the liver, lung, breast, colon, stomach,
esophagus and testis, exhibited a ubiquitous expression pattern
while its expression was upregulated in tumor tissues compared with
corresponding normal tissues (30). As another study reported, both the
normal and tumor tissues of ovary were absent of SRPRB expression
(30). By contrast, in the present
study, we found that the expression level of SRPRB was
significantly downregulated in PDAC tissues, and significantly
upregulated in stromal fibroblasts. Previous studies found SRPRB
plays certain biological role in the regulation of cell
proliferation and apoptosis, SRPRB induced inhibition of HHCC
growth and cell cycle through regulating genes such as p21 and
TIMP3 (31). In vitro, we
actually observed that SRPRB overexpression obviously increased
cell apoptosis rate, and its expression level could be regulated by
SERP1. In addition, correlation analysis found that NF-κB were
significantly upregulated in PDAC tissues, and positively
correlated with SERP1 level and negatively correlated with SRPRB
level. NF-κB activation plays a critical role in regulating cell
survival (32), it has been proved
that downregulation of NF-κB mediated cell apoptosis (33). For example, inhibition of
prosurvival Akt/NF-κB signaling induced apoptosis in prostate
cancer cells (34), ovarian cancer
cells (35) and astroglioma cells
(36). Activation of NF-κB mainly
occurs via phosphorylation of NF-κB proteins, such as p65, within
their transactivation domain by a variety of kinases in response to
distinct stimuli, and then enhanced cell anti-apoptosis ability
(37). In fact, phosphorylated p65
was significantly upregulated in PDAC cell lines, and associated
with cell proliferation, cell cycle, and apoptosis (38). Moreover, specific targeted
inhibition the phosphorylation NF-κB pathways could induce cell
apoptosis (39). In this study, we
found that SRPRB overexpression obviously decreased the expression
of NF-κB and the phosphorylation NF-κB. Hence, we assumed that
downregulated SERP1 may promote PDAC cell apoptosis via inhibiting
NF-κB activation, which may be associated with SRPRB expression. We
considered that anti-apoptosis induced by SERP1 may be one of the
important mechanisms that could promote tumor progression, and
affect the prognosis of patients, which has not been previously
reported. Although we have demonstrated the relationship between
SERP1 and prognosis as well as its possible mechanism, great number
of clinical sample data are still need.
In conclusion, SERP1 is a novel potential prognostic
marker for PDAC, and downregulated SERP1 expression promoted PDAC
cell apoptosis and inhibited NF-κB activation probably by
upregulating SRPRB expression. The present study provided a novel
pathogenetic mechanism and a new treatment direction for PDAC. To
the best of our knowledge, this is the first study showing the
prognostic value of SERP1 for PDAC patients.
Abbreviations:
|
SERP1
|
stress associated endoplasmic
reticulum protein 1
|
|
SRPRB
|
SRP receptor β subunit
|
|
PDAC
|
pancreatic ductal adenocarcinoma
|
|
ER
|
endoplasmic reticulum
|
Acknowledgments
This study was supported by the Special Foundation
for Scientific Research in the Public Interest by the National
Health and Family Planning Commission of China (no. 201402001),
CAMS Innovation Fund for Medical Sciences (no. 2016-I2M-1-002), the
National Natural Science Foundation of China (no. 31471366).
References
|
1
|
Martinez-Useros J and Garcia-Foncillas J:
The role of BRCA2 mutation status as diagnostic, predictive, and
prognosis biomarker for pancreatic cancer. Biomed Res Int.
2016:18693042016. View Article : Google Scholar
|
|
2
|
Swords DS, Firpo MA, Scaife CL and
Mulvihill SJ: Biomarkers in pancreatic adenocarcinoma: Current
perspectives. Onco Targets Ther. 9:7459–7467. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pietrasz D, Pécuchet N, Garlan F, Didelot
A, Dubreuil O, Doat S, Imbert-Bismut F, Karoui M, Vaillant JC, Taly
V, et al: Plasma circulating tumor DNA in pancreatic cancer
patients is a prognostic marker. Clin Cancer Res. 23:116–123. 2017.
View Article : Google Scholar
|
|
4
|
Tanouchi A, Taniuchi K, Furihata M,
Naganuma S, Dabanaka K, Kimura M, Watanabe R, Kohsaki T, Shimizu T,
Saito M, et al: CCDC88A, a prognostic factor for human pancreatic
cancers, promotes the motility and invasiveness of pancreatic
cancer cells. J Exp Clin Cancer Res. 35:1902016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun B, Liu X, Gao Y, Li L and Dong Z:
Downregulation of miR-124 predicts poor prognosis in pancreatic
ductal adenocarcinoma patients. Br J Biomed Sci. 73:152–157. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nuzhat Z, Kinhal V, Sharma S, Rice GE,
Joshi V and Salomon C: Tumour-derived exosomes as a signature of
pancreatic cancer - liquid biopsies as indicators of tumour
progression. Oncotarget. 8:17279–17291. 2017.
|
|
7
|
Chandra Gupta S and Nandan Tripathi Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar
|
|
8
|
Yang D, Shi J, Fu H, Wei Z, Xu J, Hu Z,
Zhang Y, Yan R and Cai Q: Integrin β1 modulates tumour resistance
to gemcitabine and serves as an independent prognostic factor in
pancreatic adenocarcinomas. Tumour Biol. 37:12315–12327. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Q, Wang H, Zogopoulos G, Shao Q, Dong
K, Lv F, Nwilati K, Gui XY, Cuggia A, Liu JL, et al: Reg proteins
promote acinar-to-ductal metaplasia and act as novel diagnostic and
prognostic markers in pancreatic ductal adenocarcinoma. Oncotarget.
7:77838–77853. 2016.PubMed/NCBI
|
|
10
|
Tsuboi M, Taniuchi K, Furihata M, Naganuma
S, Kimura M, Watanabe R, Shimizu T, Saito M, Dabanaka K, Hanazaki
K, et al: Vav3 is linked to poor prognosis of pancreatic cancers
and promotes the motility and invasiveness of pancreatic cancer
cells. Pancreatology. 16:905–916. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li X, Xiao Y, Fan S, Xiao M, Wang X, Zhu
X, Chen X, Li C, Zong G, Zhou G, et al: Overexpression of DIXDC1
correlates with enhanced cell growth and poor prognosis in human
pancreatic ductal adenocarcinoma. Hum Pathol. 57:182–192. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hori O, Miyazaki M, Tamatani T, Ozawa K,
Takano K, Okabe M, Ikawa M, Hartmann E, Mai P, Stern DM, et al:
Deletion of SERP1/RAMP4, a component of the endoplasmic reticulum
(ER) translocation sites, leads to ER stress. Mol Cell Biol.
26:4257–4267. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim HS, Chang I, Kim JY, Choi KH and Lee
MS: Caspase-mediated p65 cleavage promotes TRAIL-induced apoptosis.
Cancer Res. 65:6111–6119. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guzmán EA, Maers K, Roberts J,
Kemami-Wangun HV, Harmody D and Wright AE: The marine natural
product microsclerodermin A is a novel inhibitor of the nuclear
factor kappa B and induces apoptosis in pancreatic cancer cells.
Invest New Drugs. 33:86–94. 2015. View Article : Google Scholar
|
|
15
|
Mak P, Li J, Samanta S and Mercurio AM:
ERβ regulation of NF-κB activation in prostate cancer is mediated
by HIF-1. Oncotarget. 6:40247–40254. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Simone De V, Franzè E, Ronchetti G,
Colantoni A, Fantini MC, Di Fusco D, Sica GS, Sileri P, MacDonald
TT, Pallone F, et al: Th17-type cytokines, IL-6 and TNF-α
synergistically activate STAT3 and NF-κB to promote colorectal
cancer cell growth. Oncogene. 34:3493–3503. 2015. View Article : Google Scholar
|
|
17
|
Dolcet X, Llobet D, Pallares J and
Matias-Guiu X: NF-κB in development and progression of human
cancer. Virchows Arch. 446:475–482. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ma Q, Peng Z, Wang L, Li Y, Wang K, Zheng
J, Liang Z and Liu T: miR-19a correlates with poor prognosis of
clear cell renal cell carcinoma patients via promoting cell
proliferation and suppressing PTEN/SMAD4 expression. Int J Oncol.
49:2589–2599. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kong FY, Wei X, Zhou K, Hu W, Kou YB, You
HJ, Liu XM, Zheng KY and Tang RX: Bioinformatics analysis reveals
distinct molecular characteristics of hepatitis B-related
hepatocellular carcinomas from very early to advanced Barcelona
clinic liver cancer stages. PLoS One. 11:e01582862016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen Y, Yang Y, Sun M, Yan Z, Wu L, Cui X,
Zhang G, Morris SW and Zhang Q: Inhibition of caspase-8 activity
caused by overexpression of BCL10 contributes to the pathogenesis
of high-grade MALT lymphoma. Pediatr Blood Cancer. 58:865–871.
2012. View Article : Google Scholar
|
|
21
|
Staudt LM: Oncogenic activation of
NF-kappaB. Cold Spring Harb Perspect Biol. 2:a0001092010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kuo SH, Chou CH, Cheng AL, Wang CW, Chen
YH and Chen RJ: Expression of BCL10 in cervical cancer has a role
in the regulation of cell growth through the activation of
NF-κB-dependent cyclin D1 signaling. Gynecol Oncol. 126:245–251.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Z, Zhang H, Chen Y, Fan L and Fang J:
Forkhead transcription factor FOXO3a protein activates nuclear
factor κB through B-cell lymphoma/leukemia 10 (BCL10) protein and
promotes tumor cell survival in serum deprivation. J Biol Chem.
287:17737–17745. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Martinez-Useros J and Garcia-Foncillas J:
Can molecular biomarkers change the paradigm of pancreatic cancer
prognosis? BioMed Res Int. 2016:48730892016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mucaj V, Lee SS, Skuli N, Giannoukos DN,
Qiu B, Eisinger-Mathason TS, Nakazawa MS, Shay JE, Gopal PP,
Venneti S, et al: MicroRNA-124 expression counteracts pro-survival
stress responses in glioblastoma. Oncogene. 34:2204–2214. 2015.
View Article : Google Scholar
|
|
26
|
Yamaguchi A, Hori O, Stern DM, Hartmann E,
Ogawa S and Tohyama M: Stress-associated endoplasmic reticulum
protein 1 (SERP1)/Ribosome-associated membrane protein 4 (RAMP4)
stabilizes membrane proteins during stress and facilitates
subsequent glycosylation. J Cell Biol. 147:1195–1204. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mujumdar N, Banerjee S, Chen Z, Sangwan V,
Chugh R, Dudeja V, Yamamoto M, Vickers SM and Saluja AK: Triptolide
activates unfolded protein response leading to chronic ER stress in
pancreatic cancer cells. Am J Physiol Gastrointest Liver Physiol.
306:G1011–G1020. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Faria D, Lentze N, Almaça J, Luz S,
Alessio L, Tian Y, Martins JP, Cruz P, Schreiber R, Rezwan M, et
al: Regulation of ENaC biogenesis by the stress response protein
SERP1. Pflugers Arch. 463:819–827. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yan W, Wang WL, Zhu F, Chen SQ, Li QL and
Wang L: Isolation of a novel member of small G protein superfamily
and its expression in colon cancer. World J Gastroenterol.
9:1719–1724. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Y, Li Q, Zhu F, Cui J, Li K, Li Q,
Wang R, Wang W, Wang W and Yan W: Subcellular localization of
APMCF1 and its biological significance of expression pattern in
normal and malignant human tissues. J Exp Clin Cancer Res.
28:1112009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Q, Yan W, Cheng S, Guo S and Wang W,
Zhang Z, Wang L, Zhang J and Wang W: Introduction of G1 phase
arrest in human hepatocellular carcinoma cells (HHCC) by APMCF1
gene transfection through the downregulation of TIMP3 and
upregulation of the CDK inhibitors p21. Mol Biol Rep. 33:257–263.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu M, Chen Y, He Y, Podd A, Fu G, Wright
JA, Kleiman E, Khan WN, Wen R and Wang D: Critical role of B cell
lymphoma 10 in BAFF-regulated NF-κB activation and survival of
anergic B cells. J Immunol. 189:5185–5193. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kadirareddy RH, Vemuri SG and Palempalli
UM: Probiotic conjugated linoleic acid mediated apoptosis in breast
cancer cells by downregulation of NFκB. Asian Pac J Cancer Prev.
17:3395–3403. 2016.
|
|
34
|
Liu Y, Gao X, Deeb D, Zhang Y, Shaw J,
Valeriote FA and Gautam SC: Mycotoxin verrucarin A inhibits
proliferation and induces apoptosis in prostate cancer cells by
inhibiting prosurvival Akt/NF-κB/mTOR signaling. J Exp Ther Oncol.
11:251–260. 2016.PubMed/NCBI
|
|
35
|
Yang J, Li G and Zhang K: Pro-survival
effects by NF-κB, Akt and ERK(1/2) and anti-apoptosis actions by
Six1 disrupt apoptotic functions of TRAIL-Dr4/5 pathway in ovarian
cancer. Biomed Pharmacother. 84:1078–1087. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xia W, Tian H, Cai X, Kong H, Fu W, Xing
W, Wang Y, Zou M, Hu Y and Xu D: Inhibition of SUMO-specific
protease 1 induces apoptosis of astroglioma cells by regulating
NF-κB/Akt pathways. Gene. 595:175–179. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang L, Shao L, Creighton CJ, Zhang Y,
Xin L, Ittmann M and Wang J: Function of phosphorylation of NF-κB
p65 ser536 in prostate cancer oncogenesis. Oncotarget. 6:6281–6294.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hu YQ, Si LJ, Ye ZS, Lin ZH and Zhou JP:
Inhibitory effect of ARHI on pancreatic cancer cells and NF-κB
activity. Mol Med Rep. 7:1180–1184. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li W, Wu J, Li Z, Zhou Z, Zheng C, Lin L,
Tan B, Huang M and Fan M: Melatonin induces cell apoptosis in Mia
PaCa-2 cells via the suppression of nuclear factor-κB and
activation of ERK and JNK: A novel therapeutic implication for
pancreatic cancer. Oncol Rep. 36:2861–2867. 2016. View Article : Google Scholar : PubMed/NCBI
|