Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignant tumors in China and worldwide, accounting for
one-sixth of all malignant tumors, and the mortality rate of HCC
ranks second among all cancer-related deaths (1). China has a large population of HCC
patients, accounting for nearly half of the world's total, and
approximately 422,100 patients were expected to die from this
cancer in 2015 (2). Although
surgery, together with other comprehensive treatments, has
significantly improved the prognosis of patients with unresectable
HCC, effective treatments are still lacking (3). Knowledge of the pathogenic mechanisms
of HCC is therefore still in its infancy.
Long non-coding RNAs (lncRNAs), with a size >200
nucleotides, lack protein-coding ability (4). Increasing evidence has recently
suggested that lncRNAs play important roles in cancer occurrence
and progression, by regulating gene expression at the epigenetic,
transcriptional, posttranscriptional, and translational levels
(5–8). Recently, Lessard et al
reported that the lncRNA cancer susceptibility candidate 15
(CASC15) is involved in melanoma progression and phenotype
switching (9), while it functions
as a tumor suppressor in neuroblastoma (10). However, the function of CASC15 in
human HCC is still unknown. We therefore investigated the clinical
correlations and biological function of CASC15 in human HCC.
Materials and methods
Patient data and tissue samples
Eighty-two frozen HCC tumor samples and the
corresponding adjacent normal tissues were obtained from patients
who underwent HCC resection in our department (Hepatobiliary and
Pancreatic Surgery, The First Affiliated Hospital, College of
Medicine, Zhejiang University, China) from 2013 to 2015. No
patients had other liver diseases or received preoperative therapy.
All tissues were preserved at −80°C until analyzed. Clinical data
were collected and checked by two independent physicians. Overall
survival was defined as the time from the date of the surgery to
death or recurrence. The study was approved by the Human Research
Ethics Committee of The First Affiliated Hospital, College of
Medicine, Zhejiang University. Written informed consent was
obtained from all patients.
Total RNA extraction and reverse
transcription quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA was extracted from tissue samples or cell
lines using TRizol® reagent (Invitrogen, Carlsbad, CA,
USA) according to the manufacturer's instructions. The RNA
concentration and quality were determined by UV spectrophotometry.
Reverse transcription of total RNA was performed using the iScript™
cDNA Synthesis kit (Bio-Rad, Hercules, CA, USA) according to the
manufacturer's instructions. The 7500 Fast PCR instrument (Applied
Biosystems, Foster City, CA, USA) was used for quantitative PCR
amplification. The following primers were used: CASC15 forward,
5′-CACACGCATGGAAAACCCAG-3′ and reverse, 5′-GAGGACCTGAGCTGTAAGCC-3′;
and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward,
5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse, 5′-AGGGGCCATCCACAGTCTTC-3′
(9).
Cell lines and cell culture
HCC cells (SMMC7721, Hep-G2, Huh-7, HCCLM3, and
Hep3B) and human normal hepatocytes (QSG-7701) were maintained at
our institute. The cells were maintained in Dulbecco's minimal
essential medium or RPMI-1640 containing 10% fetal calf serum, 100
U/ml penicillin, and 100 µg/ml streptomycin. All cells were
maintained in a humidified incubator at 37°C containing 5%
CO2 and were passaged using standard cell culture
techniques (11).
The CASC15 knockdown model
SMMC7721 and Huh-7 cells were transfected with siRNA
(Smart Silencer; RiboBio, Guangzhou, China) against CASC15 using
Lipofectamine® 2000 (Invitrogen) according to the
manufacturer's protocols. The siRNA sequences were as follows:
locus 1, 5′-CCCTCAGGTGACTACAGAT-3′ (sense) and
5′-GCTCAACCACATCTAATTT-3′ (antisense); locus 2,
5′-GCAACATGCTTCACTGTCT-3′ (sense) and 5′-GATCGCTGGGAATTCTCCAC-3′
(antisense); and locus 3, 5′-TCAGAGCTGGCTGCCTGACA-3′ (sense) and
5′-GCCAAGAAGAGTATGCAGAG-3′ (antisense). CASC15-specific short
hairpin (sh-CASC15) RNA lentiviruses were produced using the
sequence 5′-GAGCAGATAGCTGAAGAGAGA-3′. After transfection, the cells
were treated with 3 µg/ml puromycin (Invitrogen) to
establish stable cell lines. Knockdown of CASC15 was confirmed by
RT-qPCR.
Cell proliferation assays
Cell proliferation was monitored using the Cell
Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). Cells
(1×104) were seeded into 96-well plates after
transfection and were counted daily for 3 consecutive days. The
absorbance was read at 450 nm. Cell viability was also assessed
using the 5-ethynyl-2′-deoxyuridine (EdU) assay (Cell-light EdU
Apollo 567 in vitro imaging kit; Ribobio, Guangzhou, China)
following the manufacturer's instructions. For the colony formation
assay, the cells were inoculated into 6-well plates (1,000/well)
and maintained in a humidified atmosphere at 37°C containing 5%
CO2, and the size of the colonies was observed after 2
weeks. Subsequently, the cells were fixed with methanol and stained
with 1% crystal violet.
Cell cycle analysis
The cells were fixed in 70% ethanol, stored at 4°C
for >24 h, stained with DNA Prep (Beckman Coulter, Brea, CA,
USA), and analyzed for cell cycle distribution using flow
cytometry.
Cell apoptosis analysis
The cells were collected 48 h after transfection,
and then stained with fluorescein isothiocyanate (FITC)-Annexin V
and propidium iodide using the FITC Annexin V Apoptosis Detection
kit (BD Biosciences, San Jose, CA, USA) according to the
manufacturer's protocol. The stained cells were analyzed using a
flow cytometer (Cytomics FC 500; Beckman Coulter, Miami, FL,
USA).
Cell migration and invasion assays
To assess cell migration, 4×104
transfected cells in 200 µl serum-free medium were added to
the upper chamber of a Transwell® and incubated for 48
h. The bottom chamber contained 10% fetal bovine serum. After
incubation, the membranes were isolated and stained using
Diff-Quick (Polyscience, Warrington, PA, USA) according to the
manufacturer's protocols. The cells from 10 fields were counted
using an inverted microscope (Leica, Malvern, PA, USA). For the
cell invasion assay, 40 µl diluted Matrigel™ (1:8) was added
to the upper chamber of the Transwell. After incubation at 37°C for
30 min, 4×104 transfected cells in 200 µl
serum-free medium were added to the upper chamber, and subsequent
procedures were performed as described above.
Xenograft model
Five-week-old female BALB⁄c nude mice were purchased
from the Shanghai Sippr/BK Laboratory Animal Co. (Shanghai, China).
SMMC7721 cells were collected 48 h after transfecting with
Sh-CASC15 or the control (Shanghai Genechem, Shanghai, China).
SMMC7721 cells were suspended in phosphate-buffered saline and
injected subcutaneously into the left flank of mice
(5×106 cells/mouse). The mice were euthanized after 21
days, and relative data were collected. The tumor volume was
calculated using the following formula: volume = (L ×
W2)/2, where L and W were the longest and shortest
diameters of the tumors, respectively. All research involving
animals complied with protocols approved by the Zhejiang Medical
Experimental Animal Care Commission.
Immunohistochemistry (IHC)
Tumors from mice were immunostained with a primary
antibody targeting Ki-67 (1:400; Abcam, Cambridge, UK).
Western blot analysis
Western blot analysis was performed as described
previously (11). Equal protein
loading was monitored using an anti-GAPDH antibody (60004-1-IG;
Proteintech, Rosemount, IL, USA). Primary antibodies against cyclin
D1 (ab134175; Abcam), cyclin E1 (ab88259; Abcam), CDK2 (ab32146;
Abcam), CDK4 (ab137675; Abcam), CDK6 (ab124821; Abcam), caspase 3
(ab17868; Abcam), polyADP-ribose polymerase (PARP) (ab32064;
Abcam), Bax (ab32503; Abcam), matrix metalloprotein (MMP)-3,
E-cadherin, and N-cadherin (Cell Signaling Technology, Danvers, MA,
USA) were used.
Statistical analysis
Data were analyzed using SPSS statistical software
for Windows, version 19.0 (IBM, Chicago, IL, USA). The results are
presented as means ± SD. All experiments were performed in
triplicate. A value of P<0.05 was considered statistically
significant.
Results
CASC15 is overexpressed in HCC tumor
specimens and associated with metastasis, tumor size, TNM stage,
and poor prognosis
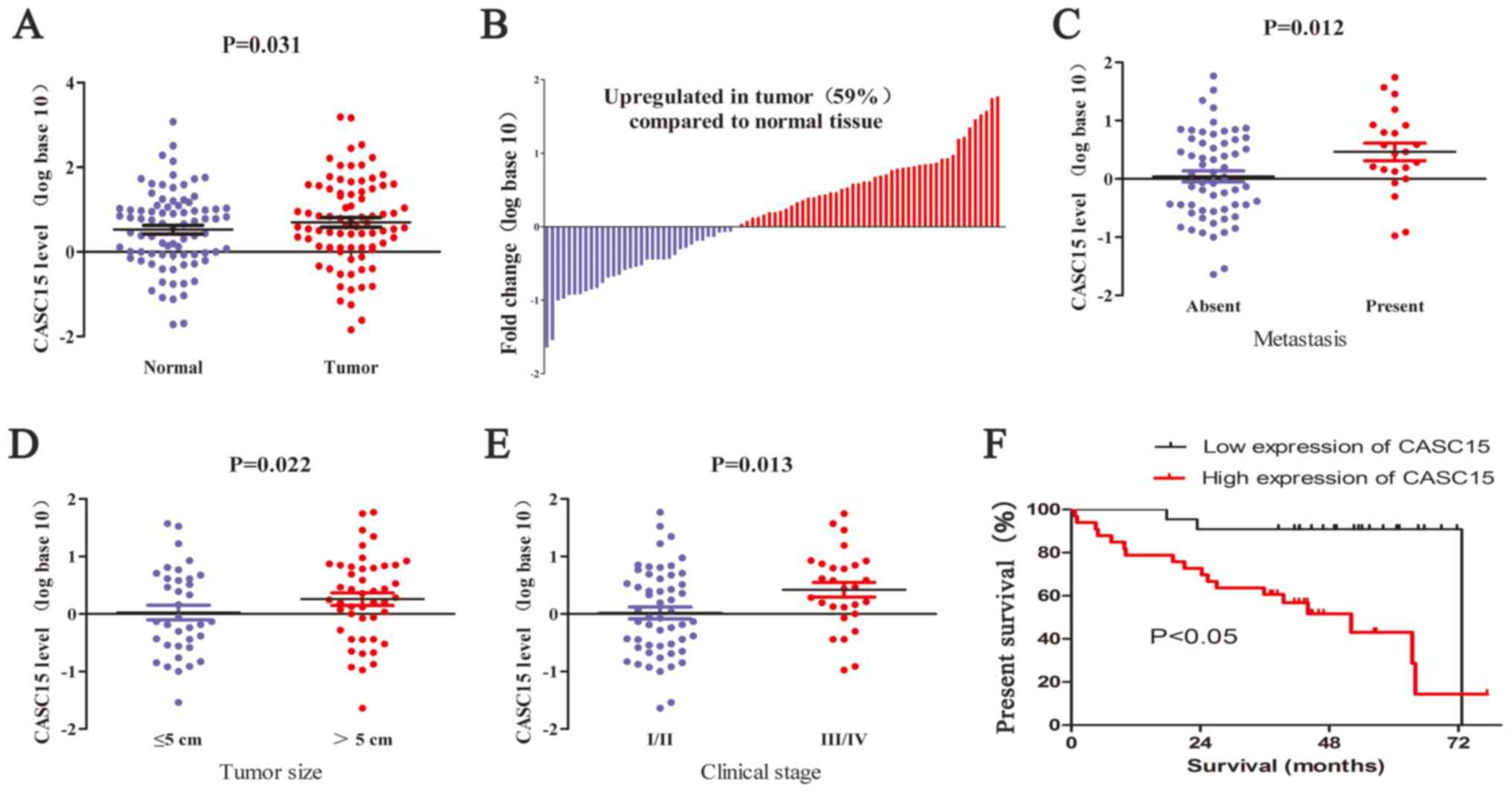
We determined the expression levels of CASC15 in HCC
tissues using RT-qPCR and found that the expression of CASC15 was
upregulated in 59% (48 of 82) of HCC tissues compared with that in
paired adjacent normal tissues (Fig.
1A and B). Analyses of potential correlations between CASC15
expression and clinicopathological features in the 82-patient
cohort showed that a high expression of CASC15 was significantly
correlated with metastasis, large tumor size, advanced and tumor
stages (Fig. 1D–F and Table I). To assess an association between
CASC15 expression and overall survival rate, the 82 patients with
HCC after hepatectomy were divided into two groups: high CASC15
expression (n=48) and low CASC15 expression (n=34). Kaplan-Meier
survival curves showed that high CASC15 expression was associated
with poor prognoses in patients with HCC (P<0.05).
 | Table ICorrelation between CASC15 expression
and clinicopathological characteristics with HCC. |
Table I
Correlation between CASC15 expression
and clinicopathological characteristics with HCC.
| Clinical
parameter | CASC-15
| P value |
|---|
| Low no. cases | High no. cases |
|---|
| Age (years) | | | 0.027 |
| <50 | 5 | 18 | |
| ≥50 | 29 | 30 | |
| Sex | | | 0.302 |
| Male | 31 | 47 | |
| Female | 3 | 1 | |
| HBV | | | 0.342 |
| Yes | 31 | 39 | |
| No | 3 | 9 | |
| Cirrhosis | | | 1.000 |
| Yes | 26 | 36 | |
| No | 8 | 12 | |
| AFP | | | 0.122 |
| ≤20 | 20 | 17 | |
| 20–400 | 3 | 7 | |
| ≥400 | 11 | 24 | |
| Tumor size | | | 0.022 |
| ≤5 cm | 20 | 16 | |
| >5 cm | 14 | 32 | |
| TNM stage | | | 0.013 |
| I–II | 28 | 27 | |
| I II–IV | 6 | 21 | |
| Tumor
differentiation | | | 0.246 |
| I–II | 14 | 26 | |
| III–IV | 20 | 22 | |
| Metastasis | | | 0.012 |
| Yes | 4 | 18 | |
| No | 30 | 30 | |
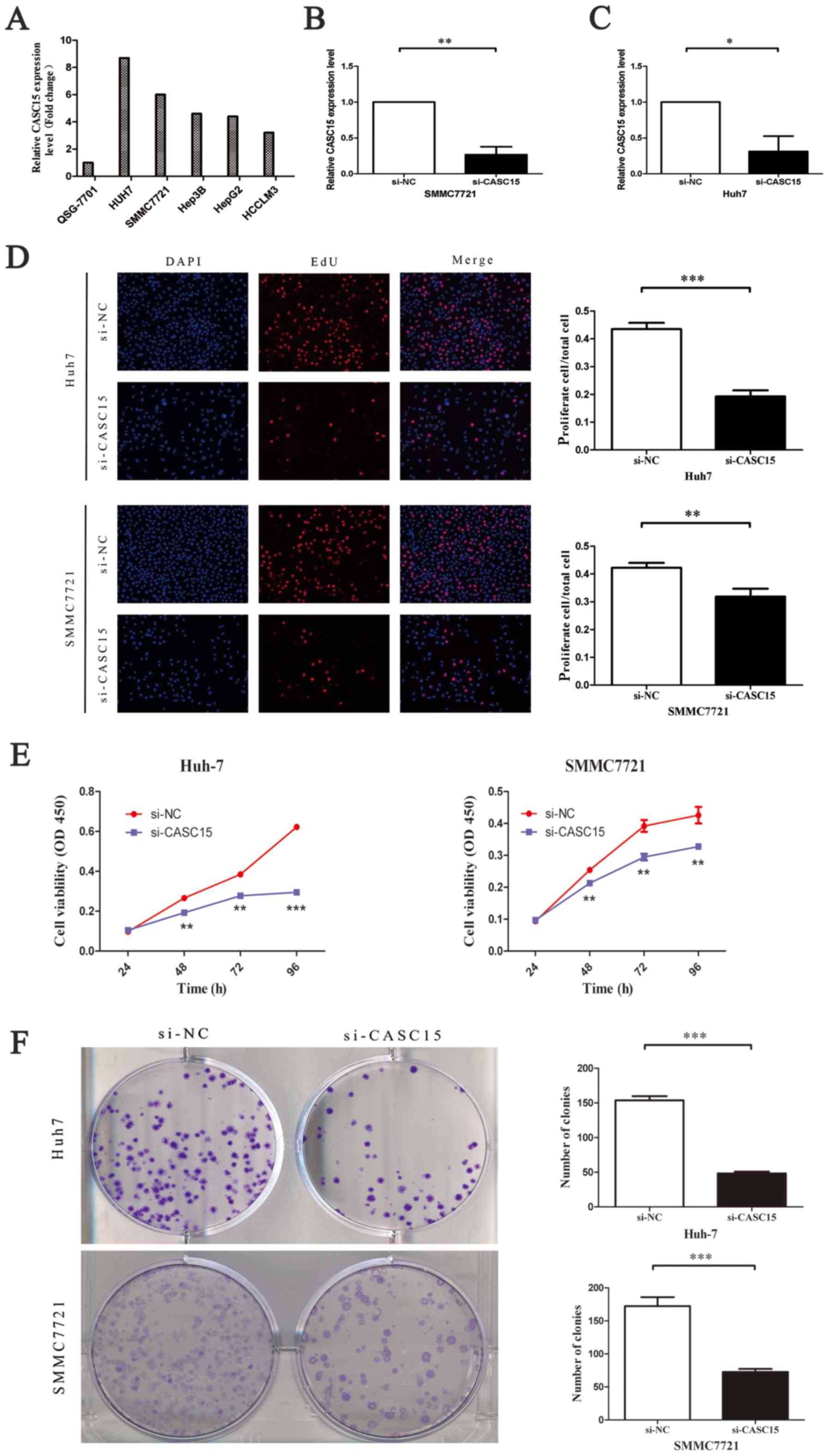
CASC15 is upregulated in HCC cell lines,
promoting tumor cell proliferation and repressing apoptosis in
vitro
The expression of CASC15 in human HCC cells
(SMMC7721, Hep-G2, Huh-7, HCCLM3 and Hep3B) and human normal
hepatocytes (QSG-7701) was detected by RT-qPCR. The mRNA level of
CASC15 was higher in the HCC cells than in QSG-7701 cells (Fig. 2A). Next, we transfected SMMC-7721
and Huh-7 cells with non-targeting control siRNA or CASC15 siRNA
(Fig. 2B and C). An EdU assay
performed to quantitate cell proliferation showed that cell
proliferation decreased significantly after treatment with the
CASC15 siRNA in both Huh7 and SMMC7721 cells (Fig. 2D). This was consistent with the
results of the Cell Counting Kit-8 assay (Fig. 2E). In addition, a colony formation
assay showed that downregulation of CASC15 significantly inhibited
colony formation in HCC cells (Fig.
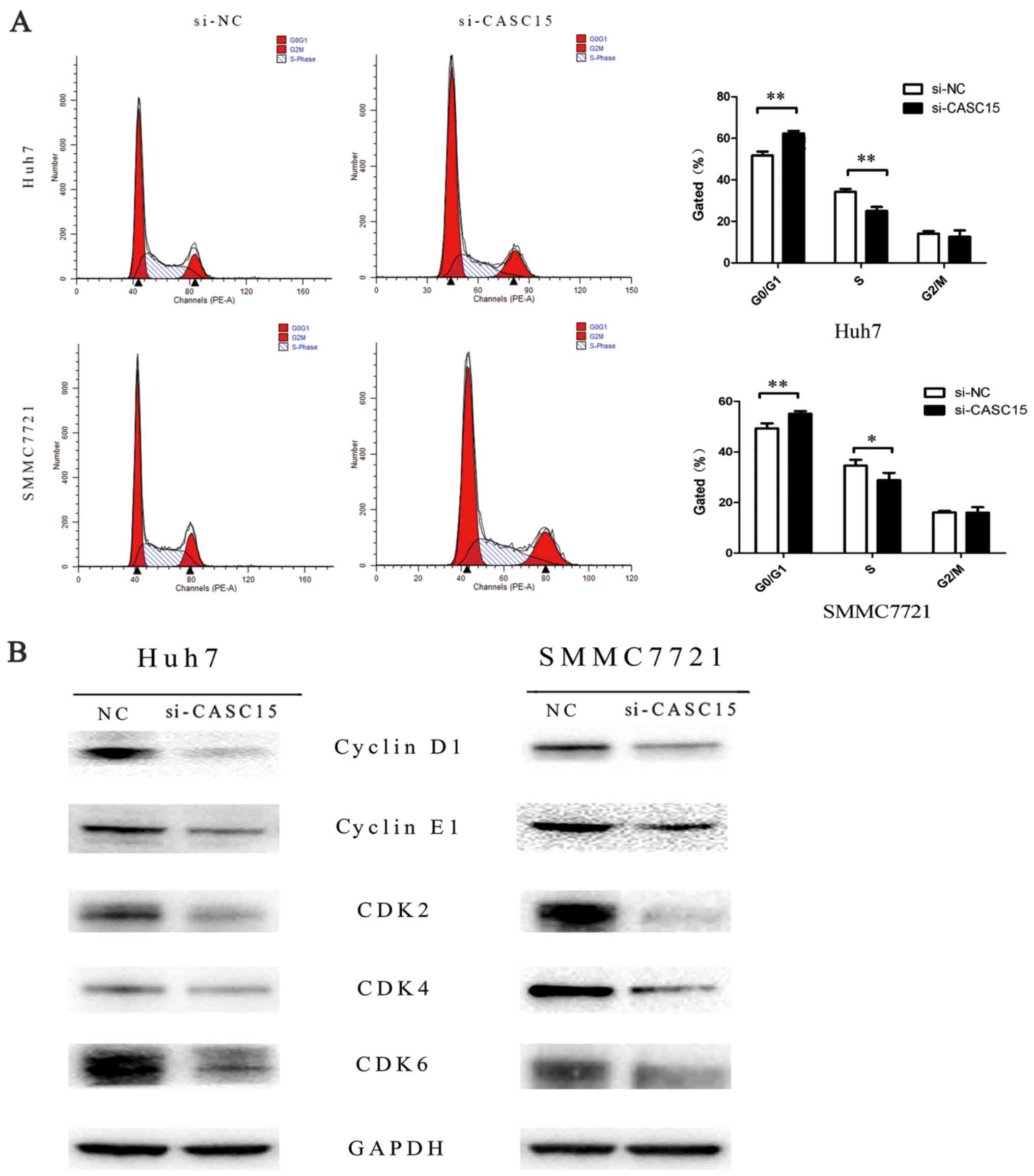
2F). To determine if the effect on cell proliferation affected
the cell cycle, we assessed cell cycle changes and found that more
cells in G1 phase arrest were detected in the CASC15
siRNA-transfected cells compared with the negative control
siRNA-transfected cells (Fig. 3A),
with decreased expression of cyclin D1, cyclin E1, CDK2, CDK4, and
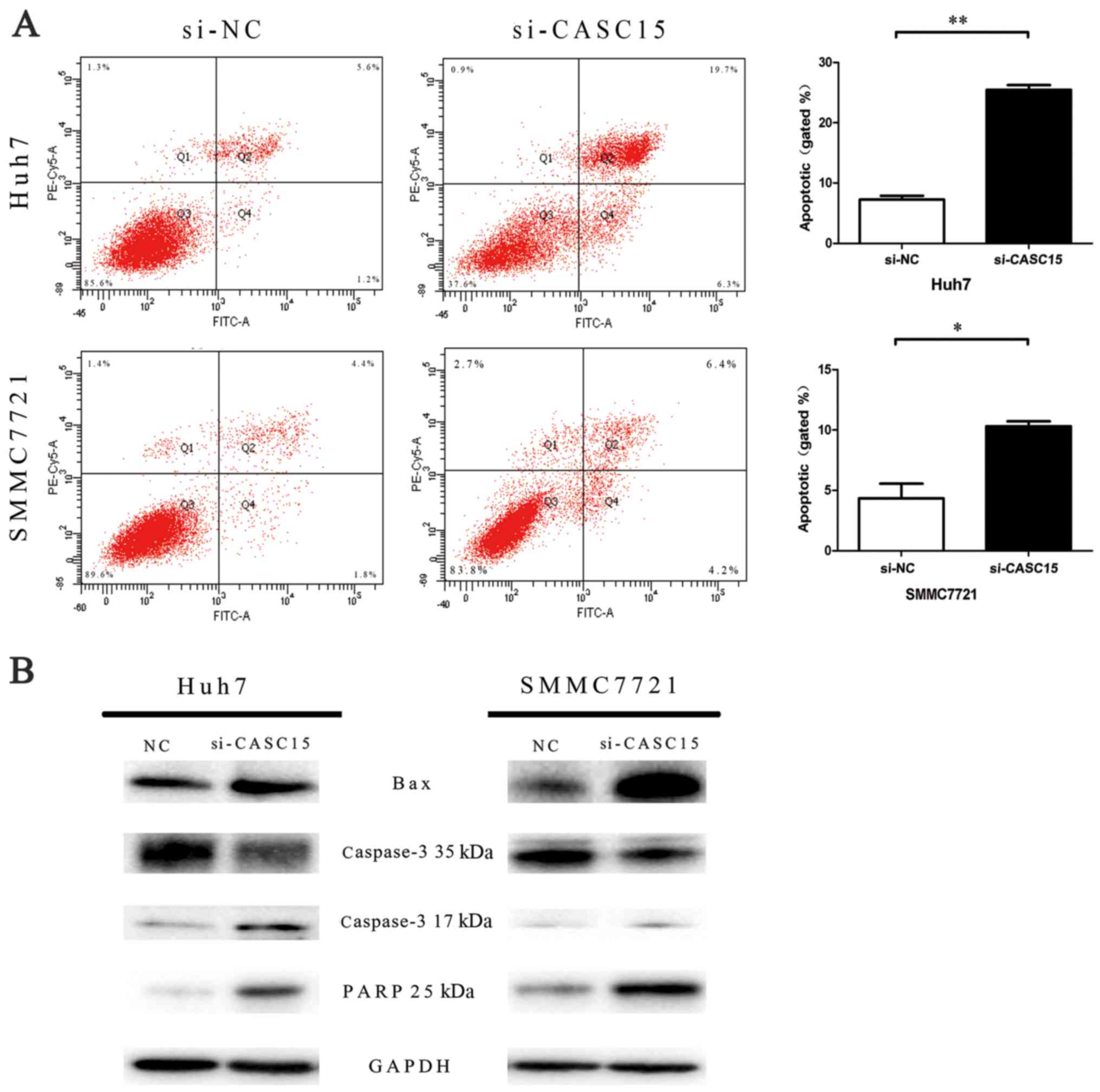
CDK6 compared with the negative control (Fig. 3B). In addition, flow cytometric
analyses indicated that knockdown of CASC15 expression increased
apoptosis (Fig. 4).
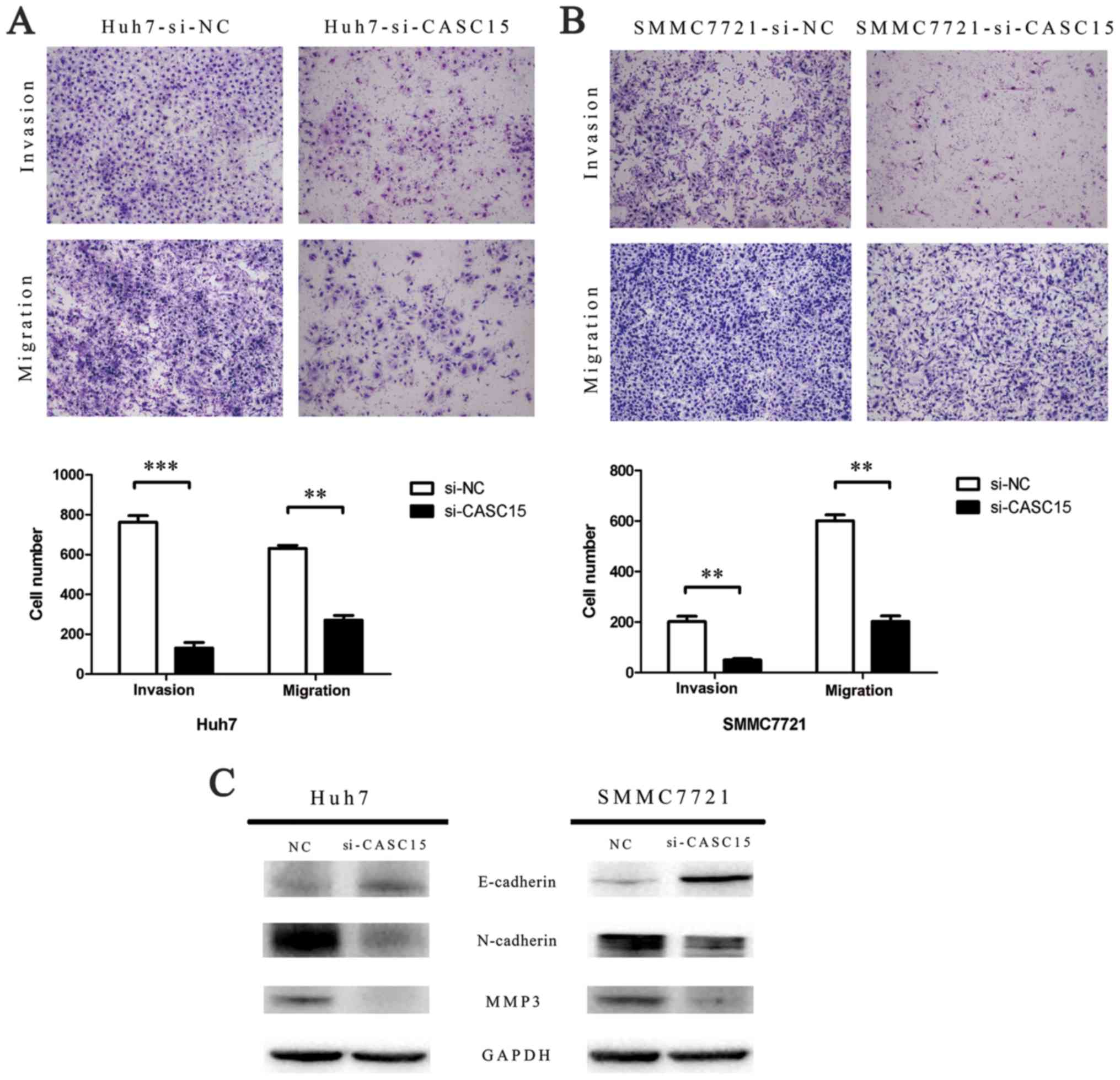
Knockdown of CASC15 inhibits HCC cell
migration and invasion in vitro
The role of CASC15 in HCC progression was assessed
by Transwell assay to measure cell migration and invasion ability.
The results showed that the migration and invasion capabilities of
CASC15 siRNA-transfected cells were reduced significantly compared
with the negative control siRNA-transfected cells (Fig. 5A and B). Furthermore, western blots
showed that E-cadherin expression was increased, while N-cadherin
and MMP-3 expression was decreased, in CASC15 siRNA-transfected
cells (Fig. 5C).
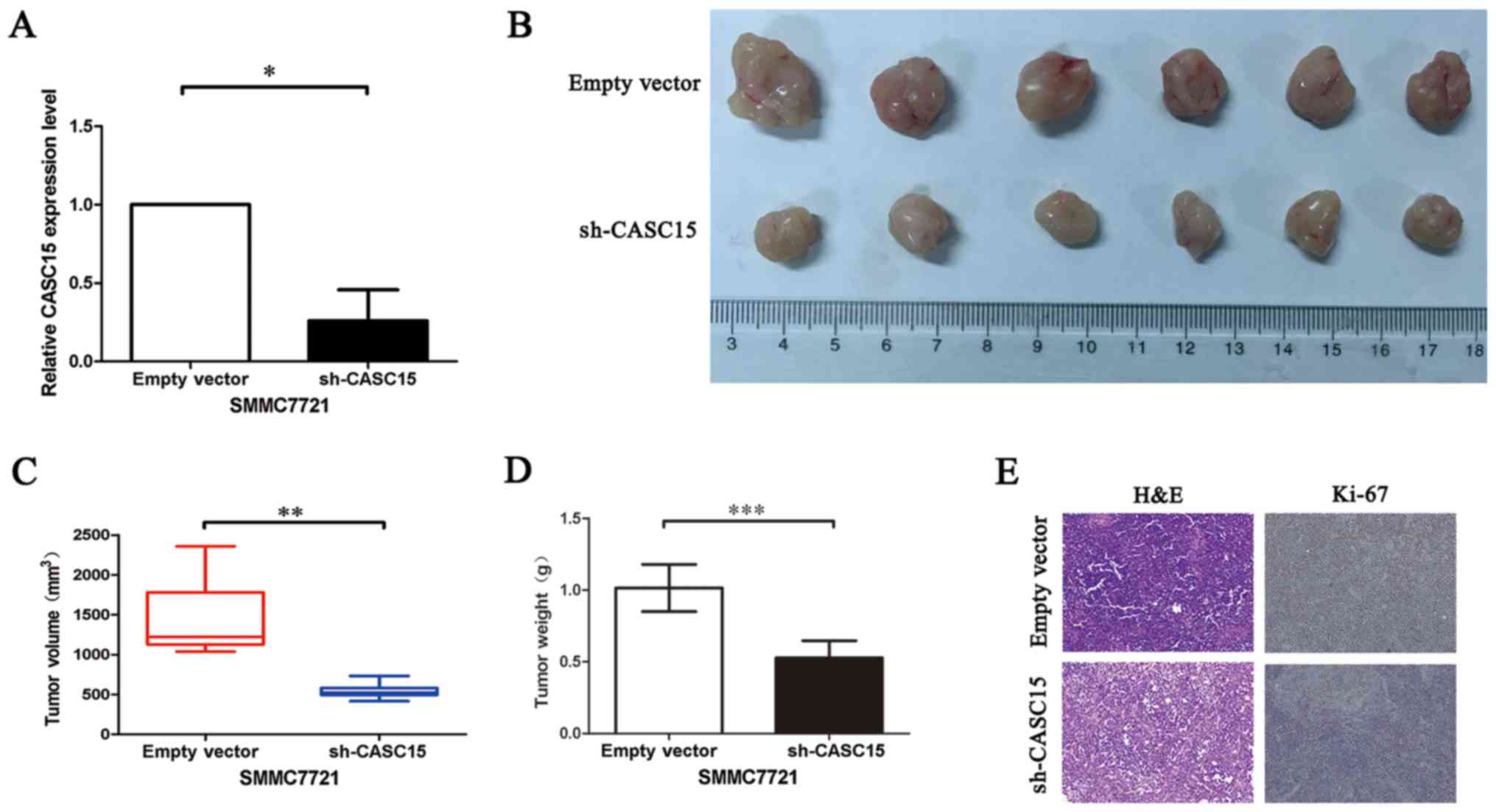
Knockdown of CASC15 suppressed HCC
progression in vivo
To confirm the in vivo oncogenic role of
CASC15, we constructed a subcutaneous xenograft model. CASC15 was
knocked down in SMMC7721 cells using sh-CASC15 (Fig. 6A). Then, sh-CASC15 and control
SMMC7721 cells were injected subcutaneously into nude mice.
Consistent with our in vitro results, the sh-CASC15 cells
showed significant inhibition of tumor growth compared with the
control cells (Fig. 6B and C). The
tumor weight of the sh-CASC15-induced tumors was also significantly
lower than that of the control tumors (Fig. 6D). Ki-67 staining confirmed these
results (Fig. 6E). Taken together,
the results indicated that knockdown of CASC15 suppressed in
vivo tumor progression.
Discussion
HCC remains one of the major causes of
cancer-related death worldwide (12), even though surgical treatment,
together with other comprehensive treatments, have significantly
improved prognosis. The difficulty in diagnosing HCC early and the
lack of effective treatments for patients with unresectable HCC
contribute to the severity of this disorder. Therefore, there is an
urgent need for novel markers for early detection and effective
molecular targeting.
Increasing numbers of studies have reported that
lncRNAs play important roles in cancer occurrence and progression,
by regulating gene expression at the epigenetic, transcriptional,
posttranscriptional, and translational levels (13–17).
For example, HULC was the first lncRNA identified as specifically
upregulated in HCC, in 2007 (18).
HULC modulates abnormal lipid metabolism to promote HCC progression
(19). Many other lncRNAs, such as
HOTTIP, HEIH, and DREH, have been reported to play important roles
in HCC (20–22).
In the present study, we identified another lncRNA
upregulated in HCC, CASC15, which was overexpressed in HCC tissues
compared with corresponding adjacent normal tissues. While CASC15
has many isoforms, which may have unique functions (9), our findings showed that their
globally upregulated expression was correlated with metastasis,
tumor size, TNM stage, and survival outcomes, suggesting that
CASC15 may be used as a predictive molecular marker for HCC
prognosis. We also found CASC15 consistently upregulated in HCC
cell lines, acting as an oncogene in HCC.
Knockdown of CASC15 in SMMC7721 and Huh7 cells
resulted in an increase in the number of cells in G1
phase arrest. Cell cycle-related proteins (cyclins E1/D1 and
CDK2/4/6) were decreased after CASC15 knockdown. In previous
studies, when CDK2 was bound to cyclin E, the complex promoted
G1/S transition (23),
and cyclin D bound to CDK4 and CDK6 led to phosphorylation and
inactivation of the retinoblastoma protein, to facilitate
E2F-dependent transcription (24).
Thus, CASC15 may facilitate proliferation via upregulation of
cyclins and CDKs.
Defects in apoptosis are closely related to tumor
development (25). Apoptosis
includes two main pathways: the extrinsic pathway and intrinsic
pathways (26). Bcl-2/Bax
activates the intrinsic pathway (27). Caspase-3 is the most important of
the executioner caspases, and cleaved caspase-3 cleaves PARP,
leading to apoptosis (28). In our
study, cell apoptosis increased with increasing expression of
apoptosis-related proteins (Bax cleaved caspase-3 and PARP) after
knockdown of CASC15 expression. Thus, CASC15 may facilitate
proliferation by inhibiting the intrinsic apoptosis pathway.
Many studies have reported that the
epithelial-mesenchymal transition (EMT) is closely associated with
the ability of tumor cells to migrate to distant organs (29,30).
Tumor cells modulate their phenotype by activation of various
EMT-associated pathways (31). In
the present study, cell migration and invasion ability were
suppressed by increased expression of an epithelial marker protein
(E-cadherin) and decreased expression of a mesenchymal marker
protein (N-cadherin), after knockdown of CASC15 expression. Thus,
CASC15 facilitates cell migration and invasion by affecting the
EMT-associated pathway.
In conclusion, this study is the first to show that
CASC15 is overexpressed in HCC tissues compared with corresponding
adjacent normal tissues. Furthermore, it showed that high
expression of CASC15 is correlated with a poor prognosis. CASC15
promoted HCC proliferation by regulation of apoptosis and the cell
cycle, both in vitro and in vivo. Moreover, CASC15
facilitated cell migration and invasion by affecting the
EMT-associated pathway. These results indicate that CASC15 acts as
an oncogene in HCC and is a potential predictive biomarker for
HCC.
Acknowledgments
This study was supported by National Natural Science
Foundation of China [grant no. 81572307(WLW)], Major Project of
Medical and Health Technology Development Program in Zhejiang
Province [grant no. 7211902 (WLW)], National Key Basic Research
Development Program (973 Program) [grant no. 2013CB531403 (WLW)]
and Science and Technology Major Project of Zhejiang Province
[grant no. 2014C13G2010059 (WLW)].
References
|
1
|
Stewart BW and Wild CP: World Cancer
Report. Organization World Health; 2014, https://shop.iarc.fr/products/wcr2014.
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Llovet JM and Bruix J: Systematic review
of randomized trials for unresectable hepatocellular carcinoma:
Chemoembolization improves survival. Hepatology. 37:429–442. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long non-coding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang ZZ, Shen ZY, Shen YY, Zhao EH, Wang
M, Wang CJ, Cao H and Xu J: HOTAIR long non-coding RNA promotes
gastric cancer metastasis through suppression of poly r(C)-binding
protein (PCBP) 1. Mol Cancer Ther. 14:1162–1170. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia
R, Liu YW, Liu XH, Zhang EB, Lu KH, et al: Long non-coding RNA
ANRIL promotes non-small cell lung cancer cell proliferation and
inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer
Ther. 14:268–277. 2015. View Article : Google Scholar
|
|
7
|
Li L, Chen H, Gao Y, Wang YW, Zhang GQ,
Pan SH, Ji L, Kong R, Wang G, Jia YH, et al: Long non-coding RNA
MALAT1 promotes aggressive pancreatic cancer proliferation and
metastasis via the stimulation of autophagy. Mol Cancer Ther.
15:2232–2243. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wilusz JE, Sunwoo H and Spector DL: Long
non-coding RNAs: Functional surprises from the RNA world. Genes
Dev. 23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lessard L, Liu M, Marzese DM, Wang H,
Chong K, Kawas N, Donovan NC, Kiyohara E, Hsu S, Nelson N, et al:
The CASC15 long Intergenic non-coding RNA locus is involved in
melanoma progression and phenotype switching. J Invest Dermatol.
135:2464–2474. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Russell MR, Penikis A, Oldridge DA,
Alvarez-Dominguez JR, McDaniel L, Diamond M, Padovan O, Raman P, Li
Y and Wei JS: CASC15-S is a tumor suppressor lncRNA at the 6p22
neuroblastoma susceptibility locus. Cancer Res. 75:3155–3166. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang W, Xie Q, Zhou X, Yao J, Zhu X, Huang
P, Zhang L, Wei J, Xie H, Zhou L, et al: Mitofusin-2 triggers
mitochondria Ca2+ influx from the endoplasmic reticulum
to induce apoptosis in hepatocellular carcinoma cells. Cancer Lett.
358:47–58. 2015. View Article : Google Scholar
|
|
12
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang KC, Yang YW, Liu B, Sanyal A,
Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta
RA, et al: A long non-coding RNA maintains active chromatin to
coordinate homeotic gene expression. Nature. 472:120–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang J, Jiang R, Deng L, Zhang X, Wang K
and Sun B: Circulation long non-coding RNAs act as biomarkers for
predicting tumorigenesis and metastasis in hepatocellular
carcinoma. Oncotarget. 6:4505–4515. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y,
Chen N, Sun F and Fan Q: CREB up-regulates long non-coding RNA,
HULC expression through interaction with microRNA-372 in liver
cancer. Nucleic Acids Res. 38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tu ZQ, Li RJ, Mei JZ and Li XH:
Down-regulation of long non-coding RNA GAS5 is associated with the
prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol.
7:4303–4309. 2014.PubMed/NCBI
|
|
17
|
Huang MD, Chen WM, Qi FZ, Xia R, Sun M, Xu
TP, Yin L, Zhang EB, De W and Shu YQ: Long non-coding RNA ANRIL is
upregulated in hepatocellular carcinoma and regulates cell
apoptosis by epigenetic silencing of KLF2. J Hematol oncol.
8:502015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M, et al: Characterization of HULC, a novel gene with
striking up-regulation in hepatocellular carcinoma, as non-coding
RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cui M, Xiao Z, Wang Y, Zheng M, Song T,
Cai X, Sun B, Ye L and Zhang X: Long non-coding RNA HULC modulates
abnormal lipid metabolism in hepatoma cells through an
miR-9-mediated RXRA signaling pathway. Cancer Res. 75:846–857.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang
Y, Tang GN, Zhou WP and Sun SH: Hepatitis B virus X protein
(HBx)-related long non-coding RNA (lncRNA) down-regulated
expression by HBx (Dreh) inhibits hepatocellular carcinoma
metastasis by targeting the intermediate filament protein vimentin.
Hepatology. 57:1882–1892. 2013. View Article : Google Scholar
|
|
21
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long non-coding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Quagliata L, Matter MS, Piscuoglio S,
Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z,
Boldanova T, et al: Long non-coding RNA HoTTiP/HoXA13 expression is
associated with disease progression and predicts outcome in
hepatocellular carcinoma patients. Hepatology. 59:911–923. 2014.
View Article : Google Scholar :
|
|
23
|
Tsai LH, Harlow E and Meyerson M:
Isolation of the human cdk2 gene that encodes the cyclin A- and
adenovirus E1A-associated p33 kinase. Nature. 353:174–177. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Massagué J: G1 cell-cycle control and
cancer. Nature. 432:298–306. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kerr JF, Wyllie AH and Currie AR:
Apoptosis: A basic biological phenomenon with wide-ranging
implications in tissue kinetics. Br J Cancer. 26:239–257. 1972.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Elmore S: Apoptosis: A review of
programmed cell death. Toxicol Pathol. 35:495–516. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gu W, Li C, Yin W, Guo Z, Hou X and Zhang
D: Shen-fu injection reduces postresuscitation myocardial
dysfunction in a porcine model of cardiac arrest by modulating
apoptosis. Shock. 38:301–306. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lazebnik YA, Kaufmann SH, Desnoyers S,
Poirier GG and Earnshaw WC: Cleavage of poly(ADP-ribose) polymerase
by a proteinase with properties like ICE. Nature. 371:346–347.
1994. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Klymkowsky MW and Savagner P:
Epithelial-mesenchymal transition: A cancer researcher's conceptual
friend and foe. Am J Pathol. 174:1588–1593. 2009. View Article : Google Scholar : PubMed/NCBI
|