Introduction
Lung cancer is the leading cause of cancer-related
deaths world-wide, accounting for 12.7% of new cancer cases and
18.2% of new cancer deaths (1,2).
Lung cancer is a major health concern and pose heavy burden on
family and society (2). Based on
histology, lung cancer can be classified into non-small cell lung
cancer (NSCLC) and small cell lung cancer (SCLC) (3). In general, NSCLC accounts for >80%
of all lung cancers, and can be further classified into squamous
carcinoma, adenocarcinoma, and large cell carcinoma (4,5).
Despite the significant improvement in surgery and chemotherapy,
the prognosis of advanced lung cancer remains poor (6,7). The
overall 5-year survival of lung cancer remains ~17% (8). Tumor recurrence and distant
metastasis are the leading causes of death in advanced lung cancer
patients (9). Thus, it is
important to understand the underlying molecular mechanism of tumor
progression and metastasis in order to design new therapeutic
agents for lung cancer patients.
LIM and SH3 protein 1 (LASP-1), also called
metastatic lymph node gene 50 proteins (MLN50), were identified
from a cDNA library of breast cancer (7,10).
LASP-1 encodes a putative protein of 261 amino acids, with an
N-terminal LIM domain and a C-terminal SRC homology 3 (SH3) domain
(10,11). The LIM domain is followed by two
actin-binding domains in the core of LASP-1 protein, which
interacts with various binding partners within the cytoskeleton and
transmit signals from the cytoplasm into the nucleus (11,12).
The SH3 domain is involved in protein-protein interactions, binding
proline-rich sequences, such as zyxin and pallidin (13). Owing to its ubiquitous expression
in many tissues, LASP-1 exhibits wide range of biological
functions, including cell morphology, signal transduction, and cell
motility (14,15). LASP-1 has been reported to be
overexpressed in several types of human cancers, such as breast
cancer (16,17), ovarian cancer (18), gastric cancer (19), hepatocellular cancer (20), colorectal cancer (21), lung cancer (7,22),
and renal cell cancer (23),
suggesting LASP-1 as a potential biomarker for the treatment of
cancer. Several studies have shown that LASP-1 can promote cell
proliferation, migration, and invasion in a wide variety of tumors
both in vitro and in vivo (11,17).
In this study, we demonstrated the role of LASP-1 as potential
biomarker in cancer; however, the underlying mechanism of how
LASP-1 mediates oncogenesis in lung cancer remains unclear
(7).
SOX9, a transcription factor and member of the SOX
family, is a key regulatory protein, which is involved in
developmental processes, including male sex determination,
chondrogenesis, neurogenesis, and neural crest development
(24–26). Recent cogent evidence has provided
a link between SOX9 and cancer progression. SOX9 overexpression has
been reported in breast cancer (27), prostate cancer (28), colorectal cancer (29), and lung cancer (30–32),
where its expression is correlated with malignancy and overall
survival. Although an association between upregulation of SOX9 and
lung cancer progression has been reported (33), the potential regulatory effect of
SOX9 and LASP-1 in lung cancer needs further validation.
In the present study, we investigated the expression
of LASP-1 in lung cancer using human tissue samples and assessed
the potential role of LASP-1 in lung cancer cell-lines. The results
have shown that knockdown of LASP-1 expression by siRNA reduced
cell proliferation and increased cell apoptosis. Moreover,
mechanistic investigation showed that LASP-1 was a critical
downstream target of SOX9. Taken together, these findings suggested
that SOX9-LASP1 axis plays an important role in cell proliferation,
migration, and invasion.
Materials and methods
Ethics statement
The Institutional Ethics Committee at the Ningbo
First Hospital, China, approved this study, and written informed
consent was obtained from all the patients prior to their
participation.
Cell culture
Human lung cancer cell lines, A549, NCI-H838,
NCI-H1299, and NCI-H1650 were obtained from the American Type
Culture Collection (ATCC, Rockville, MD, USA) and cultured in
RPMI-1640 (Hyclone) supplemented with 10% fetal bovine serum (FBS)
(Gibco) and 1% penicillin/streptomycin. The immortalized human
bronchial epithelial cell line, BEAS-2B (ATCC), was maintained in
BEGM medium (Lonza, Switzerland) supplemented with 10% FBS and 1%
penicillin/streptomycin. The cells were cultured at 37°C in
water-saturated 5% CO2 atmosphere.
Tissue samples
Thirteen human lung cancer and paired tumor-adjacent
normal tissues were obtained from Ningbo First Hospital, China. All
patients were pathologically and clinically diagnosed as lung
cancer patients. None of the patients had undergone radiotherapy or
chemotherapy before surgical resection. The histological diagnosis
of melanoma was evaluated according to the World Health
Organization (WHO). The tissues, >5 cm away from the cancer
lesions, were defined as tumor-adjacent normal tissues.
Plasmid
Twist2, Nkx2-5, and Sox9 were amplified using PCR
from NCI-1650 and cloned into the pGL3-basic vector (Promega,
Madison, WI, USA). Mutations of the Twist2, Nkx2-5, and Sox9
binding site was performed by Quik Change Site Mutagenesis kit
(Stratagene, La Jolla, CA, USA) according to the manufacturer's
instructions. The primers used for cloning are as follows: Twist2
KpnI: 5′-GGGGTACCTCTGAACACAATGATTGGGT-3′ (forward), Twist2
XhoI: 5′-CCCTCGAGGAAGTTCACAGGGCAGAGTC-3′ (reverse); Twist2
mut: 5′-CCCGGGTGGCAGATCAGTTCTAACTCATTGTCATTCAACA-3′ (forward),
Twist2 mut: 5′-TGTTGAATGACAATGAGTTAGAACTGATCTGCCACCCGGG-3′
(reverse); Nkx2-5 KpnI: 5′-GGGGTACCATAAAACATTCATTAGGCTCC-3′
(forward), Nkx2-5 XhoI: 5′-CCCTCGAGTCGTATCTATGGAAAGGGTAT-3′
(reverse); Nkx2-5 mut:
5′-TAGTTAGGAAAAAATGATAACCCGTTCTTTTTTGTGTAC-3′ (forward), Nkx2-5
mut: 5′-GTACACAAAAAAGAACGGGTTATCATTTTTTCCTAACTA-3′ (reverse); Sox9
KpnI: 5′-GGGGTACCCAATCTTAGACAAATCACCA-3′ (forward), Sox9
XhoI: 5′-CCCTCGAGGCTAGTCTTGAACTTCTGGT-3′ (reverse); Sox9
mut: 5′-TCAATTCCATACAAATGTCACAGGCTGAATGTATATGGC-3′ (forward), Sox9
mut: 5′-GCCATATACATTCAGCCTGTGACATTTGTATGGAATTGA-3′ (reverse).
Immunohistochemistry (IHC)
The immunohistochemical analysis was performed to
determine the expression of LASP-1 protein in 13 pairs of human
lung cancer and paired non-tumor tissues, using
avidin-biotin-peroxidase complex method, with anti LASP-1 antibody.
The tissue sections were incubated with polyclonal antibody against
LASP-1 (1:200; Millipore, USA) overnight at 4°C.
RNA extraction and real-time PCR
Total RNA was extracted using TRIzol regent
(Invitrogen) and reverse transcribed using the transcriptase cDNA
synthesis kit (Takara, Dalian, China) according to the
manufacturer's instructions. Real-time PCR analysis was performed
using SYBR-Green (Takara) in ABI 7500 fast fluorescence temperature
cycler. The primers were used at a concentration of 0.5 µM
to generate single PCR product. The primers used are as follows:
LASP-1: sense, 5′-GGTGCGGCAAGATCGTGTA-3′; antisense,
5′-TGCAGGTCTCGCAATGGAA-3′. GAPDH sense, 5′-ACGGATTTGGTCGTATTGGG-3′;
antisense, 5′-CGCTCCTGGAAGATGGTGAT-3′.
Small interfering RNAs
Small interfering RNAs (siRNA) were used to
knockdown the expression of LASP-1 and SOX9 in lung cancer cells.
All the siRNA duplexes were purchased from GenePharma (GenePharma,
Shanghai, China). The specific siRNAs (siLASP-1:
5′-TGTAGTTCTTCATGTTCAGTG-3′ and siSOX9:
5′-TCTTCATGAAGGGGTCCAGGA-3′) were used for RNA interference and the
non-specific scramble siRNA duplexes (5′-TTCTCCGAACGTGTCACGTTT-3′)
were used as normal control. The siRNA duplexes were transduced
into NCI-H1650 cells at a final concentration of 50 nM using
Lipofectamine™ 2000 (Invitrogen Corp., Carlsbad, CA, USA).
Cell proliferation
The cells were seeded in 96-well plates, and the
proliferation of the cells was assayed at 0, 24, 48, and 72 h using
CCK-8 kit (Dojindo Laboratories, Japan) according to the
manufacturer's instructions. Cell viability was assessed by
measuring of absorbance at 450 nm using a microplate reader.
Cell migration and invasion assays
The cell migration ability was determined using
wound healing assay. The migration was assessed by determining the
movement of cells into a scraped area created by pipette tip. After
scratching, the cells were cultured in media supplemented with 0.1%
FBS to eliminate the effect of cell proliferation. The cell
invasive ability of lung cancer cells was determined using
Transwell chamber (8 µm, Corning). Cells (5×104)
in serum-free media were placed in the top chambers, and complete
media was added to the bottom chambers. The chambers were incubated
for 24 h at 37°C. After incubation, the medium was removed from
both the wells, and the chambers were fixed with methanol for 30
min and stained with crystal violet for 30 min.
Flow cytometry
Cell apoptosis was evaluated using FITC-Annexin V
Apoptosis Detection kit. Briefly, the cells were harvested and
washed with cold PBS, and then incubated with 5 µl of
FITC-conjugated Annexin V and 5 µl PI for 10 min at room
temperature in the dark. The samples were analyzed by flow
cytometry. For cell cycle assay, the cells were harvested and fixed
in 70% ethanol for 48 h. The nuclei were stained with 50
µg/ml PI in 1% Triton X-100 containing 100 µg/ml
DNase-free RNase, and the DNA content was analyzed by flow
cytometry.
Luciferase reporter assays
The LASP-1 promoter region −2,500/+1 construct was
amplified from genomic DNA of NCI-H1650 cells. The WT and mutated
LASP-1 promoter constructs were cloned into the pGL3-Basic reporter
gene vector and verified by sequencing. HEK293T cells were
transfected by Lipofectamine 2000 (Invitrogen) in 6-well plates.
Co-transfection of Renilla luciferase plasmid was used as
the internal control for transfection efficiency. Luciferase
activities were measured using the Dual luciferase assay kit (cat.
no. E1960, Promega), with a Berthold Detection system GmbH
chemiluminometer. The results were expressed as ratio of firefly
luciferase activity to Renilla luciferase activity.
Chromatin immunoprecipitation (ChIP)
ChIP was performed according to the manufacturer's
instructions (Active Motif, Carlsbad, CA, USA) in NCI-H1650 cells,
with anti-SOX9 antibody. Real-time PCR analysis was performed using
SYBR-Green (Takara) in ABI 7500 fast fluorescence temperature
cycler. Normal rabbit IgG was used as negative control. ChIP-qPCR
assay was performed in triplicates. The primers used for ChIP are
as follows: LASP-1-F, AAGCTACCTTGGCCAGTCG; R,
AGGGGTCTCACTATGTTGCC.
Statistical analysis
Student's t-test or one-way analysis of variance
(ANOVA) was used to evaluate the significance between groups, using
the statistical package SPSS 17.0 in all the experiments. A value
of P<0.05 was considered to indicate statistical
significance.
Results
LASP-1 expression in lung cancer
Increasing evidence has showed that expression of
LASP-1 is increased in various human cancers, such as breast
cancer. To elucidate the expression level of LASP-1 in lung cancer,
we compared LASP-1 expression in 13 lung cancer tissue samples and
adjacent normal tissues. Patient characteristics, such as age, sex,
histopathological diagnosis, clinical stage, and metastasis status
are shown in Table I. Real-time
PCR and IHC were performed to determine SOX9 and LASP-1 expression
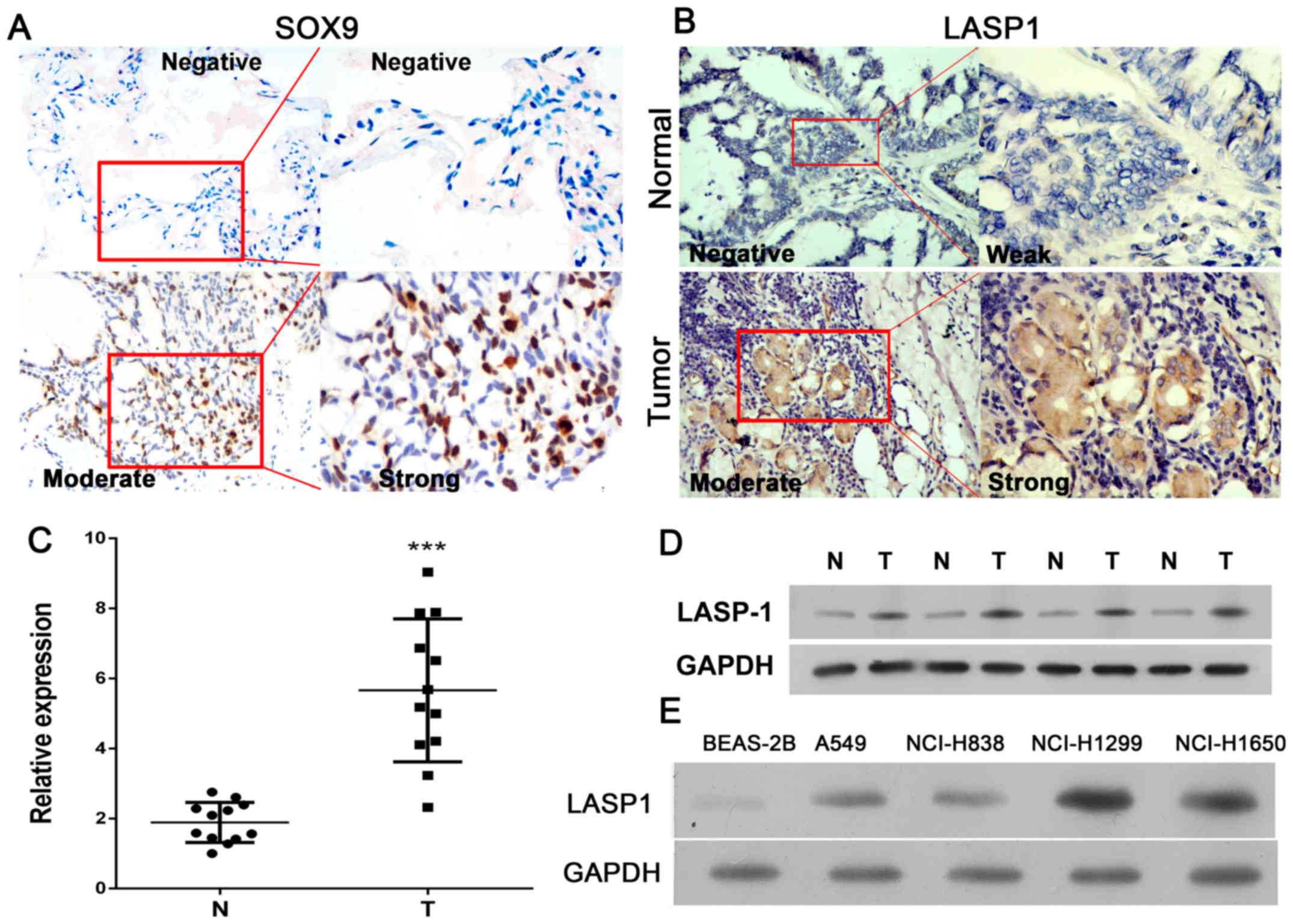
in tissues. As shown in Fig. 1A,
SOX9 expression was measured by IHC staining, and elevated
expression presented in tumor tissues than normal. The results
showed that expression of LASP-1 was increased in lung cancer
tissues as compared to normal tissues (Fig. 1B and C). To confirm the increase in
expression, western blotting was performed to determine LASP-1
expression level in lung cancer tissue and paired tumor-adjacent
normal tissue samples. As illustrated in Fig. 1D, LASP-1 expression was upregulated
in tumor tissues as compared to normal tissues. Next, we determined
the expression of LASP-1 in lung cancer cell lines. As shown in
Fig. 1E, LASP-1 expression was
increased in lung cancer cell-lines, NCI-H1299 and NCI-H1650,
compared with LASP-1 expression level in immortalized human
bronchial epithelial BEAS-2B cells.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Clinical
features | No. (%) |
|---|
| Age, average
(range, years) | 70 (48–91) |
| Tumor size, average
(range, cm) | 3.2 (1–7) |
| Sex | |
| Male | 7 (70) |
| Female | 3 (30) |
| Cigarette | |
| Yes | 8 (80) |
| No | 2 (20) |
| pT stage | |
| T1-T2 | 5 |
| T3-T4 | 5 |
| Histology | |
| Squamous | 5 |
| Adenoma | 5 |
| Metastasis
status | |
| Negative | 6 |
| Positive | 4 |
| Pro-operation
radiation | |
| Yes | 2 |
| No | 8 |
| Lasp-1 expression
(IHC) | |
| Negative | 0 |
| Weak | 2 |
| Moderate | 5 |
| Strong | 3 |
Functional analysis after knockdown of
LASP-1 expression
Based on our findings that LASP-1 was overexpressed
in lung cancer cells (Fig. 1), we
utilized siRNAs approach to knockdown LASP-1 expression in lung
cancer NCI-H1650 cells. Western blot assay was performed to
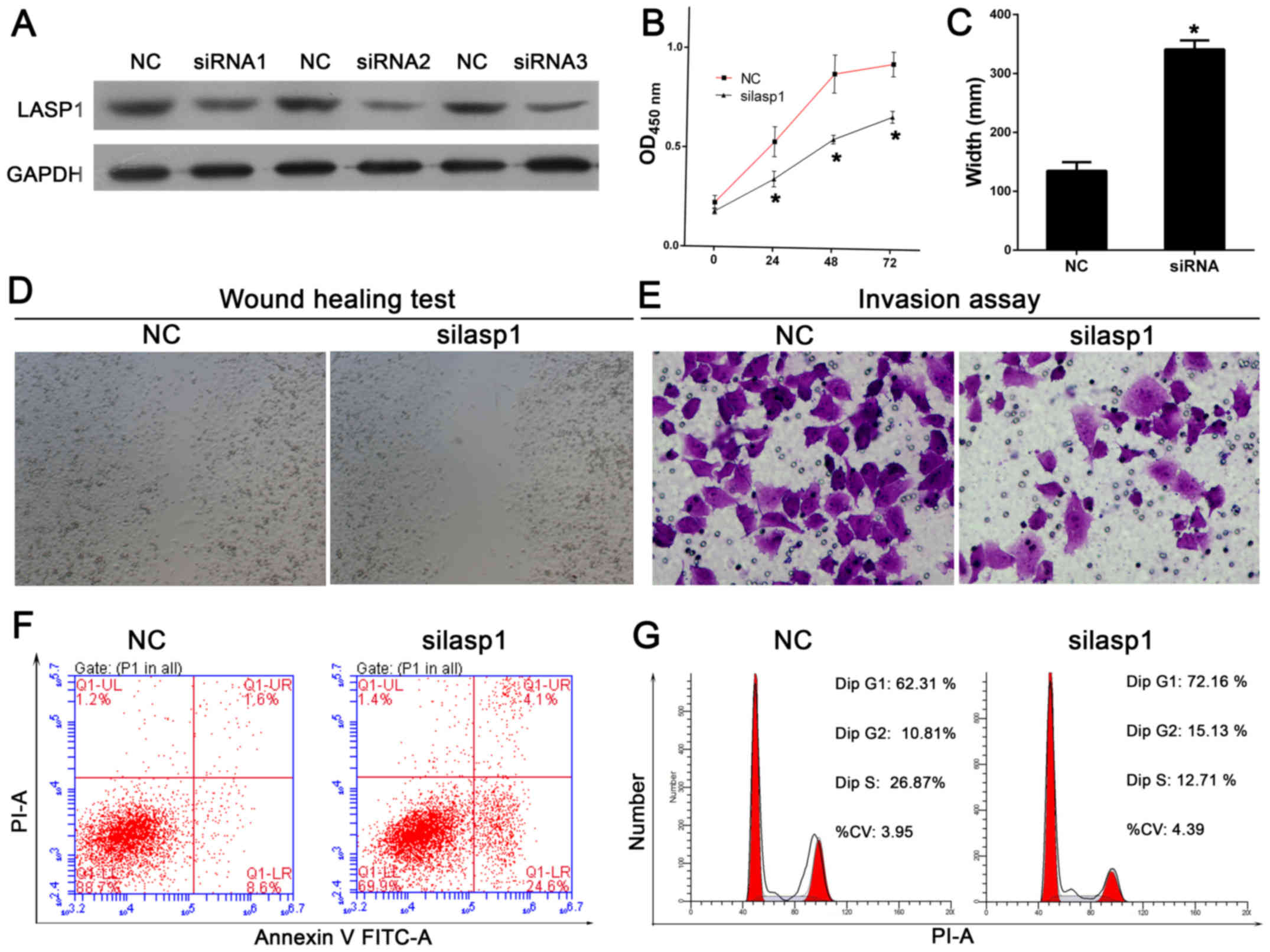
determine the expression level of LASP-1. The results showed that
LASP-1 was significantly silenced by siRNAs (Fig. 2A). siRNA2 was selected for further
experiments. The cell proliferation ability of LASP-1 was
determined using CCK-8 assay after knockdown of the LASP-1
expression. The results showed that cell proliferation was
significantly inhibited at 24, 48, and 72 h in LASP-1 knockdown
cells as compared to control cells (Fig. 2B). The role of LASP-1 in the
regulation of cell migration and invasion was studied by wound
healing assay, using reconstituted extracellular matrices in porous
culture chambers. As shown in Fig. 2C
and D, wound closure occurred gradually after scratching in
control cells, whereas this effect was significantly reduced in
siLASP-1 cells. In line with this finding, siLASP-1 cells showed
reduced invasive capacity as compared to control cells (Fig. 2E). Cell apoptosis was determined by
staining the cells for Annexin V and PI. siRNA-mediated silencing
of LASP-1 led to cell apoptosis of lung cancer cells (Fig. 2F), based on the observations of the
percentage of the Annexin V-positive cells (increase from 9.2 to
17.1% in NCI-1650). It was also observed that the proportion of
cells in S phase was decreased, whereas the proportion of cells in
G1 phase was significantly increased as compared to control
(Fig. 2G). Moreover,
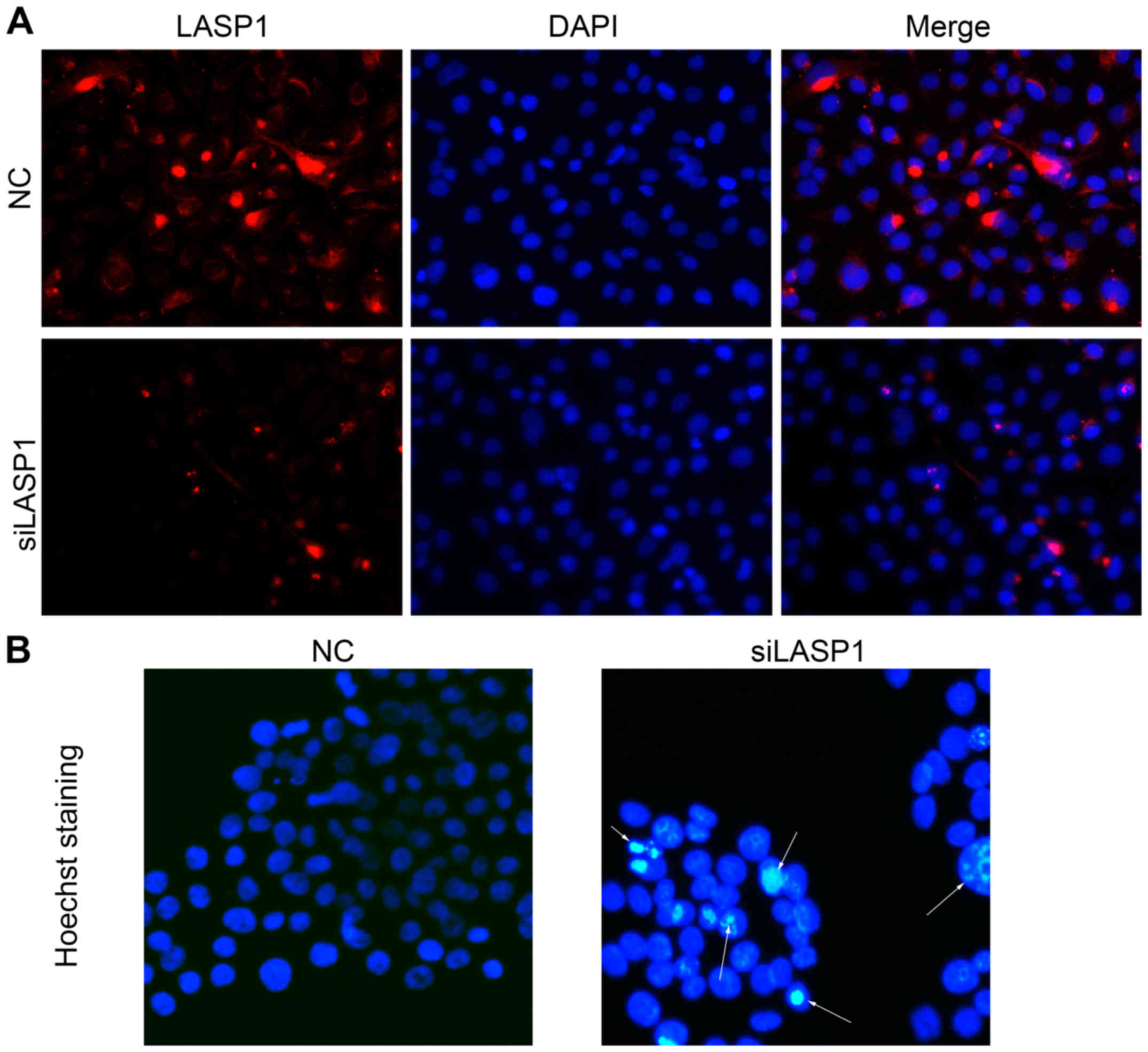
immunofluorescence images of lung cancer cells transfected with
siRNA and stained with antibody against LASP-1. The results showed
that LASP-1 expression was decreased in NCI-1650 cells transfected
with LASP-1 siRNAs as compared to NC group (Fig. 3A). Hoechst staining also showed
that cell apoptosis was increased after knockdown of the expression
of LASP-1 in NCI-1650 cells (Fig.
3B), which was consistent with the results obtained from
Annexin V and PI staining (Fig.
2F). Together, these results suggested that LASP-1 may function
as an oncogene in lung cancer progression.
LASP-1 is a direct target gene of
SOX9
Although the function of LASP-1 in cancer
progression has been well demonstrated, little is known about the
underlying molecular mechanism of LASP-1 in mediating the
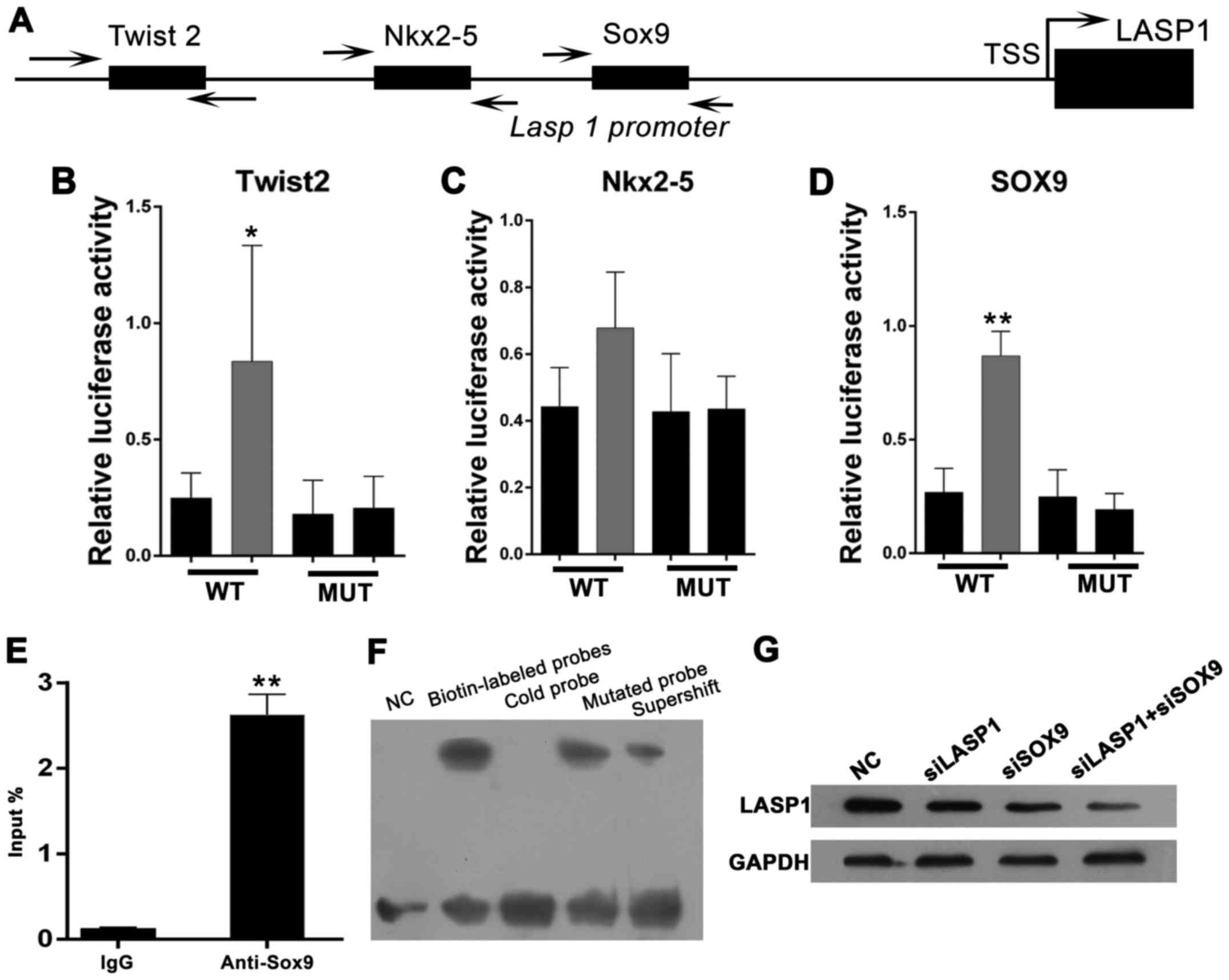
progression of lung cancer. Through functional analysis of the
proximal promoter region of LASP-1, some putative binding sites of
transcriptional factors, including Twist2, Nkx2-5 and SOX9
(Fig. 4A), were observed. We
amplified the promoter region of LASP-1 (range, −2,500 to +1) and
cloned into pGL3-Basic plasmid. Next, the putative binding sites of
Twist2, Nkx2-5 and SOX9 were removed, pGL3-LASP1-mut1,
pGL3-LASP1-mut2, and pGL3-LASP1-mut3, respectively. Then, the WT
and pGL3-LASP1-mut1 constructs were transfected alone or with the
pcR-Twist2 expression vector into HEK293T cells to determine the
promoter activities in the absence or presence of Twist2 (Fig. 4B). The data showed that the
promoter activity of LASP-1 was markedly increased after transient
transfection of HEK293T cells with Twist2 (Fig. 4B, P<0.05). However, the mutated
construct significantly decreased the promoting effect of Twist2 as
compared to the control (Fig. 4B).
Similar results were observed in Nkx2-5; however, the difference
did not reach the statistical significance threshold (Fig. 4C). Furthermore, co-transfection
with SOX9 expression vector also showed significantly increased
promoter activity of LASP-1 (Fig.
4D, P<0.01). The promoting effect, however, can be reversed
by deleting the putative binding sites of SOX9 (Fig. 4D). Among the three transcription
factors, we focused on SOX9 due to our luciferase reporter assay
results. To determine whether LASP-1 is a direct target gene of
SOX9, we performed ChIP assays using a monoclonal antibody against
SOX9 and amplified the pull-down DNA by real-time PCR. The primers
were designed to amplify the region mediating the promoting effects
of SOX9 on the LASP-1 promoter. To determine whether SOX9 could
directly bind to the LASP-1 element in the promoter region, the
electrophoretic mobility shift assay (EMSA) and chromatin
immunoprecipitation (ChIP) assays were performed. As shown in
Fig. 4E, LASP-1 promoter region
was markedly amplified from the SOX9-immunoprecipitated NCI-1650
chromatins, but almost absent from chromatin immunoprecipitated by
the control IgG. The EMSA showed a DNA/protein band of expected
mobility, which reflected the interaction between the probe
containing SOX9 and the nuclear extract of NCI-H1650 cells. The
interaction was increased after treatment with biotin-labeled
probes, but competitively inhibited by high concentration of cold
(unlabeled) probe (not by cold mutated probe; Fig. 4F). In addition, NCI-1650 cells were
transfected with negative control (NC), siSOX9, siLASP-1, siSOX9,
and siLASP-1. Western blot assay was performed to detect the
expression levels of LASP-1. The results showed that LASP-1
expression was decreased in siSOX9 and siLASP-1 groups as compared
to NC group; and the combination of siSOX9 and siLASP-1 has a
synergistic effect (Fig. 4G).
Taken together, these results indicated that LASP-1 is a direct
target gene of SOX9.
Role of LASP-1 in SOX9-induced cell
proliferation and invasion
As a transcription activator of LASP-1, SOX9 has
been found to be overexpressed in different types of human cancers,
including breast cancer and colorectal cancer. Although the
association between the upregulation of SOX9 and lung cancer
progression has been reported, the role of SOX9 in regulating cell
proliferation and invasion of cancer cells remains in need of
further elucidation. To investigate the effect of SOX9, siRNAs were
used to silence the expression of SOX9. CCK-8 assays showed that
cell proliferation was markedly inhibited in siSOX9 cells as
compared to control group. Furthermore, combined use of siRNAs
targeting SOX9 and LASP-1 significantly inhibited cell
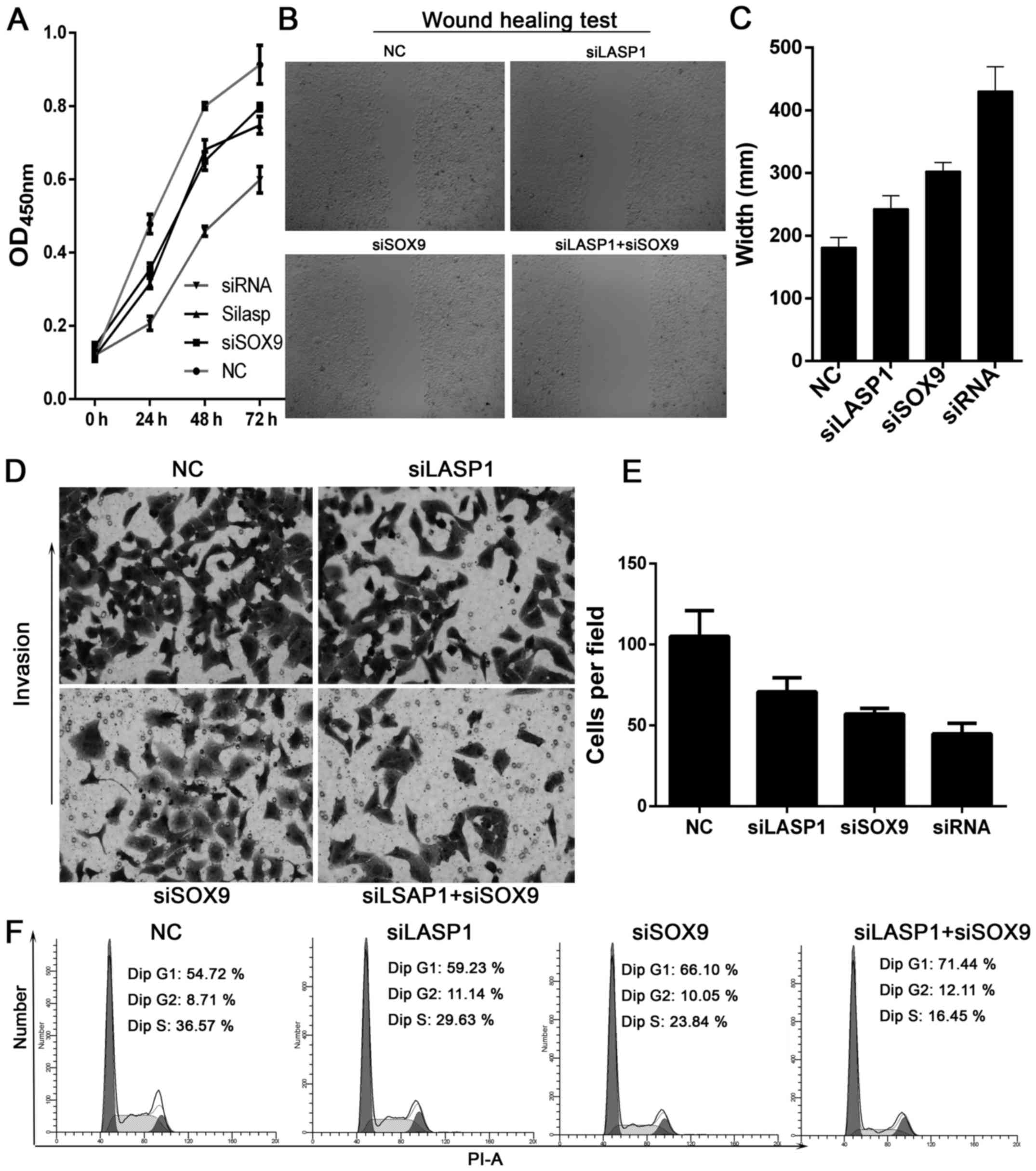
proliferation in NCI-1650 cells than either used alone (Fig. 5A). The scratch assay was used to
study the effect of SOX9 on cell migration. The results showed that
knockdown of the expression of SOX9 reduced cell migration
(Fig. 5B and C). Similarly, the
invasive capability of NCI-H1650 cells was decreased after
silencing the expression of SOX9. Thus, combined use of siRNAs
targeting SOX9 and LASP-1 notably reduced invasive ability as
compared with NC-transfected cells (Fig. 5D and 5E). Next, we examined cell cycle
distribution using flow cytometry. SOX9 knockdown decreased the
proportion of cells in the S phase (29.63% of siSOX9 cells vs.
36.57% of control cells, Fig. 5F),
and increased the proportion of cells in the G1 phase as compared
with control (Fig. 5F). In
addition, combined use of siRNAs targeting SOX9 and LASP-1 induced
apoptosis of NCI-H1650 cells as compared with control (33.1 vs.
8.3%, Fig. 6A and B). Hoechst
staining also showed that cell apoptosis was increased after
silencing the expression of SOX9 and LASP-1 (Fig. 6C). These results showed that
SOX9-LASP-1 axis was involved in the progression of lung
cancer.
Discussion
Various studies have shown that LASP-1 is
overexpressed in many cancers, such as metastatic breast cancer,
ovarian cancer, and colorectal cancer (18,34,35).
Its expression is strongly associated with lymph node metastasis
and poor clinical prognosis. However, until now, little is known
about the role of LASP-1 in lung cancer progression. In this study,
results from real-time PCR and immunohistochemical assays
demonstrated that LASP-1 was not expressed in tumor-adjacent normal
tissues; however, significant expression was observed in lung
cancer tissues, which was consistent with the results reported by
Zheng et al (7).
In this study, the effect of LASP-1 on lung cancer
pathogenesis was evaluated in vitro using NCI-H1650 cells.
Silencing LASP-1 by siRNA transfection significantly inhibited cell
proliferation and induced cell apoptosis. Furthermore, the results
of wound healing and Transwell assays indicated that LASP-1
silencing suppressed the migration and invasion of lung cancer
cells. These findings are in line with several other studies
demonstrating reduced cell motility after LASP-1 silencing
(17–19,36).
However, the studies focused on the underlying molecular mechanism
of LASP-1 in promoting tumor progression are very rare. Grunewald
et al and Shimizu et al reported that downregulation
of LASP-1 induced G2/M phase accumulation of breast cancer and
ovarian cancer cells (17,37). In addition, silencing LASP-1
expression resulted in an increase in number of cells in G1 phase.
In addition, our study showed that knockdown of LASP-1 expression
induced G2 phase accumulation; besides, knockdown of LASP-1 leads
to decreased expression of cyclin A and cyclin B, and increased
phospho-cdc2 (Tyr15) expression (37). Our study demonstrated that
silencing of LASP1 significantly inhibited prostate cancer cell
growth by decreasing cyclin D1 and increasing p21 and p27 (38). Migration and invasion are key
determinants of cancer cell progression and metastasis. Some
studies have demonstrated that LASP-1 is involved in cell migration
and invasion, based on the ability of the cells to interact with a
series of focal adhesion proteins, such as F-actin, zyxin, and
CXCR2 (18,39). Epithelial-mesenchymal transition
(EMT) is a reversible process by which cancer cells can switch from
a sessile epithelial phenotype to an invasive mesenchymal state
(40). In 2016, Zhang et al
reported that knockdown of LASP-1 expression inhibited the
migration and invasion of CCA cells by inducing EMT (20). Wang et al showed that LASP-1
plays a critical role in the TGF-β-mediated EMT process in
colorectal cancer metastasis (35). While increasing number of evidence
suggests that LASP-1 plays an important role in cancer progression,
the role of LASP-1 in mediating the proliferation and metastasis of
cancer cells remains unclear and requires further study.
An additional observation further underscores the
importance of LASP-1 in cancer. Previous studies have shown that
upstream regulatory factors, HIF-1 and p53, have critical functions
in regulating LASP-1 expression (41,42).
The expression of LASP-1 in PDAC cells was activated by HIF-1α by
direct binding to hypoxia response element in the LASP-1 promoter
(42). On the contrary,
transcription factor, p53, repressed LASP-1 expression (20). Furthermore, IGF-1-induced
expression of LASP-1 gene in MCF-7 cells was described in a recent
study (43). Only few studies have
explored the regulatory mechanism of LASP-1 in lung cancer. In this
study, we demonstrated for the first time that SOX9 induced LASP-1
expression. Dual luciferase reporter assay and ChIP assays
demonstrated that LASP-1 is a direct target gene of SOX9. A series
of studies have demonstrated that SOX9 is a multifaceted
transcription factor, which is involved in the development of
numerous organ and tissues. SOX9 is reported to be upregulated in
several types of human cancers, including lung ADC (30). Depletion of LASP-1 by specific
siRNA inhibited lung cancer cell proliferation and invasion.
Moreover, combined use of siRNAs targeting SOX9 and LASP-1
significantly inhibited cell proliferation, migration, and invasion
of NCI-H1650 cells. These findings suggested that SOX9-LASP1 axis
is involved in lung cancer cell progression.
In conclusion, our results demonstrated that LASP-1
was upregulated in lung cancer and played an important role in cell
proliferation, migration, and invasion. Mechanistic analysis
identified LASP-1 as a novel direct target of SOX9. These findings
suggested that LASP-1 is a promising therapeutic target, and
targeting SOX9-LASP1 axis may be an effective method for treatment
of lung cancer.
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
3
|
Dela Cruz CS, Tanoue LT and Matthay RA:
Lung cancer: Epidemiology, etiology, and prevention. Clin Chest
Med. 32:605–644. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Steliga MA and Dresler CM: Epidemiology of
lung cancer: Smoking, secondhand smoke, and genetics. Surg Oncol
Clin N Am. 20:605–618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yano T, Haro A, Shikada Y, Maruyama R and
Maehara Y: Non-small cell lung cancer in never smokers as a
representative 'non-smoking-associated lung cancer': Epidemiology
and clinical features. Int J Clin Oncol. 16:287–293. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Villar Álvarez F, Muguruza Trueba I and
Vicente Antunes SI: Notes on recurrence and second tumors in lung
cancer. Arch Bronconeumol. 52:545–546. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng J, Wang F, Lu S and Wang X: LASP-1,
regulated by miR-203, promotes tumor proliferation and
aggressiveness in human non-small cell lung cancer. Exp Mol Pathol.
100:116–124. 2016. View Article : Google Scholar
|
|
8
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Temel JS, Greer JA, Muzikansky A,
Gallagher ER, Admane S, Jackson VA, Dahlin CM, Blinderman CD,
Jacobsen J, Pirl WF, et al: Early palliative care for patients with
metastatic non-small-cell lung cancer. N Engl J Med. 363:733–742.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tomasetto C, Moog-Lutz C, Régnier CH,
Schreiber V, Basset P and Rio MC: Lasp-1 (MLN 50) defines a new LIM
protein subfamily characterized by the association of LIM and SH3
domains. FEBS Lett. 373:245–249. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Orth MF, Cazes A, Butt E and Grunewald TG:
An update on the LIM and SH3 domain protein 1 (LASP1): A versatile
structural, signaling, and biomarker protein. Oncotarget. 6:26–42.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Segerer SE, Bartmann C, Kaspar S, Müller
N, Kapp M, Butt E and Kämmerer U: The cytoskeletal protein LASP-1
differentially regulates migratory activities of choriocarcinoma
cells. Arch Gynecol Obstet. 293:407–414. 2016. View Article : Google Scholar
|
|
13
|
Rachlin AS and Otey CA: Identification of
palladin isoforms and characterization of an isoform-specific
interaction between Lasp-1 and palladin. J Cell Sci. 119:995–1004.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Grunewald TG and Butt E: The LIM and SH3
domain protein family: Structural proteins or signal transducers or
both? Mol Cancer. 7:312008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Duvall-Noelle N, Karwandyar A, Richmond A
and Raman D: LASP-1: A nuclear hub for the UHRF1-DNMT1-G9a-Snail1
complex. Oncogene. 35:1122–1133. 2016. View Article : Google Scholar
|
|
16
|
Frietsch JJ, Grunewald TG, Jasper S,
Kammerer U, Herterich S, Kapp M, Honig A and Butt E: Nuclear
localisation of LASP-1 correlates with poor long-term survival in
female breast cancer. Br J Cancer. 102:1645–1653. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grunewald TG, Kammerer U, Schulze E,
Schindler D, Honig A, Zimmer M and Butt E: Silencing of LASP-1
influences zyxin localization, inhibits proliferation and reduces
migration in breast cancer cells. Exp Cell Res. 312:974–982. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Grunewald TG, Kammerer U, Winkler C,
Schindler D, Sickmann A, Honig A and Butt E: Overexpression of
LASP-1 mediates migration and proliferation of human ovarian cancer
cells and influences zyxin localisation. Br J Cancer. 96:296–305.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng J, Yu S, Qiao Y, Zhang H, Liang S,
Wang H, Liu Y, Zhou F, Jiang J and Lu S: LASP-1 promotes tumor
proliferation and metastasis and is an independent unfavorable
prognostic factor in gastric cancer. J Cancer Res Clin Oncol.
140:1891–1899. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang H, Li Z, Chu B, Zhang F, Zhang Y, Ke
F, Chen Y, Xu Y, Liu S, Zhao S, et al: Upregulated LASP-1
correlates with a malignant phenotype and its potential therapeutic
role in human cholangiocarcinoma. Tumour Biol. 37:8305–8315. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao L, Wang H, Liu C, Liu Y, Wang X, Wang
S, Sun X, Li J, Deng Y, Jiang Y, et al: Promotion of colorectal
cancer growth and metastasis by the LIM and SH3 domain protein 1.
Gut. 59:1226–1235. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin X, Liu X, Fang Y and Weng X: LIM and
SH3 protein 1 promotes tumor proliferation and metastasis in lung
carcinoma. Oncol Lett. 12:4756–4760. 2016.
|
|
23
|
Yang F, Zhou X, Du S, Zhao Y, Ren W, Deng
Q, Wang F and Yuan J: LIM and SH3 domain protein 1 (LASP-1)
overexpression was associated with aggressive phenotype and poor
prognosis in clear cell renal cell cancer. PLoS One. 9:e1005572014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Akiyama H, Chaboissier MC, Martin JF,
Schedl A and de Crombrugghe B: The transcription factor Sox9 has
essential roles in successive steps of the chondrocyte
differentiation pathway and is required for expression of Sox5 and
Sox6. Genes Dev. 16:2813–2828. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Spokony RF, Aoki Y, Saint-Germain N,
Magner-Fink E and Saint-Jeannet JP: The transcription factor Sox9
is required for cranial neural crest development in Xenopus.
Development. 129:421–432. 2002.PubMed/NCBI
|
|
26
|
Chaboissier MC, Kobayashi A, Vidal VI,
Lützkendorf S, van de Kant HJ, Wegner M, de Rooij DG, Behringer RR
and Schedl A: Functional analysis of Sox8 and Sox9 during sex
determination in the mouse. Development. 131:1891–1901. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Müller P, Crofts JD, Newman BS,
Bridgewater LC, Lin CY, Gustafsson JA and Ström A: SOX9 mediates
the retinoic acid-induced HES-1 gene expression in human breast
cancer cells. Breast Cancer Res Treat. 120:317–326. 2010.
View Article : Google Scholar
|
|
28
|
Qin GQ, He HC, Han ZD, Liang YX, Yang SB,
Huang YQ, Zhou L, Fu H, Li JX, Jiang FN, et al: Combined
overexpression of HIVEP3 and SOX9 predicts unfavorable biochemical
recurrence-free survival in patients with prostate cancer. Onco
Targets Ther. 7:137–146. 2014.PubMed/NCBI
|
|
29
|
Bruun J, Kolberg M, Nesland JM, Svindland
A, Nesbakken A and Lothe RA: Prognostic significance of β-catenin,
E-cadherin, and SOX9 in colorectal cancer: Results from a large
population-representative series. Front Oncol. 4:1182014.
View Article : Google Scholar
|
|
30
|
Jiang SS, Fang WT, Hou YH, Huang SF, Yen
BL, Chang JL, Li SM, Liu HP, Liu YL, Huang CT, et al: Upregulation
of SOX9 in lung adenocarcinoma and its involvement in the
regulation of cell growth and tumorigenicity. Clin Cancer Res.
16:4363–4373. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang X, Ju Y, Zhou MI, Liu X and Zhou C:
Upregulation of SOX9 promotes cell proliferation, migration and
invasion in lung adenocarcinoma. Oncol Lett. 10:990–994.
2015.PubMed/NCBI
|
|
32
|
Wang X, Liu Y, Liu X, Yang J, Teng G,
Zhang L and Zhou C: MiR-124 inhibits cell proliferation, migration
and invasion by directly targeting SOX9 in lung adenocarcinoma.
Oncol Rep. 35:3115–3121. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou CH, Ye LP, Ye SX, Li Y, Zhang XY, Xu
XY and Gong LY: Clinical significance of SOX9 in human non-small
cell lung cancer progression and overall patient survival. J Exp
Clin Cancer Res. 31:182012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Grunewald TG, Kammerer U, Kapp M, Eck M,
Dietl J, Butt E and Honig A: Nuclear localization and cytosolic
overexpression of LASP-1 correlates with tumor size and
nodal-positivity of human breast carcinoma. BMC Cancer. 7:1982007.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang H, Shi J, Luo Y, Liao Q, Niu Y, Zhang
F, Shao Z, Ding Y and Zhao L: LIM and SH3 protein 1 induces
TGFβ-mediated epithelial-mesenchymal transition in human colorectal
cancer by regulating S100A4 expression. Clin Cancer Res.
20:5835–5847. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hailer A, Grunewald TG, Orth M, Reiss C,
Kneitz B, Spahn M and Butt E: Loss of tumor suppressor mir-203
mediates overexpression of LIM and SH3 protein 1 (LASP1) in
high-risk prostate cancer thereby increasing cell proliferation and
migration. Oncotarget. 5:4144–4153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shimizu F, Shiiba M, Ogawara K, Kimura R,
Minakawa Y, Baba T, Yokota S, Nakashima D, Higo M, Kasamatsu A, et
al: Overexpression of LIM and SH3 protein 1 leading to accelerated
G2/M phase transition contributes to enhanced tumourigenesis in
oral cancer. PLoS One. 8:e831872013. View Article : Google Scholar
|
|
38
|
Dejima T, Imada K, Takeuchi A, Shiota M,
Leong J, Tombe T, Tam K, Fazli L, Naito S, Gleave ME, et al:
Suppression of LIM and SH3 domain protein 1 (LASP1) negatively
regulated by androgen receptor delays castration resistant prostate
cancer progression. Prostate. 77:309–320. 2017. View Article : Google Scholar
|
|
39
|
Raman D, Sai J, Neel NF, Chew CS and
Richmond A: LIM and SH3 protein-1 modulates CXCR2-mediated cell
migration. PLoS One. 5:e100502010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Felipe Lima J, Nofech-Mozes S, Bayani J
and Bartlett JM: EMT in breast carcinoma-A review. J Clin Med.
5:E652016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang B, Feng P, Xiao Z and Ren EC: LIM and
SH3 protein 1 (Lasp1) is a novel p53 transcriptional target
involved in hepatocellular carcinoma. J Hepatol. 50:528–537. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao T, Ren H, Li J, Chen J, Zhang H, Xin
W, Sun Y, Sun L, Yang Y, Sun J, et al: LASP1 is a HIF1α target gene
critical for metastasis of pancreatic cancer. Cancer Res.
75:111–119. 2015. View Article : Google Scholar
|
|
43
|
Loughran G, Huigsloot M, Kiely PA, Smith
LM, Floyd S, Ayllon V and O'Connor R: Gene expression profiles in
cells transformed by overexpression of the IGF-I receptor.
Oncogene. 24:6185–6193. 2005. View Article : Google Scholar : PubMed/NCBI
|