Introduction
Signal transducers and activators of transcription
(STATs), when phosphorylated at tyrosine elements form functional
dimmers with each other, translocate to the nucleus to regulate the
expression of genes by binding to specific elements within gene
promoters (1,2). The function of the interleukin-6
(IL-6)-STAT3 pathway in signalling and its control of the
expression of different genes prompted the investigation of the
pathway in cancer. Indeed, the IL-6-STAT3 pathway has been found to
be critical for tumour development through different mechanisms
that include direct effects on cell survival, migration and
proliferation, as well as non-direct effects on the
microenvironment surrounding the tumour (3–7).
Although preclinical data suggest that the IL-6-STAT3 pathway is an
important target for cancer therapy, to date, the practical
clinical utility and efficacy is limited (8,9).
This reflects the challenges in translating preclinical evidence
into relevant clinical practice and demonstrates the complexity of
cancer biology.
STAT3 plays a role in the development of breast
cancer, as shown by several studies (3,4,10–13).
However, some studies evaluating the prognostic role of
phosphorylated STAT3 (p-STAT3), have yielded conflicting results
(14–18). The aim of this study was to
evaluate the prognostic role of p-STAT3 in luminal breast cancer
patients in the adjuvant setting and in the context of a
prospective trial.
Materials and methods
Computation of p-STAT3 reverse phase
protein array (RPPA)-based signature
We analysed clinicopathological, normalised gene
expression and RPPA data from the TCGA repository using its online
bioinformatics tool (19).
Estrogen receptor (ER)-positive breast cancers were analysed based
on the RPPA proteomic levels. A total of 265 samples with available
gene expression and RPPA data were considered as luminal (166
luminal A and 99 luminal B) according to the PAM50 computed on the
cBioPortal website (20). Each of
the RPPA assay samples available was assigned to one of two sample
groups: 'Low p-STAT3 expression' samples correspond to those
samples with RPPA expression smaller than the first quartile of all
expression values for this RPPA assay, while 'high p-STAT3
expression' samples correspond to those samples with a RPPA
expression greater than the third quartile. To identify the genes
that were differentially expressed between the low- and
high-expression groups, we performed a differential gene expression
analysis using a Student's t-test comparing high vs. low p-STAT3
expression tumours using a Welch t-test with robust estimators of
the mean and the standard deviation.
To identify the genes that would optimise the
predictive power of our signatures, we evaluated using a nested
10-fold cross validation, the maximal Benjamini-Hochberg false
discovery rate and the minimal gene fold change that would optimise
the ability of the differentially expressed genes to predict the
high/low status of the RPPA in luminal A and B patients together
and separately. While the parameters were selected in a 10-fold
cross validation, the procedure itself was assessed using a nested
cross validation (21). This
nested procedure allowed us to remove those RPPA assays that could
not deliver signatures that we could further use for prediction
(ROC curves AUC <0.6 in luminal A, B or A and B). After this
process, we were left with 69 signatures presenting a relevant AUC
for proteomic status prediction. Among others, p-STAT3 achieved
significant prediction ability in both luminal A and B cancers. The
expression levels of these signatures in the gene expression
datasets were computed as previously described (22).
Patients and study design
The Breast International Group (BIG) 2–98
(ClinicalTrials.gov identifier of BIG
2–98: NCT00174655), is a multicenter, prospective, open-labelled,
phase III adjuvant trial that randomly assigned patients to either
anthracycline-based chemotherapy or taxane combinations (23). Women received definitive surgical
treatment (mastectomy or breast-conserving surgery) for invasive
breast adenocarcinoma with ≥1 positive axillary lymph nodes of ≥8
resected nodes. All patients provided written informed consent
prior to study entry. The full details and the CONSORT diagram were
previously reported (24).
Central pathology review and tissue
microarray (TMA) construction
A primary tumour sample (blocks or slides) was
required for the central pathology review. Primary tumour samples
were stored centrally at the Institut Jules Bordet (Brussels,
Belgium). The central pathology review was carried out at the
European Institute of Oncology (Milan, Italy). The tumour grade was
centrally reviewed. Immunostaining experiments for the localization
of ER and progesterone receptor (PgR) and HER2 protein were carried
out on consecutive tissue sections using an automated immunostainer
(Autostainer; Dako, Glostrup, Denmark). The following primary
antibodies were used: 1D5 monoclonal antibody (mAb) to ER (at 1/100
dilution), 1A6 mAb to PgR (1/800) and polyclonal antiserum (1/800)
(all from Dako) to HER2 protein (23).
Only nuclear reactivity was taken into account for
ER and PgR, and the results were recorded as the percentage of
immunoreactive cells over at least 2,000 neoplastic cells.
Fluorescent in situ hybridization (FISH) was carried out for
HER2 according to the manufacturer's instructions (Vysis; Abbott
Laboratories, Abbott Park, IL, USA). Positivity thresholds were ER
≥1%; PgR ≥1%; HER 2=3+ (>10% invasive tumour cells
with intense and circumferential membrane staining) and/or
FISH-positive (HER2:CEP17 ratio ≥2).
p-STAT3 staining
The biomarker protocol for the evaluation of p-STAT3
phosphorylation in association with the clinical outcome was
approved by the Institutional Review Board of Hadassah Medical
Center and the BIG2-98 Study Steering Committee. From the 2,887
patients randomised in the BIG 2–98 trial, 2,173 cases had tumour
blocks that were centrally evaluated. A TMA was constructed from
950 blocks.
Paraffin blocks were submitted to the coordinating
center and 4 cores from each tumour were collected and placed in 2
different TMAs; each TMA contains 2 cores of the same tumour. Two
laboratories performed independent staining for p-STAT3
[immunohistochemistry (IHC) and immunofluorescence (IF)]: Each
laboratory received a set of available BIG2-98 specimens in TMAs.
The BIG 2-98 TMA set contained 19 slides with ~170 tissue cores per
slide. Two slides containing ER-negative samples were of low
quality, and although stained, could not be annotated. In total,
610 and 585 ER-positive samples were interpretable for p-STAT3 by
IHC and IF, respectively.
IHC was performed experimentally. The tissue
micro-array sections slides were deparaffinised with xylene rinses
and then transferred through two changes of 100% ethanol.
Endogenous peroxidase activity was blocked by a 5-min incubation in
a 3% hydrogen peroxide buffer. Antigen retrieval was performed
using Tris-EDTA buffer (10 mM Tris Base, 1 mM EDTA solution).
Following antigen retrieval, the slides were incubated with
blocking buffers at room temperature to reduce non-specific
background staining and then incubated with primary antibody at 4˚C
overnight (1:100 dilution of p-STAT3 antibody; cat. no.
9145-D3A7-XP; Cell Signaling Technology, Beverly, MA, USA) followed
by 30 min of incubation with the secondary anti-rabbit antibody
(Histofine® Simple Stain™ MAX PO (R), cat. no. 414141F;
Nichirei Biosciences Inc., Tokyo, Japan). Staining was visualised
using DAB.
The IHC nuclear p-STAT3 staining was determined and
scored separately for each spot and specimen by two pathologists
(R.S. and G.V.E.) who were blinded to the clinical pathological
data and reached an agreed score for each spot. The staining was
analysed according to the H-score (range, 0–300; intensity 1–3 ×
percentage of positive cells 1–100). For each spot, separate scores
where provided for the tumour and stromal parts. For specimens that
were uninterpretable, a score of 'not applicable' (N/A) was
assigned. To define tumours as p-STAT3-positive, a cut-off point of
>0 H score was selected, as well as analysing the data as a
continuous variable. Specificity/sensitivity, performance and
reproducibility tests of the p-STAT3 IHC were performed. Full
slides from representative positive (n=9) and negative (n=8) TMAs
were stained processed, and evaluated similarly as the experimental
slides, for the indication of reproducibility and performance of
the TMAs. In total, 8/9 positive TMAs were found to be positive on
the full slides and 8/8 of the negative TMAs were found to be
negative on the full slides, demonstrating 100% sensitivity and 89%
specificity. As the controls for our procedure, we used HeLa cells
without treatment that served as a negative control and
serum-starved HeLa cells prepared with interferon-α (IFN-α)
treatment that served as a positive control (all control slides
were purchased from Cell Signaling Technology). The control slides
were used for qualifying the procedure and not as an interpretation
reference. The presence of a brown reaction product at the cell
nucleus was indicative of positive reactivity.
In parallel, TMAs were stained using IF at the
laboratory of J.F.B. For IF staining, the TMAs were deparaffinised
and processed using the automated Ventana deparaffinization
solution (Ventana Medical Systems, Inc., Tucson, AZ, USA) and CC1
antigen retrieval. The tissue sections were blocked for 30 min in
10% normal goat serum, 2% BSA in phosphate-buffered saline (PBS).
The incubation with the p-STAT3 antibody was carried for 2 h,
followed by 16 min of biotinylated goat anti-rabbit IgG (cat. no.
PK6101; Vector Laboratories, Inc., Burlingame, CA, USA) at a 1:200
dilution (=7.5 µg/ml). Streptavidin-HRP (Ventana Medical Systems,
Inc.) was applied for 12 min followed by incubation with
Tyramide-Alexa Fluor 488 (cat. no. T20922; Invitrogen, Carlsbad,
CA, USA) for 16 min. p-STAT3 antibody was from Cell Signaling
Technology (cat. no. 9145-D3A7-XP) a rabbit monoclonal has been
purchased in large amounts (5 ml), mixed and re-aliquoted to avoid
batch-batch variation. All slides were in the same run.
The IF results were analysed using a semi-supervised
free-scoring tool by a digital imaging system. Specifically, each
slide was scanned using Pannoramic Flash (3DHistech Ltd., Budapest,
Hungary) and each tumour/stromal section was exported into a tif.
image by Pannoramic Viewer (3DHistech Ltd.). Image analysis was
performed using Metamorph (Molecular Devices, Sunnyvale CA, USA) in
which the number of nuceli was counted using the DAPI channel, and
the level of green fluorescence (p-STAT3)/nuclei was
quantified.
Tissue microarray construction, the determination of
the proteomic status, patient selection, assay performance and data
analysis are reported according to the Recommendations for Tumour
Marker Prognostic Studies (REMARK) criteria (25).
Statistical analysis
In total, 39 gene expression datasets of expression
profiles from >7,000 tumours were retrieved from public
databases or authors' websites [36 previously described (26) and another 3 sets: PNC, METABRIC and
TCGA (19,27,28)]. To ensure the comparability of the
expression values across multiple data sets, we performed a 0.95
quantile normalization. Differences in the p-STAT3 expression
signatures according to subtype were examined using the
Kruskal-Wallis test. Distant metastasis-free survival was the
primary survival end-point, which is defined as the time elapsing
between breast cancer diagnosis and the date of systemic relapse.
When distant metastasis-free survival data were not reported,
relapse-free survival information was used if available. Survival
plots according to the p-STAT3 signatures tertiles were drawn using
the Kaplan-Meier method, and the significance of the survival
differences were evaluated using the log-rank P-test. The
association of the signatures with good or bad prognosis were
computed using uni- or multivariate Cox regression analyses. All
analyses were performed using the genefu package of R
(v3.2)/Bioconductor (v1.18) statistical suite.
For the BIG 2–98 outcome analysis, patients were
classified according to the presence of p-STAT3. The primary
outcomes were disease-free survival (DFS) and overall survival
(OS). DFS was defined as the interval from the date of
randomization to the date of local, regional or metastatic relapse
or second primary cancer or death for any cause. OS was calculated
from the date of randomization to last follow-up or death from any
cause. Univariate and multivariate models were computed with the
use of Cox proportional-hazards regression. The Chi-square test for
categorical data and unpaired Student's t-test for continuous
variables were used in order to determine an association between
p-STAT3 and clinical pathological parameters, and P-values <0.05
were considered to indicate statistically significant
differences.
Results
Association of the p-STAT3 and p-STAT3
gene expression signature with clinical parameters and outcome in
patients with ER-positive breast cancer
To capture the clinical importance of p-STAT3 in
ER-positive breast cancer, we analysed breast cancer samples from
the TCGA repository (19). Using
the PAM50 classification model, patients were assigned to the main
luminal breast cancer molecular subtypes: Luminal A and luminal B.
We first assessed whether p-STAT3 expression (by RPPA) was
associated with a particular subtype. Our analysis revealed that
luminal A-type cancers were more likely to possess p-STAT3 high
levels in comparison to luminal B-type cancers (Wilcoxon
P=3e−6) (Fig. 1A).
We then derived a gene signature whose expression
levels could predict adequately the p-STAT3 RPPA levels by
computing the differentially expressed genes between tumour samples
with high (upper quartile) and low (lower quartile) RPPA levels of
p-STAT3. To capture the clinical relevance of the p-STAT3
expression signature in ER-positive breast cancer, we combined 39
publically available microarray datasets comprising over 7,000
breast cancer patients to build a pooled set of gene-expression
profiles with available outcome data. Using the PAM50
classification model, patients were assigned to the main breast
cancer molecular subtypes, namely luminal A, luminal B,
HER2-enriched, basal-like and normal-like breast cancers. We first
assessed whether the p-STAT3 expression signatures were associated
with any particular luminal subtype. As expected, in the pooled set
analysis, the p-STAT3 expression signature was significantly
associated will luminal A-type cancers (P<0.01−10)
(Fig. 1B). We then assessed
whether p-STAT3 correlated with survival in patients with
ER-positive breast cancer in cohorts where relapse data was
available. As shown in Fig. 2 and
Table I, the p-STAT3 expression
signature was significantly associated with a good prognosis in all
patients with luminal tumours (log-rank, P<0.01−10)
(Fig. 2A). Similar results were
found with patients treated with hormonal therapy only (2D
log-rank, P=4.58e−9). Of interest, the p-STAT3
expression signature was able to identify patients with a good
prognosis irrespective of the luminal subtypes (luminal A,
log-rank, P=0.027; luminal B, log-rank, P=0.006) (Fig. 2B and C). Cox multivariate analysis
confirmed the independent prognostic value of p-STAT3 (Table I).
 | Table ICox univariate and multivariate
analysis of relapse data from the pooled analysis according to the
p-STAT3 gene expression signature status. |
Table I
Cox univariate and multivariate
analysis of relapse data from the pooled analysis according to the
p-STAT3 gene expression signature status.
| Subtype | Treatment | Hazard Cox
univariate | P-value | Hazard Cox
multivariate | P-value |
|---|
| All luminal | Treated and not
treated | 0.52 |
<0.01−10 | 0.58 |
1.27E−09 |
| All luminal | Hormonotherapy
treatment only | 0.43 |
3.37E−10 | 0.55 |
3.48E−05 |
| Luminal A | Treated and not
treated | 0.64 | 0.004 | 0.67 | 0.059 |
| Luminal A | Hormonotherapy
treatment only | 0.46 | 0.02 | 0.64 | 0.23 |
| Luminal B | Treated and not
treated | 0.72 | 0.002 | 0.78 | 0.045 |
| Luminal B | Hormonotherapy
treatment only | 0.64 | 0.018 | 0.76 | 0.16 |
Association of p-STAT3 with
clinicopathological characteristics and outcome in the BIG 2-98
randomised trial
We then confirmed our observations at the proteomic
level in the context of prospective data from the BIG 2–98
repository. There were 610 and 585 ER-positive tumour TMAs
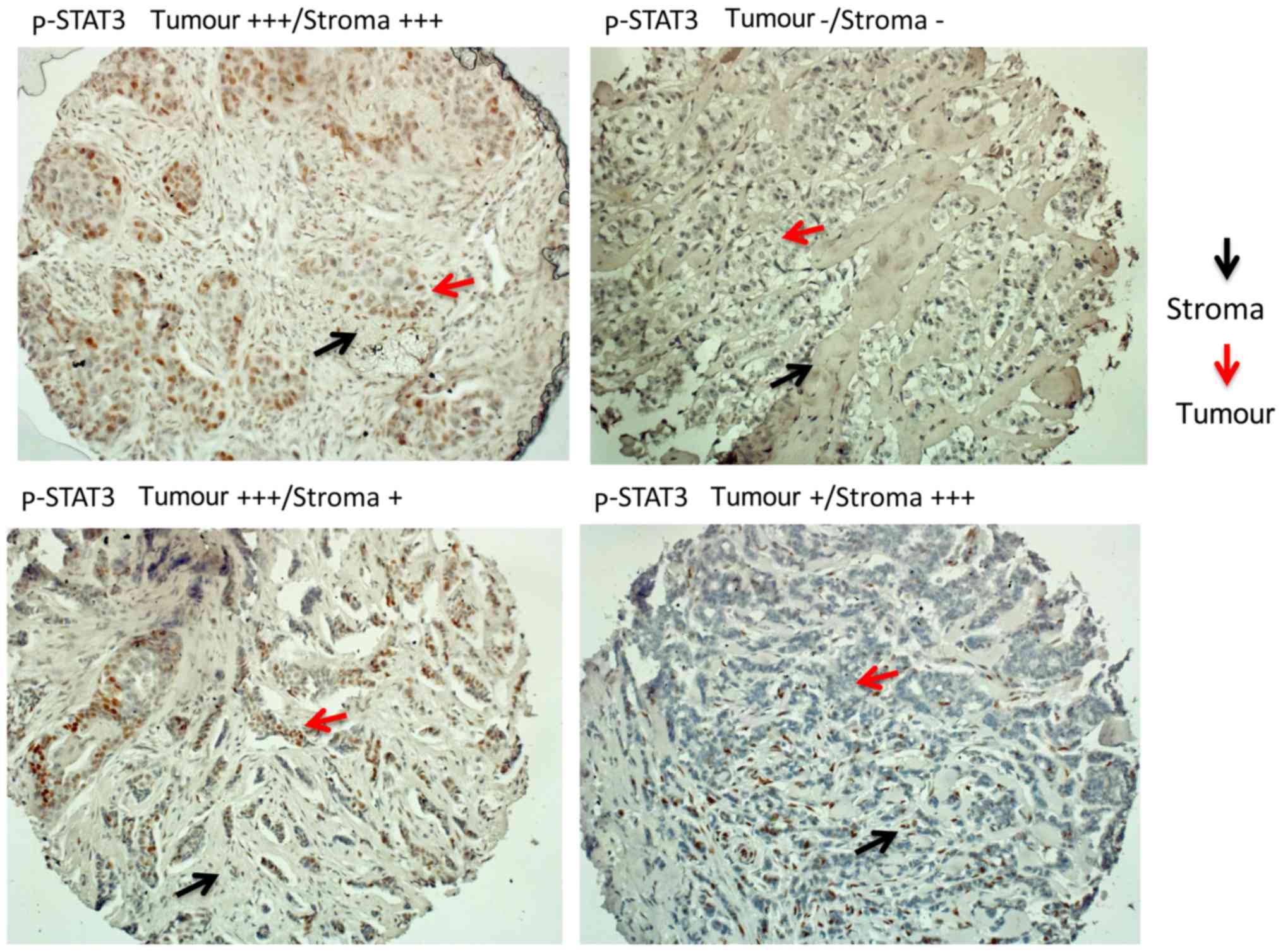
available for the evaluation of p-STAT3 by IHC and IF,
respectively. Evaluation was performed for the tumoral and stromal
component separately (Fig. 3). Any
level of p-STAT3 staining by IHC in the tumour or stroma was
detected in 174 out of the 610 samples (28.5%). p-STAT3 (in the
tumour or stroma) was associated with a smaller tumour size when
analysed by IHC (Table II) or IF
(data not shown). p-STAT3 expression between the tumour and stroma
strongly correlated using IHC (R=0.67; 95% CI, 0.63–0.71) or IF
(R=0.86; 95% CI, 0.84–0.88) as evaluated by Spearman's correlation.
For the IF analysis, two patterns of staining were found (patchy
and diffuse) that were not associated with different clinical or
pathological parameters (data not shown).
 | Table IIAssociation of p-STAT3 (by
immunohistochemistry) expression with clinicopathological
parameters in ER-positive breast cancer. |
Table II
Association of p-STAT3 (by
immunohistochemistry) expression with clinicopathological
parameters in ER-positive breast cancer.
|
Characteristics | p-STAT3-negative in
both tumour and stroma
(n=436) | p-STAT3-positive in
tumour or stroma
(n=174) | P-value |
|---|
| Age at
randomization, years | | | |
| Mean ± SD | 48.6±9.2 | 48.4±8.5 | 0.80 |
| Median
(range) | 49 (20–69) | 48 (27–69) | |
| No. of involved
nodes, no (%) | | | |
| 1–3 | 229 (52.5) | 95 (54.6) | 0.70 |
| 4–10 | 153 (35.1) | 58 (33.3) | |
| >10 | 54 (12.4) | 21 (12.1) | |
| Tumor size, no
(%) | | | |
| ≤2 cm | 133 (30.8) | 72 (41.6) | 0.01 |
| >2 cm | 299 (69.2) | 101 (58.4) | |
| pTx | 4 | 1 | |
| Tumor grade, no
(%) | | | |
| G1–G2 | 234 (55.9) | 103 (61.7) | 0.20 |
| G3 | 185 (44.2) | 64 (38.3) | |
| Gx | 17 | 7 | |
| PR, no (%) | | | |
|
PR− | 41 (9.6) | 13 (7.6) | 0.44 |
|
PR+ | 388 (90.4) | 158 (92.4) | |
| Missing info | 7 | 3 | |
| HER2, no (%) | | | |
|
HER2− | 364 (84.5) | 152 (88.4) | 0.21 |
|
HER2+ | 67 (15.6) | 20 (11.6) | |
| Missing info | 5 | 2 | |
| Taxane, no (%) | | | |
| No | 131 (30.1) | 59 (33.9) | 0.35 |
| Yes | 305 (70.0) | 115 (66.1) | |
| Seq/combined, no
(%) | | | |
| Sequential | 213 (48.9) | 104 (59.8) | 0.01 |
| Combined | 223 (51.2) | 70 (40.2) | |
| Median follow-up,
years (95% CI) | 10.40
(10.15–10.61) | 10.45
(10.11–10.70) | 0.51 | |
| No. of deaths | 98 | 34 | |
| No. of events (BCR,
SPM, death) | 159 | 51 | |
For the prognostic evaluations, all treatment arms
were pooled. We examined p-STAT3 in the tumour or stroma using IHC
and their association with DFS and OS end points. As summarised in
Table III, there was no
significant prognostic effect in the global population (all
ER-positive), although a trend for an improved DFS was observed
(HR, 0.72; 95% CI, 0.51–1.04; P=0.08). For the ER-positive/HER2
negative group, p-STAT3 was found to be prognostic using univariate
(positive vs. negative) analysis for DFS (Cox univariate HR, 0.66;
95% CI, 0.44–0.98; P=0.04), but not OS (Cox univariate HR, 0.70;
95% CI, 0.43–1.15; P=0.16). Multivariate analysis, adjusting for
the grade and number of positive lymph nodes, did not reach
significance (Cox multivariate, HR, 0.76; 95% CI, 0.51–1.15;
P=0.19). The analysis of ER-positive/HER2 negative samples from IF
staining with diffuse distribution demonstrated the same trend of
improved outcome in p-STAT3-positive tumours (DFS: HR, 0.59; 95%
CI, 0.35–1.01, P=0.056; OS: HR, 0.53; 95% CI, 0.26–1.05, P=0.068)
(data not shown).
 | Table IIIPrognostic value of p-STAT3
(immunohistochemistry) using univariate analysis. |
Table III
Prognostic value of p-STAT3
(immunohistochemistry) using univariate analysis.
| DFS
| OS
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| All | | | | |
| Tumour p-STAT3
ordinala | 0.84
(0.68–1.04) | 0.10 | 0.83
(0.63–1.08) | 0.16 |
| Tumour p-STAT3
binary (positive vs. negative) | 0.72
(0.51–1.04) | 0.08 | 0.75
(0.48–1.18) | 0.21 |
| Stroma p-STAT3
ordinala | 0.85
(0.68–1.05) | 0.13 | 0.88
(0.67–1.15) | 0.34 |
| Stroma p-STAT3
binary (positive vs. negative) | 0.78
(0.55–1.08) | 0.14 | 0.79
(0.52–1.21) | 0.28 |
|
ER+/HER2− | | | | |
| Tumour p-STAT3
ordinala | 0.80
(0.63–1.01) | 0.06 | 0.83
(0.62–1.11) | 0.22 |
| Tumour p-STAT3
binary (positive vs. negative) | 0.66
(0.44–0.98) | 0.04 | 0.70
(0.43–1.15) | 0.16 |
| Stroma p-STAT3
ordinala | 0.86
(0.67–1.09) | 0.20 | 0.92
(0.68–1.22) | 0.55 |
| Stroma p-STAT3
binary (positive vs. negative) | 0.75
(0.52–1.09) | 0.13 | 0.82
(0.52–1.30) | 0.39 |
|
ER+/HER2+ | | | | |
| Tumor p-STAT3
ordinala | 1.09
(0.66–1.80) | 0.73 | 0.86
(0.40–1.85) | 0.71 |
| Tumor p-STAT3
binary (positive vs. negative) | 1.33
(0.56–3.21) | 0.52 | 1.34
(0.45–3.93) | 0.60 |
| Stroma p-STAT3
ordinala | 0.86
(0.49–1.52) | 0.61 | 0.78
(0.36–1.67) | 0.52 |
| Stroma p-STAT3
binary (positive vs. negative) | 0.93
(0.41–2.12) | 0.86 | 0.79
(0.27–2.34) | 0.67 |
Discussion
In the present study, we sought to better understand
the associations between p-STAT3 the luminal phenotype and outcome
of breast cancer, using an approach that integrated RPPA and
microarrays gene expression data. Specifically, we were interested
in identifying a p-STAT3 expression signature in luminal breast
cancers which could be used for prognostic purposes. Although
significant data exist on p-STAT3 target genes/signatures in
triple-negative breast cancer models and datasets, relatively few
have been identified for luminal breast cancers (29). Notably, Walker et al
determined that ER-positive tumours co-express both p-STAT5 and
p-STAT3 and identified a STAT3/STAT5 expression signature (albeit
using a small number of samples) distinct from STAT3 which
correlated with an improved outcome (18). In this study, we did not analyse
p-STAT5 levels, as they are not represented in the TCGA RPPA data
set. In addition to the p-STAT3 levels, total STAT3 is considered a
'surrogate' for p-STAT3, as STAT3 is positively regulated by
p-STAT3. Unfortunately, we could not perform this analysis, as the
total STAT3 protein levels were similarly not available from the
TCGA RPPA dataset.
We also validated the observation that p-STAT3 is
associated with a better outcome in ER-positive patients randomised
in the BIG 2-98 trial. We found that tumours highly expressing
p-STAT3 were associated with a smaller tumour size, luminal A
phenotype and a good clinical outcome in comparison to those
expressing low levels of p-STAT3. In a recent study, it was
suggested that patients in the luminal A population were much more
likely to possess a p-STAT3 high phenotype, but this was not
associated with specific genes that were differentially expressed
between the p-STAT3 high vs. low groups (29). Therefore, it was hypothesised that
although STAT3 is activated variably between tumour subtypes, the
overall mechanisms through which STAT3 affects gene expression are
multifactorial and may not entirely correlate with STAT3
phosphorylation.
In a recent study, hormonal treatment did not
improve outcome in patients with increased levels of protein
inhibitor of activated STAT3 (PIAS3). It was suggested that an
increased PIAS3 expression attenuates the effectiveness of
tamoxifen (30). This is in
support of our observation that STAT3 activation was associated
with an improved outcome in ER-positive tumours.
Although our data strongly suggest that a high
p-STAT3 and a p-STAT3 expression signature correlate with an
improved outcome, these observations do not allow us to propose
that targeting this molecule or pathway would have deleterious
effects. For example, activating mutations in the PI3K pathway
found predominantly in luminal breast cancers are associated with
an improved clinical outcome (31). Despite this correlation, targeting
this pathway with small molecule inhibitors in conjunction with
anti-estrogens has led to improved outcomes in patients with
metastatic disease (32).
As regards the IL-6/STAT3 signalling pathway, it was
recently shown in pre-clinical models of luminal breast cancer that
reversing its activity with an IL-6R blocking antibody
(tocilizumab) restored a dependence on anti-estrogens through
increased ER expression and elimination of CD133 stem cells
(33). The potential benefits of
targeting this pathway in endocrine-therapy resistant metastatic
disease are presently being tested. Specifically, a clinical trial
combining the JAK1/2 inhibitor ruxolitinib with endocrine therapy
is ongoing (NCT01594216). Notably, the selection of inhibitor used
could affect the outcome. Specifically, a JAK1/2 inhibitor can also
target STAT5 and STAT1 which could influence response and prevent
one from concluding that the outcome is due to targeting only
STAT3.
In conclusion, this study provides evidence that the
p-STAT3 status is a marker of favorable outcome in ER-positive
breast cancer.
Acknowledgments
This study was funded by a Clinical Research Career
Development Award from the Israel Cancer Research Fund grants
(16-116-CRCDA) and from the Israeli Cancer Research Association
(2017-0140).
References
|
1
|
Darnell JE Jr, Kerr IM and Stark GR:
Jak-STAT pathways and transcriptional activation in response to
IFNs and other extracellular signaling proteins. Science.
264:1415–1421. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhong Z, Wen Z and Darnell JE Jr: Stat3: A
STAT family member activated by tyrosine phosphorylation in
response to epidermal growth factor and interleukin-6. Science.
264:95–98. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Garcia R, Bowman TL, Niu G, Yu H, Minton
S, Muro-Cacho CA, Cox CE, Falcone R, Fairclough R, Parsons S, et
al: Constitutive activation of Stat3 by the Src and JAK tyrosine
kinases participates in growth regulation of human breast carcinoma
cells. Oncogene. 20:2499–2513. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Niu G, Wright KL, Huang M, Song L, Haura
E, Turkson J, Zhang S, Wang T, Sinibaldi D, Coppola D, et al:
Constitutive Stat3 activity up-regulates VEGF expression and tumor
angiogenesis. Oncogene. 21:2000–2008. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu H, Lee H, Herrmann A, Buettner R and
Jove R: Revisiting STAT3 signalling in cancer: New and unexpected
biological functions. Nat Rev Cancer. 14:736–746. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Knüpfer H and Preiss R: Significance of
interleukin-6 (IL-6) in breast cancer (Review). Breast Cancer Res
Treat. 102:129–135. 2007. View Article : Google Scholar
|
|
7
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: A leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yue P and Turkson J: Targeting STAT3 in
cancer: How successful are we? Expert Opin Investig Drugs.
18:45–56. 2009. View Article : Google Scholar :
|
|
9
|
Sansone P and Bromberg J: Targeting the
interleukin-6/Jak/stat pathway in human malignancies. J Clin Oncol.
30:1005–1014. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chang Q, Bournazou E, Sansone P, Berishaj
M, Gao SP, Daly L, Wels J, Theilen T, Granitto S, Zhang X, et al:
The IL-6/JAK/Stat3 feed-forward loop drives tumorigenesis and
metastasis. Neoplasia. 15:848–862. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Azare J, Doane A, Leslie K, Chang Q,
Berishaj M, Nnoli J, Mark K, Al-Ahmadie H, Gerald W, Hassimi M, et
al: Stat3 mediates expression of autotaxin in breast cancer. PLoS
One. 6:e278512011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gritsko T, Williams A, Turkson J, Kaneko
S, Bowman T, Huang M, Nam S, Eweis I, Diaz N, Sullivan D, et al:
Persistent activation of stat3 signaling induces survivin gene
expression and confers resistance to apoptosis in human breast
cancer cells. Clin Cancer Res. 12:11–19. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Burke WM, Jin X, Lin HJ, Huang M, Liu R,
Reynolds RK and Lin J: Inhibition of constitutively active Stat3
suppresses growth of human ovarian and breast cancer cells.
Oncogene. 20:7925–7934. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sonnenblick A, Uziely B, Nechushtan H,
Kadouri L, Galun E, Axelrod JH, Katz D, Daum H, Hamburger T, Maly
B, et al: Tumor STAT3 tyrosine phosphorylation status, as a
predictor of benefit from adjuvant chemotherapy for breast cancer.
Breast Cancer Res Treat. 138:407–413. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sonnenblick A, Shriki A, Galun E, Axelrod
JH, Daum H, Rottenberg Y, Hamburger T, Mali B and Peretz T: Tissue
microarray-based study of patients with lymph node-positive breast
cancer shows tyrosine phosphorylation of signal transducer and
activator of transcription 3 (tyrosine705-STAT3) is a marker of
good prognosis. Clin Transl Oncol. 14:232–236. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Walker SR, Nelson EA, Yeh JE, Pinello L,
Yuan GC and Frank DA: STAT5 outcompetes STAT3 to regulate the
expression of the oncogenic transcriptional modulator BCL6. Mol
Cell Biol. 33:2879–2890. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dolled-Filhart M, Camp RL, Kowalski DP,
Smith BL and Rimm DL: Tissue microarray analysis of signal
transducers and activators of transcription 3 (Stat3) and
phospho-Stat3 (Tyr705) in node-negative breast cancer shows nuclear
localization is associated with a better prognosis. Clin Cancer
Res. 9:594–600. 2003.PubMed/NCBI
|
|
18
|
Walker SR, Xiang M and Frank DA: Distinct
roles of STAT3 and STAT5 in the pathogenesis and targeted therapy
of breast cancer. Mol Cell Endocrinol. 382:616–621. 2014.
View Article : Google Scholar
|
|
19
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Varma S and Simon R: Bias in error
estimation when using cross-validation for model selection. BMC
Bioinformatics. 7:912006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sonnenblick A, Brohée S, Fumagalli D,
Rothé F, Vincent D, Ignatiadis M, Desmedt C, Salgado R, Sirtaine N,
Loi S, et al: Integrative proteomic and gene expression analysis
identify potential biomarkers for adjuvant trastuzumab resistance:
Analysis from the Fin-her phase III randomized trial. Oncotarget.
6:30306–30316. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sonnenblick A, Francis PA, Azim HA Jr, de
Azambuja E, Nordenskjöld B, Gutiérez J, Quinaux E, Mastropasqua MG,
Ameye L, Anderson M, et al: Final 10-year results of the Breast
International Group 2-98 phase III trial and the role of Ki67 in
predicting benefit of adjuvant docetaxel in patients with oestrogen
receptor positive breast cancer. Eur J Cancer. 51:1481–1489. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Francis P, Crown J, Di Leo A, Buyse M,
Balil A, Andersson M, Nordenskjöld B, Lang I, Jakesz R, Vorobiof D,
et al: BIG 02-98 Collaborative Group: Adjuvant chemotherapy with
sequential or concurrent anthracycline and docetaxel: Breast
International Group 02-98 randomized trial. J Natl Cancer Inst.
100:121–133. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
McShane LM, Altman DG, Sauerbrei W, Taube
SE, Gion M and Clark GM; Statistics Subcommittee of NCI-EORTC
Working Group on Cancer Diagnostics : REporting recommendations for
tumor MARKer prognostic studies (REMARK). Breast Cancer Res Treat.
100:229–235. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Haibe-Kains B, Desmedt C, Loi S, Culhane
AC, Bontempi G, Quackenbush J and Sotiriou C: A three-gene model to
robustly identify breast cancer molecular subtypes. J Natl Cancer
Inst. 104:311–325. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al METABRIC Group: The genomic and transcriptomic architecture of
2,000 breast tumours reveals novel subgroups. Nature. 486:346–352.
2012.PubMed/NCBI
|
|
28
|
Dedeurwaerder S, Desmedt C, Calonne E,
Singhal SK, Haibe-Kains B, Defrance M, Michiels S, Volkmar M,
Deplus R, Luciani J, et al: DNA methylation profiling reveals a
predominant immune component in breast cancers. EMBO Mol Med.
3:726–741. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tell RW and Horvath CM: Bioinformatic
analysis reveals a pattern of STAT3-associated gene expression
specific to basal-like breast cancers in human tumors. Proc Natl
Acad Sci USA. 111:12787–12792. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang SF, Hou MF, Chen FM, Ou-Yang F, Wu
YC, Chai CY and Yeh YT: Prognostic value of protein inhibitor of
activated STAT3 in breast cancer patients receiving hormone
therapy. BMC Cancer. 16:202016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Loi S, Haibe-Kains B, Majjaj S, Lallemand
F, Durbecq V, Larsimont D, Gonzalez-Angulo AM, Pusztai L, Symmans
WF, Bardelli A, et al: PIK3CA mutations associated with gene
signature of low mTORC1 signaling and better outcomes in estrogen
receptor-positive breast cancer. Proc Natl Acad Sci USA.
107:10208–10213. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Baselga J, Campone M, Piccart M, Burris HA
III, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun
F, et al: Everolimus in postmenopausal hormone-receptor-positive
advanced breast cancer. N Engl J Med. 366:520–529. 2012. View Article : Google Scholar
|
|
33
|
Sansone P, Ceccarelli C, Berishaj M, Chang
Q, Rajasekhar VK, Perna F, Bowman RL, Vidone M, Daly L, Nnoli J, et
al: Self-renewal of CD133(hi) cells by IL6/Notch3 signalling
regulates endocrine resistance in metastatic breast cancer. Nat
Commun. 7:104422016. View Article : Google Scholar : PubMed/NCBI
|