Introduction
Glioblastoma (GBM), the most malignant form of
glioma, is highly aggressive, and most patients diagnosed with this
form of cancer often quickly succumb to the disease outcome
(1). Despite the introduction of
several modern therapeutic methods, including surgical resection,
radiation therapy, and chemotherapy, the median survival time of
patients diagnosed with GBM is only 14.6 months, with a 2-year
survival rate of only 26% (2,3).
Therefore, a better understanding of the molecular and cellular
changes that occur in GBM cells is essential to identify effective
therapeutic targets for the treatment of GBM.
MicroRNAs (miRNAs) regulate the expression of their
target mRNAs via partial or complete complementarity to the 3′
untranslated region (UTR) of their target gene (4), and have been shown to play a critical
role in tumor pathogenesis (5,6).
Studies have shown that some miRNAs have tumor suppressive
functions, and many are aberrantly expressed in variety of cancers
(7). Furthermore, several miRNAs
have been shown to be involved in glioblastoma tumorigenesis by
targeting oncogenes and tumor suppressor genes (8–10).
For example, miR-204 has been shown to suppress glioma cell
proliferation, migration, and invasion via the inhibition of
activating transcription factor (11), and miR-92b-I has been reported to
inhibit proliferation, invasion, and migration, and to stimulate
apoptosis of glioma cells via the regulation of the PTEN/Akt
signaling pathway (12). Another
study showed that miR-423-5p functions as a tumor suppresser in
glioma tissues by targeting the inhibitor of growth 4 gene
(13). Recently, several studies
have reported that miR-205 is dysregulated in clinical samples from
solid tumors and glioma cell lines (14–17).
However, to date, the role of miR-205 in the development of GBM has
not been elucidated.
The epithelial-mesenchymal transition (EMT) is a
vital morphogenic process during embryonic development, and is
crucial for epithelial cancer cell acquisition of an invasive
phenotype (18). EMT is regulated
by several transcription factors, including zinc finger E-box
binding homeobox 1 (ZEB1) and Twist1, which have been shown to be
transcriptional repressors of E-cadherin (19). ZEB1 plays a key role in the
regulation of tumor metastasis by inducing EMT (20). Furthermore, studies have indicated
that ZEB1 is upregulated in GBM cells and may repress genes
involved in cancer cell adhesion and polarity (21). Therefore, inhibition of ZEB1
expression may be beneficial for the therapeutic management of
GBM.
In this study, we show that miR-205 expression is
reduced in GBM tissues and cell lines, and that miR-205 expression
is negatively associated with ZEB1 expression in GBM tissues.
Furthermore, we show miR-205 downregulates ZEB1, causing
suppression of GBM cell migration and invasion, and reverses EMT
via the Akt/mTOR signaling pathway. These findings elucidate the
role of miR-205 in GBM, and suggest that miR-205 may be an
effective therapeutic target for the treatment of patients with
GBM.
Materials and methods
Tissue specimens
Tissue specimens from 76 GBM and corresponding
normal brain tissues were obtained from the Guangzhou Women and
Children's Medical Center and Sun Yat-Sen Memorial Hospital between
January 2013 and October 2015. Each sample was frozen in liquid
nitrogen within 2 h of extraction. The medical history of each
patient was recorded as shown in Table
I. GBM diagnosis was made according to the revised WHO
classification system (22).
Patients who had received treatment, including chemotherapy and
radiation therapy, before surgery, were excluded. All experiments
were performed in accordance with the guidelines approved by the
Ethics Committee of Sun YatSen Memorial Hospital, and informed
consent was obtained from each patient.
 | Table ICharacteristics of glioma
patients. |
Table I
Characteristics of glioma
patients.
|
Characteristics | Patients
(n=76) |
|---|
| Median age
(years) | 50.2 |
| Range | 34–75 |
| Male | 39 (51.3%) |
| Smoking
status | |
| Ever and
current | 23 (30.3%) |
| Never | 53 (69.7%) |
| Alcohol
consumption | |
| Ever and
current | 25 (32.9%) |
| Never | 51 (67.1%) |
| KPS score | |
| ≥70 | 29 (38.2%) |
| <70 | 51 (61.8%) |
| Extent of
resection | |
| Gross total
resection | 53 (69.7%) |
| Subtotal
resection | 23 (30.3%) |
| WHO | |
| I+II | 29 (38.2%) |
| III+IV | 47 (61.8%) |
Cell culture and transfection
The human glial HEB and glioblastoma cell lines
U87MG, SHG-44, U251, and A172 were maintained in Dulbecco's
modified Eagle's medium (DMEM) with low glucose, supplemented with
10% fetal bovine serum (Gibco, MD, USA). Cells were incubated at
37°C with 5% CO2. Control RNA mimics and miR-205 mimics
were obtained from Ribobio (Guangzhou, China), and transfected into
cells at a working concentration of 50 nM using Lipofectamine 2000
reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer's instructions.
ZEB1 gene expression rescue assay
The full length ZEB1 gene open reading frame (ORF)
was amplified via PCR and cloned into pcDNA-3.1 (Invitrogen) to
generate a pcDNA-3.1+ZEB1 construct (hereinafter, ZEB1). The empty
pcDNA-3.1 served as the control (hereinafter, vector). SHG-44 and
A172 cells were transfected with the miR-205 mimic (50 nM), in
6-well plates, followed by co-transfection with 2.0 µg of
either pcDNA-3.1+ZEB1 or control vector for 48 h.
RNA extraction and quantitative real-time
PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent
(Invitrogen) according to the manufacturer's protocol, and
reverse-transcription was conducted using PrimeScript™ RT-PCR kit
(Takara, Otsu, Shiga, Japan). qRT-PCR was performed to quantify
expression levels using the SYBR Green PCR Master mix (Applied
Biosystems, Foster City, CA, USA) according to the manufacturer's
instructions. qRT-PCR was performed at 94°C for 2 min followed by
40 cycles at 94°C for 10 sec, 60°C for 1 min, and 30 sec at 72°C on
an ABI PRISM 7500 Real-Time PCR system (Applied Biosystems).
Expression of mature miR-205 was assayed using the Bulge-Loop™
miRNA qRT-PCR Primer Set and the miRNA qRT-PCR Control Primer Set
(RiboBio, Guangdong, China). GAPDH and U6 were used as internal
controls. All sequences used are shown in Table II. Each reaction was performed in
triplicate. Relative quantification of gene expression levels was
expressed as fold change, normalized against internal controls,
using the ΔΔCq method. Each sample was detected in triplicate.
 | Table IIThe forward and reverse primers for
real-time PCR. |
Table II
The forward and reverse primers for
real-time PCR.
| Name | Sequence |
|---|
| Hsa-miR-205 | Forward
GTGACCAACATACCACCGG |
| Reverse
TGGTGTCGTGGAGTCG |
| ZEB1 | Forward
CAGCTTGATACCTGTGAATGGG |
| Reverse
TATCTGTGGTCGTGTGGGACT |
| E-cadherin | Forward
AAAGGCCCATTTCCTAAAAACCT |
| Reverse
TGCGTTCTCTATCCAGAGGCT |
| N-cadherin | Forward
AGCCAACCTTAACTGAGGAGT |
| Reverse
GGCAAGTTGATTGGAGGGATG |
| Vimentin | Forward
AGTCCACTGAGTACCGGAGAC |
| Reverse
CATTTCACGCATCTGGCGTTC |
| U6 | Forward
CTCGCTTCGGCAGCACA |
| Reverse
AACGCTTCACGAATTTGCGT |
| GAPDH | Forward
ACAACTTTGGTATCGTGGAAGG |
| Reverse
GCCATCACGCCACAGTTTC |
Western blotting
Total protein was extracted from cells using 1% RIPA
lysis buffer (Beyotime, Jiangsu, China), and the BCA method was
used for protein quantitation. Equal concentrations of protein were
separated by SDS-PAGE, transferred onto PVDF membranes, and probed
with ZEB1, mTOR, phospho-mTOR (Abcam, MA, USA), Akt, phospho-Akt,
E-cadherin, N-cadherin, vimentin (Cell Signaling Technology,
Beverly, MA, USA) and GAPDH (Abcam) antibodies overnight at 4°C.
The membranes were incubated with horseradish peroxidase
(HRP)-conjugated secondary antibodies (Abcam) for 2 h.
Immunocomplexes were visualized using the ECL detection reagent
(Beyotime) and the intensities of the signals were quantified using
ImageJ software 6.0.
Cell migration and invasion assay
Approximately 106 cells were seeded into
each well of 6-well plates. A cell scratch spatula was used to
scratch the cell after transfection. Plates were washed using
warmed phosphate-buffered saline three times to remove cellular
debris, and the cells were incubated at 37°C for 48 h, examined and
photographed under a microscope (Olympus Corp., Tokyo, Japan) 0 and
48 h after wounding. Each cell condition was assayed in
triplicate.
Transwell chambers (8-µm pore size;
Millipore, Billerica, MA, USA) coated with Matrigel (BD
Biosciences, San Jose, CA, USA) were used for the invasion assays.
Following trans-fection, cells were seeded onto the upper wells,
and 500 µl DMEM containing 10% fetal bovine serum (FBS) was
used as a chemoattractant in the lower chambers. The cells were
allowed to migrate for 24 h, the Matrigel and the cells in the
upper surface were removed, and the cells that had migrated to the
lower surface were fixed in 4% paraformaldehyde and stained with
0.1% crystal violet (Sigma, St. Louis, MO, USA). The number of
invaded cells were counted under a light microscope (Axiovert 200
inverted microscope; Zeiss, Germany) in five fields, and presented
as the average number of cells per field of view.
Luciferase reporter assay
Potential targets of miR-205 were predicted using
miRbase, and finally the putative complementary sequence of miR-205
was identified in the 3′UTR of ZEB-1 mRNA. Luciferase reporter
vectors were constructed using the full 3′UTR of ZEB-1 and inserted
into the pEZX-MT01 vector (GeneCopia, Labomics S.A, Nivelles,
Belgium). Empty vectors were used as a negative control.
Renilla luciferase, encoded by the same vector, served as an
internal control. Each luciferase reporter construct, including the
Luc+miR205, Luc+ZEB1 3′UTR, and negative control vectors, was
co-transfected into SHG-44 cells and A172 cells using Lipofectamine
2000 (Invitrogen). After 24 h of incubation, the cells were
re-seeded in 96-well plates and then Firefly and Renilla
luciferase activities were determined using Luc-Pair™ miR
Luciferase assay kits (GeneCopia) according to the manufacturer's
instructions. All transfection experiments were performed in
triplicate and repeated three times.
Immunofluorescence analysis
To analyze immuno-fluorescence, SHG-44 and A172
cells were transfected with miRNA-NC or miRNA-205 mimics for 48 h
and cultured on circular coverslips (BD Biosciences) in 6-well
plates, then washed and fixed with 4% paraformaldehyde for 15 min.
The cells were then permeabilized with 0.1% Triton X for 5 min and
blocked for 30 min with 3% bovine serum albumin (BSA)-PBS at room
temperature. The cells were incubated overnight at 4°C with
anti-ZEB1 primary antibodies, then incubated with a fluorescent
secondary antibody for 1 h at room temperature. To visualize the
nuclei, the wells were stained with DAPI (Sigma) for 5 min.
Fluorescent images (at ×200 magnification) were acquired using a
fluorescence inverted microscope (Olympus) and images were
processed with Image-ProPlus 6.0 (Media Cybernetics, USA).
Statistical analysis
Data were expressed as the mean ± standard deviation
(SD). Student's t-test was used to compare differences between two
groups. One-way analysis of variance (ANOVA) was used to compare
the differences between more than two groups, and significance was
determined using the LSD t-test. Pearson correlation was used to
analyze the relationship between miR-205 and ZEB1 expression. A
P-value of <0.05 was considered statistically significant. All
statistical analyses were performed using SPSS 20.0 (SPSS, Inc.,
Chicago, IL, USA), and GraphPad Prism 6.0 (GraphPad Software, Inc.,
CA, USA) was used to generate graphs.
Results
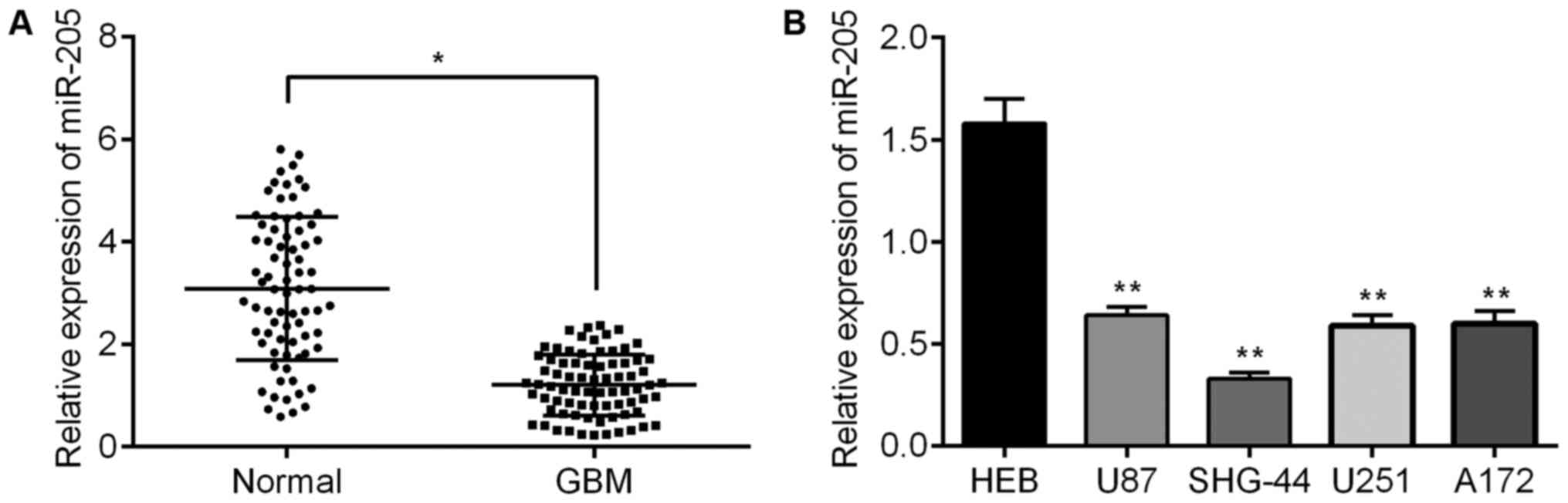
miR-205 is downregulated in GBM tissues
and cell lines
Previous studies have shown miR-205 expression is
reduced in GBM tissues and cell lines (14,15).
Therefore, we investigated miR-205 expression, via qRT-PCR, from
surgically excised specimens. Our results showed that miR-205 was
significantly downregulated in GBM tissues, as compared to the
corresponding non-neoplastic tissues (Fig. 1A). Furthermore, we found miR-205
expression was also substantially decreased in several glioma cell
lines, as compared to human glial HEB cells (Fig. 1B).
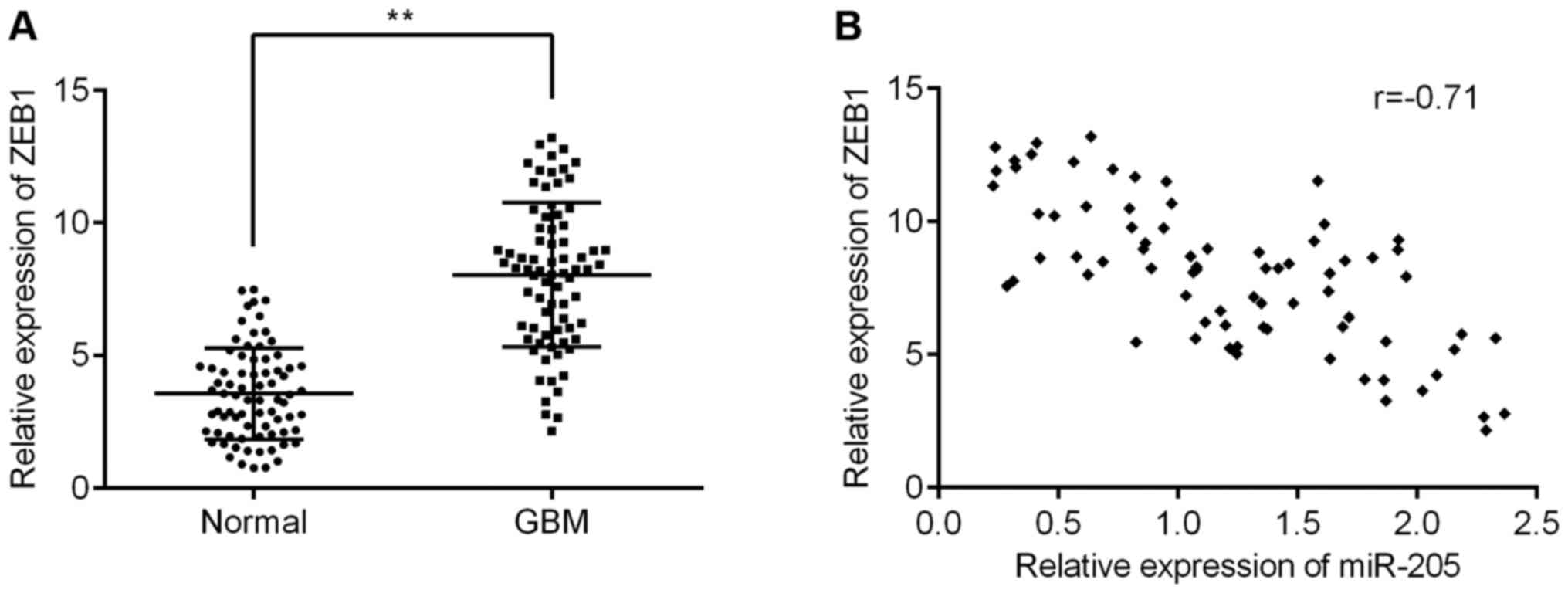
miR-205 directly targets the 3′UTR of
ZEB1 in GBM
To determine the downstream targets of miR-205 and
its role in GBM, we queried candidate target genes from the human
microRNA database, and found ZEB1 was a putative target of miR-205.
Therefore, we decided to investigate ZEB1 expression in GBM tissues
and glioma cell lines. As shown in Fig. 2A, the levels of ZEB1 in GMB tissues
were lower than those in the non-tumor tissues. A subsequent
correlation analysis found ZEB1 was negatively correlated with
miR-205 expression (Fig. 2B). Our
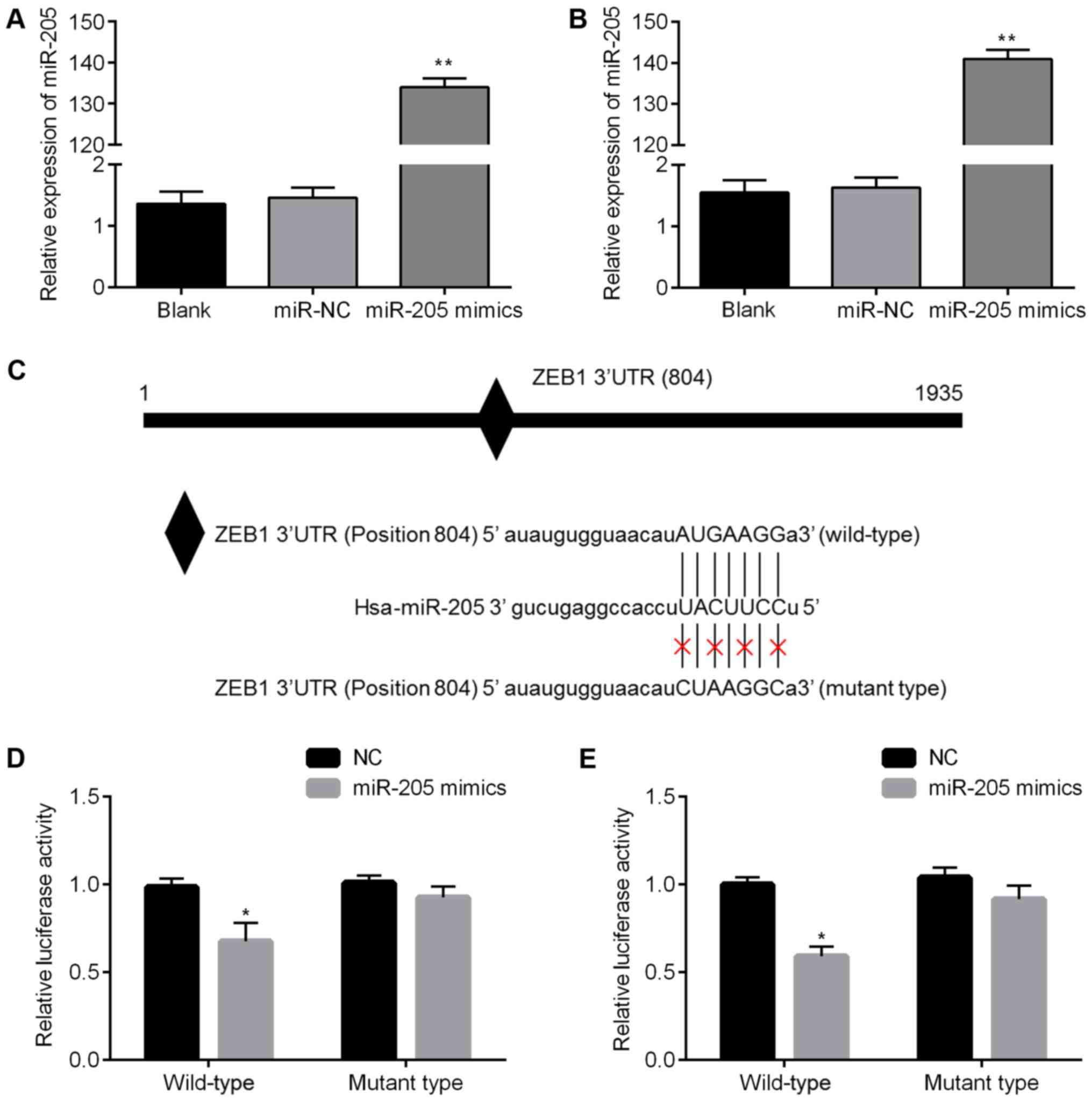
qRT-PCR results showed that miR-205 expression in SHG-44 and A172
cells increased more than 130-fold in the miR-205 mimics group
compared to miR-NC group (Fig. 3A and
B). Fig. 3C shows the putative
position of the miR-205 target site in the 3′UTR of ZEB1 mRNA. To
confirm these findings, we performed a luciferase assay, and found
that in SHG-44 and A172 cells overexpression of miR-205 suppressed
luciferase activity, while transfection of a scrambled sequence had
no effect (Fig. 3D and E). These
results suggest that miR-205 regulates ZEB1 expression by targeting
its 3′UTR.
miR-205 regulates the Akt/mTOR signaling
pathway by targeting ZEB1
In order to further elucidate the molecular
mechanisms of miR-205 in GBM metastasis, we examined the downstream
signaling pathways activated by ZEB1. We found ZEB1 protein
expression in SHG-44 and A172 cells transfected with the miR-205
mimics were significantly inhibited, as detected by western
blotting (Fig. 4A–D). Along with
the decrease in ZEB1 levels, overexpression of miR-205 also
inhibited p-Akt and p-mTOR expression levels, but had no effect on
total Akt and mTOR expression levels (Fig. 4A–D). These results indicate that
miR-205 regulates the Akt/mTOR signaling pathway by targeting ZEB1.
We also performed immunofluorescent staining on SHG-44 and A172
cells transfected with miR-205 mimics or miR-NC mimics. As shown in
Fig. 4E, the ZEB1 protein is
expressed in the cytoplasm of the SHG-44 and A172 cells, and
Fig. 4F shows its level is reduced
in the miR-205 mimics group as compared with the miR-NC group,
which corresponds with our western blotting results.
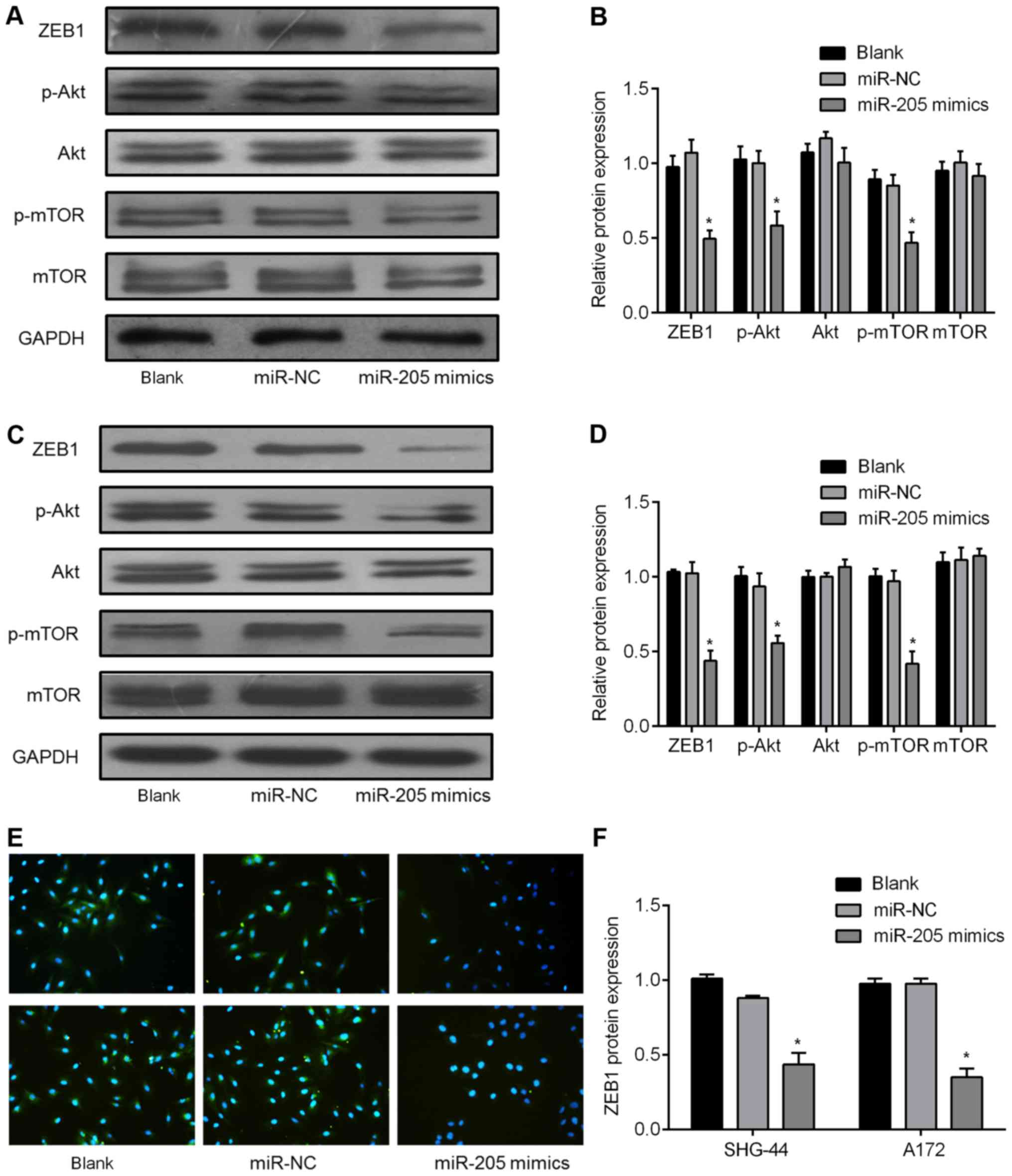 | Figure 4miR-205 regulates Akt/mTOR signaling
pathways by targeting ZEB1. (A) SHG-44 and (C) A172 cells
transfected with a miR-205 mimic or negative control mimics
(miR-NC) showing total protein used to detect ZEB1, p-Akt,
total-Akt, p-mTOR, total-mTOR, and GAPDH expression by western
blotting. (B) SHG-44 and (D) A172 cell relative protein levels
presented as mean ± SD from three experiments. The asterisk
indicates expression levels of ZEB1, p-Akt, and p-mTOR were
significantly lower in the miR-205 mimic group than that in miR-NC
group (*P<0.05). (E, F) SHG-44 (upper) or A172 cells
(lower) stained for ZEB1 by immunofluorescence. Red, ZEB1; blue,
DAPI nuclear staining. |
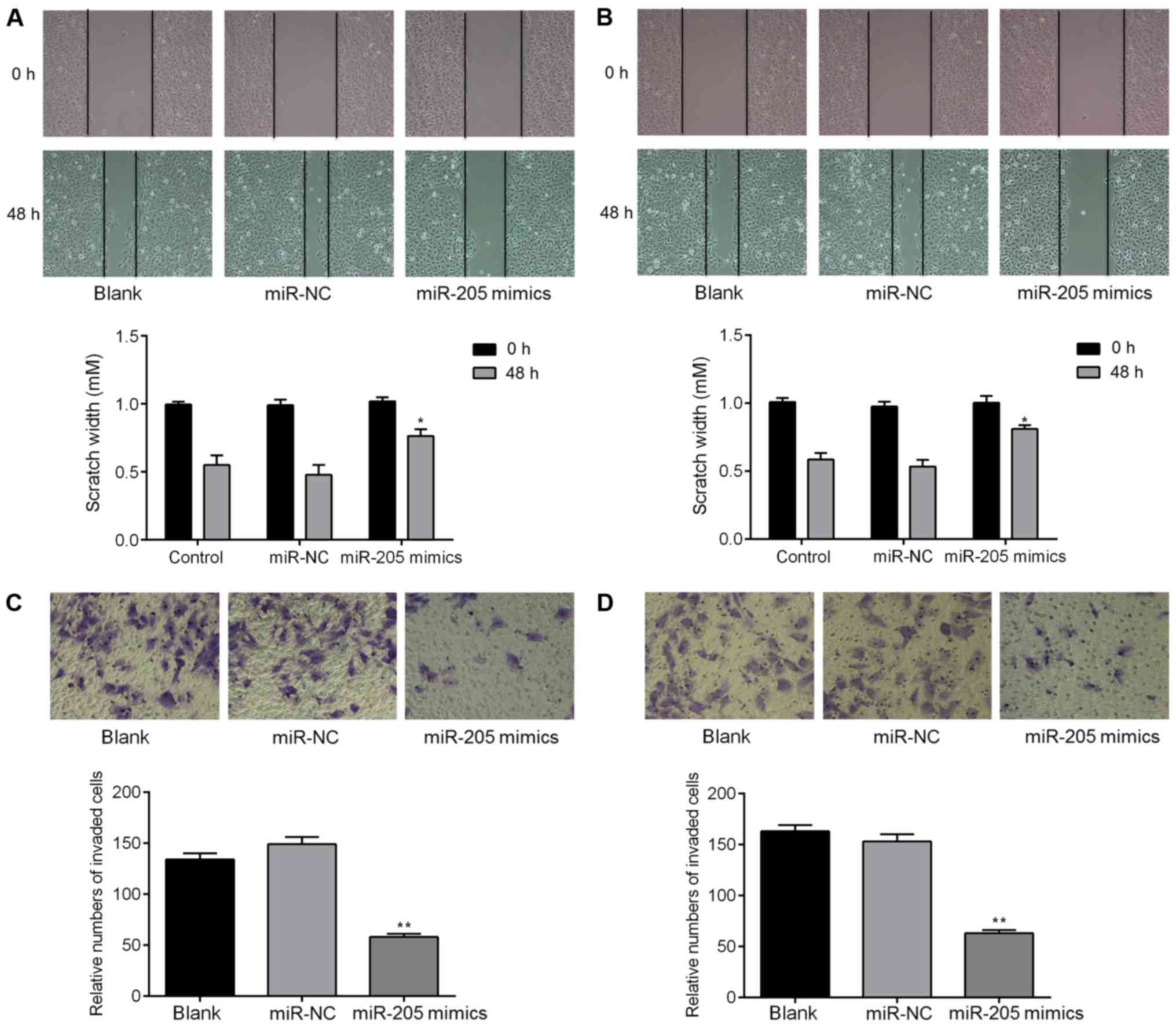
miR-205 inhibits cell migration and
invasion and reverses EMT in GBM
We also tested the effects of miR-205 on the
migration and invasion properties of glioma cells. Our wound
healing assay showed that the cell migration was suppressed when
miR-205 was overexpressed in both SHG-44 and A172 cells (Fig. 5A and B). Furthermore, our Transwell
invasion assay indicated that ectopic expression of miR-205
significantly inhibited the invasive response of both SHG-44 and
A172 cells (Fig. 5C and D). To
determine the regulatory effect of miR-205 on EMT, we performed
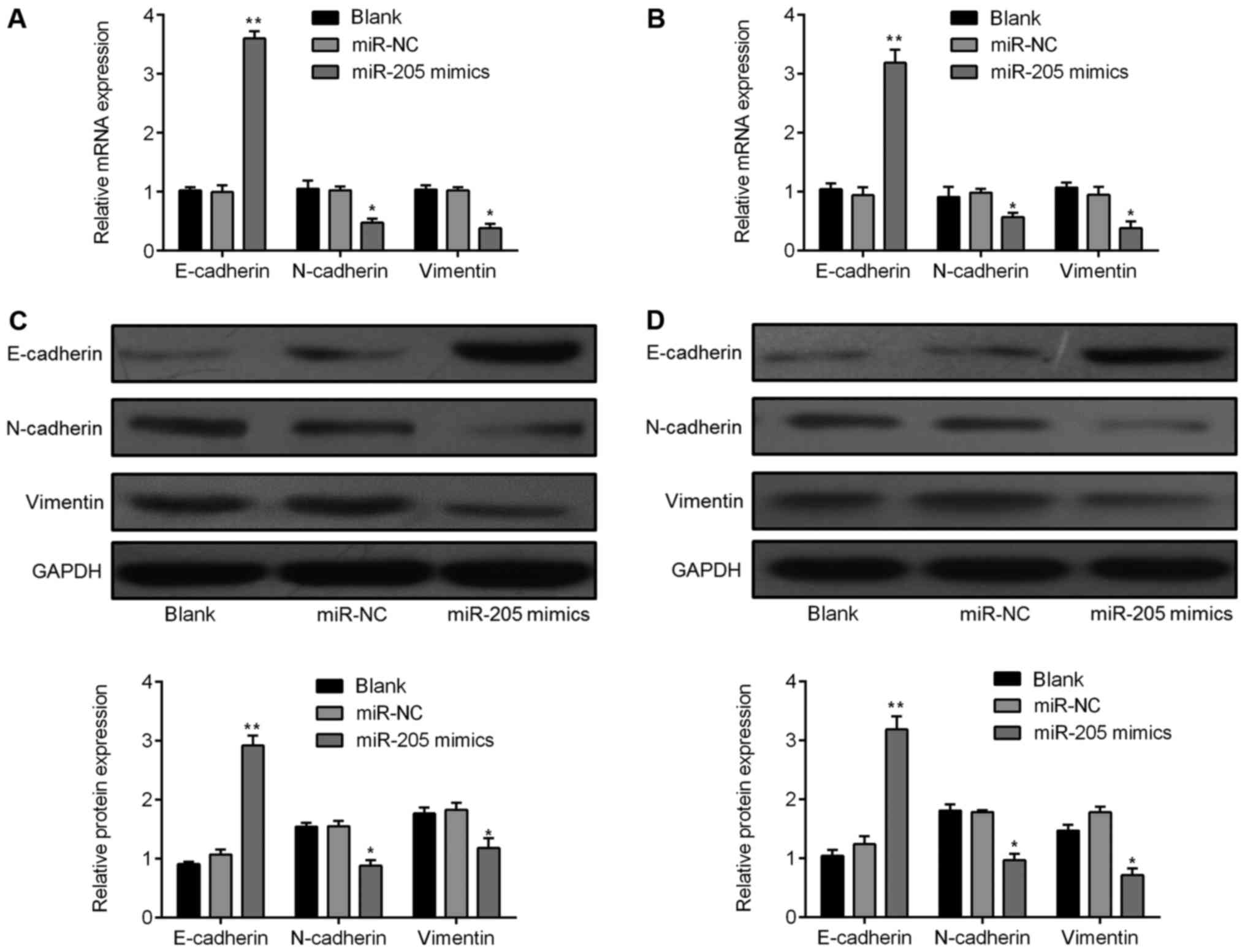
qRT-PCR and western blot analyses. The E-cadherin mRNA expression
level was markedly increased in the miR-205 mimics group, while
N-cadherin and vimentin were decreased in both SHG-44 and A172
cells (Fig. 6A and B). Consistent
with the qRT-PCR results, E-cadherin, N-cadherin, and vimentin
protein expression showed similar changes (Fig. 6C and D).
ZEB1 is a key regulator of the tumor
regulatory function of miR-205
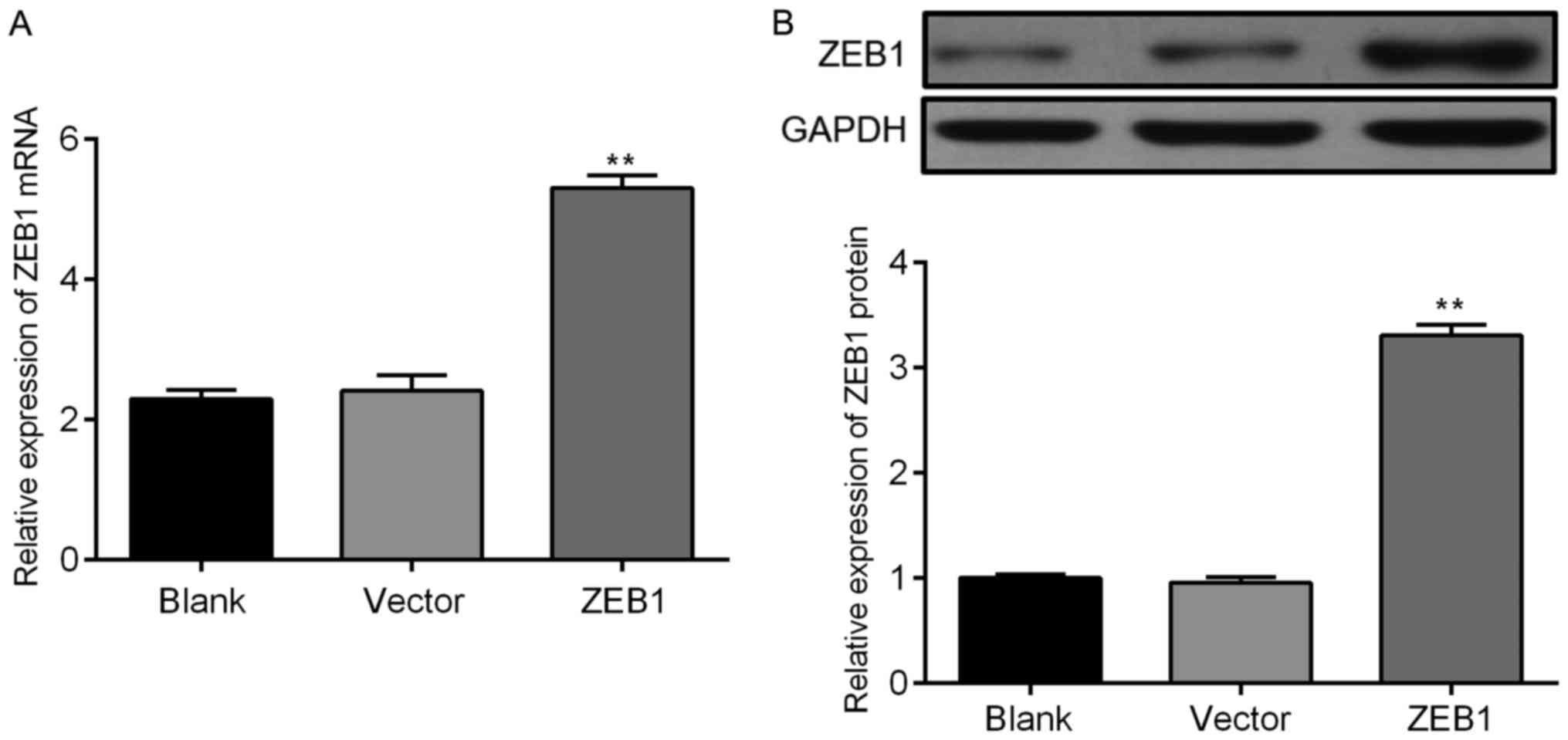
We performed a rescue assay to investigate if ZEB1
is involved in the miR-205-mediated regulation of glioma cells
(23–25). Following transfection, the mRNA and
protein levels of ZEB1 increased in glioma cells transfected with
pcDNA-3.1+ZEB1 (Fig. 7). We also
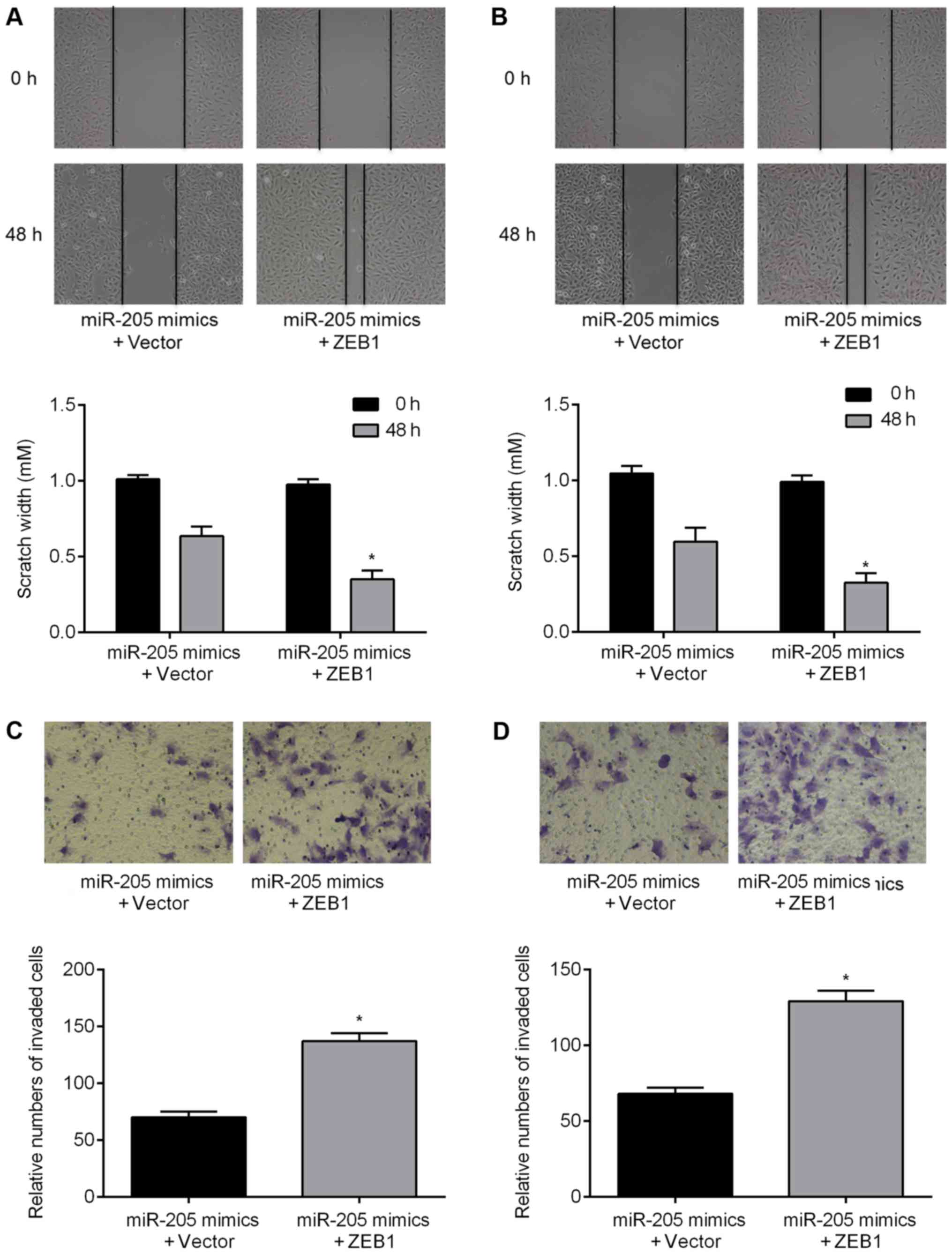
found an increase in the migratory ability of glioma cells
transfected with the ZEB1 construct following treatment with the
miR-205 mimics (Fig. 8A and B). In
addition, upregulation of ZEB1 partially reversed the effects of
miR-205 on cell invasion ability (Fig.
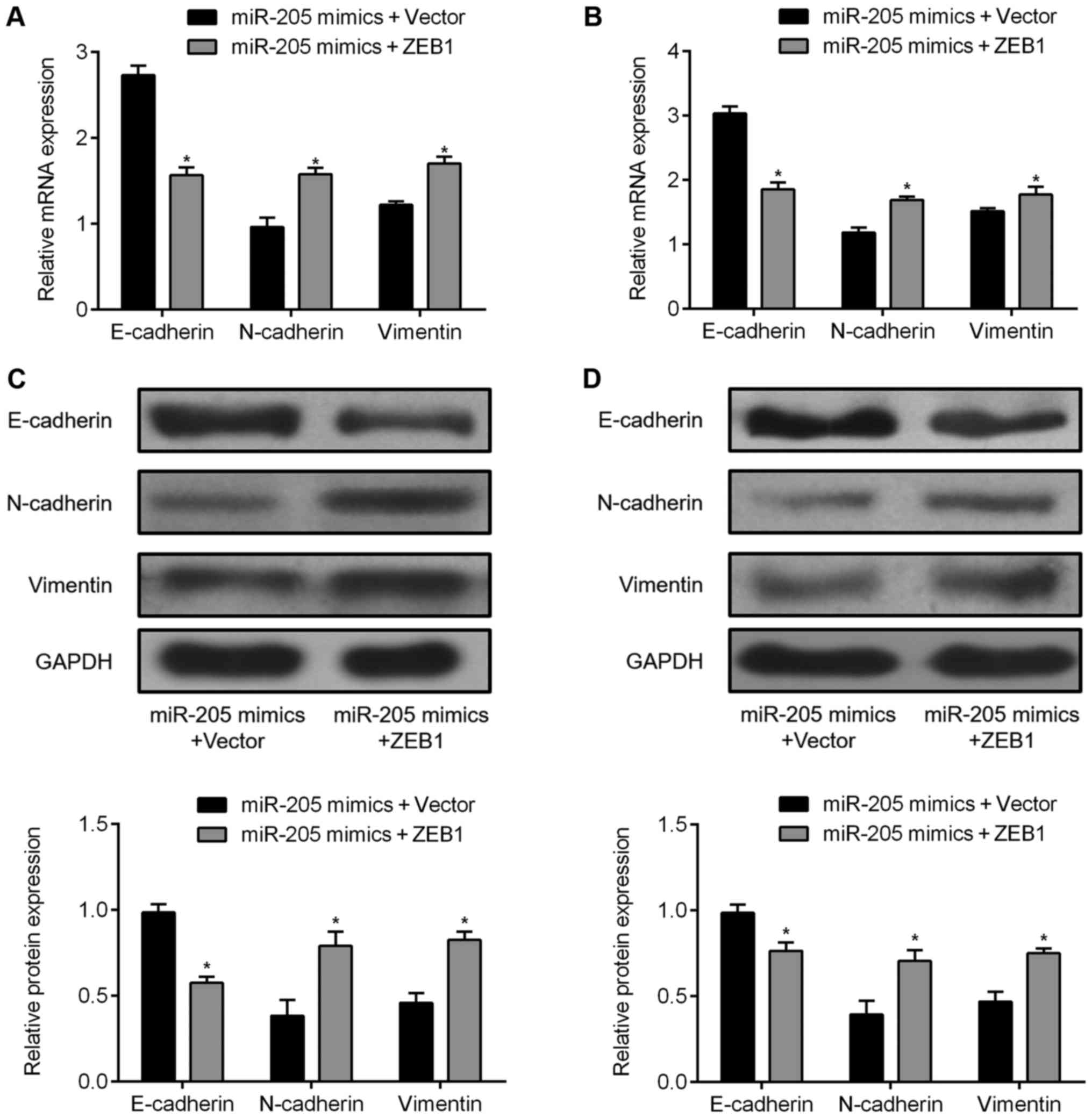
8C and D). Overexpression of ZEB1 inhibited E-cadherin
expression and induced the expression of N-cadherin and vimentin as
detected by qRT-PCR (Fig. 9A and
B) and western blotting (Fig. 9C
and D). These data indicate that miR-205 inhibits cell
migration and invasion, and reverses EMT by downregulating ZEB1 in
GBM cells.
Discussion
MicroRNAs can be either upregulated or
downregulated, and function as oncogenes or tumor suppressors
depending on their target genes (26). Accumulating evidence reveals the
vital role of miRNAs involved in processes such as proliferation
and invasion and in prognosis of GBM (27–29).
It is well established that ZEB1 is a powerful regulator of EMT,
and numerous miRNAs have been reported to be either direct or
indirect regulators of EMT (30–32).
In this study, we examined the function of miR-205
in malignant glioma cells. We showed that miR-205 expression is
significantly decreased in GBM tissues and cell lines. We also
found overexpression of miR-205 inhibits migration and invasion,
and reverses EMT in glioma cells (Figs. 5 and 6). To determine the potential mechanisms
of miR-205 function, we identified downstream targets, and found
the 3′UTR of ZEB1 contained a sequence complementary to miR-205
(Fig. 3). Furthermore, we found
upregulation of ZEB1 could abolish the inhibitory effects of
miR-205 in glioma cell lines (Figs.
8 and 9). These results
suggest an underlying mechanism whereby miR-205 suppression might
contribute to the malignancy of GBM.
Previous studies have demonstrated a relationship
between miR-205 and metastasis in several cancers. miR-205
suppresses proliferation, migration, and invasion of osteosarcoma
cells by binding to the 3′UTR of the vascular endothelial growth
factor A gene (VEGFA) (33). Liu
et al determined miRNA expression levels in samples of
melanoma tissues and found significantly reduced miR-205 levels.
Overexpression of miR-205 suppressed cell migration and
proliferation in vitro and in vivo (17). In addition, miR-205 plays a crucial
role in EMT as a silencer of the ZEB1/2 transcriptional repressor
(34,35). Xu et al previously indicated
that miR-205 prevents EMT by targeting ZEB1 in gastric cancer
cells, suggesting that miR-205 may be applied in the treatment of
human gastric cancer (36). In the
present study, we identified the loss of miR-205 in GBM tissues and
cell lines, and the following experiments strongly suggested that
ZEB1 is a direct downstream target of miR-205. Subsequent
investigation found that enforced expression of miR-205 could
promote Akt/mTOR activation, by targeting ZEB1. Akt controls
multiple biological processes including cell proliferation,
differentiation, apoptosis, and tumorigenesis (37). miR-200c has been reported to
enhance non-small cell lung cancer cell sensitivity in response to
gefitinib and induce apoptosis through the PI3K/Akt signaling
pathway via targeting ZEB1 (38).
Another study suggested that miR-205 promotes EMT in nasopharyngeal
carcinoma cells via the regulation of PTEN and Akt signaling
(39). Accordingly, we found
miR-205 moderated cell metastasis and EMT through activating the
Akt/mTOR signaling pathway by targeting ZEB1 in GBM cell lines.
Furthermore, we found that overexpression of ZEB1 could partially
abolish miR-205-mediated suppression of migration and invasion of
these cells.
ZEB1 is a transcriptional factor that was originally
shown to suppress interleukin (IL)-2 gene expression (40). Numerous studies have reported that
ZEB1 is expressed in a variety of cancers and is an important
regulator of EMT, which plays an important role in cancer
metastasis (41–43). Qu et al indicated that
miR-33b suppresses tumor cell growth and EMT via directly targeting
ZEB1 (44), and Sun et al
found that miR-431 suppresses ZEB1-induced EMT in hepatocellular
carcinoma (45). Moreover, a
recent study reported that over-expression of ZEB1 in GBM could
also induce EMT, which was regulated by miR-590-3p (46). However, this finding does not
clarify the potential signaling pathway of ZEB1-induced EMT. In the
present study, we focused on the importance of the Akt signaling
pathway and hypothesized that this pathway underlies the function
of ZEB1 in EMT. Furthermore, our in vitro studies suggested
ZEB1 might regulate EMT in GBM via the Akt/mTOR signaling
pathway.
In conclusion, our integrated approach demonstrates
the biological functions of miR-205 in GBM. We also show that
overexpression of miR-205 downregulates ZEB1, inhibits GBM cell
migration and invasion, and prevents EMT in GBM cells via
regulation of the Akt/mTOR signaling pathway, which has important
implications for further understanding the mechanisms involved in
modulating tumorigenesis in GBM.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (nos. 81703011, 81272774 and 81572497)
and the Science Foundation of Guangzhou Women and Children's
Medical Center (nos. 5001-3001023 and 5001-2150010).
Abbreviations:
|
GBM
|
glioblastoma
|
|
miRNAs
|
microRNAs
|
|
ZEB1
|
zinc finger E-box binding homeobox
1
|
|
EMT
|
epithelial-mesenchymal transition
|
References
|
1
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer Genome Atlas Research Network:
Comprehensive genomic characterization defines human glioblastoma
genes and core pathways. Nature. 455:1061–1068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dai C, Lv S, Shi R, Ding J, Zhong X, Song
H, Ma X, Fan J, Sun B, Wang R, et al: Nuclear protein C23 on the
cell surface plays an important role in activation of CXCR4
signaling in glioblastoma. Mol Neurobiol. 52:1521–1526. 2015.
View Article : Google Scholar
|
|
4
|
Janga SC and Vallabhaneni S: MicroRNAs as
post-transcriptional machines and their interplay with cellular
networks. Adv Exp Med Biol. 722:59–74. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ong CA, Lao-Sirieix P and Fitzgerald RC:
Biomarkers in Barrett's esophagus and esophageal adenocarcinoma:
Predictors of progression and prognosis. World J Gastroenterol.
16:5669–5681. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Du M, Shi D, Yuan L, Li P, Chu H, Qin C,
Yin C, Zhang Z and Wang M: Circulating miR-497 and miR-663b in
plasma are potential novel biomarkers for bladder cancer. Sci Rep.
5:104372015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu X, Lei Q, Yu Z, Xu G, Tang H, Wang W,
Wang Z, Li G and Wu M: MiR-101 reverses the hypomethylation of the
LMO3 promoter in glioma cells. Oncotarget. 6:7930–7943. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
She X, Yu Z, Cui Y, Lei Q, Wang Z, Xu G,
Xiang J, Wu M and Li G: miR-128 and miR-149 enhance the
chemosensitivity of temozolomide by Rap1B-mediated cytoskeletal
remodeling in glioblastoma. Oncol Rep. 32:957–964. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tang H, Liu X, Wang Z, She X, Zeng X, Deng
M, Liao Q, Guo X, Wang R, Li X, et al: Interaction of hsa-miR-381
and glioma suppressor LRRC4 is involved in glioma growth. Brain
Res. 1390:21–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Song S, Fajol A, Tu X, Ren B and Shi S:
miR-204 suppresses the development and progression of human
glioblastoma by targeting ATF2. Oncotarget. 7:70058–70065.
2016.PubMed/NCBI
|
|
12
|
Song H, Zhang Y, Liu N, Wan C, Zhang D,
Zhao S, Kong Y and Yuan L: miR-92b regulates glioma cells
proliferation, migration, invasion, and apoptosis via PTEN/Akt
signaling pathway. J Physiol Biochem. 72:201–211. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li S, Zeng A, Hu Q, Yan W, Liu Y and You
Y: miR-423-5p contributes to a malignant phenotype and temozolomide
chemo-resistance in glioblastomas. Neurooncol. 19:55–65. 2017.
|
|
14
|
Song H and Bu G: MicroRNA-205 inhibits
tumor cell migration through down-regulating the expression of the
LDL receptor-related protein 1. Biochem Biophys Res Commun.
388:400–405. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hou SX, Ding BJ, Li HZ, Wang L, Xia F, Du
F, Liu LJ, Liu YH, Liu XD, Jia JF, et al: Identification of
microRNA-205 as a potential prognostic indicator for human glioma.
J Clin Neurosci. 20:933–937. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Boll K, Reiche K, Kasack K, Mörbt N,
Kretzschmar AK, Tomm JM, Verhaegh G, Schalken J, von Bergen M, Horn
F, et al: MiR-130a, miR-203 and miR-205 jointly repress key
oncogenic pathways and are downregulated in prostate carcinoma.
Oncogene. 32:277–285. 2013. View Article : Google Scholar
|
|
17
|
Liu S, Tetzlaff MT, Liu A, Liegl-Atzwanger
B, Guo J and Xu X: Loss of microRNA-205 expression is associated
with melanoma progression. Lab Invest. 92:1084–1096. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu H, Wang H, Liu X and Yu T: miR-1271
inhibits migration, invasion and epithelial-mesenchymal transition
by targeting ZEB1 and TWIST1 in pancreatic cancer cells. Biochem
Biophys Res Commun. 472:346–352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Siebzehnrubl FA, Silver DJ, Tugertimur B,
Deleyrolle LP, Siebzehnrubl D, Sarkisian MR, Devers KG, Yachnis AT,
Kupper MD, Neal D, et al: The ZEB1 pathway links glioblastoma
initiation, invasion and chemoresistance. EMBO Mol Med.
5:1196–1212. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aigner K, Dampier B, Descovich L, Mikula
M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist
P, et al: The transcription factor ZEB1 (deltaEF1) promotes tumour
cell dedifferentiation by repressing master regulators of
epithelial polarity. Oncogene. 26:6979–6988. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun X, Li Y, Yu J, Pei H, Luo P and Zhang
J: miR-128 modulates chemosensitivity and invasion of prostate
cancer cells through targeting ZEB1. Jpn J Clin Oncol. 45:474–482.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang H, Cao H, Xu D and Zhu K:
MicroRNA-92a promotes metastasis of nasopharyngeal carcinoma by
targeting the PTEN/ AKT pathway. Onco Targets Ther. 9:3579–3588.
2016.
|
|
25
|
Guo K, Zheng S, Xu Y, Xu A, Chen B and Wen
Y: Loss of miR-26a-5p promotes proliferation, migration, and
invasion in prostate cancer through negatively regulating SERBP1.
Tumour Biol. 37:12843–12854. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu S, Huang S, Ding J, Zhao Y, Liang L,
Liu T, Zhan R and He X: Multiple microRNAs modulate p21Cip1/Waf1
expression by directly targeting its 3′ untranslated region.
Oncogene. 29:2302–2308. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
LeBlanc VC and Morin P Jr: Exploring
miRNA-associated signatures with diagnostic relevance in
glioblastoma multiforme and breast cancer patients. J Clin Med.
4:1612–1630. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shea A, Harish V, Afzal Z, Chijioke J,
Kedir H, Dusmatova S, Roy A, Ramalinga M, Harris B, Blancato J, et
al: MicroRNAs in glioblastoma multiforme pathogenesis and
therapeutics. Cancer Med. 5:1917–1946. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luo JW, Wang X, Yang Y and Mao Q: Role of
micro-RNA (miRNA) in pathogenesis of glioblastoma. Eur Rev Med
Pharmacol Sci. 19:1630–1639. 2015.PubMed/NCBI
|
|
30
|
Wang H, Tao T, Yan W, Feng Y, Wang Y, Cai
J, You Y, Jiang T and Jiang C: Upregulation of miR-181s reverses
mesenchymal transition by targeting KPNA4 in glioblastoma. Sci Rep.
5:130722015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yan Y, Wu J, Wu M, Xia Y, Tang W and Liao
Z: MiR-143 suppresses the epithelial-mesenchymal transition of
spinal glioblastoma through down-regulation of ERK5. Oncotarget:
oncotarget. pp. 12977
|
|
32
|
Puhr M, Hoefer J, Schäfer G, Erb HH, Oh
SJ, Klocker H, Heidegger I, Neuwirt H and Culig Z:
Epithelial-to-mesenchymal transition leads to docetaxel resistance
in prostate cancer and is mediated by reduced expression of
miR-200c and miR-205. Am J Pathol. 181:2188–2201. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang L, Shan M, Liu Y, Yang F, Qi H, Zhou
L, Qiu L and Li Y: miR-205 suppresses the proliferative and
migratory capacity of human osteosarcoma Mg-63 cells by targeting
VEGFA. Onco Targets Ther. 8:2635–2642. 2015.PubMed/NCBI
|
|
34
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Matsushima K, Isomoto H, Yamaguchi N,
Inoue N, Machida H, Nakayama T, Hayashi T, Kunizaki M, Hidaka S,
Nagayasu T, et al: MiRNA-205 modulates cellular invasion and
migration via regulating zinc finger E-box binding homeobox 2
expression in esophageal squamous cell carcinoma cells. J Transl
Med. 9:302011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu C, Li M, Zhang L, Bi Y, Wang P, Li J
and Jiang X: MicroRNA-205 suppresses the invasion and
epithelial-mesen-chymal transition of human gastric cancer cells.
Mol Med Rep. 13:4767–4773. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Manning BD and Cantley LC: AKT/PKB
signaling: Navigating downstream. Cell. 129:1261–1274. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou G, Zhang F, Guo Y, Huang J, Xie Y,
Yue S, Chen M, Jiang H and Li M: miR-200c enhances sensitivity of
drug-resistant non-small cell lung cancer to gefitinib by
suppression of PI3K/Akt signaling pathway and inhibites cell
migration via targeting ZEB1. Biomed Pharmacother. 85:113–119.
2017. View Article : Google Scholar
|
|
39
|
Mao Y, Wu S, Zhao R and Deng Q: MiR-205
promotes proliferation, migration and invasion of nasopharyngeal
carcinoma cells by activation of AKT signalling. J Int Med Res.
44:231–240. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Williams TM, Moolten D, Burlein J, Romano
J, Bhaerman R, Godillot A, Mellon M, Rauscher FJ III and Kant JA:
Identification of a zinc finger protein that inhibits IL-2 gene
expression. Science. 254:1791–1794. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang Y, Wen M, Kwon Y, Xu Y, Liu Y, Zhang
P, He X, Wang Q, Huang Y, Jen KY, et al: CUL4A induces
epithelial-mesenchymal transition and promotes cancer metastasis by
regulating ZEB1 expression. Cancer Res. 74:520–531. 2014.
View Article : Google Scholar :
|
|
42
|
Ohashi S, Natsuizaka M, Naganuma S, Kagawa
S, Kimura S, Itoh H, Kalman RA, Nakagawa M, Darling DS, Basu D, et
al: A NOTCH3-mediated squamous cell differentiation program limits
expansion of EMT-competent cells that express the ZEB transcription
factors. Cancer Res. 71:6836–6847. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Eger A, Aigner K, Sonderegger S, Dampier
B, Oehler S, Schreiber M, Berx G, Cano A, Beug H and Foisner R:
DeltaEF1 is a transcriptional repressor of E-cadherin and regulates
epithelial plasticity in breast cancer cells. Oncogene.
24:2375–2385. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Qu J, Li M, An J, Zhao B, Zhong W, Gu Q,
Cao L, Yang H and Hu C: MicroRNA-33b inhibits lung adenocarcinoma
cell growth, invasion, and epithelial-mesenchymal transition by
suppressing Wnt/β-catenin/ZEB1 signaling. Int J Oncol.
47:2141–2152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sun K, Zeng T, Huang D, Liu Z, Huang S,
Liu J and Qu Z: MicroRNA-431 inhibits migration and invasion of
hepatocellular carcinoma cells by targeting the ZEB1-mediated
epithelial-mensenchymal transition. FEBS Open Bio. 5:900–907. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pang H, Zheng Y, Zhao Y, Xiu X and Wang J:
miR-590-3p suppresses cancer cell migration, invasion and
epithelial-mesenchymal transition in glioblastoma multiforme by
targeting ZEB1 and ZEB2. Biochem Biophys Res Commun. 468:739–745.
2015. View Article : Google Scholar : PubMed/NCBI
|