Introduction
Breast cancer remains a leading cause of
cancer-related mortality in women worldwide (1). Distant metastasis is a major cause of
cancer-related mortality (2).
Clinical studies have shown that breast cancer frequently
metastasizes to the brain, lungs, bone, liver and lymph nodes
(3). Compared to the cancer cells
in the primary site, metastatic cells are less sensitive to breast
cancer treatments (4). Therefore,
the investigation of the factors which are involved in the
development of a pre-metastatic niche is of utmost importance.
Inflammatory molecules, including chemokines,
cytokines and other secreted molecules are a driving force that
affects the microenvironment of extracellular matrix, and
extracellular matrix remodeling is an important process for the
establishment of a pre-metastatic niche (5). The S100 calcium-binding protein
family are inflammatory molecules and contribute to develop an
inflammatory tumor microenvironment (6). There are >20 genes in the S100
family and some of these are considered as tumor markers (7). A recent study suggested that S100A4
and S100P, which are members of the S100 family, serve as
pro-metastatic factors (8). S100B
(also known as S100beta) also belongs to the S100 family. A
previous study also indicated that S100B overexpression led to the
enhanced migratory and invasive capacity of a lung cancer cell
line, promoting brain metastasis (9). Serum S100B and S100B autoantibodies
are biomarkers of brain metastasis in lung cancer (10). In melanoma, the serum S100B
concentration positively correlates with the tumor stage and
negatively correlates with the survival rate (11,12).
Furthermore, elevated serum S100B levels are associated with the
metastasis of melanoma, and decreased serum S100B levels are
associated with an increased survival rate (13). In addition, S100B downregulates
tumor suppressor p53 expression in melanoma. The knockdown of S100
expression restores p53-mediated apoptosis (14,15).
These data suggest that S100B is a molecule associated with tumor
metastasis and progression.
However, serum concentrations of S100B and HER2 may
not be biomarkers of brain metastasis in breast cancer (16). By contrast, a recent study revealed
that the serum levels of S100B were an independent prognostic
determinant for patients with brain metastasis (17). In the serum of breast cancer
patients subjected to endocrine therapy treatment, elevated levels
of S100B are associated with a poor disease-free survival (18). Currently, the role of S100B in
breast cancer is not well known and the association between the
cell metastatic capacity and S100B has not yet been determined.
Thus, in the present study, we aimed to investigate whether S100B
directly regulates migration by performing in vitro
experiments and using bioinformatics databases.
Materials and methods
Cells and cell culture
The human breast cancer cell lines, MCF-7
(ATCC® HTB-22™), Hs578T (ATCC® HTB-126™) and
MDA-MB-231 (ATCC® HTB-26™), were purchased from the
American Type Culture Collection (ATCC, Manassas, VA, USA). The
MCF-7, Hs578T and MDA-MB-231 cells were respectively cultured in
Eagle's minimum Essential medium (EMEM), Dulbecco's modified
Eagle's medium (DMEM) and L-15 medium supplemented with 10% fetal
bovine serum (FBS) and 1% penicillin-streptomycin (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The MCF-7 and Hs578T cells
were cultured at 37°C with 5% CO2 and the MDA-MB-231
cells were cultured at 37°C in a CO2-free incubator.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from the MCF-7, MDA-MB-231
and Hs578T cells using TRIzol reagent (Invitrogen, Carlsbad, CA,
USA). The PrimeScript RT reagent kit (Clontech Laboratories, Inc.,
Kusatsu, Japan) was used for the reverse transcription of 500 ng of
total RNA to complementary DNA. The relative expression of S100B
was determined using a Real-Time PCR system (StepOnePlus Real-Time
PCT system; Applied Biosystems, Foster City, CA, USA) using Fast
SYBR-Green Master Mix (Applied Biosystems) with the specific
primer: S100B forward, 5′-AGGGAGACAAGCACAAGCTG-3′ and reverse,
5′-CGTGGCAGGCAGTAGTAACC-3′. The expression of
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was detected using
the primers: forward, 5′-GAGTCAACGGATTTGGTCGT-3′ and reverse,
5′-TTGATTTTGGAGGGATCTCG-3′. Relative mRNA expression was normalized
to the expression level of GAPDH and calculated using the
2−ΔΔCt method (19).
Cell migration assay
The migration ability was analyzed by wound healing
assay and Transwell migration assay. For wound healing assay,
2.4×105 MDA-MB-231 cells were seeded into 24-well
plates. At 24 h after seeding, a scratch was made using a 200-ml
pipette tip. After scratching, cell debris was removed by washing
twice with phosphate-buffered saline (PBS) and the cells were then
incubated in serum-free L-15 medium containing 0, 0.1 and 1 nM
recombinant human S100B protein (R&D Systems, Minneapolis, MN,
USA). Images were acquired at 0 and 12 h following treatment with
S100B. Quantification was performed using AxioVs40 V4.8.2.0
microimaging software (MicroImaging GmbH, Gottingen, Germany). QCM™
24-well Cell Migration assay and Invasion System uncoated
8-µm pore size polycarbonate membranes (Millipore,
Billerica, MA, USA) were used for Transwell migration assay
according to the manufacturer's instructions. A total of
4×104 MDA-MB-231 or Hs578T cells were seeded in 300 ml
of serum-free medium in 24-well upper inserts and 500 ml medium
with 10% FBS was placed in the lower chamber. After 12 h, the
bottom surface of the membrane was fixed in 5% formaldehyde
solution, followed by 0.4 g/l crystal violet (Sigma-Aldrich, St.
Louis, MO, USA) staining for 2 h. The cells on the upper surface
were removed using a cotton swab after washing the crystal
violet-stained membrane. The bottom of the membrane was then
visualized using an inverted light microscope (Carl Zeiss Primo
Vert Microscope; Carl Zeiss, Munich, Germany) at x100
magnification. In total, 4 random fields of view were counted and
the relative fold of migration in each group was compared to the 0
nM S100B treatment group.
Western blot analysis
The cells were lysed on ice for 30 min in
radioimmunoprecipitation lysis buffer (RIPA) buffer (Millipore)
which was supplemented with protease inhibitor cocktail (Millipore)
at a 100:1 ratio. The total cell lysate was collected following
centrifugation at 4°C, 12,000 × g for 15 min. The protein
concentration was determined using a bicinchoninic acid (BCA)
protein assay kit (Thermo Fisher Scientific, Rockford, IL, USA).
Equal amounts of protein were loaded and separated by 10% sodium
dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The
proteins were then transferred onto polyvinylidene difluoride
membranes (Millipore). After 1 h of blocking [5% skim milk in
Tris-buffered saline with Tween-20 (TBST) buffer], the membranes
were incubated with the following primary antibodies: N-cadherin
(1:1,000, cat. no. 2167184; Millipore), E-cadherin (1:1,000, cat.
no. 610182; BD Biosciences, San Jose, CA, USA), vimentin (1:1,000,
cat. no. MAB3400; Millipore) and GAPDH (1:5,000, cat. no. MAB374;
Millipore) at 4°C overnight. After washing with TBST, the membranes
were incubated with secondary antibodies, including peroxidase
conjugated goat anti-rabbit IgG (1:5,000; cat. no. AP132P;
Millipore) and peroxidase conjugated goat anti-mouse IgG (1:5,000;
cat. no. AP124P; Millipore) for 1 h at room temperature. The
results were acquired on Alpha Innotech FluorChem FC2 Imaging
system (ProteinSimple; Bio-Techne, Minneapolis, MN, USA).
Evaluation of S100B expression from
public microarray datasets
The mRNA expression of S100B in breast cancer
samples was obtained from the Oncomine Research Edition (Thermo
Fisher Scientific; http://www.oncomine.org, v4.5) which includes the TCGA
breast dataset of Ma et al (20). The clinical data of TP53 mutation
and the expression levels of S100B in the breast cancer samples
were downloaded as z-scores from the cBioPortal (http://www.cbioportal.org, Breast cancer, TCGA, Cell
2015) (21). The expression of
S100B in different subtypes (PAM50) of breast cancer was evaluated
using the GOBO database (http://co.bmc.lu.se/gobo) (22).
Assessment of the patient survival
rate
The distant metastasis-free survival (DMFS) analysis
of the patients with breast cancer with different expression levels
of S100B was performed using Kaplan-Meier plotter (KM Plotter)
database (http://kmplot.com/analysis/)
(23). Briefly, the prognostic
value of each gene was analyzed by splitting the patient samples
into 2 groups according to the median value. To investigate the
association between S100B expression and DMFS in different subtypes
of breast cancer patients, the DMFS was determined in the subtype
of estrogen receptor (ER)-positive (n=664) patients, ER-negative
(n=218) patients, or patients subjected to endocrine therapy
(n=645). The hazard ratio with 95% confidence intervals and
log-rank P-value are shown according to the KM plotter database. To
further examine whether the expression of S100B is associated with
the TP53 status, the subtype of breast cancer was restricted to
TP53 status 'mutated' (n=188) or 'wild-type' (n=273) and the DMFS
of the patients was analyzed. The overall survival was accessed in
the dataset 'Breast Invasive Carcinoma TCGA' from the SurvExpress
database (http://bioinformatica.mty.itesm.mx:8080/Biomatec/Survivax.jsp,
Interface v2.0, Database Update: November 5, 2017) (24,25).
The censored was set to 'Survival_months', groups were divided by
'maximize risk groups' and stratification was set to 'class: ER'.
To determine the distant recurrence rate, the dataset 'Vincent
Darbon Breast' (Accession no. GSE9893 in the Gene Expression
Omnibus database) in the SurvExpress database. The censored was set
'Distant recurrence months', and the raw data of the distant
recurrence-free survival curve and mRNA expression levels were
sequentially obtained. The low- and high-risk groups were divided
according to the median risk index on the SurvExpress website. The
survival curve and mRNA expression were re-drawn using GraphPad
Prism 7 software (GraphPad Software, Inc., San Diego, CA, USA).
Statistical analysis
All graphs were generated using GraphPad Prism 7
software (GraphPad Software). The Student's t-test was used for the
analysis of differences between 2 groups and one-way analysis of
variance (ANOVA) with a Tukey's post hoc test was used for the
analysis of differences among 3 or more groups. A P-value <0.05
was considered to indicate a statistically significant
difference.
Results
Relatively low S100B expression was
observed in breast cancer
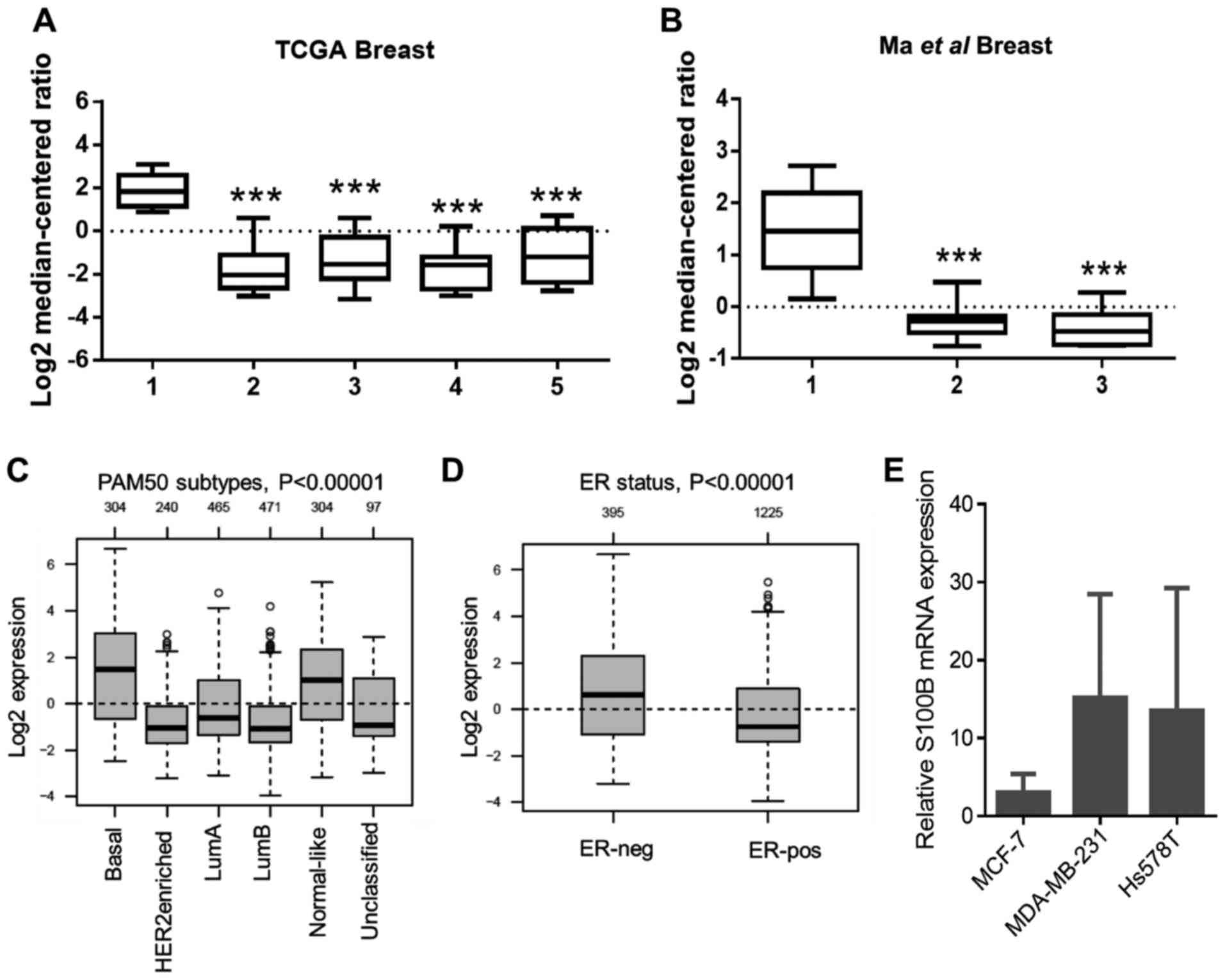
To investigate the expression levels of S100B in the
breast cancer samples, the Oncomine database (http://www.oncomine.org) was used. Compared to the
normal breast tissue, the expression of S100B in various types of
breast cancer tissue was significantly low in 2 independent
data-sets (Fig. 1A and B). To
further examine S100B expression in different subtypes of breast
cancer according to PAM50 subtypes, S100B expression was evaluated
using the GOBO database (http://co.bmc.lu.se/gobo). As shown in Fig. 1C, a relatively high S100B
expression level was observed in the basal-like type when compared
to the luminal A (LumA), LumB and HER2-enriched groups. Moreover,
S100B expression in the ER-negative group was higher than that in
the ER-positive group (Fig. 1D).
Based on these results, we hypothesized that S100B may directly
regulate the behaviors of the breast cancer cells, particularly in
basal-like or ER-negative breast cancer. mRNA expression levels in
3 cancer cell lines, including MCF-7 (luminal A, ER-positive),
MDA-MB-231 (basal-like, ER-negative) and Hs578T (basal like,
ER-negative) were then determined. As shown in Fig. 1E, the mean S100B expression in the
MDA-MB-231 and Hs578T cells was higher than that in the MCF-7
cells; however, there was no statistical difference among these 3
cell lines.
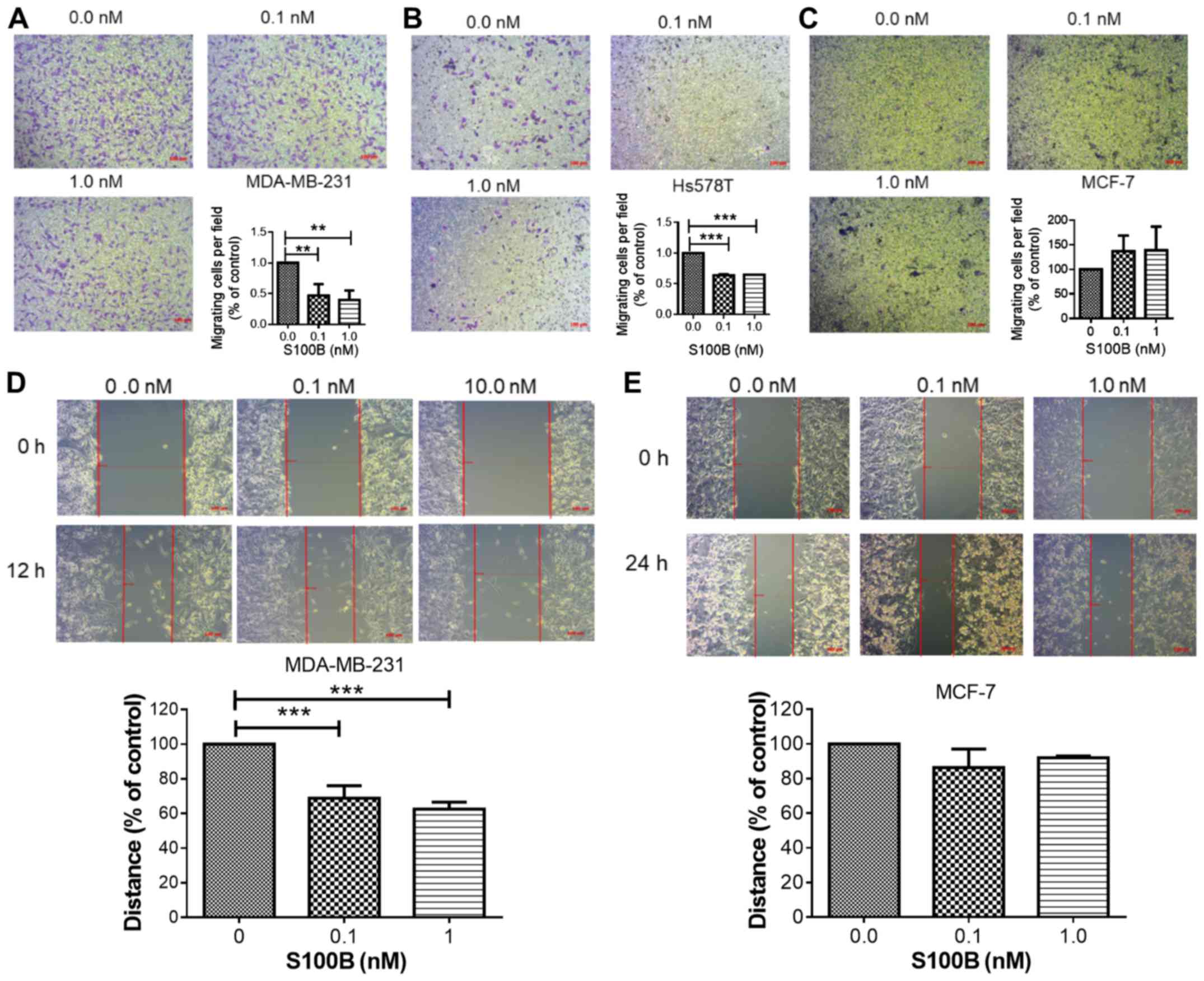
Treatment with recombinant S100B inhibits
cell migration and promotes the epithelial-phenotype
The effects of extracellular S100B were further
examined in the MCF-7, MDA-MB-231 and Hs578T cells. The cells were
treated with various concentrations of human recombinant S100B
protein. In the Transwell migration assay, S100B treatment
significantly inhibited the migration of the two basal-like cell
lines (Fig. 2A and B); however, it
did not significantly affect the migration of the MCF-7 cells
(Fig. 2C). In addition, the
inhibitory effects of S100B treatment were observed in the
MDA-MB-231 cells (Fig. 2D), but
not in the MCF-7 (Fig. 2E) cells,
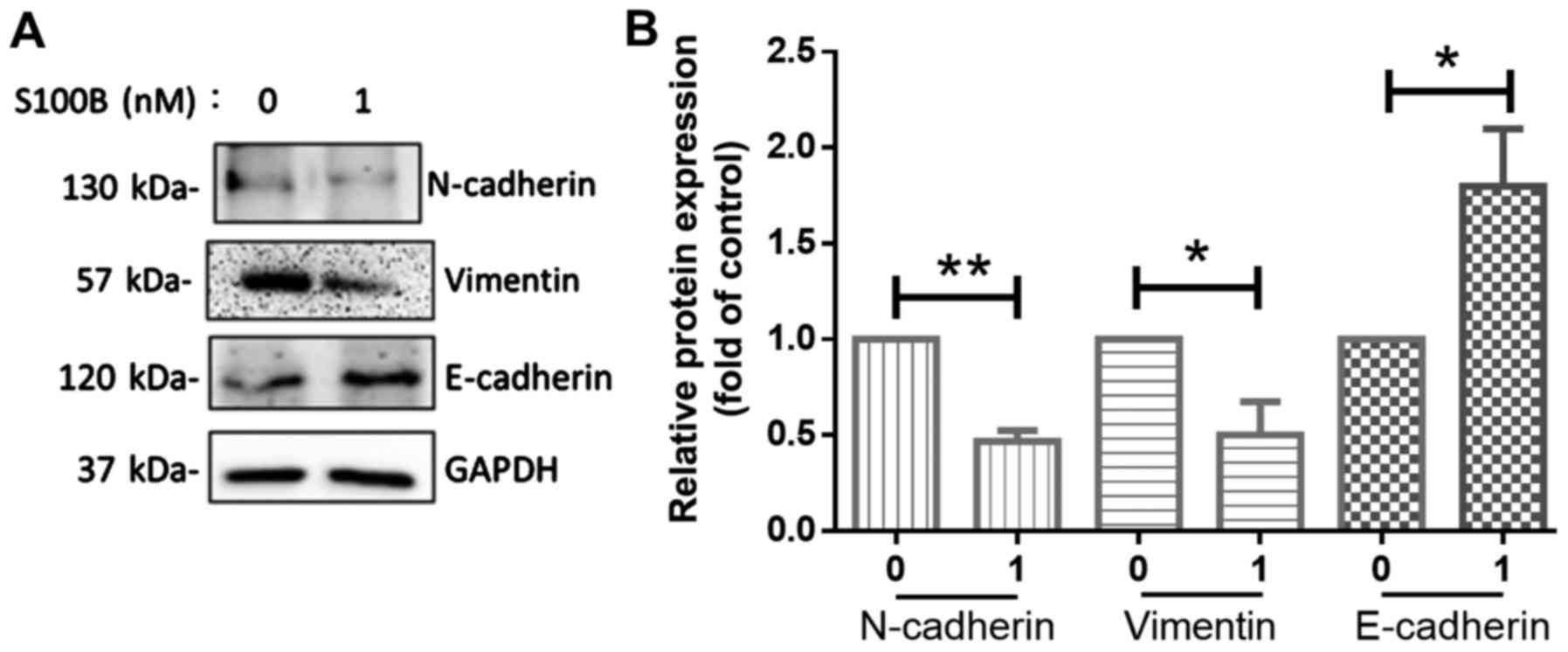
in wound healing assay. The process of epithelial-mesenchymal
transition (EMT) in cancer cells is associated with metastatic
potential in breast cancer and EMT is related to the basal-like
type of breast cancer (26). The
MDA-MB-231 and Hs578T cells are classified as the basal B type
(27). In order to determine
whether S100B affects the mesenchymal or epithelial phenotype
transition, the markers of the mesenchymal phenotype, N-cadherin
and vimentin, and the marker of the epithelial phenotype,
E-cadherin (28), were determined
in the MDA-MB-231 cells. The results of western blot analysis
revealed that S100B treatment induced the expression of E-cadherin.
Thus, S100B treatment led to the acquirement of the epithelial
phenotype (Fig. 3).
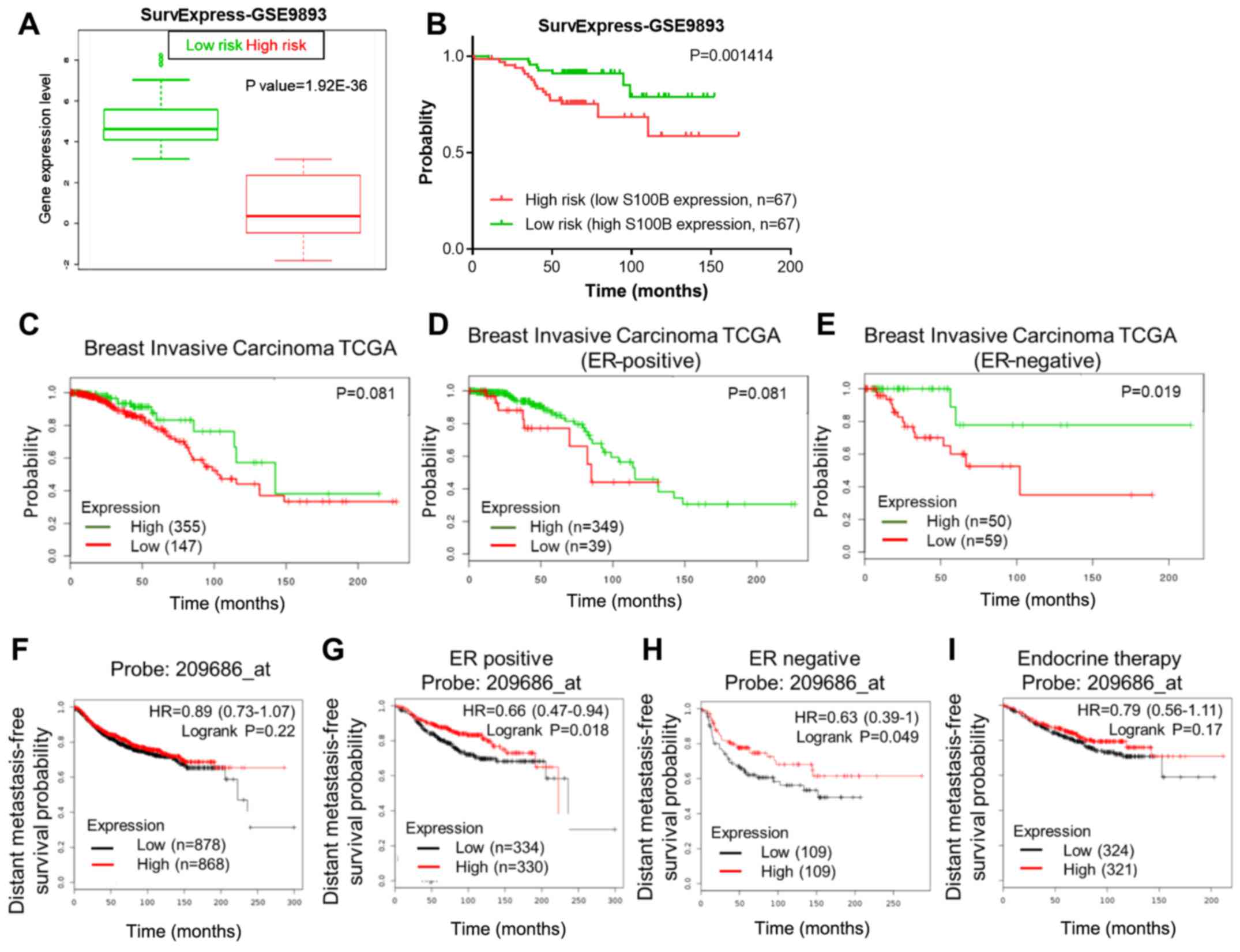
High S100B expression is associated with
a good prognosis
As S100B treatment inhibited cell metastasis and the
acquisition of the epithelial phenotype, we further investigated
whether S100B expression could be a prognostic marker for
metastasis in patients with breast cancer, particularly in patients
with metastatic breast cancer via the online databases, SurvExpress
(http://bioinformatica.mty.itesm.mx:8080/Biomatec/Survivax.jsp)
and KM plotter (http://kmplot.com/analysis/). In a dataset from the
SurvExpress database, patients with a low S100B expression
(high-risk group) had a good survival rate in a distant
recurrence-free survival analysis (Fig. 4A and B). Furthermore, we examined
whether the ER status was an important factor in the S100B-induced
inhibitory effect on cell migration. In breast invasive carcinoma,
the TCGA dataset from the SurvExpress database, a high S100B
expression was associated with a good overall survival rate in only
ER-negative breast cancer patients, but not in ER-positive breast
cancer patients and in breast cancer patients as a whole (Fig. 4C–E). As S100B treatment resulted in
the inhibition of the migration of ER-negative breast cancer cell
lines, we further investigated the DMFS using the KM plotter
database. S100B expression was not associated with DMFS in all
breast cancer patients as a whole (Fig. 4F). By contrast, S100B expression
was associated with a good DMFS in ER-positive (P=0.018) and
ER-negative (P=0.049) breast cancer patients (Fig. 4G and H). Of note, S100B expression
was a marker of prediction of DMFS in ER-positive breast cancer
patients who received endocrine therapy (Fig. 4I).
High S100B expression is associated with
good prognosis
In melanoma, S100B decreases p53 activity and
expression (14,15). In this study, the association
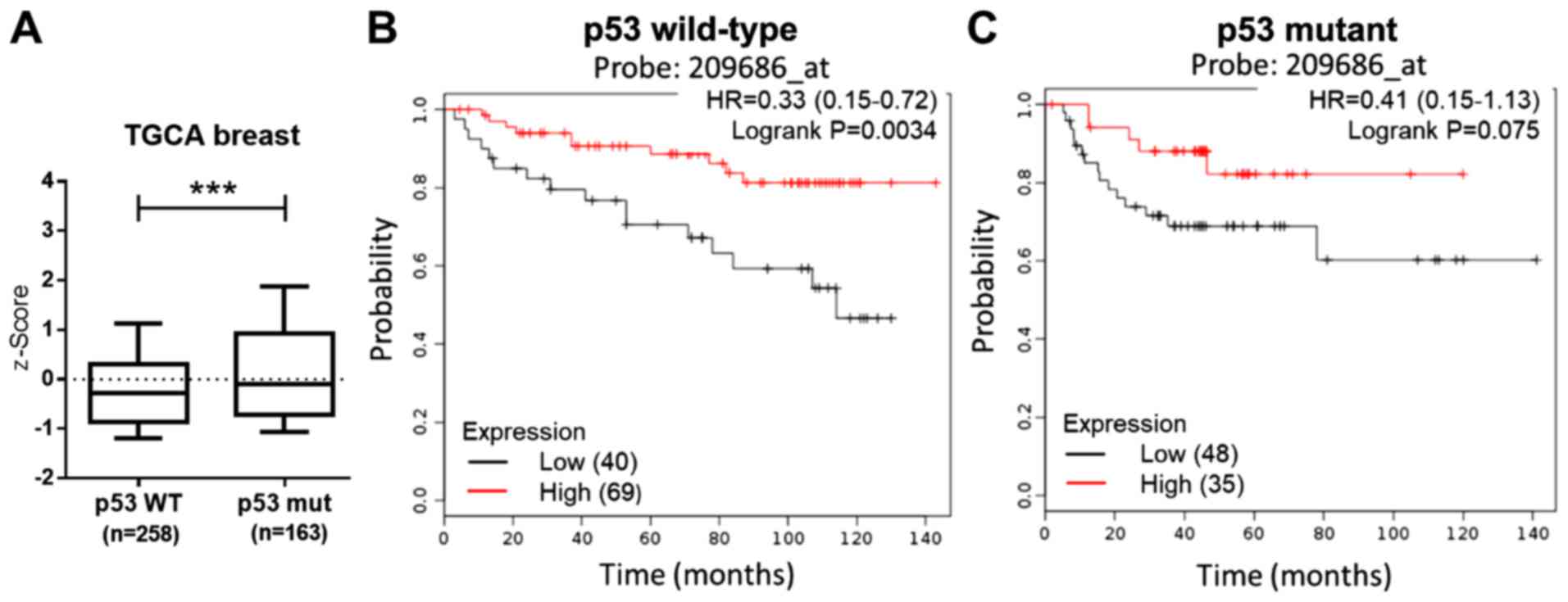
between S100B and p53 expression was examined using the TCGA breast
dataset from cBioPortal (http://www.cbioportal.org). Compared to the p53
wild-type group, a significantly higher p53 expression was observed
in the p53 mutant group (Fig. 5A).
Both the MDA-MB-231 and Hs578T cells exhibited p53 mutation
(27). However, in this study,
S100B treatment resulted in the inhibition of cell migration.
Therefore, the association between the p53 status, S100B expression
and the prognosis of breast cancer patients was evaluated. The DMFS
in breast cancer patients with mutant p53 and wild-type p53 was
respectively accessed in the KM plotter database. The results
revealed that a high S100B expression was associated with a good
outcome (Fig. 5B and C). These
findings suggest that a low S100B expression is associated with a
high metastatic rate irrespective of the p53 status in breast
cancer.
Discussion
Inflammation is a critical to create a
pre-metastatic niche in cancer (5). The interaction of inflammatory
molecules which are secreted from tumor cells, adipocytes and
tumor-infiltrating macrophages, neutrophils, myeloid-derived
suppressor cells and other types of immune cells regulate the
behaviors of tumor cells (29).
Metastasis-related pathways, such as the transition between the
epithelial and mesenchymal phenotype, were altered by inflammatory
molecules (29). Tumor cells
secrete interleukin (IL)-6 and IL-8, which are important mediators
of EMT in breast cancer (30). In
addition, tumor cells secrete IL-6 and IL-8, which affect the
phenotypes in an autocrine manner and IL-8 promotes the mesenchymal
phenotype (31,32). S100B is an inflammatory mediator
and secreted from several types of cells, such as cancer cells and
neuronal cells. Previous studies have suggested that
S100B-expressing neuronal cells exert an autocrine/paracrine effect
under physiological conditions (32,33).
We thus hypothesized that S100B may directly regulate the phenotype
of breast cancer in an autocrine or paracrine manner. Therefore,
this study focused on S100B expression in breast cancer cells and
on the effects of S100B treatment. Of note, a previous study
suggested that S100B was associated with brain metastasis in lung
cancer (10). Thus, the tumor
microenvironment may be a key factor which results in controversial
effects in breast cancer. Compared to lung cancer, breast cancer
grows near adipose tissue. Adipocytes secrete many molecules which
regulate tumorigenesis and metastasis (34). In addition, adipocytes are an
important source of serum S100B (35). The interaction of soluble factors
and S100B in the tumor microenvironment exert differential effects
on lung cancer cells and breast cancer cells.
There are >20 molecules in the S100
calcium-binding protein family. Different molecules have specific
functions and regulatory mechanisms in different types of cancer.
For example, S100A8 and S100A9 are associated with metastasis in
breast cancer (36). By contrast,
the overexpression of S100A6 inhibits metastasis and invasion in
human osteosarcoma (37). Our
results revealed a relatively high S100B expression in normal
breast tissue, as observed by bioinformatics analysis when compared
to breast cancer tissue. Furthermore, recombinant S100B treatment
significantly suppressed cell migration and promoted the epithelial
phenotype in ER-negative breast cancer cell lines. Our results thus
suggest that S100B treatment can trigger the activation of
anti-metastatic signaling pathways in ER-negative breast cancer. As
the serum S100B level is not correlated with the number of
metastatic sites of brain metastases in breast cancer (17), and S100B may affect the behavior of
breast cancer cells in an autocrine or paracrine manner, we
hypothesized that the expression S100B in breast cancer was not
associated with the serum S100B levels. The detailed regulatory
mechanisms of S100B expression warrant further investigation in
breast cancer.
Our results revealed that S100B expression in
ER-negative group was higher than that in the ER-positive group, as
shown by bioinformatics analysis (Fig.
1D). RT-qPCR analysis revealed a similar trend in breast cancer
cell lines (Fig. 1E). S100B
expression was associated with a good overall survival rate in
patients with ER-negative breast cancer, and a good DMFS in breast
cancer patients as a whole. However, current endocrine
therapy-treated patients with a high S100B expression and
ER-positive breast cancer reveals poor disease-free survival
(18). Based on these findings, it
is suggested that ER signaling pathways may affect S100B expression
and function. The anti-migratory effect of S100B was not observed
in ER-positive MCF-7 cells. In microglia, extracellular S100B,
IL-1β, and tumor necrosis factor-α activate the NF-κB and AP-1
transcriptional activity and then results in the upregulation of
cyclo-oxygenase 2 (38). The
interaction between S100B-mediated signaling pathways and ER
signaling pathways warrants further investigation in breast
cancer.
Previous studies have demonstrated that S100B binds
to the c-terminal of p53 protein, which is an important tumor
suppressor gene (14,15). The interaction of S100B and p53
results in the inhibition of p53-mediated growth suppression and
apoptosis in melanoma, lung cancer and ovarian cancer (10,14,39).
In this study, S100B treatment inhibited the migration in of 2
p53-mutated breast cancer cells and S100B expression was associated
with a good prognosis. Although we did not examine whether S100B
binds to mutant p53 in the MDA-MB-231 and Hs578T cells, and did not
assess the effects of S100B on breast cancer cell lines carrying
wild-type p53, the current results revealed that S100B was
associated with an anti-migratory effect. The interaction between
p53 and S100B may play a minor role in breast cancer. The
association between S100B, wild-type p53, mutant p53 and distant
metastasis in breast cancer warrants further investigation.
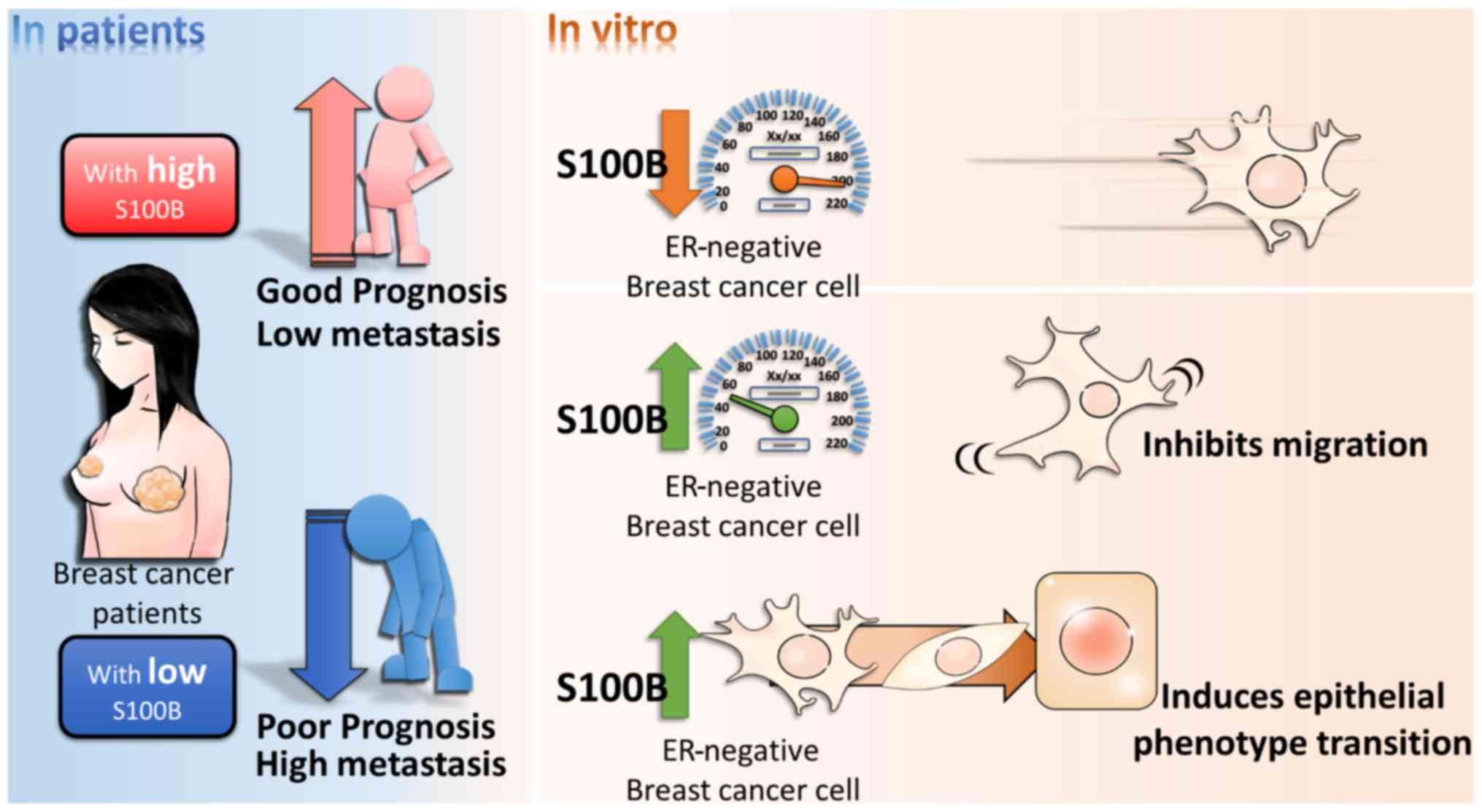
In conclusion, this study demonstrated a novel role
of S100B in the migration of breast cancer cells. S100B expression
in breast cancer tissue was lower than that in normal breast
tissue, and S100 exerted an inhibitory effect on cell migration and
the mesenchymal phenotype. The summarized model is shown in
Fig. 6. These results suggest that
the regulatory mechanisms of action of S100B in breast cancer
differ from those in melanoma, lung cancer and ovarian cancer, and
that S100B expression may provide a predictive marker for
metastasis in breast cancer.
Acknowledgments
The present study was supported by grants from the
Ministry of Science and Technology (MOST 104-2314-B-037-053-MY4;
MOST 105-2314-B-037-037-MY3; MOST 106-2314-B-037- 046) and the
Kaohsiung Medical University Hospital Research Foundation
(KMUHS10601; KMUH105-5M23).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Scully OJ, Bay BH, Yip G and Yu Y: Breast
cancer metastasis. Cancer Genomics Proteomics. 9:311–320.
2012.PubMed/NCBI
|
|
3
|
Chiang AC and Massagué J: Molecular basis
of metastasis. N Engl J Med. 359:2814–2823. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vanharanta S and Massagué J: Origins of
metastatic traits. Cancer Cell. 24:410–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Peinado H, Zhang H, Matei IR, Costa-Silva
B, Hoshino A, Rodrigues G, Psaila B, Kaplan RN, Bromberg JF, Kang
Y, et al: Pre-metastatic niches: Organ-specific homes for
metastases. Nat Rev Cancer. 17:302–317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lukanidin E and Sleeman JP: Building the
niche: The role of the S100 proteins in metastatic growth. Semin
Cancer Biol. 22:216–225. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Salama I, Malone PS, Mihaimeed F and Jones
JL: A review of the S100 proteins in cancer. Eur J Surg Oncol.
34:357–364. 2008. View Article : Google Scholar
|
|
8
|
Mei Y, Yang JP and Qian CN: For robust big
data analyses: A collection of 150 important pro-metastatic genes.
Chin J Cancer. 36:162017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pang X, Min J, Liu L, Liu Y, Ma N and
Zhang H: S100B protein as a possible participant in the brain
metastasis of NSCLC. Med Oncol. 29:2626–2632. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Choi H, Puvenna V, Brennan C, Mahmoud S,
Wang XF, Phillips M, Janigro D and Mazzone P: S100B and S100B
auto-antibody as biomarkers for early detection of brain metastases
in lung cancer. Transl Lung Cancer Res. 5:413–419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bonfrer JM, Korse CM, Nieweg OE and Rankin
EM: The luminescence immunoassay S-100: a sensitive test to measure
circulating S-100B: its prognostic value in malignant melanoma. Br
J Cancer. 77:2210–2214. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Heizmann CW: S100B protein in clinical
diagnostics: Assay specificity. Clin Chem. 50:249–251. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schlagenhauff B, Schittek B, Ellwanger U,
Stroebel W, Blum A, Schwarz M, Rassner G and Garbe C: Significance
of serum protein S100 levels in screening for melanoma metastasis:
Does protein S100 enable early detection of melanoma recurrence?
Melanoma Res. 10:451–459. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin J, Yang Q, Wilder PT, Carrier F and
Weber DJ: The calcium-binding protein S100B down-regulates p53 and
apoptosis in malignant melanoma. J Biol Chem. 285:27487–27498.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yoshimura C, Miyafusa T and Tsumoto K:
Identification of small-molecule inhibitors of the human S100B-p53
interaction and evaluation of their activity in human melanoma
cells. Bioorg Med Chem. 21:1109–1115. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bechmann T, Madsen JS, Brandslund I, Lund
ED, Ormstrup T, Jakobsen EH, Jylling AM, Steffensen KD and Jakobsen
A: Predicting brain metastases of breast cancer based on serum
S100B and serum HER2. Oncol Lett. 6:1265–1270. 2013.PubMed/NCBI
|
|
17
|
Darlix A, Lamy PJ, Lopez-Crapez E,
Braccini AL, Firmin N, Romieu G, Thézenas S and Jacot W: Serum NSE,
MMP-9 and HER2 extracellular domain are associated with brain
metastases in metastatic breast cancer patients: Predictive
biomarkers for brain metastases? Int J Cancer. 139:2299–2311. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Charmsaz S, Hughes É, Bane FT, Tibbitts P,
McIlroy M, Byrne C, Cocchiglia S, McBryan J, Hennessy BT, Dwyer RM,
et al: S100β as a serum marker in endocrine resistant breast
cancer. BMC Med. 15:792017. View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Ma XJ, Dahiya S, Richardson E, Erlander M
and Sgroi DC: Gene expression profiling of the tumor
microenvironment during breast cancer progression. Breast Cancer
Res. 11:R72009. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ciriello G, Gatza ML, Beck AH, Wilkerson
MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et
al: TCGA Research Network: Comprehensive molecular portraits of
invasive lobular breast cancer. Cell. 163:506–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ringnér M, Fredlund E, Häkkinen J, Borg Å
and Staaf J: GOBO: Gene expression-based outcome for breast cancer
online. PLoS One. 6:e179112011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar
|
|
24
|
Aguirre-Gamboa R, Gomez-Rueda H,
Martínez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R,
Rodriguez-Barrientos A, Tamez-Peña JG and Treviño V: SurvExpress:
An online biomarker validation tool and database for cancer gene
expression data using survival analysis. PLoS One. 8:e742502013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chanrion M, Negre V, Fontaine H, Salvetat
N, Bibeau F, Mac Grogan G, Mauriac L, Katsaros D, Molina F,
Theillet C, et al: A gene expression signature that can predict the
recurrence of tamoxifen-treated primary breast cancer. Clin Cancer
Res. 14:1744–1752. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sarrió D, Rodriguez-Pinilla SM, Hardisson
D, Cano A, Moreno-Bueno G and Palacios J: Epithelial-mesenchymal
transition in breast cancer relates to the basal-like phenotype.
Cancer Res. 68:989–997. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chavez KJ, Garimella SV and Lipkowitz S:
Triple negative breast cancer cell lines: One tool in the search
for better treatment of triple negative breast cancer. Breast Dis.
32:35–48. 2010. View Article : Google Scholar
|
|
28
|
Lamouille S, xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dominguez C, David JM and Palena C:
Epithelial-mesenchymal transition and inflammation at the site of
the primary tumor. Semin Cancer Biol. 47:177–184. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Demirkan B: The roles of
epithelial-to-mesenchymal transition (EMT) and
mesenchymal-to-epithelial transition (MET) in breast cancer bone
metastasis: Potential targets for prevention and treatment. J Clin
Med. 2:264–282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Giri D, Ozen M and Ittmann M:
Interleukin-6 is an autocrine growth factor in human prostate
cancer. Am J Pathol. 159:2159–2165. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Horiguchi K, Fujiwara K, Higuchi M,
Yoshida S, Tsukada T, Ueharu H, Chen M, Hasegawa R, Takigami S,
Ohsako S, et al: Expression of chemokine CxCL10 in
dendritic-cell-like S100β-positive cells in rat anterior pituitary
gland. Cell Tissue Res. 357:757–765. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Villarreal A, Seoane R, González Torres A,
Rosciszewski G, Angelo MF, Rossi A, Barker PA and Ramos AJ: S100B
protein activates a RAGE-dependent autocrine loop in astrocytes:
Implications for its role in the propagation of reactive gliosis. J
Neurochem. 131:190–205. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nieman KM, Romero IL, Van Houten B and
Lengyel E: Adipose tissue and adipocytes support tumorigenesis and
metastasis. Biochim Biophys Acta. 1831:1533–1541. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gonçalves CA, Leite MC and Guerra MC:
Adipocytes as an important source of serum S100B and possible roles
of this protein in adipose tissue. Cardiovasc Psychiatry Neurol.
2010:7904312010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lim SY, Yuzhalin AE, Gordon-Weeks AN and
Muschel RJ: Tumor-infiltrating monocytes/macrophages promote tumor
invasion and migration by upregulating S100A8 and S100A9 expression
in cancer cells. Oncogene. 35:5735–5745. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Luo X, Sharff KA, Chen J, He TC and Luu
HH: S100A6 expression and function in human osteosarcoma. Clin
Orthop Relat Res. 466:2060–2070. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bianchi R, Giambanco I and Donato R:
S100B/RAGE-dependent activation of microglia via NF-kappaB and AP-1
Co-regulation of COx-2 expression by S100B, IL-1beta and TNF-alpha.
Neurobiol Aging. 31:665–677. 2010. View Article : Google Scholar
|
|
39
|
Yang T, Cheng J, Yang Y, Qi W, Zhao Y,
Long H, xie R and Zhu B: S100B mediates stemness of ovarian cancer
stem-like cells through inhibiting p53. Stem Cells. 35:325–336.
2017. View Article : Google Scholar
|