Introduction
Globally, gastric cancer is the fourth most common
malignancy and the second leading cause of cancer-related
mortality, affecting approximately one million individuals each
(1,2). Chemotherapy has been applied widely
in the treatment of gastric cancer at different stages (3). However, a major issue in the
treatment of gastric cancer is the development of resistance to
multiple chemotherapeutic agents in tumor cells (4). Multidrug resistance (MDR) in cancer
cells is an acquired resistance to multiple drugs, which may be
structurally and functionally different (5). Various mechanisms may lead to the
development of MDR in cancer cells, including the altered
expression of drug influx/efflux transporters, aberrant DNA repair
and impairment, the prevention of apoptosis, the mutation of drug
targets in targeted therapy, alterations in the cell cycle and
checkpoints and an altered tumor microenvironment (5,6). The
signaling pathways involved include, in some cancers,
Wnt/β-catenin, NOTCH and PI3K/AKT, among others, leading to
increased resistance to drug treatment with both chemotherapy and
targeted therapy (7–10). Interfering with these signaling
pathways may be a novel antitumor strategy with which to
prevent/inhibit MDR in clinical therapies.
Isobaric tags for the relative and absolute
quantification (iTRAQ) analysis is an emerging quantitative
proteomics technology that utilizes peptides labeled with
isotope-coded covalent tags for the analysis of changes in protein
expression in different samples (11). In the present study, the
iTRAQ-based proteomic approach was applied to identify
differentially expressed proteins in the SGC7901 and SGC7901/DDP
cell lines. Among the proteins screened by this approach,
tetraspanin-8 (TSPAN8) expression was found to be significantly
increased in the SGC7901/DDP cells.
The TSPAN8 gene encodes a cell surface
glycoprotein characterized by 4 transmembrane domains and
well-conserved cysteine residues in a large extracellular loop, and
is expressed in gastric, colon, rectal and pancreatic carcinomas,
but not in the majority of normal tissues (12–15).
Within the Tetraspanin-enriched microdomain (TEM), TSPAN8 acts as a
molecular facilitator (16), being
involved in tissue differentiation (17), tumor-cell metastasis (18), and cell motility and cell fusion
(18,19). TSPAN8 has been shown to be
overexpressed in gastric cancer and to promote cancer cell
proliferation, migration and invasion (20). However, the role of TSPAN8 in MDR
gastric cancer cells remains unknown. Thus, in the present study,
we identified TSPAN8 as a pro-drug resistance protein using
iTRAQ-based quantitative proteomics. The silencing of TSPAN8
enhanced the sensitivity of the SGC7901/DDP cells to
chemotherapeutic drugs. Additionally, TSPAN8 mediated the
activation of the Wnt/β-catenin pathway by binding to NOTCH2. These
results indicate that TSPAN8 increases the MDR of gastric cancer
cells. The inhibition of TSPAN8 may reduce drug resistance and may
prove to be a strategy for the clinical treatment of patients with
gastric cancer.
Materials and methods
Cell culture, transfection and drug
treatment
The cell lines used in this study were purchased
from the China Center for Type Culture Collection (Wuhan, China).
SGC7901/DDP is an MDR gastric cancer cell line in which resistance
was induced by cisplatin and it is derived from the human gastric
cancer cell line, SGC7901. The cells were cultured in RPMI-1640
medium containing 10% fetal bovine serum (FBS) (both from Gibco,
Grand Island, NY, USA) and 1% penicillin-streptomycin solution. The
biological characteristics of MDR of the SGC7901/DDP cell line were
maintained by the addition of 1 µg/ml cisplatin
(Sigma-Aldrich, St. Louis, MO, USA) to the complete medium. The
cells were incubated in an atmosphere with 5% carbon dioxide at
37°C. Three small interfering RNA (siRNA) duplexes targeting human
TSPAN8 and a control siRNA were synthesized by GenePharma Co., Ltd.
(Shanghai, China). The sequences of the siRNA-TSPAN8 were as
follows: Sequence 1 forward, 5′-GUAUCUUGAUCCUAGCAUUdTdT-` and
reverse, 5′-AAUGCUAGGAUCAAGAUACdTdT-3′; sequence 2 forward,
5′-GUCUGAUCGCAUUGUGAAUdTdT-3′ and reverse,
5′-AUUCACAAUGCGAUCAGACdTdT-3′; sequence 3 forward,
5′-GAGUUUAAAUGCUGCGGUUd TdT-3′ and reverse,
5′-AACCGCAGCAUUUAAACUCdTdT-3′; and siRNA-NC forward,
5′-UUCUUCGAAGGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′. The SGC7901/DDP cells were transfected
with the siRNA using siRNA-Mate (GenePharma Co., Ltd.) following
the manufacturer's instructions. The inhibitors of the Wnt pathway
(CCT036477 and XAV939) and the inhibitor of NOTCH2 (DAPT) were
purchased from Santa Cruz Biotechnology, (Santa Cruz, CA, USA). All
these drugs were suspended in dimethyl sulfoxide (DMSO; Sangon
Biotech, Shanghai, China) at a stock concentration according to the
manufacturer's instructions and stored at -80°C. Following siRNA
transfection for 48 h, the cells were exposed to the inhibitors,
which were diluted into the culture medium (10 µM CCT036447,
10 µM XAV939, 20 µM DAPT or DMSO alone as a control)
for 48 h, respectively.
Determination of half maximal inhibitory
concentration (IC50)
The cytotoxic effects of the cisplatin,
5-fluorouracil and adriamycin (both from Sangon Biotech) on the
SGC7901 and SGC7901/DDP cells were measured by cell counting kit-8
(CCK-8) assay (21). The cells
were counted using the Neubauer cell-counting chamber (BRAND GMBH +
CO KG, Wertheim, Germany) following the manufacturer's
instructions. The cells were then seeded in 96-wells at a density
of 5×103 cells/well, and cultured in an incubator at
37°C for 24 h before being treated with the chemotherapeutic drugs.
Cisplatin, 5-fluorouracil (5-Fu) and adriamycin in graded
concentrations were added to the cells. Following treatment for 48
h, the medium was replaced with fresh medium containing 10% CCK-8
reagent (Dojindo, Kumamoto, Japan), and the cells were incubated
for an additional 1–4 h. The optical density was then measured by
Thermo Scientific Varioskan Flash spectral scanning multimode
reader (Thermo Fisher Scientific, Waltham, MA, USA) at 450 nm. The
IC50 values obtained following treatment of the SGC7901
and SGC7901/DDP cells with each drug were analyzed using IBM SPSS
Statistics v21 software (SPSS Inc., Chicago, IL, USA) via probit
analysis (22).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted using a High Purity Total
RNA Rapid Extraction kit (RP1201; BioTeke, Beijing, China)
according to the manufacturer's instructions. cDNA was synthesized
using a iSCRIPT cDNA Synthesis kit (GeneCopoeia Co., Ltd.,
Guangzhou, China). The primers used for the amplification of
TSPAN8, β-catenin, NOTCH2 and glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) were synthesized by GeneCopoeia Co., Ltd.
GAPDH was used as an internal standard, and the relative expression
of each gene was normalized to GAPDH. The real-time PCR kit was
purchased from GeneCopoeia Co., Ltd. PCR cycling conditions were as
follows: 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec,
60°C for 20 sec and 72°C for 10 sec. The relative quantification of
gene expression was analyzed using the 2−ΔΔCt method
(23). Each sample was analyzed in
triplicate.
Western blot analysis
Protein was extracted from the cells using RIPA
lysis buffer (Beyotime, Shanghai, China) and the concentration was
determined using the 2D Quantification kit (Amersham Biosciences,
Little Chalfont, UK). The protein samples were separated on a 10%
polyacrylamide gel, and electrotransferred onto polyvinylidene
fluoride membranes (Millipore Corp., Billerica, MA, USA). The
membranes were then blocked with 5% non-fat dried milk for 1 h at
room temperature. This was followed by the addition of the primary
antibodies: anti-TSPAN8 antibody (ab70007), anti-β-catenin antibody
(ab16051) (both from Abcam, Cambridge, MA, USA), anti-cellular
retinoic acid-binding protein 2 (CRABP2) antibody (10225-1-AP),
anti-voltage-dependent anion-selective channel protein 2 (VDAC2)
antibody (11663-1-AP), anti-Bcl-2 antibody (12789-1-AP) (all from
Proteintech, Wuhan, China), anti-heat shock protein 90 (HSP90)
antibody (bs-0889R), anti-erythrocyte membrane protein band 4.1
(EPB41) antibody (bs-13080R), anti-tumor protein D54 (TPD54)
antibody (bs-6743R), anti-mucin 13 (MUC13) antibody (bs-10074R),
anti-GAPDH antibody (bs-10900R), anti-caspase-3 antibody (bs-0081R)
and anti-Bax antibody (bs-0127R) (all from Bioss, Beijing, China)
and overnight incubation at 4°C. All primer antibodies were diluted
(1:1,000) by Tris-buffered saline containing 0.1% Tween-20 (TBS-T)
After washing 3 times with TBS-T, the membranes were incubated with
a horseradish peroxidase-conjugated goat anti-rabbit IgG as a
secondary antibody (1:5,000, ab6721; Abcam) for 2 h at room
temperature. After washing 3 times with TBS-T buffer, the membranes
were visualized with an ECL detection system (KeyGen Biotech Inc.,
Nanjing, China). All western blot analyses were repeated at least 3
times.
Luciferase reporter assay
The cells were seeded in a 6-well plate and
transfected with siRNA according to the protocol of siRNA-Mate
(GenePharma Co., Ltd.). TOP-flash reporter plasmid was purchased
from Shanghai Qcbio Science and Technologies Co., Ltd. (Shanghai,
China) and was transfected into the cells by Endofectin™-Plus
(GeneCopoeia) according to the manufacturer's instructions 48 h
after siRNA transfection. The reporter gene assay was performed 48
h post-plasmid-transfection using the Dual Luciferase Assay System
(Promega, Madison, WI, USA). Firefly luciferase activity was
normalized for transfection efficiency using the corresponding
Renilla luciferase activity. All experiments were performed
at least in triplicate.
Immunoprecipitation
The plasmids (HA-TSPAN8, Flag-TSPAN8, HA-NOTCH2 and
Flag-NOTCH2) used for exogenous co-immunoprecipitation were
synthesized by GeneCopoeia Co., Ltd. The cells were lysed by
sonication and centrifugation at 4°C, 16,000 × g, 10 min (TDZ4-WS
centrifuge; Thermo Fisher Scientific) in IP lysis buffer (Beyotime,
Beijing, China) supplemented with phosphatase/protease inhibitor
cocktail and 1 mM PMSF. The supernatant was transferred to a
separate microfuge tube, pre-cleared with protein A/G agarose beads
(Yanji Biotechnology, Shanghai, China) and centrifuged at 4°C,
16,000 × g, 5 min (TDZ4-WS centrifuge; Thermo Fisher Scientific) to
pellet the beads and remove protein impurities. The supernatant was
collected and incubated with rabbit IgG (bs-0295P; Bioss) overnight
at 4°C. The beads were collected by centrifugation at 4°C, 16,000 ×
g, 10 min (TDZ4-WS centrifuge; Thermo Fisher Scientific), washed 3
times with IP lysis buffer and resuspended with 2× sodium dodecyl
sulfate (SDS) loading buffer. Bound protein was eluted off the
beads by boiling and examined by western blot analysis as described
above.
Immunofluorescence
The cells were incubated with 4.0% paraformaldehyde
for 15 min at room temperature. The cells were then washed 3 times
with phosphate-buffered saline (PBS). To increase permeability,
0.1% Triton X-100 was added to the cells for 10 min. The cells were
then washed again thrice with PBS. The anti-β-catenin antibody
(ab16051; 1:100 diluted by PBS; Abcam) was added to the wells
followed by incubation overnight at 4°C. The cells were then washed
and incubated in Alexa Fluor-conjugated secondary antibody (1:100
diluted with Bioss antifade mounting medium; Bioss). DAPI
(Invitrogen, Carlsbad, CA, USA) was used to dye the nuclei. The
cells were incubated with DAPI for 20 min at room temperature.
After being washed 3 times with PBS, the cells were imaged under a
microscope (Ci-L; Nikon, Tokyo, Japan).
Protein extraction and iTRAQ
labelling
Total protein extracts were prepared in lysis buffer
[7 M urea, 1 mg/ml DNase I, 1 mM Na3VO4 (all
from Sangon Biotech), and 1 mM PMSF (Bioss, Beijing, China)] using
the Sample Grinding kit from Amersham Biosciences. Following being
centrifuged at 17,000 × g for 15 min at 4°C, the supernatant was
collected and the protein concentrations were quantified with a 2-D
Quantification kit (Amersham Biosciences).
From each sample, 100 µg of protein was
precipitated, denatured, cysteine-blocked and digested with
sequencing-grade modified trypsin, according to the manufacturer's
instructions (iTRAQ Reagent 8 Plex Multi-plex; Applied Biosystems,
Foster City, CA, USA). The samples were then labeled with the iTRAQ
tags (SGC7901, 113, 115 tags; SGC7901/DDP 114, 116 tags; Applied
Biosystems). The labeled samples were pooled prior to further
analysis.
Fractionation of peptides
The iTRAQ-labeled samples were solubilized in 300
µl of 1% Pharmalyte (Amersham Biosciences) and 8 M urea
solution. The samples were rehydrated on IPG gel strips (pH
3.0–10.0; Amersham Biosciences) at 30 V for 14 h. The peptides were
subsequently focused successively at 500 V for 1 h, 1,000 V for 1
h, 3,000 V for 1 h and 8,000 V for 8.5 h. Following
electrofocusing, the peptides were extracted from the gel using a
solution containing 0.1% formic acid and 2% acetonitrile for 1 h.
The fractions were then purified and concentrated on a C18
Discovery DSC-18 SPE column (Sigma-Aldrich), lyophilized and
maintained at -20°C.
Mass spectrometry
The samples were analyzed using a QStar Elite hybrid
mass spectrometer (Applied Biosystems) coupled with a liquid
chromatography system (Amersham Biosciences, Little Chalfont,
UK).
The mass spectrometer was set to perform
information-dependent acquisition (IDA) in the positive ion mode at
a mass range of 300–1800 m/z. Peptides with +2 to +4 charge states
were selected for tandem mass spectrometry, and the time of
summation of MS/MS events was set to 3 sec. We selected the two
most abundantly charged peptides above a 20-count threshold for
MS/MS and dynamic exclusion was set to 30 sec with a 50 mDa mass
tolerance. Data were processed using ProteinPilot version 2.0
software (Applied Biosystems) and searched against the UnitProt
(http://www.uniprot.org/) human protein database
(v3.77). Protein identification was based on selection thresholds
of ProtScore >1.3 or ProtScore <0.77, and false discovery
rate P-values <0.05.
Bioinformatics analysis
The results obtained by iTRAQ-labeled proteomics
were analyzed using by protein analysis using the evolutionary
relationships (PANTHER) classification system (www.pantherdb.org) following the instructions
available online (24). STRING
10.5 (http://string.embl.de/) was used to
predict the interaction between proteins following the instruction
online (25).
Statistical analysis
The in vitro experiments were repeated at
least 3 times. Data are presented as the means ± standard deviation
(SD). Significance between groups from in vitro experiments
was determined using the Student's t-test or Dunnett's T3 test. A
value of P<0.05 was considered to indicate a statistically
significant difference.
Results
iTRAQ-coupled 2D LC-MS/MS analysis of
differentially expressed proteins
To identify potential proteins associated with
resistance to cisplatin, iTRAQ-based quantification was performed
on proteins isolated from cisplatin-sensitive gastric cancer cells
(SGC7901) and from DDP-resistant gastric cancer cells
(SGC7901/DDP). The specimens were iTRAQ-labeled in duplicate in
order to verify the results. Protein samples were labeled as
follows: SGC7901, tags 113 and 114; SGC7901/DDP, tags 115 and 116.
The relative abundance of protein from the SGC7901/DDP cells with
respect to proteins from SGC7901 cells was calculated as the iTRAQ
ratios 115:113 and 116:114. These fractions were analyzed by
LC/MS/MS. The workflow of the iTRAQ proteomics approach is
presented in Fig. 1. ProteinPilot
2.0 software was used for protein quantification and
identification. Considering the technical variations of the method
and statistical analysis in the relative quantification analysis,
and in order to reduce false-positives and increase accuracy, a
1.3-fold cut-off for all iTRAQ ratios was used (26,27).
Therefore, proteins with iTRAQ ratios <0.77- or >1.3-fold
cut-off (P<0.05) were considered to be downregulated or
upregulated, respectively. A total of 1,324 differentially
expressed proteins were identified, regardless of whether or not
there was a significant P-value in the iTRAQ ratios. Of these, 112
proteins were differentially expressed in the SCG7901/ddp cells
compared to the SGC7901 cells (64 upregulated and 48 downregulated
proteins). The top 30 downregulated and upregulated proteins are
shown in Table I.
 | Table IPartial list of proteins
differentially expressed between the SGC7901and SGC7901/DDP
cells. |
Table I
Partial list of proteins
differentially expressed between the SGC7901and SGC7901/DDP
cells.
| No. | UniProtKB accession
ID | UniProtKB ID | Protein name | Peptides
(95%) | 115:113 | pvAl 115:113 | 116:114 | pvAl 116:114 |
|---|
| Top 30
downregulated proteins in the multidrug-resistant SGC7901/DDP
gastric cancer cells |
| 1 | B4DQE1 | B4DQE1_HUMAN | Annexin | 48 | 0.622325778 | 0.02227027 | 0.634360694 | 7.79E-05 |
| 2 | P11171 | 41_HUMAN | Protein 4.1 | 72 | 0.624904871 | 0.03530645 | 0.710090963 | 4.57E-06 |
| 3 | G3V4C1 | G3V4C1_HUMAN | Heterogeneous
nuclear ribonucleoproteins C1/C2 | 31 | 0.628175676 | 0.000760296 | 0.725739842 | 4.02E-05 |
| 4 | P22626 | ROA2_HUMAN | Heterogeneous
nuclear ribonucleoproteins A2/B1 | 10 | 0.655659914 | 0.00013268 | 0.639985561 | 0.001628224 |
| 5 | A8K2H4 | A8K2H4_HUMAN | cDNA FFJ78235 | 15 | 0.660123587 | 0.005608639 | 0.673961346 | 0.00055461 |
| 6 | Q9NZ23 | Q9NZ23_HUMAN | Drug-sensitive
protein 1 | 9 | 0.663049996 | 0.01362959 | 0.715051212 | 0.007643274 |
| 7 | PI4866 | HNRPF_HUMAN | Heterogeneous
nuclear ribonucleoprotein F | 32 | 0.663075626 | 4.51E-05 | 0.614491193 | 2.93E-08 |
| 8 | 043390 | HNRPR_HUMAN | Heterogeneous
nuclear ribonucleoprotein R | 16 | 0.675710618 | 7.89E-07 | 0.617349922 | 0.01043923 |
| 9 | PI0253 | FYAG_HUMAN | Fysosomal
α-glucosidase | 7 | 0.677277982 | 0.004195308 | 0.622946714 | 0.001522158 |
| 10 | B4DVA7 | B4DVA7_HUMAN |
β-hexosaminidase | 25 | 0.690114915 | 5.21E-05 | 0.728313433 | 0.003678592 |
| 11 | Q6RFH5 | WDR74_HUMAN | WD
repeat-containing protein 74 | 8 | 0.703277528 | 0.038117841 | 0.727350508 | 0.01727649 |
| 12 | B3KM89 | B3KM89_HUMAN | cDNA FUJI0528 hs,
clone NT2RP2000943, highly similar to protein transport protein
Sec24D | 36 | 0.706753314 | 0.024660509 | 0.737365516 | 0.000550256 |
| 13 | D3DQU2 | D3DQU2_HUMAN | Tripeptidyl
peptidase I, isoform CRA_a | 22 | 0.707172573 | 0.003796435 | 0.658462612 | 1.84E-05 |
| 14 | P07900 | HS90A_HUMAN | Heat shock protein
HSP 90-α | 7 | 0.71462971 | 8.03E-08 | 0.703796549 | 0.002079676 |
| 15 | Q53GL6 | Q53GF6_HUMAN | RNA binding protein
(autoantigenic, hnRNP-associated with lethal yellow) long isoform
variant (Fragment) | 5 | 0.729635179 | 0.002883428 | 0.721853618 | 0.024822449 |
| 16 | P17050 | NAGAB_HUMAN | α
-N-acetylgalactosaminidase | 3 | 0.736388922 | 0.016721571 | 0.683589351 | 0.0137383 |
| 17 | Q15334 | F2GF1_HUMAN | Fethal(2) giant larvae protein homolog 1 | 23 | 0.741260827 | 0.01128356 | 0.676991196 | 8.27E-05 |
| 18 | P08238 | HS90B_HUMAN | Heat shock protein
HSP 90-β | 31 | 0.745372176 | 4.53E-05 | 0.736570076 | 3.96E-05 |
| 19 | Q9UHL4 | DPP2_HUMAN | Dipeptidyl
peptidase 2 | 15 | 0.745664179 | 0.01644465 | 0.723860763 | 0.001684257 |
| 20 | B3KXS5 | B3KXS5_HUMAN | Eon protease
homolog, mitochondrial | 9 | 0.750456929 | 1.18E-07 | 0.623211837 | 0.02029554 |
| 21 | P29373 | RABP2_HUMAN | Cellular retinoic
acid-binding protein 2 | 6 | 0.752079427 | 0.000808526 | 0.647008409 | 0.0207015 |
| 22 | B3KQS9 | B3KQS9_HUMAN | cDNA PSEC0141 hs,
clone PFACE1005913, highly similar to deoxyribonuclease-2- α (EC
3.1.22.1) | 17 | 0.753284812 | 0.02952897 | 0.736696043 | 0.01036579 |
| 23 | A8K9X5 | A8K9X5_HUMAN | cDNA FFJ76472,
highly similar to Homo sapiens Fas (TNFRSF6)associated
factor 1 (FAF1), transcript variant 1, mRNA | 12 | 0.754278779 | 0.000863104 | 0.61996146 | 0.009885758 |
| 24 | Q53FG3 | Q53FG3_HUMAN | Interleukin
enhancer binding factor 2 variant (Fragment) | 12 | 0.760721684 | 0.000109113 | 0.713951109 | 0.006046766 |
| 25 | F8W1F5 | F8W1F5_HUMAN | Formin-like protein
3 | 2 | 0.762245297 | 0.04186723 | 0.695746288 | 0.009152975 |
| 26 | 060826 | CCD22_HUMAN | Coiled-coil
domain-containing protein 22 | 19 | 0.766084313 | 0.031637449 | 0.680582983 | 7.48E-05 |
| 27 | D6RD18 | D6RD18_HUMAN | Heterogeneous
nuclear ribonucleoprotein A/B | 11 | 0.768395722 | 0.001917408 | 0.630256551 | 0.003946933 |
| 28 | Q9HB71 | CYBP_HUMAN | Calcyclin-binding
protein | 12 | 0.768992722 | 0.01458319 | 0.735216274 | 0.03016348 |
| 29 | 043175 | SERA_HUMAN |
D-3-phosphoglycerate dehydrogenase | 8 | 0.769186914 | 1.83E-05 | 0.709687752 | 0.005728934 |
| 30 | Q53FB6 | Q53FB6_HUMAN | Mitochondrial
aldehyde dehydrogenase 2 variant (Fragment) | 62 | 0.7699821 | 2.35E-06 | 0.695289527 | 1.13E-10 |
| Top 30 upregulated
proteins in the multidrug-resistant SGC7901/DDP gastric cancer
cells |
| 1 | P02768 | ALBU_HUMAN | Serum albumin | 21 | 2.153665066 | 0.004250416 | 1.802739155 | 0.003726222 |
| 2 | P05787 | K2C8_HUMAN | Keratin, type II
cytoskeletal 8 | 188 | 1.95599401 | 1.16E-14 | 1.636557607 | 0.043917108 |
| 3 | P08727 | K1C19_HUMAN | Keratin, type I
cytoskeletal 19 | 105 | 1.943524003 | 0.003219714 | 1.795400163 | 0.001931372 |
| 4 | Q9GZL9 | Q9GZL9_HUMAN | β-globin
(Fragment) | 5 | 1.916771054 | 0.01608417 | 1.928079502 | 0.004268234 |
| 5 | 075348 | VATG1_HUMAN | V-type proton
ATPase subunit G1 | 4 | 1.765862942 | 0.02341911 | 1.686415416 | 0.021376461 |
| 6 | B7Z8Q2 | B7Z8Q2_HUMAN | cDNA FFJ55606,
highly similar to α-2-HS-glycoprotein | 5 | 1.763658047 | 0.0266499 | 1.5277603 | 0.02854233 |
| 7 | B2RA03 | B2RA03_HUMAN | cDNA, FFJ94640,
highly similar to Homo sapiens keratin 18 (KRT18), mRNA | 139 | 1.743123055 | 1.17E-11 | 1.530218785 | 0.037320711 |
| 8 | B2RAU8 | B2RAU8_HUMAN | cDNA, FFJ95131,
highly similar to Homo sapiens nucleolar and coiled-body
phosphoprotein 1 (NOFC1), mRNA | 33 | 1.705428004 | 0.002081443 | 1.580844317 | 0.03559231 |
| 9 | P45880 | VDAC2_HUMAN | Voltage-dependent
anion-selective channel protein 2 | 32 | 1.693142056 | 8.20E-08 | 1.963505651 | 0.035407521 |
| 10 | Q9NY12 | GAR1_HUMAN | H/ACA
ribonucleoprotein complex subunit 1 | 4 | 1.674811006 | 0.033414129 | 1.769748098 | 0.003661653 |
| 11 | I1VZV6 | I1VZV6_HUMAN | Hemoglobin α1 | 5 | 1.66385603 | 0.00999979 | 1.995862361 | 0.03718495 |
| 12 | D3DV26 | D3DV26_HUMAN | S100 calcium
binding protein A10 [Annexin II ligand, calpactin I, light
polypeptide (PI 1)], isoform CRA_b (Fragment) | 5 | 1.651872993 | 0.024565291 | 1.546541111 | 0.039673839 |
| 13 | A0A024RD07 |
A0A024RD07_HUMAN | Trinucleotide
repeat containing 5, isoform CRA_c | 4 | 1.56981504 | 0.04436332 | 1.891481629 | 0.01262225 |
| 14 | B2R6W1 | B2R6W1_HUMAN | cDNA, FFJ93143,
highly similar to Homo sapiens complement component 7 (C7),
mRNA | 6 | 1.568076968 | 0.02081763 | 1.911388803 | 0.042174641 |
| 15 | B4DRB6 | B4DRB6_HUMAN | cDNA FFJ59394,
highly similar to Homo sapiens ubiquitin associated protein
2 (UBAP2), transcript variant 1, mRNA | 7 | 1.550289989 | 0.00021213 | 2.001291157 | 0.016664799 |
| 16 | B2RAW0 | B2RAW0_HUMAN | cDNA, FFJ95154,
highly similar to Homo sapiens disabled homolog 2,
mitogen-responsive phosphoprotein (Drosophila) (DAB2),
mRNA | 3 | 1.548756003 | 0.002253909 | 1.844182745 | 0.04310175 |
| 17 | Q96AG4 | LRC59_HUMAN | Feucine-rich
repeat-containing protein 59 | 28 | 1.539394975 | 1.52E-06 | 2.009917718 | 0.045030121 |
Cellular and molecular functional
characteristics of the proteins
The 112 proteins, which were potentially
differentially expressed between the SGC7901/DDP cells and SGC7901
cells, were classified into 5 functional categories using the
Protein Analysis through Evolutionary Relationships (PANTHER)
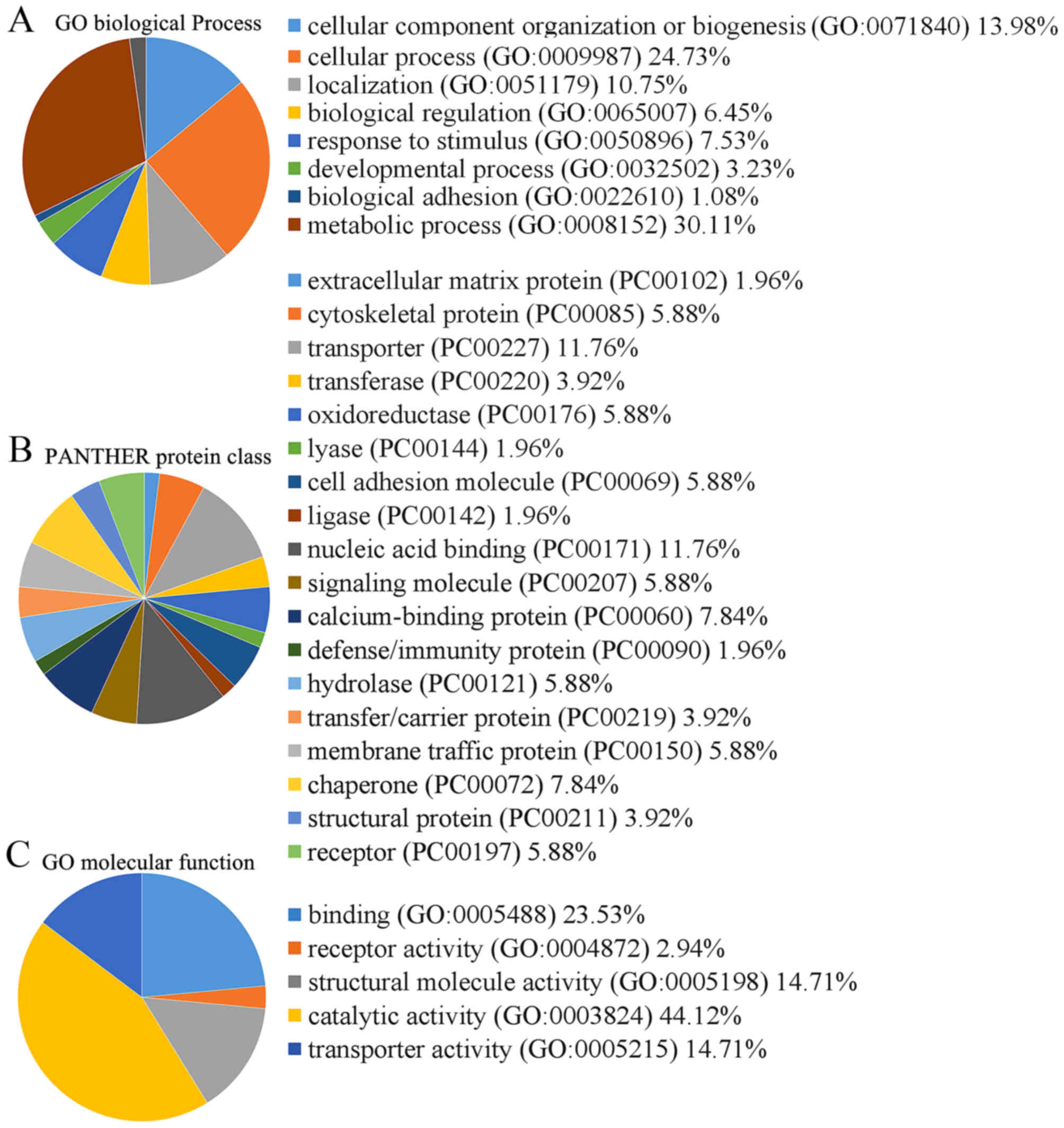
classification system (Fig. 2).
The molecular function categories were binding (23.5%), receptor
activity (2.9%), structural molecule activity (14.7%), catalytic
activity (44.1%) and transporter activity (14.7%) (Fig. 2).
Validation of differentially expressed
proteins
The differentially expressed proteins identified by
iTRAQ were validated by RT-qPCR and western blot analysis. The
proteins selected for validation were the ones most significantly
dysregulated according to protein classification or the ones
closely related to multidrug resistance. TSPAN8 has been reported
to promote the proliferation and metastasis of SGC7901 cells. The
results from iTRAQ-coupled 2D LC-MS/MS revealed that TSPAN8 was
potentially related to drug resistance in the SGC7901/DDP cells.
Thus, it was selected as the object of the following analysis. The
mRNA levels of HSP90, drug-sensitive protein 1 (YA61), EPB41 and
CRABP2 were decreased in the SGC7901/DDP cells when compared with
those in the SGC7901 cells, whereas the mRNA levels of TSPAN8,
VDAC2, TPD54 and MUC13 were increased (Fig. 3A). The results of western blot
analysis revealed that the protein expression levels of HSP90,
YA61, EPB41 and CRABP2 were downregulated in the SGC7901/DDP cells
when compared to those in the SGC7901 cells, whereas the levels of
TSPAN8, VDAC2, TPD54 and MUC13 were upregulated (Fig. 3B). These results were consistent
with the trend observed in iTRAQ analysis.
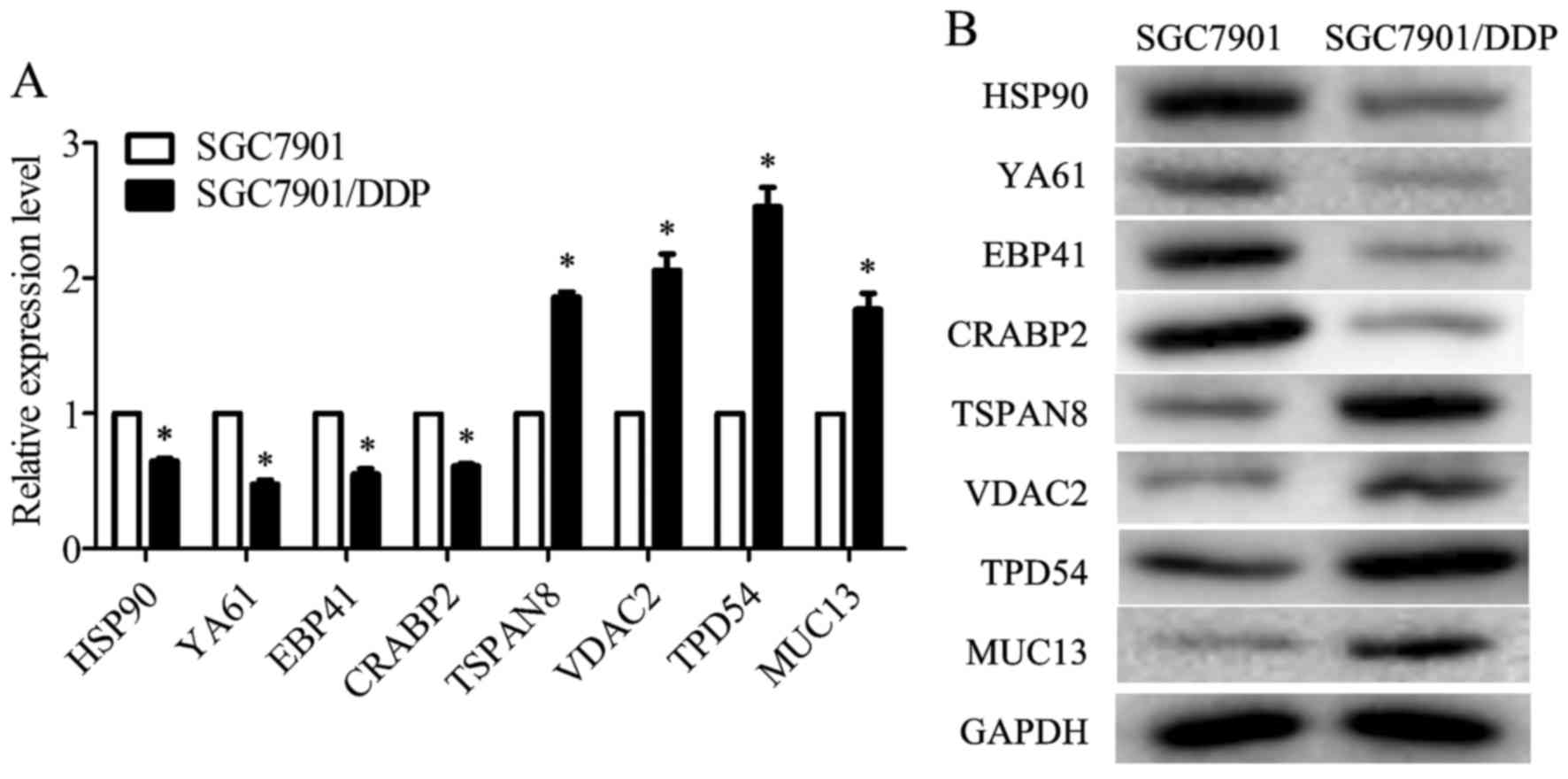 | Figure 3Validation of heat shock protein 90
(HSP90), drug-sensitive protein 1 (YA61), erythrocyte membrane
protein band 4.1 (EPB41), cellular retinoic acid-binding protein 2
(CRABP2), tetraspanin-8 (TSPAN8), voltage-dependent anion-selective
channel protein 2 (VDAC2), tumor protein D54 (TPD54) and mucin 13
(MUC13) expression in the SGC7901 and SGC7901/DDP cells. (A)
RT-qPCR was used to detect the relative mRNA expression levels of
HSP90, YA61, EPB41, CRABP2, TSPAN8, VDAC2, TPD54 and MUC13, as
normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
(P<0.05). (B) Representative western blot analyses for HSP90,
YA61, EPB41, CRABP2, TSPAN8, VDAC2, TPD54 and MUC13 expression in
cells. GAPDH was used as the normalization standard. Data are the
means ± SD; *P<0.05 vs. negative control (NC). |
Silencing of TSPAN8 in SGC7901/DDP cells
reduces MDR
TSPAN8 has been reported to be an oncoprotein in
gastric cancer, enhancing gastric cancer cell proliferation and
metastasis (20). However, the
role of TSPAN8 in gastric cancer cell drug resistance remains
unclear. In the present study, TSPAN8 was knocked down by siRNA.
RT-qPCR and western blot analysis confirmed the efficacy of the
silencing of TSPAN8. As shown in Fig.
4A, the relative mRNA level of TSPAN8 was significantly
decreased following transfection with siRNA against TSPAN8. The
results of western blot analysis revealed that TSPAN8 protein
expression in specific siRNA-transfected SGC7901/DDP cells was
effectively suppressed (Fig. 4B).
Of the 3 siRNA sequences, sequence 1 was found to be the most
suitable for our purposes (Fig. 4A and
B), and was thus used in all subsequent experiments.
MDR is the main cause of chemotherapy failure in
gastric cancer treatment. Thus, in this study, to assess the
association between TSPAN8 and MDR, the siRNA-transfected
SCG7901/DDP cells were treated with cisplatin, 5-Fu and adriamycin
(the most commonly used drugs in clinical practice for the
chemotherapeutic treatment of gastric cancer), for 2 days and the
IC50 values were determined. The IC50 values
of cisplatin, 5-Fu and adriamycin were significantly decreased in
the TSPAN8-silenced SGC7901/DDP cells compared with the negative
controls (Table II). This result
suggested that the silencing of TSPAN8 reduced the resistance of
the SGC7901/DDP cells to the aforementioned drugs, which, in turn,
indicated that TSPAN8 may contribute to the MDR of this cell line.
In the following experiments, only cisplatin was used to maintain
the drug resistance of the SGC7901/DDP cells.
 | Table IIIC50 values (mg/l) for
selected reagents after siRNA transfection. |
Table II
IC50 values (mg/l) for
selected reagents after siRNA transfection.
| Treatment | NC | siTSPAN8 |
|---|
| Cisplatin | 8.25±0.57 | 3.89±0.27a |
| 5-Fu | 4.43±0.22 | 2.41±0.16a |
| Adriamycin | 2.48±0.19 | 1.54±0.10a |
Furthermore, compared with the negative control
SGC7901/DDP cells, apoptosis was increased in the TSPAN8-silenced
cells (Fig. 4C). Moreover, the
levels of apoptosis-related proteins (caspase-3, Bax and Bcl-2)
were examined by western blot analysis. The results (Fig. 4D) revealed that the levels of
caspase-3 and Bax were upregulated, while those of Bcl-2, an
anti-apoptotic protein, were downregulated in the TSPAN8-silenced
SGC7901/DDP cells. These results indicated that the silencing of
TSPAN8 promoted SGC7901/DDP cell apoptosis.
Silencing of TSPAN8 sensitizes
SGC7901/DDP cells to chemotherapy by mediating Wnt/β-catenin
Thus far, our findings suggested that TSPAN8 plays a
critical role in the drug resistance of SGC7901/DDP cells. It is
believed that metastasis is the persistence of cancer stem cells
(CSCs), which are highly resistant to chemotherapy (28). The Wnt/β-catenin signaling pathway
has been reported to increase gastric cancer cell migration and
invasion (29). Therefore, in this
study, we investigated whether TSPAN8-mediated gastric cancer cell
drug resistance is also related to the Wnt/β-catenin pathway. The
Wnt/β-catenin pathway activity was detected using a TOP-flash
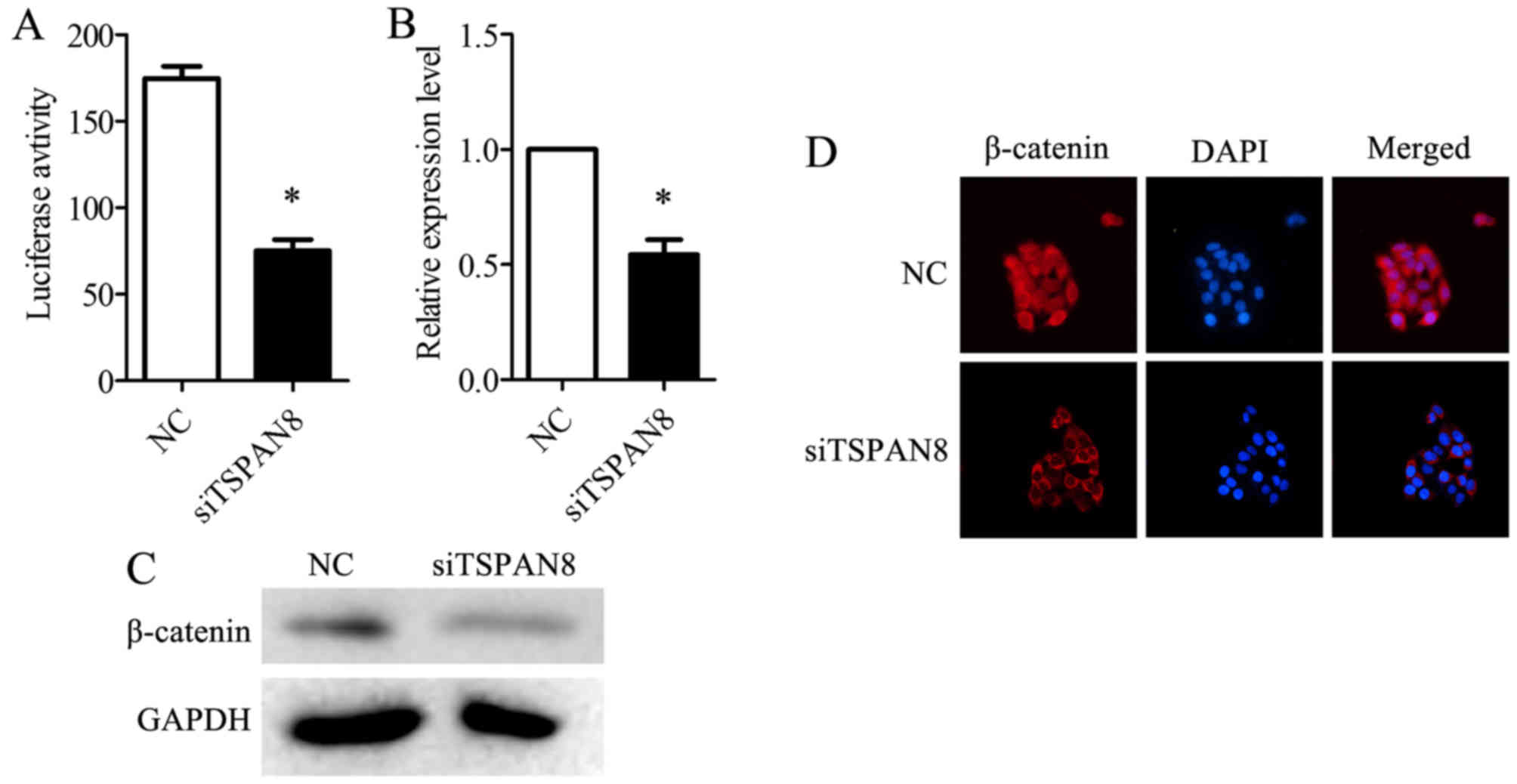
luciferase reporter. The silencing of TSPAN8 in the SGC7901/DDP
cells significantly decreased TOP-flash luciferase activity
(Fig. 5A). The TSPAN8-silenced
cells displayed a decreased expression of β-catenin at both the
mRNA (Fig. 5B) and protein level
(Fig. 5C), compared to negative
control (NC)-infected SGC7901/DDP cells. Additionally, the
accumulation of β-catenin in the nucleus was impaired in the
TSPAN8-silenced SGC7901/DDP cells (Fig. 5D). The cells were treated with
CCT036477 (CCT) and XAV939 (inhibitors of the Wnt-β-catenin
pathway) (30). The reduced
IC50 value caused by TSPAN8 silencing was partially
reversed when the Wnt-β-catenin pathway inhibitors were added
(Table III). These data
indicated that TSPAN8 enhanced the resistance of the SGC7901/DDP
cells to chemotherapy through the activation of the Wnt/β-catenin
pathway and by increasing β-catenin expression and accumulation in
the nucleus. However, compared to the NC group, the inhibitors of
the Wnt pathway still decreased the IC50 values
(Table III).
 | Table IIITSPAN8-silencing meditated reduction
of IC50 could be partially reversed by Wnt/β-catenin
inhibitors. |
Table III
TSPAN8-silencing meditated reduction
of IC50 could be partially reversed by Wnt/β-catenin
inhibitors.
| Treatment | NC | siTSPAN8 | siTSPAN8 +
CCT036477 | siTSPAN8 +
XAV939 |
|---|
| Cisplatin | 7.97±0.62 | 3.77±0.41a | 5.92±0.51a,b | 5.84±0.48a,b |
TSPAN8 mediated Wnt/β-catenin through
binding to NOTCH2
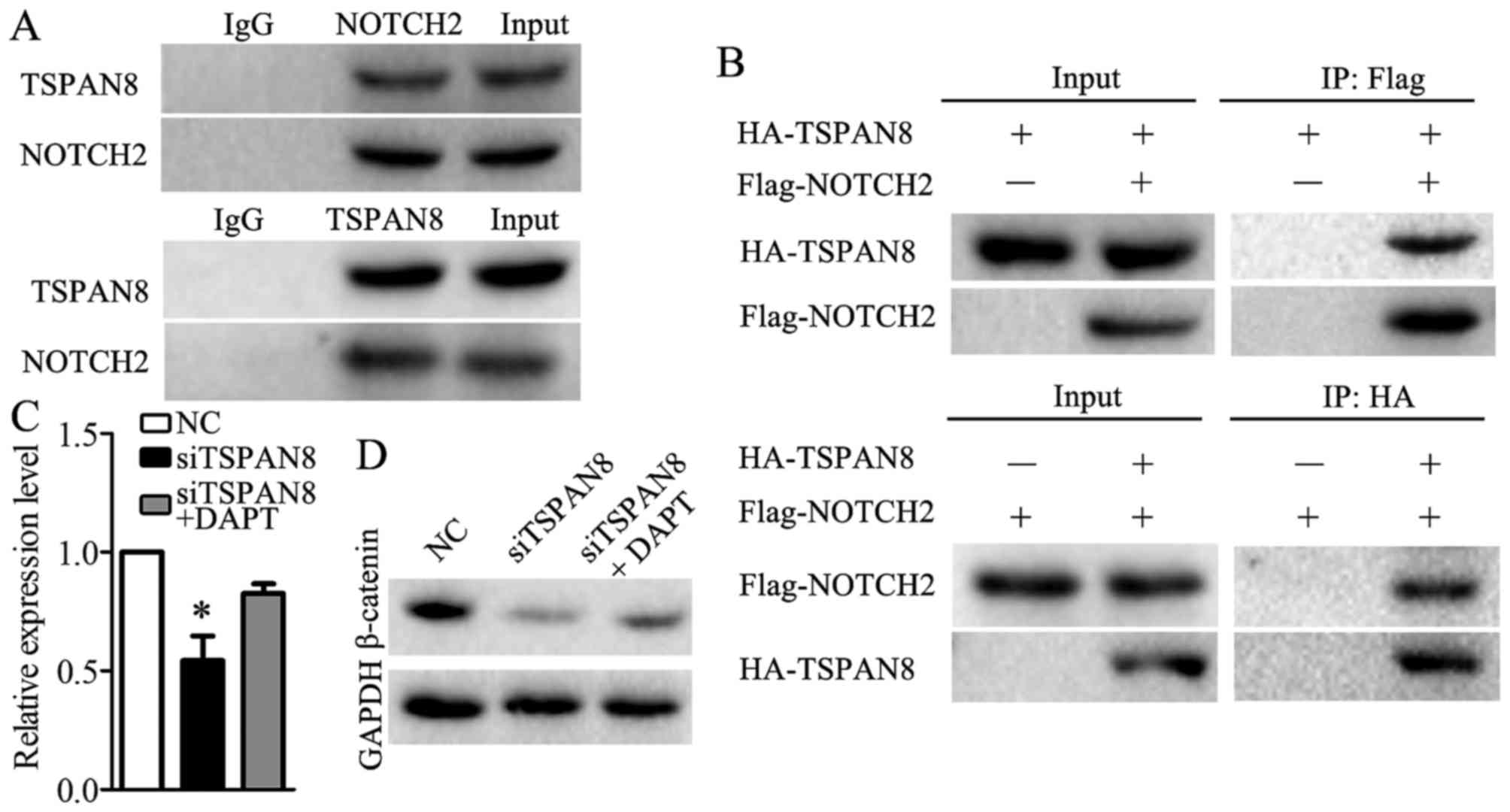
To identify which protein or proteins interact with
TSPAN8, we utilized STRING 10.5. NOTCH2 was predicted to interact
with TSPAN8. Co-immunoprecipitation was used to validate the
association between TSPAN8 and NOTCH2. Endogenous
co-immunoprecipitation assays revealed that TSPAN8 interacted with
NOTCH2 in the SGC7901/DDP cells (Fig.
6A). Consistent with this result, the exogenous interaction
between TSPAN8 and NOTCH2 was also observed in the SGC7901/DDP
cells that were co-transfected with HA-TSPAN8 and Flag-NOTCH2
(Fig. 6B). These findings revealed
that TSPAN8 acts in combination with NOTCH2 in gastric cancer
cells. Furthermore, we found that the impairment of β-catenin
expression was partially compensated when DAPT (30), a NOTCH2 inhibitor, was used in the
TSPAN8-silenced SGC7901/DDP cells (Fig. 6C and D). The results data indicated
that TSPAN8 mediated the activation of the Wnt/β-catenin pathway by
binding to NOTCH2.
Discussion
Gastric cancer is one of the most common malignant
tumors worldwide (31), and is the
leading cause of morbidity and mortality among malignant tumors in
East Asia (32). Unfortunately,
the majority of patients are diagnosed at the advanced stages of
the disease, when chemotherapy is regarded as an important
component of multimodal treatment (33). Platinum- or fluorouracil-based
chemotherapy is established as the first-line treatment for
patients with advanced gastric cancer (34). Cisplatin and other platinum-based
cancer drugs destroy tumor cells by binding to DNA strands and
interfering with DNA replication (33,34).
While cisplatin is often effective when first administered,
clinical drug resistance to cisplatin-based chemotherapy is
considered a major impediment in the treatment of patients with
gastric cancer (3,34). Drug resistance in cancer patients
includes the development of intrinsic or acquired drug resistance
against chemotherapeutic agents (35). The resistance phenotype is
associated with cancer cells gaining a cross-resistance to a large
range of drugs that are structurally and functionally different,
referred to as MDR (36). The
mechanisms of MDR in cancer remain understood on only a limited
basis. A wide range of mechanisms contribute to MDR, including drug
efflux mediation by ATP-binding cassette (ABC) transporter, the
prevention of apoptosis, alterations in drug targets, the aberrant
activation of cell signaling pathways, altered cell cycle events,
cancer stem cells (CSC), epigenetic regulation, tumor
microenvironment and many other causes (8,36).
MDR results in treatment failure or even death in patients with
gastric cancer (4,37) and, as such, strategies to reverse
MDR have been a high priority goal in cancer research.
In the present study, we searched for proteins
possibly related to drug resistance in the human gastric cancer
cell lines, SGC7901 and SCG7901/DDP, using iTRAQ-based quantitative
proteomics. In total, 64 proteins were found to be increased, while
48 proteins were found to be decreased, in the SGC7901/DDP cancer
cells, compared with the drug-sensitive SGC7901 cells. VDAC2,
TPD53, MUC13 and HSP90 (38–41)
have been previously reported to be closely associated with MDR.
Thus, these proteins and another 4 of the mostly dysregulated
proteins were selected for validation. Western blot analysis
revealed that the expression levels of TSPAN8, VDAC2, TPD54, MUC13,
HSP90, YA61, EPB41 and CRABP2 were validated at the same levels as
those obtained from the results of the quantitative proteomic
analysis, confirming that the iTRAQ-based quantitative proteomics
is an efficient and powerful method for the analysis of MDR-related
proteins. TSPAN8 expression was found to be significantly increased
in the SGC7901/DDP cells, the drug-resistant gastric cancer cell
line. The overexpression of TSPAN8 has been reported in many types
of cancer, including hepatocellular carcinoma, pancreatic cancer,
colon carcinoma and gastric cancer (14–15,20,42).
TSPAN8 has been implicated as increasing the proliferation,
migration and invasion of many types of cancer cells, including
gastric cancer cells (20).
However, the role of TSPAN8 in the MDR of gastric cancer cells
remains unknown. In this study, the iTRAQ-based quantitative
proteomics data indicated that TSPAN8 contributed to MDR in the
SGC7901/DDP cells. To confirm this, we silenced TSPAN8 in the
SGC7901/DDP cells via RNA interference. The IC50 results
revealed that the silencing of TSPAN8 increased the response of the
gastric cancer cells to the anticancer drugs. The silencing of
TSPAN8 also increased cell apoptosis. These results indicated that
TSPAN8 facilitates the MDR of SGC7901/DDP cells by suppressing
apoptosis.
The aberrant activation of the Wnt/β-catenin pathway
leads to cancer cell invasion, migration and MDR (27,43).
Thus, in this study, Wnt/β-catenin activity was monitored in the
TSPAN8-silenced cells. The results revealed that silencing TSPAN8
significantly decreased Wnt activity and β-catenin expression in
the SGC7901/DDP cells. We also found that the IC50 of
the SGC7901/DDP cells treated with cisplatin was decreased when
TSPAN8 was silenced; however, this effect of TSPAN8 silencing was
partially reversed when Wnt/β-catenin pathway inhibitors were used.
All these data indicated that TSPAN8 enhanced the resistance of
SGC7901/DDP cells to chemotherapy through the activation of the
Wnt/β-catenin pathway and by increasing β-catenin expression and
accumulation in the nucleus. When the Wnt/β-catenin pathway is
aberrantly activated, the transcription of downstream genes
mediated by Wnt signaling increases. A number of Wnt targeting
genes, such as LEF1 and c-MYC, induce drug resistance in cancer
cells (44,45). This explains how TSPAN8 increases
the MDR of SGC7901/DDP cells by mediating the Wnt/β-catenin
pathway.
To further explore the mechanisms of action of
TSPAN8 as regards MDR, we searched the biological database. It was
predicted that TSPAN8 may interact with NOTCH2 (46), which has been reported to
participate in Wnt/β-catenin-based MDR in osteosarcoma (31). We hypothesized that TSPAN8 mediated
the activation of the Wnt/β-catenin pathway by binding to NOTCH2 in
SGC7901/DDP cells. Co-immunoprecipitation revealed that TSPAN8
bound to NOTCH2. The impairment of β-catenin expression was
partially compensated when DAPT, a NOTCH2 inhibitor, was used in
TSPAN8-silenced SGC7901/DDP cells. These data indicated that TSPAN8
mediated Wnt/β-catenin pathway activation by binding to NOTCH2.
However, further studies are warranted in order to elucidate the
mechanisms through which TSPAN8 interacts with NOTCH2 in MDR. Taken
together, our study indicates that the inhibition of TSPAN8
sensitizes gastric cancer cells to chemotherapeutic drugs. However,
to obtain a more complete picture of the molecular mechanisms
involved in the regulation of the MDR of SGC7901/DDP by TSPAN8,
further studies are required in the future.
In conclusion, the present study demonstrates that
TSPAN8 impairs the sensitivity of SGC7901/DDP gastric cancer cells
to chemotherapeutic agents by mediating Wnt/β-catenin activity.
TSPAN8 also mediates β-catenin expression and accumulation by
binding to NOTCH2. This study provides novel insight for drug
designs that overcome cisplatin resistance in gastric cancer
cells.
Acknowledgments
This study was supported by the Foundation for Young
Scientists of Guizhou Provincial People's Hospital [grant no.
GZSYQN (2016) 19] and the Foundation of Health and Family Planning
Commission of Guizhou Province (grant no. GZWJKT2015-1-022).
References
|
1
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ajani JA, Ota DM, Jessup JM, Ames FC,
McBride C, Boddie A, Levin B, Jackson DE, Roh M and Hohn D:
Resectable gastric carcinoma. An evaluation of preoperative and
postoperative chemotherapy. Cancer. 68:1501–1506. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang D and Fan D: Multidrug resistance in
gastric cancer: Recent research advances and ongoing therapeutic
challenges. Expert Rev Anticancer Ther. 7:1369–1378. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kartal-Yandim M, Adan-Gokbulut A and Baran
Y: Molecular mechanisms of drug resistance and its reversal in
cancer. Crit Rev Biotechnol. 36:716–726. 2016.
|
|
6
|
Borst P: Genetic mechanisms of drug
resistance. A review Acta Oncol. 30:87–105. 1991. View Article : Google Scholar
|
|
7
|
Cui J, Jiang W, Wang S, Wang L and Xie K:
Role of Wnt/β-catenin signaling in drug resistance of pancreatic
cancer. Curr Pharm Des :. 2464–2471. 2012. View Article : Google Scholar
|
|
8
|
Abdi J, Chen G and Chang H: Drug
resistance in multiple myeloma: Latest findings and new concepts on
molecular mechanisms. Oncotarget. 4:2186–2207. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang Z, Guo L, Liu D, Sun L, Chen H, Deng
Q, Liu Y, Yu M, Ma Y, Guo N, et al: Acquisition of resistance to
trastuzumab in gastric cancer cells is associated with activation
of IL-6/STAT3/ Jagged-1/Notch positive feedback loop. Oncotarget.
6:5072–5087. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
McAuliffe SM, Morgan SL, Wyant GA, Tran
LT, Muto KW, Chen YS, Chin KT, Partridge JC, Poole BB, Cheng KH, et
al: Targeting Notch, a key pathway for ovarian cancer stem cells,
sensitizes tumors to platinum therapy. Proc Natl Acad Sci USA.
109:E2939–E2948. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ren T, Lin S, Wang Z and Shang A:
Differential proteomics analysis of low- and high-grade of
astrocytoma using iTRAQ quantification. Onco Targets Ther.
9:5883–5895. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Berthier-Vergnes O, El Kharbili M, de la
Fouchardière A, Pointecouteau T, Verrando P, Wierinckx A, Lachuer
J, Le Naour F and Lamartine J: Gene expression profiles of human
melanoma cells with different invasive potential reveal TSPAN8 as a
novel mediator of invasion. Br J Cancer. 104:155–165. 2011.
View Article : Google Scholar :
|
|
13
|
Yue S, Mu W, Erb U and Zöller M: The
tetraspanins CD151 and Tspan8 are essential exosome components for
the crosstalk between cancer initiating cells and their
surrounding. Oncotarget. 6:2366–2384. 2015. View Article : Google Scholar :
|
|
14
|
Kanetaka K, Sakamoto M, Yamamoto Y,
Yamasaki S, Lanza F, Kanematsu T and Hirohashi S: Overexpression of
tetraspanin CO-029 in hepatocellular carcinoma. J Hepatol.
35:637–642. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gesierich S, Paret C, Hildebrand D, Weitz
J, Zgraggen K, Schmitz-Winnenthal F H, Horejsi V, Yoshie O, Herlyn
D, Ashman LK, et al: Colocalization of the tetraspanins, CO-029 and
CD151, with integrins in human pancreatic adenocarcinoma: Impact on
cell motility. Clin Cancer Res. 11:2840–2852. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hemler ME: Tetraspanin functions and
associated microdomains. Nat Rev Mol Cell Biol. 6:801–811. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yáñez-Mó M, Barreiro O, Gordon-Alonso M,
Sala-Valdés M and Sánchez-Madrid F: Tetraspanin-enriched
microdomains: A functional unit in cell plasma membranes. Trends
Cell Biol. 19:434–446. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zöller M: Tetraspanins: Push and pull in
suppressing and promoting metastasis. Nat Rev Cancer. 9:40–55.
2009. View
Article : Google Scholar
|
|
19
|
Nazarenko I, Rana S, Baumann A, McAlear J,
Hellwig A, Trendelenburg M, Lochnit G, Preissner KT and Zöller M:
Cell surface tetraspanin Tspan8 contributes to molecular pathways
of exosome-induced endothelial cell activation. Cancer Res.
70:1668–1678. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wei L, Li Y and Suo Z: TSPAN8 promotes
gastric cancer growth and metastasis via ERK MAPK pathway. Int J
Clin Exp Med. 8:8599–8607. 2015.PubMed/NCBI
|
|
21
|
Mao ZL, He SB, Sheng WH, Dong XQ and Yang
JC: Adenovirusmediated ING4 expression reduces multidrug resistance
of human gastric carcinoma cells in vitro and in vivo. Oncol Rep.
30:2187–2194. 2013.PubMed/NCBI
|
|
22
|
Soothill JS, Ward R and Girling AJ: The
IC50: An exactly defined measure of antibiotic sensitivity. J
Antimicrob Chemother. 29:137–139. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
24
|
Mi H, Muruganujan A and Thomas PD: PANTHER
in 2013: Modeling the evolution of gene function, and other gene
attributes, in the context of phylogenetic trees. Nucleic Acids
Res. 41:D377–D386. 2013. View Article : Google Scholar :
|
|
25
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar
|
|
26
|
Gan CS, Chong PK, Pham TK and Wright PC:
Technical, experimental, and biological variations in isobaric tags
for relative and absolute quantitation (iTRAQ). J Proteome Res.
6:821–827. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gong J, Shen S, Yang Y, Qin S, Huang L,
Zhang H, Chen L, Chen Y, Li S, She S, et al: Inhibition of FASN
suppresses migration, invasion and growth in hepatoma carcinoma
cells by deregulating the HIF-1α/IGFBP1 pathway. Int J Oncol.
50:883–892. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mitra A, Mishra L and Li S: EMT, CTCs and
CSCs in tumor relapse and drug-resistance. Oncotarget.
6:10697–10711. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Clements WM, Wang J, Sarnaik A, Kim OJ,
MacDonald J, Fenoglio-Preiser C, Groden J and Lowy AM: β-Catenin
mutation is a frequent cause of Wnt pathway activation in gastric
cancer. Cancer Res. 62:3503–3506. 2002.PubMed/NCBI
|
|
30
|
Ma Y, Ren Y, Han EQ, Li H, Chen D, Jacobs
JJ, Gitelis S, O'Keefe RJ, Konttinen YT, Yin G, et al: Inhibition
of the Wnt-β-catenin and Notch signaling pathways sensitizes
osteosarcoma cells to chemotherapy. Biochem Biophys Res Commun.
431:274–279. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hartgrink HH, Jansen EPM, van Grieken NCT
and van de Velde CJ: Gastric cancer. Lancet. 374:477–490. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Loomis D, Huang W and Chen G: The
International Agency for Research on Cancer (IARC) evaluation of
the carcinogenicity of outdoor air pollution: Focus on China. Chin
J Cancer. 33:189–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wagner AD, Grothe W, Haerting J, Kleber G,
Grothey A and Fleig WE: Chemotherapy in advanced gastric cancer: A
systematic review and meta-analysis based on aggregate data. J Clin
Oncol. 24:2903–2909. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim DY, Kim JH, Lee SH, Kim TY, Heo DS,
Bang YJ and Kim NK: Phase II study of oxaliplatin, 5-fluorouracil
and leucovorin in previously platinum-treated patients with
advanced gastric cancer. Ann Oncol. 14:383–387. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shen DW, Pouliot LM, Hall MD and Gottesman
MM: Cisplatin resistance: A cellular self-defense mechanism
resulting from multiple epigenetic and genetic changes. Pharmacol
Rev. 64:706–721. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Krishna R and Mayer LD: Multidrug
resistance (MDR) in cancer. Mechanisms, reversal using modulators
of MDR and the role of MDR modulators in influencing the
pharmacokinetics of anticancer drugs Eur J Pharm Sci. 11:265–283.
2000.
|
|
37
|
Endo K, Maehara Y, Kusumoto T, Ichiyoshi
Y, Kuwano M and Sugimachi K: Expression of
multidrug-resistance-associated protein (MRP) and chemosensitivity
in human gastric cancer. Int J Cancer. 68:372–377. 1996.
View Article : Google Scholar
|
|
38
|
Yang YX, Xiao ZQ, Chen ZC, Zhang GY, Yi H
and Zhang PF: Proteome analysis of multidrug resistance in
vincristine-resistant human gastric cancer cell line SGC7901/ VCR.
Proteomics. 6:2009–2021. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
McCubrey JA, Lertpiriyapong K, Fitzgerald
TL, Martelli AM, Cocco L, Rakus D, Gizak A, Libra M, Cervello M,
Montalto G, et al: Roles of TP53 in determining therapeutic
sensitivity, growth, cellular senescence, invasion and metastasis.
Adv Biol Regul. 63:32–48. 2017. View Article : Google Scholar
|
|
40
|
Sheng Y, Ng CP, Lourie R, Shah ET, He Y,
Wong KY, Seim I, Oancea I, Morais C, Jeffery PL, et al: MUC13
overexpression in renal cell carcinoma plays a central role in
tumor progression and drug resistance. Int J Cancer. 140:2351–2363.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang J, Zhang Y, Liu T, Guo CH and Wan YF:
Reversal effect of inhibition of HSP90 activity on adriamycin
resistance of human hepatocellular carcinoma HepG2/ADR cells.
Tumor. 36:414–423. 2016.
|
|
42
|
Ren YP, Song C and Meng QK: Expression of
Tspan8 in colon cancer cell lines and effects of siRNA-mediated
Tspan8 gene silencing on the cell function of SW620. Zhongguo Yike
Daxue Xuebao. 44:34–37. 2015.In Chinese.
|
|
43
|
Castillo V, Valenzuela R, Huidobro C,
Contreras HR and Castellon EA: Functional characteristics of cancer
stem cells and their role in drug resistance of prostate cancer.
Int J Oncol. 45:985–994. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Niimi S, Nakagawa K, Yokota J, Tsunokawa
Y, Nishio K, Terashima Y, Shibuya M, Terada M and Saijo N:
Resistance to anticancer drugs in NIH3T3 cells transfected with
c-myc and/or c-H-ras genes. Br J Cancer. 63:237–241. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hayward P, Brennan K, Sanders P, Balayo T,
DasGupta R, Perrimon N and Martinez Arias A: Notch modulates Wnt
signalling by associating with Armadillo/β-catenin and regulating
its transcriptional activity. Development. 132:1819–1830. 2005.
View Article : Google Scholar : PubMed/NCBI
|