Introduction
Approximately 15% of breast cancers are classified
as triple-negative breast cancer (TNBC), lacking expression of the
estrogen receptor (ER) and the progesterone receptor (PR), and
which is characterized by absence or low expression or no
amplification on the gene level, of the human epidermal growth
factor receptor 2 (HER2) (1,2).
Thus, unfortunately, breast cancer patients suffering from TNBC do
not benefit from cancer therapeutics targeting these receptors.
Gene expression profiling revealed that TNBC shows overlapping
characteristics with the basal-like breast cancer type, including
various subtypes. Patients afflicted with this malignancy are known
for early disease onset with high aggressiveness, poor clinical
outcome and high nuclear grade (3–9).
Often, TNBCs show BRCAness, characterized by clinicopathological
features normally found with BRCA1-mutated tumors (10–14).
One has to admit that little progress has been made
in the last decade regarding novel, suitable druggable targets and
targeted drugs for TNBC patients (15), subsequently, chemotherapy remains
the essential therapeutic tool in TNBC, both in the adjuvant and
the neoadjuvant setting (3,13,15,16).
Clinical data do suggest that the addition of platinum to
anthracycline- and taxane-based chemotherapy regimens is an
additional option in the treatment of both early-stage and advanced
TNBC (4,17–25).
Breast cancer patients who are undergoing
chemotherapy have an increased risk of developing cardiovascular
complications, and anthracyclines (e.g. doxorubicin, daunorubicin,
idarubicin and epirubicin), are some of the most frequently used
agents. The administration of non-anthracycline agents, that also
may cause cardiotoxicity, frequently results in synergistic
toxicity when anthracyclines are given concurrently (26,27).
Therefore, the identification of additional molecular biomarkers to
predict response and/or potential cytotoxic side-effects to
specific chemotherapeutics is still of high unmet medical need to
further improve strategies to treat TNBC patients (3,7,19).
Epigenetic DNA-methylation plays an important role
in controlling gene activity and nucleus architecture (28–31).
DNA-methylation markers were shown to have prognostic and/or
predictive value, thus, being considered valuable, additive tools
for physicians to choose the appropriate therapy regimen for the
cancer patient (32–34). The PITX2 gene (paired-like
homeodomain transcription factor 2), a member of the paired-like
homeodomain transcription factor family, which in the healthy
organism is known to play an important role during embryogenesis
and organogenesis, might serve as a prime example (35,36).
Recent data strongly suggest that methylation of
certain CpG island promoters of the PITX2 gene may play an
essential role in the very early stages of breast cancer
pathogenesis and its methylation status being associated with
response to adjuvant chemotherapy of a breast cancer patient
(37–42). Thus, unexpectedly, in breast cancer
patients, PITX2 emerged to be a key molecule in breast cancer
pathophysiology, but not only associated with the course of the
disease but also with response to adjuvant systemic endocrine or
anthracycline-based chemotherapy (39–41).
The aim of this retrospective pilot study was to
demonstrate that PITX2 DNA-methylation is a potential predictive
breast cancer biomarker in the triple-negative breast cancer
subgroup (TNBC), treated with adjuvant anthracycline-based
chemotherapy regimens. Our present results, for the first time
indicate that quantitative determination of the PITX2
DNA-methylation status in primary TNBC breast cancer tissues will
allow selection of those TNBC patients who most probably will
benefit from anthracycline-based chemotherapy or not. Consequently,
TNBC patients who possibly will not respond should be spared the
potentially toxic burden of such chemotherapy, but could be
allocated to alternative treatment modalities (23,43–45).
Materials and methods
Materials
Unless otherwise stated, all the reagents applied in
the present study were obtained from Qiagen (Hilden, Germany),
Sigma-Aldrich (Taufkirchen, Germany), or Merck KGaA (Darmstadt,
Germany).
Patients
Inclusion criteria for the retrospective study were
breast cancer patients with histologically confirmed invasive
triple-negative breast cancer (n=56), no signs of distant
metastasis at time of diagnosis, availability of frozen tumor
tissue specimens for DNA extraction, follow-up data and signed
informed patient consent. All patients were treated between 1991
and 2006 at the Department of Obstetrics and Gynecology, Klinikum
rechts der Isar, Technical University of Munich, Munich, Germany.
Study approval was obtained from the Ethics Committee of the
Medical Faculty of the Technical University of Munich. Clinical and
histomorphological patient-related data are summarized in Table I. Histopathologic tumor grade was
determined according to the Nottingham modification of the
Scarff-Bloom-Richardson grading scheme. Absence of estrogen
receptor (ER) and/or progesterone receptor (PR) protein expression
was confirmed either by the dextran-coated charcoal method, by
enzyme immunoassay, or immunohistochemistry, whereby positive
staining of either receptor denoted receptor positivity. Presence
of human epidermal growth factor receptor 2 (HER2)
expression/amplification was demonstrated by immunohistochemistry
using a semi-quantitative scoring system or by fluorescent in
situ hybridization analysis (40). Twenty patients were younger than 50
years at the time of diagnosis. Median time of follow-up was 74
months (range, 8–179). Fifteen patients were treated with breast
conserving therapy, 41 patients with mastectomy and 51 patients
received radiotherapy. TNBC patients were allocated to various
types of adjuvant anthracycline-based polychemotherapy regimens
including FEC (n=19), EC (n=13); EC+CMF (n=3), anthracycline plus
taxane (n=20), or idarubicin-based therapeutics (n=1).
 | Table IClinical and histomorphological
characteristics of TNBC patients treated with adjuvant
anthracycline-based chemotherapy. |
Table I
Clinical and histomorphological
characteristics of TNBC patients treated with adjuvant
anthracycline-based chemotherapy.
|
Characteristics | (n=56) (%) |
|---|
| Age at time of
diagnosis (years) | |
| <50 | 20 (35.7) |
| ≥50 | 36 (64.3) |
| Type of
surgery | |
| Mastectomy | 15 (26.8) |
| Breast
conserving | 41 (73.2) |
| Tumor size
(cm) | |
| ≤2 | 18 (32.1) |
| >2 | 36 (64.3) |
| Not available | 2 (3.6) |
| Histological
subtype | |
| Invasive
ductal | 42 (75.0) |
| Others | 14 (25.0) |
| Tumor grade | |
| G2 | 4 (7.1) |
| G3 | 50 (89.3) |
| Not available | 2 (3.6) |
| Nodal status | |
| Negative | 27 (48.2) |
| Positive | 28 (50.0) |
| Not available | 1 (1.8) |
| Radiotherapy | |
| Yes | 51 (91.1) |
| No | 5 (8.9) |
| Adjuvant
chemotherapy | |
| FEC | 19 (33.9) |
| EC | 13 (23.2) |
| Anthracycline plus
taxane | 20 (35.7) |
| EC plus CMF | 3 (5.4) |
| Idarubicin-based
therapeutics | 1 (1.8) |
| Disease
recurrence | |
| Yes | 18 (32.1) |
| No | 38 (67.9) |
| Deceased | |
| Yes | 16 (28.6) |
| No | 40 (71.4) |
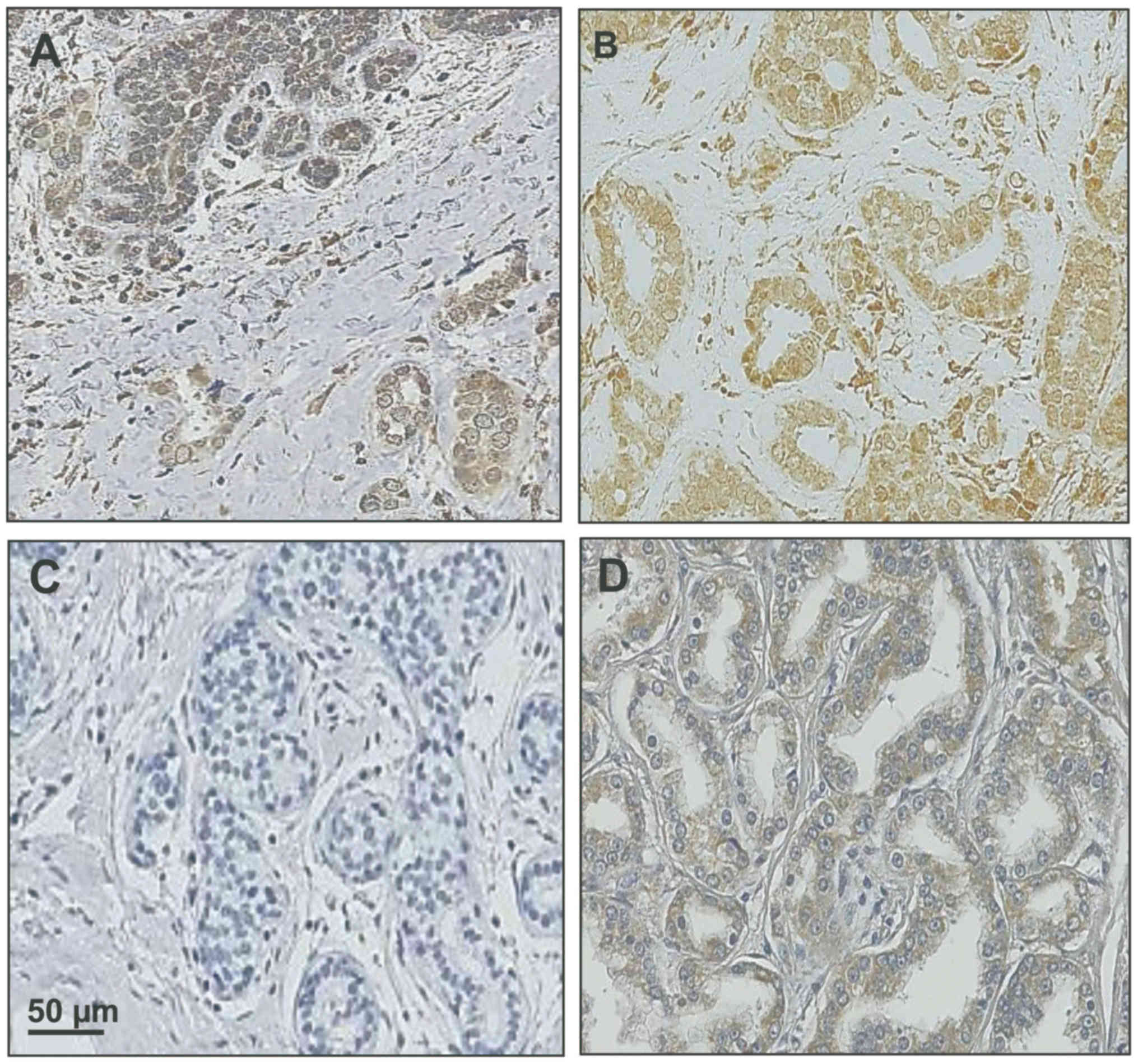
Immunohistochemistry
2–4 µm thick sections were cut from FFPE
blocks of TNBC breast cancer patients and mounted on microscope
slides (R. Langenbrinck GmbH, Emmendingen, Germany). Sections were
deparaffinized by xylene (2 × 10 min) and then rehydrated in a
series of graded ethanol, followed by washing in TBS, pH 7.6, 5
min, as previously described (46). All steps were performed at room
temperature. Antigen retrieval was accomplished by exposing the
slides to 4 min of pressure cooking (WMF, Geislingen an der Steige,
Germany). Since a peroxidase-dependent antibody-binding system was
applied (Dako EnVision + Dual Link System; Dako Deutschland GmbH,
Hamburg, Germany), endogenous peroxidase activities were blocked by
use of the peroxidase/alkaline phosphatase blocking reagent (Dako
Deutschland GmbH). For immunohistochemical staining of PITX2
protein expressed in tumor tissues, the polyclonal rabbit antibody
PITX2-484 to the human PITX2 molecule was added (1:50) in antibody
diluent (Dako Deutschland GmbH). Sections were stored overnight at
4°C to allow solid interaction of antibody PITX2-484 with its
target molecule PITX2 expressed in the tumor sections. After
washing with TBS, the secondary horseradish peroxidase-conjugated
polymer antibody to the Fc-region of rabbit immunoglobulin G was
added according to the manufacturer's recommendation (30 min, room
temperature). After another washing step with TBS, the peroxidase
detection solution (3,3′-diaminobenzidine; Dako Deutschland GmbH)
was added (8 min, room temperature). The sections were then washed
with TBS, nuclei of the tissue sections counterstained, and then
sealed with Pertex embedding medium as previously described
(46). Stained sections were
scanned and digitized using the NanoZoomer Digital Pathology RS
(NDP) scanner (Hamamatsu Photonics Deutschland GmbH, Herrsching am
Ammersee, Germany), utilizing the NDP scan 2.2 software. A
selection of TNBC primary tumors ± hematoxylin counterstain plus a
prostate cancer specimen for comparison are shown in Fig. 1.
Generation of PITX2-directed antibody
PITX2-484
Polyclonal antibody PITX2-484 was produced by Pineda
Antibody Service (Berlin, Germany), after two PITX2-peptides were
selected common for all known three PITX2 variants, by generation
in rabbits after combined immunization with the PITX2-derived
peptides Y (aa 154-170: NGFGPQFNGLMQPYDDM) and Z (aa 243-260:
NNLNNLSSPSLNSAVPTP). Peptides Y and Z relate to PITX2-B, which is
referred to by UniProt as canonical sequence of PITX2. These
peptides were synthesized plus an additional N-terminal Cys-residue
which was used for S-S-based linkage to the carrier protein KLH.
Sera of the immunized rabbits were purified by affinity
chromatography on vIDR-pHis (aa 153-261 of PITX2B).
DNA extraction
Immediately after excision of the primary breast
tumor tissue at the Department of Obstetrics and Gynecology,
Klinikum rechts der Isar, Technical University of Munich, Germany,
the removed tissues were placed on ice and transported to the
university's nearby pathologist to examine the removed tissue for
the presence of malignant cells. Approved malignant tissue was
snap-frozen and stored in the liquid nitrogen tumor bank of the
Klinikum rechts der Isar of the Technical University of Munich
until further use. On demand, tissue was removed from the liquid
nitrogen storage container and the still-frozen tumor tissue
pulverized by use of the Mikro-Dismembrator S (Sartorius Stedim
Biotech, Göttingen, Germany), the powder was then suspended in
Tris-buffered saline (0.02 M Tris-HCl/0.125 M NaCl, pH 8.5)
containing 0.1% of the non-ionic detergent Triton X-100, to be
centrifuged at 100,000 × g (60 min, 4°C) (47). The supernatant and the cellular
debris, containing the DNA-containing nuclei, were aliquoted
separately and stored in liquid nitrogen until further use.
Aliquots of cellular debris representing ~30 mg of breast cancer
tissue were used for DNA extraction by following the QIAamp DNA
Mini and Blood Mini Handbook protocol, employing the semi-automated
QIAcube system (Qiagen, Hilden, Germany). Extracted genomic DNA was
aliquoted and stored at −80°C until further use. DNA concentration
was determined by use of the NanoDrop 2000c spectrophotometer
(Thermo Fisher Scientific, Wilmington, DE, USA). Breast cancer cell
lines MCF-7 and MDA-MB-231 (CLS Cell Lines Service GmbH, Eppelheim,
Germany), genomic unconverted DNA, and water as no template
control, as well as fully methylated bisulfite-converted DNA
(EpiTect PCR Control DNA Set; Qiagen) served as controls for PITX2
DNA-methylation status.
PITX2-probe and primer system
specifications (according to patent EP1561821)
Entrez gene ID: 5308. Amplicon length 144. Reference
sequence (RefSeq) ID: NT_016354.18. Detected CpG in RefSeq: 3 CpG
in 36106573 - 36106600 (39).
DNA-methylation-specific quantitative
real-time PCR
For PITX2 DNA-methylation status determination, for
each specimen, 310 ng of DNA was applied in the subsequent
bisulfite conversion step which was performed following the EpiTect
Bisulfite Handbook protocol (Qiagen) employing an ABI PCR Cycler
(Applied Biosystems, Darmstadt, Germany). Program details: 1st: 5
min at 99°C. 2nd: 25 min at 60°C. 3rd: 5 min at 99°C. 4th: 85 min
at 60°C. 5th: 5 min at 99°C. 6th: 175 min at 60°C. Clean-up of the
bisulfite-converted DNA was carried out following the EpiTect
Bisulfite kit protocol. Primers and probes for the methylated and
unmethylated PITX2 DNA-methylation status were applied in a duplex
probe system combined in a 10× ready to use primer and probe Master
Mix; qPCR was performed according to the provider protocol (EpiTect
MethyLight Assay Hs_PITX2; Qiagen) using the ABI 7000 Taqman system
(Applied Biosystems). Run details: 1st: 15 min at 95°C. 2nd: 48
cycles comprising of each 15 sec at 95°C and of 1 min at 60°C,
including 2 µl primer and probe Master Mix, 2 µl
bisulfite converted DNA (7.5 ng) and 10 µl QuantiTect 2×
QPCR Master Mix (Qiagen), supplemented with water to a final volume
of 20 µl. Each specimen was assessed in triplicates. A total
of 5 ng fully methylated bisulfite-converted human control DNA
(Qiagen) and 7.75 ng MCF-7 bisulfite-converted DNA served as
positive controls, RNAse-free water as the negative control.
Statistics
Reporting of this study was carried out respecting
the REMARK criteria (48,49). For calculation of the PITX2
DNA-methylation status, the modified ΔΔCT-method as described by
Harbeck et al (40) was
employed. Mean values of triplicates were calculated for the
methylated and the unmethylated PITX2 DNA-methylation status,
respectively, which were then used for calculation of the
individual PITX2 DNA-methylation scores. CT-values (methylated or
unmethylated) obtained with >38 cycles were disregarded. Mean
values, standard deviation, and coefficient of variation of the
different qPCR runs were calculated. Only values with a coefficient
of variation <0.3 were considered for the statistical
evaluation. The relationship between PITX2 DNA-methylation score
and established clinical factors to the primary endpoints
disease-free (DFS) and overall survival (OS) was calculated
applying univariate and multivariate Cox proportional hazard
models. The date of surgery was considered as the follow-up index
date. In order to discriminate between low- and high-risk patients
with regards to DFS and OS, optimized cut-off values were
calculated with the 'maximum-selected log-rank statistic' using the
maxstat.test function as implemented from the program library
'maxstat' of the program 'R' (R Development Core Team 2012)
(50,51). Death before incidence of distant
recurrence was considered censoring event. Survival curves were
calculated according to the Kaplan-Meier method (40). The log-rank test was used for
calculating the respective P-values. Cox regression models were
employed for univariate risk estimation (hazard ratios, HR) for DFS
and OS. Due to the limited numbers of events (disease recurrence,
deaths), multivariate analyses were carried out in an exploratory
fashion. For this, covariates (tumor size, tumor grading and age)
were added stepwise to the variable PITX2 DNA-methylation (high vs.
low) and the according hazard ratios and 95% confidence intervals
depicted in forest plot diagrams in order to test whether the PITX2
DNA-methylation status adds statistically independent additional
information to DFS and OS.
Results
In breast cancer, the methylation status of the
PITX2 gene has previously been shown to be a clinically
relevant outcome predictor in early stage breast cancer patients,
either treated with endocrine (tamoxifen) therapy or
anthracycline-containing chemotherapy (39–41).
Applying the same technical approach as described in these
publications, we now for the first time present data demonstrating
the additional potential clinical utility of PITX2 DNA-methylation
as a suitable biomarker to predict response to
anthracycline-containing adjuvant chemotherapy in the aggressive
subgroup of triple-negative breast cancer (TNBC) patients.
PITX2 DNA-methylation as a predictive
candidate biomarker in TNBC patients treated with
anthracycline-based adjuvant chemotherapy
Rational
An initial study investigating high-risk breast
cancer patients with pN>1 (>3 affected lymph nodes) showed a
clinical significance of PITX2 DNA-methylation assessment to
predict response to anthracycline-containing chemotherapy (41). For this quantitative assessment of
the methylation status of the PITX2 gene in primary tumor tissue
samples of breast cancer patients, a quantitative
methylation-specific duplex-probe qPCR system was developed, as a
follow-up technology to the microarray-based screening system
described in the study by Maier et al (39) and tested for reliability both for
ER- and PR-positive breast cancer FFPE samples (40).
With this qPCR test system, valid PITX2
DNA-methylation scores were obtained for all of the 56 TNBC
patients included in this explorative biomarker study. The median
PITX2 DNA-methylation score was 10.05. An optimized cut-off value
of 6.35 percent methylation ratio (PMR) for clinical impact on DFS
and OS was established with the 'maximum-selected log-rank
statistic' using the maxstat.test function as implemented from the
program library 'maxstat' of the program 'R'. To analyze whether
PITX2 DNA-methylation might constitute a predictive marker in TNBC,
at first, univariate Cox regression analyses were performed for DFS
and OS, then multivariate Cox regression analyses were carried out
for DFS and OS by stepwise inclusion of the established
clinicopathological factors age, tumor grading, tumor size and
nodal status, as covariates for testing whether PITX2
DNA-methylation (<6.35 vs. ≥6.35%) constitutes a statistically
significant independent variable.
PIT X2 DNA-methylation assay
stability
PITX2 DNA-methylation assay stability and
reproducibility were demonstrated by qPCR through gradual and
serial dilution series of 7.75 ng bisulfite-converted DNA obtained
from the MCF-7 breast cancer cell line and two breast cancer
tissues. The low coefficients of variation (CV ≤0.06) obtained
indicate that even with input amounts of bisulfite-converted DNA as
low as 1 ng into the qPCR reaction, PITX2 DNA-methylation scores
can be determined reliably. For quality assurance and validation of
the different qPCR runs, various PITX2 expression-positive and
negative controls were included in each qPCR run [DNA extracted
from tumor cell line MCF7, genomic unconverted DNA, fully
methylated bisulfite converted DNA (EpiTect Control DNA Set;
Qiagen), no-template control (water)]. The resulting low CVs
(≤0.08) evidence that stable scores could be obtained throughout
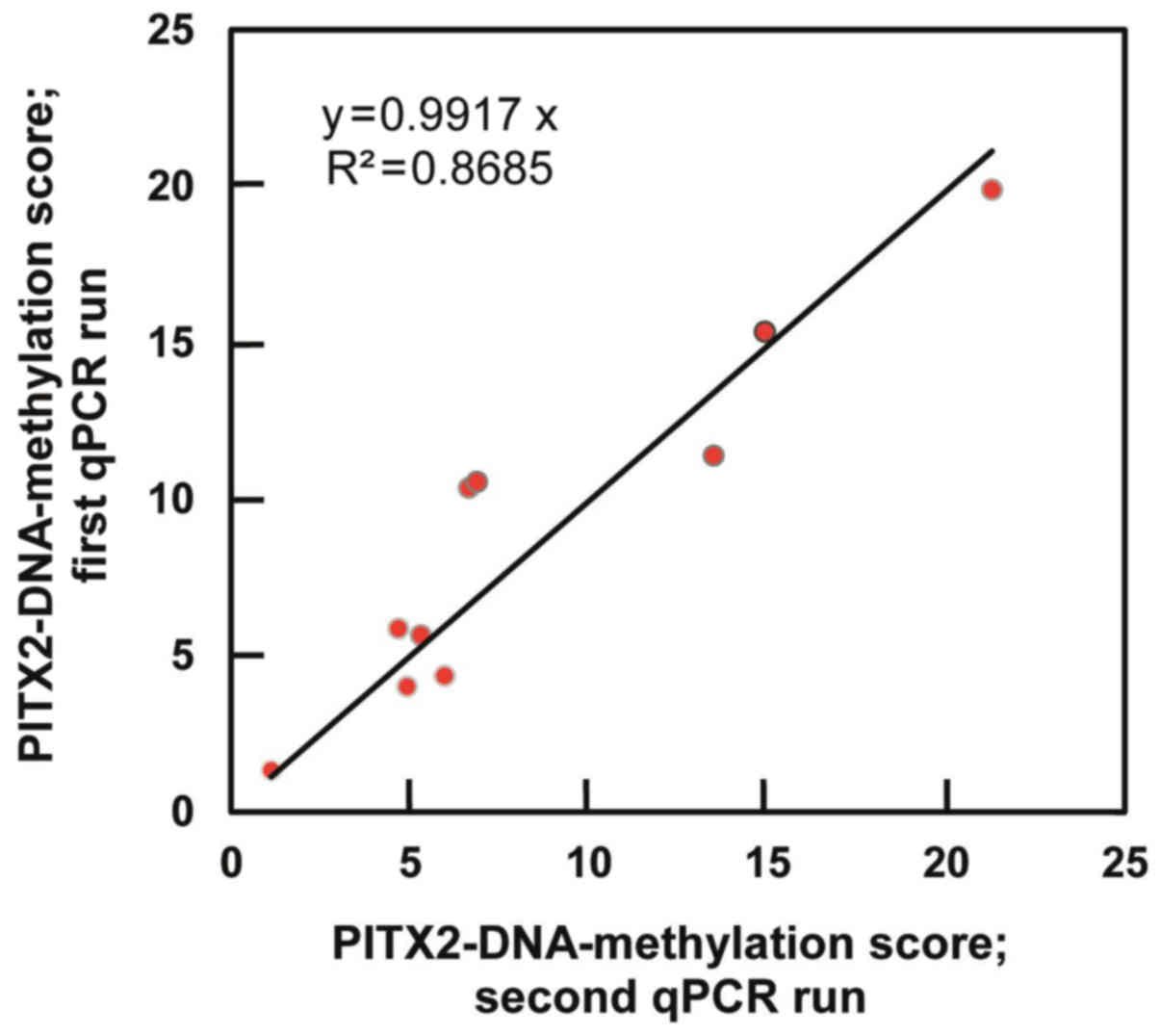
the different qPCR runs. Comparison of the DNA-methylation scores
of ten randomly chosen triple-negative breast cancer tumor tissue
specimens, which were processed in two different qPCR runs,
revealed a low median CV of 0.10 between the calculated PMR values
of two runs with high correlation of results (r>0.93) (Fig. 2).
Allocation of PITX2 DNA-methylation
scores (low vs. high) to various anthracycline-based chemotherapy
regimens
Correlation with clinical outcome
The TNBC patients included in our explorative
clinical study were not stratified for adjuvant anthracycline-based
chemotherapy by respecting their PITX2 DNA-methylation status, but
treated with anthracycline-containing adjuvant chemotherapy
following the actual German AGO (Arbeitsgemeinschaft
Gynaekologischer Onkologen) guidelines effective at the time of
treatment (3).
Connecting clinical outcome (DFS and OS) of the 56
TNBC-patients with their treatment modalities and their respective
PITX2 DNA-methylation status, we noted that the patients with low
PITX2 DNA-methylation scores <6.35 experienced a poor clinical
outcome for both DFS and OS (Table
II). Remarkably, this was not only true for the EC and FEC
groups, but also for the group of TNBC patients treated with
anthracycline-based chemotherapy followed by taxanes. The TNBC
patients allocated to the low-methylation group (n=17) thus
experienced ten of the disease recurrences and eight of the
deaths.
 | Table IIAllocation of PITX2 DNA-methylation
scores (low versus high) to various anthracycline-based
chemotherapy regimens. |
Table II
Allocation of PITX2 DNA-methylation
scores (low versus high) to various anthracycline-based
chemotherapy regimens.
| A, Allocations of
PITX2 DNA-methylation scores (low) to various anthracycline-based
chemotherapy regimens. |
|---|
|
|---|
Adjuvant
chemotherapy schedule PITX2 DNA-methylation PMR values listed
| Events Low PITX2
(PMR <6.35)
|
|---|
| Patient | FEC | EC | FEC + CMF | Anthracycline +
taxane | Other combination
of anthracycline-based therapeutics | Disease recurrence
(months) | Death (months) |
|---|
| 1 | 1.56 | | | | | 22 | 24 |
| 2 | 1.78 | | | | | | |
| 3 | 4.11 | | | | | 18 | 26 |
| 4 | 4.31 | | | | | 12 | 14 |
| 5 | 4.53 | | | | | | |
| 6 | | 2.30 | | | | | |
| 7 | | 2.38 | | | | 18 | 20 |
| 8 | | 2.72 | | | | | |
| 9 | | 5.00 | | | | 21 | 31 |
| 10 | | 6.12 | | | | 27 | |
| 11 | | | 3.65 | | | | |
| 12 | | | | 2.39 | | 29 | |
| 13 | | | | 3.52 | | | |
| 14 | | | | 3.81 | | 34 | |
| 15 | | | | 5.09 | | 5 | 8 |
| 16 | | | | 5.52 | | 4 | 15 |
| 17 | | | | | 1.19 | | 10 |
| B, Allocations of
PITX2 DNA-methylation scores (high) to various anthracycline-based
chemotherapy regimens. |
|---|
|
|---|
Adjuvant
chemotherapy schedule PITX2 DNA-methylation PMR values listed
| Events High PITX2
(PMR ≥6.35)
|
|---|
| Patient | FEC | EC | FEC + CMF | Anthracycline +
taxane | Disease recurrence
(months) | Death (months) |
|---|
| 18 | 6.95 | | | | 31 | 42 |
| 19 | 7.70 | | | | | |
| 20 | 8.70 | | | | | |
| 21 | 9.17 | | | | | |
| 22 | 10.05 | | | | | |
| 23 | 10.53 | | | | 9 | 10 |
| 24 | 12.44 | | | | | |
| 25 | 20.22 | | | | 72 | |
| 26 | 22.59 | | | | | |
| 27 | 26.11 | | | | | |
| 28 | 40.52 | | | | | |
| 29 | 60.83 | | | | 59 | |
| 30 | 61.78 | | | | | |
| 31 | 63.65 | | | | | 60 |
| 32 | | 6.35 | | | 13 | 13 |
| 33 | | 6.52 | | | | |
| 34 | | 11.96 | | | | |
| 35 | | 20.59 | | | | |
| 36 | | 21.68 | | | | |
| 37 | | 29.10 | | | | |
| 38 | | 40.85 | | | | |
| 39 | | 47.45 | | | | 49 |
| 40 | | | 7.57 | | | |
| 41 | | | 15.14 | | | |
| 42 | | | | 6.64 | | |
| 43 | | | | 8.06 | | |
| 44 | | | | 8.72 | | |
| 45 | | | | 8.78 | | |
| 46 | | | | 9.33 | | |
| 47 | | | | 11.20 | | |
| 48 | | | | 13.18 | | |
| 49 | | | | 13.19 | | |
| 50 | | | | 14.10 | 27 | 30 |
| 51 | | | | 15.18 | | |
| 52 | | | | 16.91 | | |
| 53 | | | | 25.60 | 29 | 35 |
| 54 | | | | 38.36 | 28 | 43 |
| 55 | | | | 44.10 | | |
| 56 | | | | 59.47 | | |
| C, Allocation of
TNBC patients to treatment groups in relation to DFS and OS
events. |
|---|
|
|---|
| Patients (n) | Adjuvant
treatment | PITX2
DNA-methylation status (Cut-off: 6.35 PMR) | DFS: Events
(%) | OS: Events (%) |
|---|
| 12 | Anthracycline, no
taxane | Low | 6 (50) | 6 (50) |
| 5 | Anthracycline plus
taxane | Low | 4 (80) | 2 (40) |
| 24 | Anthracycline, no
taxane | High | 5 (20.8) | 5 (20.8) |
| 15 | Anthracycline plus
taxane | High | 3 (20) | 3 (20) |
In contrast, in the high-methylation group
(methylation score ≥6.35; n=39), only eight disease recurrences and
eight of the deaths were noted in this larger group of patients,
reflecting a better clinical outcome of the anthracycline-treated
TNBC patients, for both DFS and OS, independent of the different
anthracycline-based chemotherapy regimens.
Univariate and multivariate Cox
regression analyses to assess the clinical impact of PITX2 DNA
methylation status to predict response of TNBC patients to adjuvant
anthracycline-based chemotherapy
The 'maxstat.test' R-function was applied in order
to search for an optimized PITX2 DNA-methylation cut-off value to
distinguish TNBC patients who will respond to anthracycline-based
adjuvant chemotherapy from those who will not. Using this
statistical test which already accounts for multiple testing, a
cut-off value of 6.35 (percentage methylation rate) was defined in
order to estimate and graphically display empirical survival
tendencies for DFS and OS.
Both by univariate and multivariate Cox regression
analyses, the PITX2 DNA-methylation status was found to contribute
significant information regarding DFS and OS (Table III). Univariate analysis: DFS:
HR, 5.36, 95% CI, 2.06–13.95; OS: HR, 3.78, 95% CI, 1.40–10.20.
Multivariate analysis: DFS: HR, 6.40, 95% CI, 1.96–20.88; OS: HR,
3.62, 95% CI, 1.03–12.72. Concerning the established prognostic
factors, only age contributed statistically significant but weak
information as assessed by univariate analysis of OS (HR, 1.05, 95%
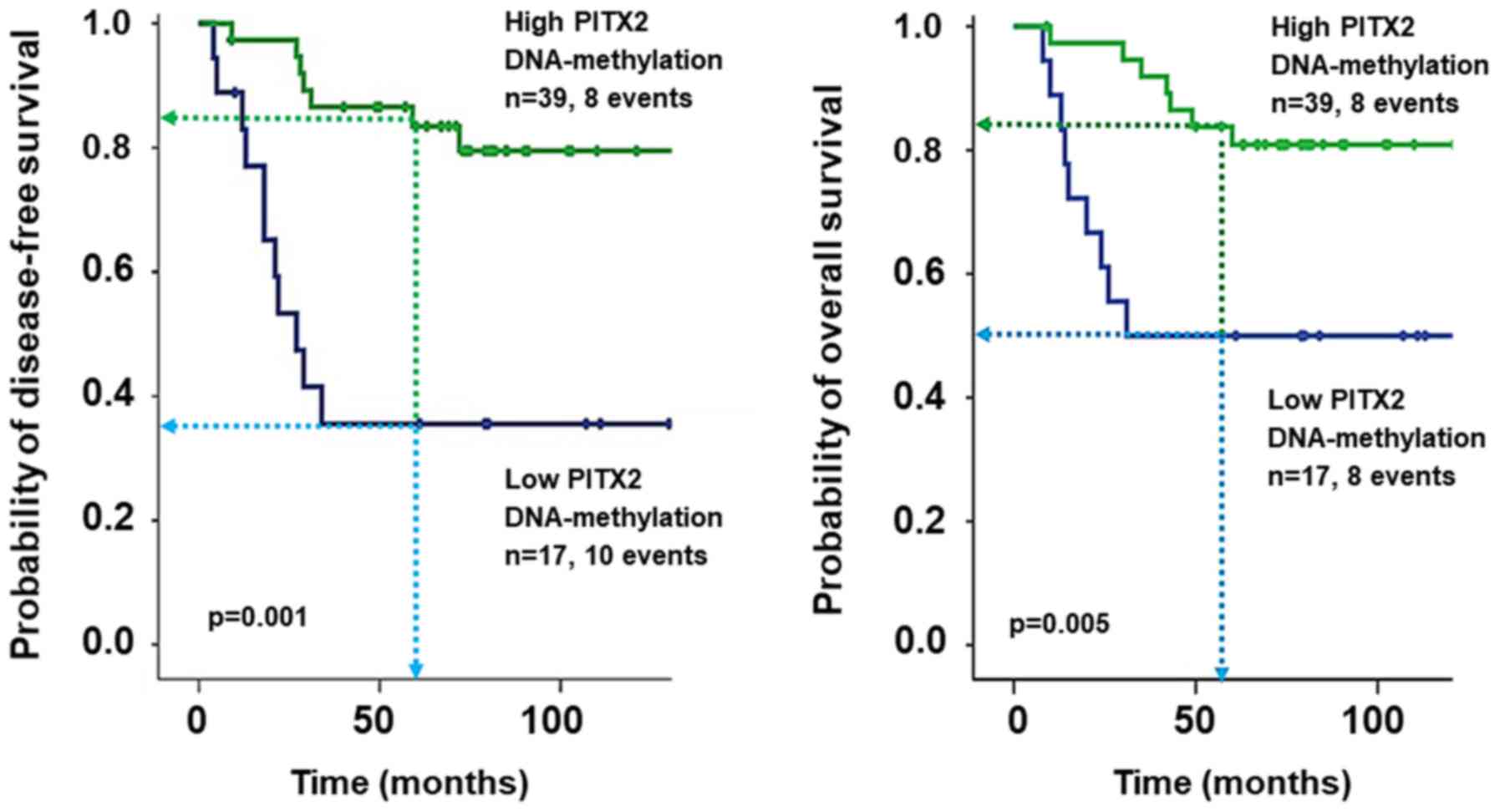
CI, 1.01–1.10). Kaplan-Meier analyses were carried out employing
the same cut-off, regarding DFS and OS, low PITX2 DNA-methylation
was associated with poor clinical outcome of the patients (DFS:
P<0.001, 5-year DFS 35.6 vs. 83.5%; OS: P=0.005, 5-year OS 50.0
vs. 80.9%) (Fig. 3).
 | Table IIIUnivariate and multivariate Cox
regression analyses to assess the clinical impact of PITX2
DNA-methylation status to predict response of TNBC patients to
adjuvant anthracycline-based chemotherapy. |
Table III
Univariate and multivariate Cox
regression analyses to assess the clinical impact of PITX2
DNA-methylation status to predict response of TNBC patients to
adjuvant anthracycline-based chemotherapy.
| Variables | Disease-free
survival
|
|---|
Univariate analysis
| Multivariate
analysis
|
|---|
| n | HR | 95% CI | P-value | n | HR | 95% CI | P-value |
|---|
| Tumor size | 54 | 2.75 | 0.80–9.52 | 0.109 | 51 | 1.13 | 0.26–5.01 | 0.871 |
| Nodal status | 55 | 1.55 | 0.60–4.00 | 0.365 | 51 | 2.04 | 0.61–6.84 | 0.249 |
| Grading | 54 | 1.30 | 0.17–9.83 | 0.798 | 51 | 0.86 | 0.10–7.78 | 0.893 |
| Age | 56 | 1.02 | 0.98–1.06 | 0.415 | 51 | 1.00 | 0.96–1.05 | 0.981 |
| PITX2
DNA-methylation | 56 | 5.36 | 2.06–13.95 | 0.001 | 51 | 6.40 | 1.96–20.88 | 0.002 |
|
|
Variables | Overall
survival
|
Univariate
analysis
| Multivariate
analysis
|
| n | HR | 95% CI | P-value | n | HR | 95% CI | P-value |
|
| Tumor size | 54 | 1.62 | 0.52–5.02 | 0.404 | 56 | 1.12 | 0.23–5.58 | 0.886 |
| Nodal status | 55 | 1.46 | 0.52–4.11 | 0.471 | 56 | 0.99 | 0.30–3.21 | 0.982 |
| Grading | 54 | 1.09 | 0.14–8.30 | 0.933 | 56 | 0.54 | 0.06–5.23 | 0.594 |
| Age | 56 | 1.05 | 1.01–1.10 | 0.032 | 56 | 1.05 | 0.99–1.11 | 0.092 |
| PITX2
DNA-methylation | 56 | 3.78 | 1.40–10.20 | 0.009 | 56 | 3.62 | 1.03–12.72 | 0.045 |
Evaluation of the association between
PITX2-antigen expression and PITX2-DNA-methylation status in
primary TNBC breast cancer tumor tissues (FFPE)
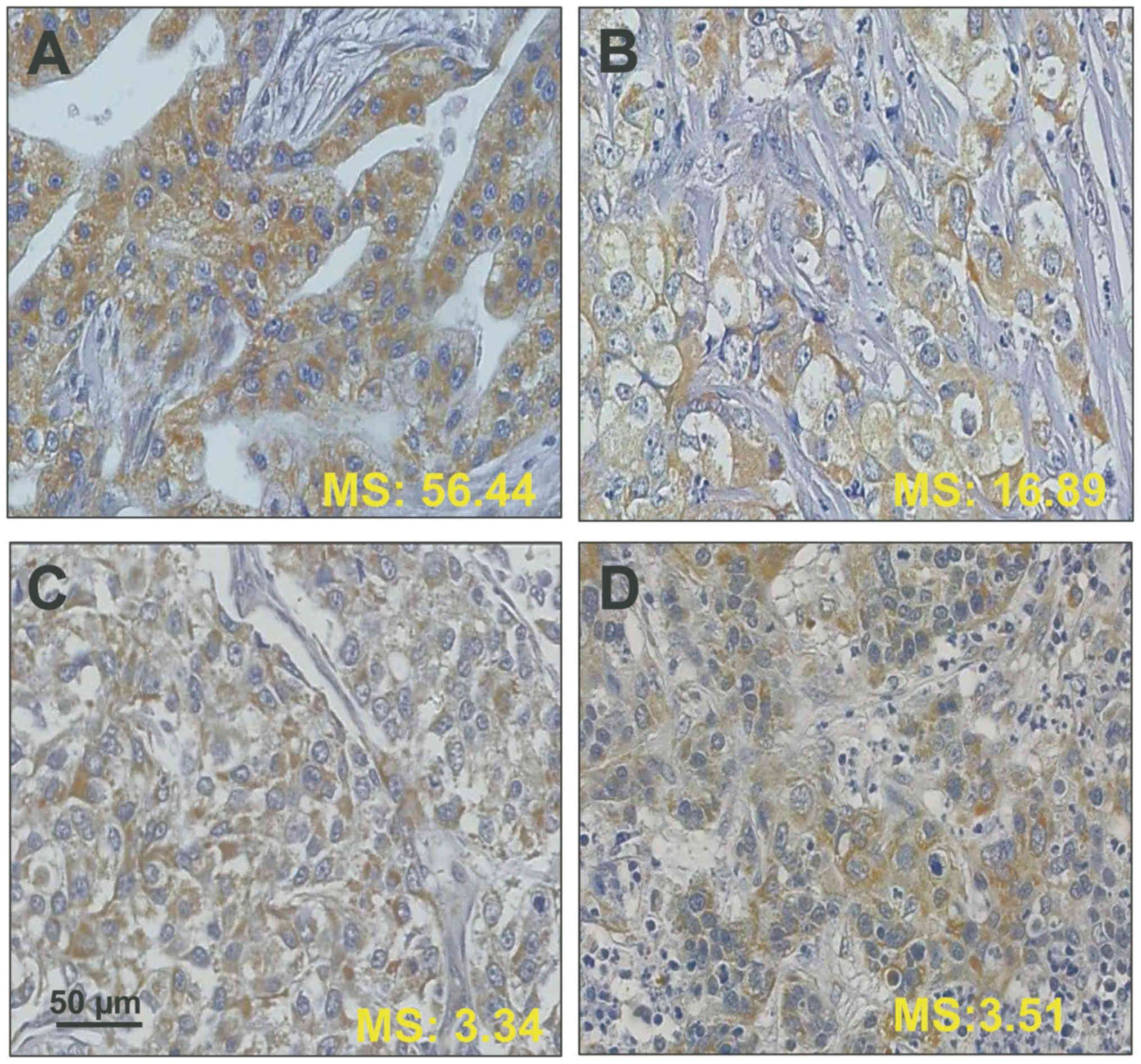
PITX2 DNA-methylation score measurements and
immunohistochemical staining against PITX2 applying
affinity-purified antibody PITX2-484 was performed for a few
randomly selected TNBC breast cancer tissues (Fig. 4). In general, in the tissues shown,
immunostaining for PITX2-antigen does not match with its respective
DNA-methylation status, e.g. high PITX2 DNA-methylation scores do
not reflect a substantial decrease in immunostaining intensity.
Discussion
During embryonic development, expression of PITX2, a
bicoid-like developmental transcription factor, determines the
left-right symmetry of the body and tightly controls the correct
placement of various internal organs (36,52–54).
In breast cancer, the methylation status of the PITX2 gene has been
reported to be both a prognostic and a predictive biomarker for
response of patients to endocrine therapy or anthracycline-based
chemotherapy (39–41). In the present study, a test system
was applied which determines the PITX2 DNA-methylation status in
FFPE-breast cancer tumor tissues, first described by Harbeck et
al (40), resulting from a
multicenter trans-European cooperation of European academic and
commercial partners supported by the European Union Framework
Program FP6. It is a real-time quantitative methylation-specific
PCR-based (qMSP) assay; the sample type is bisDNA, i.e.
bisulfite-converted gDNA. For this, gDNA is extracted from FFPE
breast cancer tumor tissues, then exposed to bisulfite treatment to
distinguish between the methylated and unmethylated PITX2 status
(39,40).
DNA methylation changes in routinely available FFPE
tissues or fresh-frozen tissues not only can serve as biomarkers
for the detection of malignant disease and for the assessment of
the clinical course of cancer disease but also as specific
biomarkers to predict whether a cancer patient will respond to
systemic drug therapy or not (39–41,55–61).
Tests to assess promoter methylation patterns of some genes (e.g.
MGMT, GSTP1, SHOX2, SEPT9, ASTN1 and ZNF671), in malignancies other
than breast cancer, have already been transformed into commercially
available clinical assays (e.g. by MDxHealth, Epigenomics and
Oncgnostics).
Increasing evidence suggests that aberrant PITX2 DNA
methylation is not only prominent in breast cancer but is also
associated with other malignant diseases, e.g. cancers of the
urogenital and gastrointestinal tract, that of the thyroid and of
head and neck, and of leukemia (62–71).
Yet, the regulatory role that PITX2 DNA methylation plays in these
diseases is still not fully explored. Further clinical studies are
warranted to determine whether the results available are
transferable to larger patient cohorts.
In a recent Clinical Practice Guideline published on
behalf of the American Society of Clinical Oncology (ASCO),
evidenced-based recommendations were published on the appropriate
use of breast tumor biomarker assay results to guide decisions on
adjuvant systemic therapy for women with early-stage invasive
breast cancer with known ER/PR/HER2 status (72). One focal point of the guideline was
to provide recommendations to physicians and patients on the
potential ability of predictive biomarkers to indicate benefit of a
certain chemotherapeutic for a respective class of
ER/PR/HER2-positive and negative breast cancer patients. In
summary, in view of the guideline panel, sufficient clinical
evidence was provided in the scientific literature to recommend the
use of multiplex biomarker assays to manage treatment of
ER/PR-positive breast cancer patients. Such recommendation could
not be given for the group of TNBC patients, which means that there
is still a fundamental lack to guide decision on adjuvant systemic
therapy for this group of patients (72).
This assessment supports the idea that the PITX2
assay should be considered as a novel, valuable additive clinically
useful test, different from the established multiplex gene
signatures as depicted by Harris et al (72). The PITX2 assay may assist the
physician to guide decisions on adjuvant systemic therapy, both for
the ER/PR-positive and negative (TNBC) breast cancer subsets
(39–42).
The present clinical investigation centered on the
question whether breast cancer patients afflicted with the
aggressive TNBC subtype would benefit from anthracycline-containing
adjuvant chemotherapy by grouping the respective TNBC patients to
their low or high PITX2 gene methylation status. Different from
that, previously published investigations described the relation of
PITX2 DNA-methylation status with estrogen/progesterone
receptor-positive breast cancer disease, not including the TNBC
phenotype (39–42).
PITX2 DNA-methylation profiles in TNBC patients'
tumor tissues apply to the methylation status of particular CpG
sites in this gene; such changes can be used clinically as a
prognostic and/or predictive marker for this kind of malignant
disease (39–42,55).
Thus, the aim of the present study was to explore a possible
statistical correlation between methylation in the promoter region
of the PITX2 gene and clinical outcome of TNBC patients treated
with anthracycline-based chemotherapy.
In addition, we also analyzed PITX2 protein
expression in a limited number of randomly selected TNBC primary
breast cancer tumor tissue specimens by immunohistochemistry, by
employing the proprietary antibody PITX2-484, but did not observe
any lack of PITX2-expression in the breast cancer tumor tissue
specimens looked at, demonstrating that with respect to PITX2
protein expression, partial methylation of CpG islands is not
associated with complete gene silencing.
In our TNBC cases, the antibody applied (PITX2-484
to two PITX2-peptides located on canonical PITX2-variant PITX2-B)
reacted predominantly with cytoplasmic PITX2 protein but
occasionally nuclear and perinuclear staining was observed as well.
Since PITX2 is a transcription factor, nuclear localization of
PITX2 was expected. Yet, in the immunohistochemical assessment of
PITX2 in breast cancer tissues presented by Wan Abdul Rahman et
al (73), the authors claimed
that in invasive ductal breast cancer, including TNBC, PITX2
protein expression is preferentially associated with the cytoplasm
of the tumor cells. Other but single clinically relevant studies
assessed cytoplasmic PITX2 protein expression in malignant diseases
as well, e.g. in odontogenic tumors, thyroid and esophageal cancer
(69,74,75).
Another study was related to PITX2-protein expression in ovarian
cancer. The authors observed both cytoplasmic and nuclear
localization of PITX2, depending on the malignant stage (62).
TNBC is characterized by large-scale
transcriptional, mutational, and copy number heterogeneity; thus,
in the past, most targeted chemotherapeutic agents have
demonstrated low overall activity in unselected TNBC patients since
various biological TNBC subgroups are overlapping and so far cannot
be combined into a 'one-fits-all' model of TNBC biology (61,76–78).
Oppositely, this molecular heterogeneity has allowed to categorize
TNBC for different novel targeted therapeutic interventions, having
led to ongoing innovative clinical strategies for early-stage and
advanced TNBC, including immunotherapy and modified chemotherapy
(76,79). Besides that, TNBC is typically
treated with various combinations of chemotherapy; a sequential
anthracycline-taxane combination is the standard of care for TNBC
(3). Yet, systemic chemotoxic
treatment of TNBC patients needs to be personalized for a specific
patient to suit her best, preferentially depending on the molecular
characteristics of her disease, since at least ten different
molecular TNBC subtypes have been identified using gene copy number
and expression analyses (3,77).
Many clinical studies have demonstrated that TNBC is
sensitive to anthracycline-containing adjuvant chemotherapy
regimens (3,80,81);
anthracyclines are considered to be among the most active drugs for
the treatment of breast cancer by destabilizing the DNA through
intercalation (3,80,81).
Otherwise, anthracyclines may cause severe side-effects, including
cardiac toxicity, which can lead to heart failure and which
therefore may hamper their optimal use in treatment of TNBC
(82). The risk of TNBC patients
to experience severe side-effects caused by systemic cancer
treatment can be higher when other treatments, e.g. taxanes or
platinum-based therapeutics are used in combination with an
anthracycline (3,83).
There is general consensus that for TNBC systemic
adjuvant therapy anthracycline-containing regimens are the standard
approach for patients after primary surgery. This is also the
opinion of the American Society of Clinical Oncology (ASCO), it
recommends chemotherapy treatment for TNBC patients based on the
combination of an anthracycline with a taxane but does not
currently recommend tailoring therapy for TNBC patients by
stratifying treatment by implementing non-validated results for
TNBC biomarkers, such as tumor cell surface receptors (EGFR, IGFBP,
C-kit and PD-L1) which potentially could serve as novel target
molecules to block tumor cell proliferation and dissemination
(16). If comorbidities forbid the
use of anthracyclines, treatment with taxane-based regimens and
cyclophosphamide are recommended as alternative adjuvant treatment;
treatments using paclitaxel or CMF (cyclophosphamide, methotrexate
and 5-fluorouracil) should be considered as well (45,84–87).
Only scarce data are available in the scientific
literature naming biomarkers predictive for response of TNBC
patients to anthracycline-based adjuvant chemotherapy. For example,
Mori et al (88) speculated
on the predictive value of BRCAness as a predictive factor for the
effectiveness of anthracycline-based adjuvant chemotherapy for
patients with TNBCs. Different from that analysis, Bouchalova et
al (89) demonstrated the
usefulness of the biomarker BCL2 to predict the level of
recurrence-free and overall survival in TNBC patients treated with
anthracycline-based adjuvant chemotherapy.
More information is available for non-breast cancer
patients. For these patients, especially concerning association of
PITX2 DNA-methylation status with other biomarkers, several
epigenetic studies were published, demonstrating co-expression of
hypo- or hypermethylated biomarkers in conjunction with PITX2
DNA-methylation profiles (37,38,90–92).
None of these studies, however, specifically addressed the issue of
the potential benefit of adjuvant anthracycline treatment with
regard to PITX2 expression or its DNA-methylation status, except
the work performed by Hartmann et al (41). Obviously, there is still a strong
need to address the question of anthracycline efficacy in TNBC and
associated biomarkers to predict sensitivity or resistance to
adjuvant anthracycline-based chemotherapy and for new cellular
targets for individualized TNBC patients.
On this line, accumulated substantial evidence was
presented in the past by a European EU-FP6-framework consortium
that among all genes analyzed, PITX2 DNA-methylation analysis
performed on routinely available FFPE-tumor tissue specimens holds
promise as a novel practical assay for routine clinical use to
predict outcome of these patients, treated with endocrine or
anthracycline-based adjuvant therapy providing a potential link
between PITX2 expression and breast cancer progression (39–42).
Otherwise, these studies on steroid hormone
receptor-positive breast cancer were different from the present
investigation which exclusively focusses on the TNBC subgroup of
breast cancer patients, lacking both ER/PR-steroid hormone receptor
and oncoprotein HER2 expression (3). To the best of our knowledge, this is
the first clinical study examining whether determination of the
methylation status of the promoter region of the PITX2 gene in
primary tumor tissues can serve as a biomarker to predict response
of TNBC patients to adjuvant anthracycline-based chemotherapy.
Collectively, we show by statistical analyses of
TNBC patients treated with anthracycline-based adjuvant
chemotherapy that low PITX2 DNA-methylation is associated with poor
clinical outcome, demonstrating its statistically independent
nature by univariate and multivariate statistics. Notably, the
PITX2-TNBC response data indicate a reverse relationship between
PITX2 DNA-methylation and response to anthracy-cline-based TNBC
when compared to the non-TNBC breast cancer studies (39–42),
supporting the notion that TNBC reflects a selected kind of breast
cancer disease, different in many phenotypic and genomic aspects
from endocrine receptor-positive breast cancer (3). Remarkably, our results also
demonstrated that the sequential addition of taxanes to adjuvant
systemic anthracycline-based chemotherapy did not alter the
predictive value of PITX2.
Since searching for optimised cut-off values could
potentially lead to overfitting, it is important to note that this
study does not claim that the optimized cut-off value defined in
this anthracycline-receiving TNBC subgroup is the optimal one for
future application. The pilot character of the present study is
reflected by its retrospective design and the non-homogeneous
therapy regimens applied but also by a small number of cases and
number of events. Therefore, the current results of this first-time
observation study should rather serve as a clue towards validation
of the data which have to be evaluated in future clinically
relevant studies.
Abbreviations:
|
C
|
cyclophosphamide
|
|
M
|
methotrexate
|
|
F
|
5-fluo-rouracil
|
|
E
|
epirubicine
|
|
gDNA
|
genomic DNA
|
Acknowledgments
The present study was supported in part by the
Wilhelm Sander-Stiftung, Munich, Germany, contract number
2012.028.1. to M.A. and M.S. and 2016.024.1. to M.A., A.W., V.M.
and M.S. and by the German Research Foundation (DFG), contract
number DO 1772/1-1.
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Wolff AC, Hammond ME, Hicks DG, Dowsett M,
McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M,
Fitzgibbons P, et al American Society of Clinical Oncology; College
of American Pathologists: Recommendations for human epidermal
growth factor receptor 2 testing in breast cancer: American Society
of Clinical Oncology/College of American Pathologists clinical
practice guideline update. J Clin Oncol. 31:3997–4013. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bauer KR, Brown M, Cress RD, Parise CA and
Caggiano V: Descriptive analysis of estrogen receptor
(ER)-negative, progesterone receptor (PR)-negative, and
HER2-negative invasive breast cancer, the so-called triple-negative
phenotype: A population-based study from the California Cancer
Registry. Cancer. 109:1721–1728. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harbeck N and Gnant M: Breast cancer.
Lancet. 389:1134–1150. 2017. View Article : Google Scholar
|
|
4
|
Jiang T, Shi W, Wali VB, Pongor LS, Li C,
Lau R, Győrffy B, Lifton RP, Symmans WF, Pusztai L, et al:
Predictors of chemo-sensitivity in triple negative breast cancer:
An integrated genomic analysis. PLoS Med. 13:e10021932016.
View Article : Google Scholar
|
|
5
|
Liedtke C and Kiesel L: Breast cancer
molecular subtypes--modern therapeutic concepts for targeted
therapy of a heterogeneous entity. Maturitas. 73:288–294. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Irshad S, Ellis P and Tutt A: Molecular
heterogeneity of triple-negative breast cancer and its clinical
implications. Curr Opin Oncol. 23:566–577. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Karn T, Pusztai L, Holtrich U, Iwamoto T,
Shiang CY, Schmidt M, Müller V, Solbach C, Gaetje R, Hanker L, et
al: Homogeneous datasets of triple negative breast cancers enable
the identification of novel prognostic and predictive signatures.
PLoS One. 6:e284032011. View Article : Google Scholar
|
|
8
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lord CJ and Ashworth A: BRCAness
revisited. Nat Rev Cancer. 16:110–120. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Spugnesi L, Gabriele M, Scarpitta R,
Tancredi M, Maresca L, Gambino G, Collavoli A, Aretini P, Bertolini
I, Salvadori B, et al: Germline mutations in DNA repair genes may
predict neoadjuvant therapy response in triple negative breast
patients. Genes Chromosomes Cancer. 55:915–924. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mori H, Kubo M, Nishimura R, Osako T,
Arima N, Okumura Y, Okido M, Yamada M, Kai M, Kishimoto J, et al:
BRCAness as a biomarker for predicting prognosis and response to
anthracycline-based adjuvant chemotherapy for patients with
triple-negative breast cancer. PLoS One. 11:e01670162016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sharma P: Sharma. Oncologist.
21:1050–1062. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lips EH, Mulder L, Oonk A, van der Kolk
LE, Hogervorst FB, Imholz AL, Wesseling J, Rodenhuis S and Nederlof
PM: Triple-negative breast cancer: BRCAness and concordance of
clinical features with BRCA1-mutation carriers. Br J Cancer.
108:2172–2177. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cardoso F, Harbeck N, Barrios CH, Bergh J,
Cortés J, El Saghir N, Francis PA, Hudis CA, Ohno S, Partridge AH,
et al: Research needs in breast cancer. Ann Oncol Nov. 28:208–217.
2017.
|
|
16
|
Fleisher B, Clarke C and Ait-Oudhia S:
Current advances in biomarkers for targeted therapy in
triple-negative breast cancer. Breast Cancer (Dove Med Press).
8:183–197. 2016.
|
|
17
|
Telli M: Optimizing chemotherapy in
triple-negative breast cancer: The role of platinum. Am Soc Clin
Oncol Educ Book. 33:e37–e42. 2014. View Article : Google Scholar
|
|
18
|
Guan X, Ma F, Fan Y, Zhu W, Hong R and Xu
B: Platinum-based chemotherapy in triple-negative breast cancer: A
systematic review and meta-analysis of randomized-controlled
trials. Anticancer Drugs. 26:894–901. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Anders CK, Abramson V, Tan T and Dent R:
The evolution of triple-negative breast cancer: From biology to
novel therapeutics. Am Soc Clin Oncol Educ Book. 35:34–42. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gerratana L, Fanotto V, Pelizzari G,
Agostinetto E and Puglisi F: Do platinum salts fit all triple
negative breast cancers. Cancer Treat Rev. 48:34–41. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kern P, Kalisch A, Kolberg HC, Kimmig R,
Otterbach F, von Minckwitz G, Sikov WM, Pott D and Kurbacher C:
Neoadjuvant, anthracycline-free chemotherapy with carboplatin and
docetaxel in triple-negative, early-stage breast cancer: A
multicentric analysis of feasibility and rates of pathologic
complete response. Chemotherapy. 59:387–394. 2013. View Article : Google Scholar
|
|
22
|
Zheng R, Han S, Duan C, Chen K, You Z, Jia
J, Lin S, Liang L, Liu A, Long H, et al: Role of taxane and
anthracycline combination regimens in the management of advanced
breast cancer: a meta-analysis of randomized trials. Medicine.
94:e8032015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adium R and Liedtke C: Neoadjuvant therapy
for patients with triple negative breast cancer (TNBC). Rev Recent
Clin Trials. 12:73–80. 2017. View Article : Google Scholar
|
|
24
|
von Minckwitz G, Schneeweiss A, Loibl S,
Salat C, Denkert C, Rezai M, Blohmer JU, Jackisch C, Paepke S,
Gerber B, et al: Neoadjuvant carboplatin in patients with
triple-negative and HER2-positive early breast cancer (GeparSixto;
GBG 66): A randomised phase 2 trial. Lancet Oncol. 15:747–756.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nabholtz JM, Abrial C, Mouret-Reynier MA,
Dauplat MM, Weber B, Gligorov J, Forest AM, Tredan O, Vanlemmens L,
Petit T, et al: Multicentric neoadjuvant phase II study of
panitumumab combined with an anthracycline/taxane-based
chemotherapy in operable triple-negative breast cancer:
Identification of biologically defined signatures predicting
treatment impact. Ann Oncol. 25:1570–1577. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
McClendon AK, Dean JL, Rivadeneira DB, Yu
JE, Reed CA, Gao E, Farber JL, Force T, Koch WJ and Knudsen ES:
CDK4/6 inhibition antagonizes the cytotoxic response to
anthracycline therapy. Cell Cycle. 11:2747–2755. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Reinisch M, von Minckwitz G, Harbeck N,
Janni W, Kümmel S, Kaufmann M, Elling D, Nekljudova V and Loibl S:
Side effects of standard adjuvant and neoadjuvant chemotherapy
regimens according to age groups in primary breast cancer. Breast
Care (Basel). 8:60–66. 2013. View Article : Google Scholar
|
|
28
|
Baylin SB and Jones PA: Epigenetic
determinants of cancer. Cold Spring Harb Perspect Biol. 8:82016.
View Article : Google Scholar
|
|
29
|
Wu Y, Sarkissyan M and Vadgama JV:
Epigenetics in breast and prostate cancer. Methods Mol Biol.
1238:425–466. 2015. View Article : Google Scholar :
|
|
30
|
Atalay C: Atalay. Exp Oncol. 35:246–249.
2013.
|
|
31
|
Jovanovic J, Rønneberg JA, Tost J and
Kristensen V: The epigenetics of breast cancer. Mol Oncol.
4:242–254. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Basse C and Arock M: The increasing roles
of epigenetics in breast cancer: Implications for pathogenicity,
biomarkers, prevention and treatment. Int J Cancer. 137:2785–2794.
2015. View Article : Google Scholar
|
|
33
|
Yang X, Lay F, Han H and Jones PA:
Targeting DNA methylation for epigenetic therapy. Trends Pharmacol
Sci. 31:536–546. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jones PA: Jones. J Clin Invest. 124:14–16.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shiratori H, Yashiro K, Shen MM and Hamada
H: Conserved regulation and role of Pitx2 in situs-specific
morphogenesis of visceral organs. Development. 133:3015–3025. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wilting J and Hagedorn M: Left-right
asymmetry in embryonic development and breast cancer: Common
molecular determinants. Curr Med Chem. 18:5519–5527. 2011.
View Article : Google Scholar
|
|
37
|
Jezkova E, Kajo K, Zubor P, Grendar M,
Malicherova B, Mendelova A, Dokus K, Lasabova Z, Plank L and Danko
J: Methylation in promoter regions of PITX2 and RASSF1A genes in
association with clinicopathological features in breast cancer
patients. Tumour Biol. 37:15707–15718. 2016. View Article : Google Scholar
|
|
38
|
Martens JW, Margossian AL, Schmitt M,
Foekens J and Harbeck N: DNA methylation as a biomarker in breast
cancer. Future Oncol. 5:1245–1256. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Maier S, Nimmrich I, Koenig T,
Eppenberger-Castori S, Bohlmann I, Paradiso A, Spyratos F, Thomssen
C, Mueller V, Nährig J, et al European Organisation for Research
and Treatment of Cancer (EORTC) PathoBiology group: DNA-methylation
of the homeodomain transcription factor PITX2 reliably predicts
risk of distant disease recurrence in tamoxifen-treated,
node-negative breast cancer patients--Technical and clinical
validation in a multi-centre setting in collaboration with the
European Organisation for Research and Treatment of Cancer (EORTC)
PathoBiology group. Eur J Cancer. 43:1679–1686. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Harbeck N, Nimmrich I, Hartmann A, Ross
JS, Cufer T, Grützmann R, Kristiansen G, Paradiso A, Hartmann O,
Margossian A, et al: Multicenter study using paraffin-embedded
tumor tissue testing PITX2 DNA methylation as a marker for outcome
prediction in tamoxifen-treated, node-negative breast cancer
patients. J Clin Oncol. 26:5036–5042. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hartmann O, Spyratos F, Harbeck N,
Dietrich D, Fassbender A, Schmitt M, Eppenberger-Castori S,
Vuaroqueaux V, Lerebours F, Welzel K, et al: DNA methylation
markers predict outcome in node-positive, estrogen
receptor-positive breast cancer with adjuvant anthracycline-based
chemotherapy. Clin Cancer Res. 15:315–323. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nimmrich I, Sieuwerts AM, Meijer-van
Gelder ME, Schwope I, Bolt-de Vries J, Harbeck N, Koenig T,
Hartmann O, Kluth A, Dietrich D, et al: DNA hypermethylation of
PITX2 is a marker of poor prognosis in untreated lymph
node-negative hormone receptor-positive breast cancer patients.
Breast Cancer Res Treat. 111:429–437. 2008. View Article : Google Scholar
|
|
43
|
Kumar P and Aggarwal R: An overview of
triple-negative breast cancer. Arch Gynecol Obstet. 293:247–269.
2016. View Article : Google Scholar
|
|
44
|
Loibl S, Denkert C and von Minckwitz G:
Neoadjuvant treatment of breast cancer - Clinical and research
perspective. Breast. 24(Suppl 2): S73–S77. 2015. View Article : Google Scholar
|
|
45
|
Liedtke C and Rody A: New treatment
strategies for patients with triple-negative breast cancer. Curr
Opin Obstet Gynecol. 27:77–84. 2015. View Article : Google Scholar
|
|
46
|
Yfanti C, Mengele K, Gkazepis A, Weirich
G, Giersig C, Kuo WL, Tang WJ, Rosner M and Schmitt M: Expression
of metalloprotease insulin-degrading enzyme insulysin in normal and
malignant human tissues. Int J Mol Med. 22:421–431. 2008.PubMed/NCBI
|
|
47
|
Schmitt M, Mengele K, Schueren E, Sweep
FC, Foekens JA, Brünner N, Laabs J, Malik A and Harbeck N; European
Organisation for Research and Treatment of Cancer Pathobiology
Group: European Organisation for Research and Treatment of Cancer
(EORTC) Pathobiology Group standard operating procedure for the
preparation of human tumour tissue extracts suited for the
quantitative analysis of tissue-associated biomarkers. Eur J
Cancer. 43:835–844. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
McShane LM, Altman DG, Sauerbrei W, Taube
SE, Gion M and Clark GM; Statistics Subcommittee of the NCI-EORTC
Working Group on Cancer Diagnostics: Reporting recommendations for
tumor marker prognostic studies (REMARK). J Natl Cancer Inst.
97:1180–1184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Altman DG, McShane LM, Sauerbrei W and
Taube SE: Reporting recommendations for tumor marker prognostic
studies REMARK. Explanation and elaboration. PLoS Med.
9:e10012162012. View Article : Google Scholar
|
|
50
|
Hothorn T: Maxstat: maximally selected
rank statistics. R package version 0.7-14.
|
|
51
|
Team RD CR: A language and environment for
statistical computing. R Foundation for Statistical Computing
Vienna Austria: 2012
|
|
52
|
Blum M, Feistel K, Thumberger T and
Schweickert A: The evolution and conservation of left-right
patterning mechanisms. Development. 141:1603–1613. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tabin CJ: Tabin. Cell. 127:27–32. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Levin M: Levin. Mech Dev. 122:3–25. 2005.
View Article : Google Scholar
|
|
55
|
Paska AV and Hudler P: Aberrant
methylation patterns in cancer: A clinical view. Biochem Med
(Zagreb). 25:161–176. 2015. View Article : Google Scholar
|
|
56
|
Ahmed D, Danielsen SA, Aagesen TH,
Bretthauer M, Thiis-Evensen E, Hoff G, Rognum TO, Nesbakken A,
Lothe RA and Lind GE: A tissue-based comparative effectiveness
analysis of biomarkers for early detection of colorectal tumors.
Clin Transl Gastroenterol. 3:e272012. View Article : Google Scholar
|
|
57
|
Berghoff AS, Hainfellner JA, Marosi C and
Preusser M: Assessing MGMT methylation status and its current
impact on treatment in glioblastoma. CNS Oncol. 4:47–52. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Darwiche K, Zarogoulidis P, Baehner K,
Welter S, Tetzner R, Wohlschlaeger J, Theegarten D, Nakajima T and
Freitag L: Assessment of SHOX2 methylation in EBUS-TBNA specimen
improves accuracy in lung cancer staging. Ann Oncol. 24:2866–2870.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Dietrich D, Jung M, Puetzer S, Leisse A,
Holmes EE, Meller S, Uhl B, Schatz P, Ivascu C and Kristiansen G:
Diagnostic and prognostic value of SHOX2 and SEPT9 DNA methylation
and cytology in benign, paramalignant and malignant pleural
effusions. PLoS One. 8:e842252013. View Article : Google Scholar
|
|
60
|
Ilse P, Biesterfeld S, Pomjanski N, Wrobel
C and Schramm M: Analysis of SHOX2 methylation as an aid to
cytology in lung cancer diagnosis. Cancer Genomics Proteomics.
11:251–258. 2014.PubMed/NCBI
|
|
61
|
Székely B, Silber AL and Pusztai L: New
therapeutic strategies for triple-negative breast cancer. Oncology
(Williston Park). 31:130–137. 2017.
|
|
62
|
Fung FK, Chan DW, Liu VW, Leung TH, Cheung
AN and Ngan HY: Increased expression of PITX2 transcription factor
contributes to ovarian cancer progression. PLoS One. 7:e370762012.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kapoor S: Kapoor. APMIS. 121:10112013.
View Article : Google Scholar
|
|
64
|
Liu Y, Huang Y and Zhu GZ: Cyclin A1 is a
transcriptional target of PITX2 and overexpressed in papillary
thyroid carcinoma. Mol Cell Biochem. 384:221–227. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Toyota M, Kopecky KJ, Toyota MO, Jair KW,
Willman CL and Issa JP: Methylation profiling in acute myeloid
leukemia. Blood. 97:2823–2829. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Uhl B, Gevensleben H, Tolkach Y, Sailer V,
Majores M, Jung M, Meller S, Stein J, Ellinger J, Dietrich D, et
al: PITX2 DNA Methylation as biomarker for individualized risk
assessment of prostate cancer in core biopsies. J Mol Diagn.
19:107–114. 2017. View Article : Google Scholar
|
|
67
|
Wang Q, Li J, Wu W, Shen R, Jiang H, Qian
Y, Tang Y, Bai T, Wu S, Wei L, et al: Smad4-dependent suppressor
pituitary homeobox 2 promotes PPP2R2A-mediated inhibition of Akt
pathway in pancreatic cancer. Oncotarget. 7:11208–11222.
2016.PubMed/NCBI
|
|
68
|
Zhang JX, Chen ZH, Xu Y, Chen JW, Weng HW,
Yun M, Zheng ZS, Chen C, Wu BL, Li EM, et al: Downregulation of
microRNA-644a promotes esophageal squamous cell carcinoma
aggressiveness and stem cell-like phenotype via dysregulation of
PITX2. Clin Cancer Res. 23:298–310. 2017. View Article : Google Scholar
|
|
69
|
Zhang JX, Tong ZT, Yang L, Wang F, Chai
HP, Zhang F, Xie MR, Zhang AL, Wu LM, Hong H, et al: PITX2: A
promising predictive biomarker of patients' prognosis and
chemoradioresistance in esophageal squamous cell carcinoma. Int J
Cancer. 132:2567–2577. 2013. View Article : Google Scholar
|
|
70
|
Sailer V, Gevensleben H, Dietrich J, Goltz
D, Kristiansen G, Bootz F and Dietrich D: Clinical performance
validation of PITX2 DNA methylation as prognostic biomarker in
patients with head and neck squamous cell carcinoma. PLoS One.
12:e01794122017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
López JI, Angulo JC, Martín A,
Sánchez-Chapado M, González-Corpas A, Colás B and Ropero S: A DNA
hypermethylation profile reveals new potential biomarkers for the
evaluation of prognosis in urothelial bladder cancer. APMIS.
125:787–796. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Harris LN, Ismaila N, McShane LM, Andre F,
Collyar DE, Gonzalez-Angulo AM, Hammond EH, Kuderer NM, Liu MC,
Mennel RG, et al: American Society of Clinical Oncology: Use of
biomarkers to guide decisions on adjuvant systemic therapy for
women with early-stage invasive breast cancer: American Society of
Clinical Oncology Clinical Practice Guideline. J Clin Oncol.
34:1134–1150. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wan Abdul, Rahman WF, Fauzi MH and Jaafar
H: Expression of DNA methylation marker of paired-like homeodomain
transcription factor 2 and growth receptors in invasive ductal
carcinoma of the breast. Asian Pac J Cancer Prev. 15:8441–8445.
2014. View Article : Google Scholar
|
|
74
|
Bologna-Molina R, Mikami T, Pereira-Prado
V, Pires FR, Carlos-Bregni R and Mosqueda-Taylor A: Primordial
odonto-genic tumor: An immunohistochemical profile. Med Oral Patol
Oral Cir Bucal. 22:e314-e3232017.
|
|
75
|
Huang Y, Guigon CJ, Fan J, Cheng SY and
Zhu GZ: Pituitary homeobox 2 (PITX2) promotes thyroid
carcinogenesis by activation of cyclin D2. Cell Cycle. 9:1333–1341.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Denkert C, Liedtke C, Tutt A and von
Minckwitz G: Molecular alterations in triple-negative breast
cancer-the road to new treatment strategies. Lancet. 389:2430–2442.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Lehmann BD, Bauer JA, Chen X, Sanders ME,
Chakravarthy AB, Shyr Y and Pietenpol JA: Identification of human
triple-negative breast cancer subtypes and preclinical models for
selection of targeted therapies. J Clin Invest. 121:2750–2767.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ring BZ, Hout DR, Morris SW, Lawrence K,
Schweitzer BL, Bailey DB, Lehmann BD, Pietenpol JA and Seitz RS:
Generation of an algorithm based on minimal gene sets to clinically
subtype triple negative breast cancer patients. BMC Cancer.
16:1432016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Bianchini G, Balko JM, Mayer IA, Sanders
ME and Gianni L: Triple-negative breast cancer: Challenges and
opportunities of a heterogeneous disease. Nat Rev Clin Oncol.
13:674–690. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yadav BS, Sharma SC, Chanana P and Jhamb
S: Systemic treatment strategies for triple-negative breast cancer.
World J Clin Oncol. 5:125–133. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Burstein HJ: Patients with triple negative
breast cancer: Is there an optimal adjuvant treatment? Breast.
22(Suppl 2): S147–S148. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Christenson ES, James T, Agrawal V and
Park BH: Use of biomarkers for the assessment of
chemotherapy-induced cardiac toxicity. Clin Biochem. 48:223–235.
2015. View Article : Google Scholar :
|
|
83
|
Basso SM, Santeufemia DA, Fadda GM,
Tozzoli R, D'Aurizio F and Lumachi F: Advances in the treatment of
triple-negative early breast cancer. Med Chem. 12:268–272. 2016.
View Article : Google Scholar
|
|
84
|
Wahba HA and El-Hadaad HA: Current
approaches in treatment of triple-negative breast cancer. Cancer
Biol Med. 12:106–116. 2015.PubMed/NCBI
|
|
85
|
Yao H, He G, Yan S, Chen C, Song L, Rosol
TJ and Deng X: Triple-negative breast cancer: Is there a treatment
on the horizon. Oncotarget. 8:1913–1924. 2017.
|
|
86
|
Locatelli MA, Curigliano G and Eniu A:
Extended adjuvant chemotherapy in triple-negative breast cancer.
Breast Care (Basel). 12:152–158. 2017. View Article : Google Scholar
|
|
87
|
Liedtke C, Thill M, Jackisch C, Thomssen
C, Müller V and Janni W: AGO Recommendations for the diagnosis and
treatment of patients with early breast cancer: Update. Breast Care
(Basel). 12. pp. 172–183. 2017, View Article : Google Scholar
|
|
88
|
Mori H, Kubo M, Nishimura R, Osako T,
Arima N, Okumura Y, Okido M, Yamada M, Kai M, Kishimoto J, et al:
BRCAness as a biomarker for predicting prognosis and response to
anthracycline-based adjuvant chemotherapy for patients with
triple-negative breast cancer. PLoS One. 11:e01670162016.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Bouchalova K, Svoboda M, Kharaishvili G,
Vrbkova J, Bouchal J, Trojanec R, Koudelakova V, Radova L, Cwiertka
K, Hajduch M, et al: BCL2 is an independent predictor of outcome in
basal-like triple-negative breast cancers treated with adjuvant
anthracycline-based chemotherapy. Tumour Biol. 36:4243–4252. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Duffy MJ, Napieralski R, Martens JW, Span
PN, Spyratos F, Sweep FC, Brunner N, Foekens JA and Schmitt M;
EORTC PathoBiology Group: Methylated genes as new cancer
biomarkers. Eur J Cancer. 45:335–346. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Pillai SG, Dasgupta N, Siddappa CM, Watson
MA, Fleming T, Trinkaus K and Aft R: Paired-like Homeodomain
Transcription factor 2 expression by breast cancer bone marrow
disseminated tumor cells is associated with early recurrent disease
development. Breast Cancer Res Treat. 153:507–517. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Dietrich D, Hasinger O, Liebenberg V,
Field JK, Kristiansen G and Soltermann A: DNA methylation of the
homeobox genes PITX2 and SHOX2 predicts outcome in non-small-cell
lung cancer patients. Diagn Mol Pathol. 21:93–104. 2012. View Article : Google Scholar : PubMed/NCBI
|