Introduction
Over the past 30 years, lung cancer has had high
morbidity and mortality rates worldwide, with 75% of new diagnoses
being classified as non-small-cell lung cancer (NSCLC) and advanced
tumors at the first visit (1).
Chemotherapy and radiotherapy remain the most common treatment
methods for advanced cancer (2).
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)
induces apoptosis in malignant tumors and has little effect on
normal cells (3,4). Recombinant human TRAIL and its
receptor agonists are under investigation as promising therapeutic
approaches for the treatment of advanced cancer (5). There are 4 different transmembrane
receptors in the TRAIL receptor/ligand system, including 2 death
receptors (DR4 and DR5), 2 decoy receptors (DcR1 and DcR2), and a
soluble receptor, furthermore, osteoprotegerin DRs contain a
cytoplasmic death domain transducing the apoptosis signalling
pathway (6). Preclinical studies
have revealed that recombinant TRAIL and DR agonists inhibit tumor
growth in vitro and in vivo without systemic toxicity
(7–9). A percentage of tumor cells were
observed to respond to TRAIL therapy, which is called primary TRAIL
resistance, while some tumor cells obtained TRAIL resistance
following repeated treatments, called acquired TRAIL resistance
(10,11). Increasing the existing
understanding of the molecular alterations involved in acquired
resistance and the cytotoxicity of TRAIL is required to further
investigate its therapeutic potential.
A previous study reported that β-catenin and DRs
were co-expressed in colonic tumor tissues. DR expression increased
during colon carcinoma tumorigenesis, possibly due to upregulation
of β-catenin expression (12).
However, the mechanisms by which β-catenin regulates TRAIL
resistance remain unclear in NSCLC (13). It was, therefore, hypothesized that
β-catenin enhanced TRAIL sensitivity by regulating the
redistribution of DR4 and DR5.
In the present study, a TRAIL-resistant H460-TR cell
line was established to investigate the potential effects of
β-catenin on DR redistribution and TRAIL sensitivity.
Downregulation of β-catenin expression decreased the redistribution
of DR4 and DR5 on the cell surface, and was associated with TRAIL
resistance. While β-catenin-knockdown in H460 cells decreased their
TRAIL sensitivity, upregulation of β-catenin expression in H460-TR
cells rescued TRAIL sensitivity, increased DR distribution on the
cytomembrane and activated caspase-3/8. β-catenin may be used as a
biomarker to predict TRAIL sensitivity in the future. Patients
exhibiting high β-catenin expression may benefit more from TRAIL
treatment. Furthermore, the Wnt signaling pathway agonist may be
used to promote TRAIL sensitivity during chemotherapy.
Materials and methods
Cells
The human NSCLC cell line, NCI-H460, was provided by
the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). Cells were cultured in RPMI-1640 medium
supplemented with 10% (v/v) fetal bovine serum (both from HyClone;
GE Healthcare Life Sciences, Logan, UT, USA), 100 U/ml penicillin
and 100 mg/ml streptomycin at 37°C in an atmosphere containing 5%
CO2. The TRAIL-resistant H460-TR cell line was
established using a gradient ascent model (8, 16, 32, 64, 128 and
256 ng/ml TRAIL) from parental TRAIL-sensitive H460 cells. Cells
were continuously exposed to 50 ng/ml TRAIL to maintain their
resistant capability.
Reagents and plasmids
TRAIL was purchased from Shanghai Kaibao
Pharmaceutical Co., Ltd. (Shanghai, China). Recombinant Wnt-3A was
purchased from Peprotech, Inc. (Rocky Hill, NJ, USA). The
pCMV-C-flag-β-catenin (pCMV-β-catenin) overexpression plasmid was
constructed and identified in our laboratory, as previously
described (14).
β-catenin-silencing plasmids were purchased from Addgene, Inc.
(Cambridge, MA, USA; cat. nos. 19761 pLKO.1.puro
shRNA.β-catenin.1248 and 18803 pLKO.1.puro shRNA β-catenin). These
will be abbreviated as shRNA1 and shRNA2, respectively.
Cell viability assay
Cell viability was assessed by cell counting kit-8
(CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan).
Cells were seeded in 96-well plates (Corning, Inc., Corning, NY,
USA) at a density of 1×106 cells/well. When the cells
reached 70–80% confluence, the experimental group was treated with
the 0, 10, 25, 50, 100 or 200 ng/ml TRAIL for 24 h. The original
medium was discarded and replaced with a basal media mixture
containing 10% (v/v) CCK-8 for 1 h. The optical density (OD) was
measured at 450 nm using a microplate reader (Rayto Life and
Analytical Sciences Co., Ltd., Guangming, China). Cell viability
was calculated using the following formula: Cell viability (%) =
(OD value of the treated wells - OD value of the blank control
wells)/(OD value of the negative control wells - OD value of the
blank control wells). All assays contained 5 replicates and were
repeated 3 times under the same conditions.
RNA isolation, reverse transcription and
RT-qPCR
Total RNA was extracted from cells with TRIzol
(Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to
the manufacturer’s protocol, and the RNA concentration was detected
using a NanoDrop spectrophotometer (Thermo Fisher Scientific,
Inc.). Reverse transcription was performed with the SuperScript
First-Strand Synthesis system (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer’s protocol. The
primer sequences were as follows (forward and reverse,
respectively): DR4, 5′-AGAGAGAAGTCCCTGCACCA-3′ and
5′-GTCACTCCAGGGCGTACAAT-3′; DR5, 5′-CACCAG GTGTGATTCAGGTG-3′ and
5′-CCCCACTGTGCTTTGTA CCT-3′; DcR1, 5′-ACCAACGCTTCCAACAA-3′ and
5′-AGG GCACCTGCTACACTT-3′; DcR2, 5′-CCTTCTTGCCTGCT ATG-3′ and
5′-GTGGTCACTGTCTCCTCC-3′; FADD, 5′-GCGAGTCTGGAAGAATGTCG-3′ and
5′-GGCTTGTCA GGGTGTTT-3′; Cellular FADD-like interleukin-1β
converting enzyme inhibitory protein (c-FLIP), 5′-GGCTCCCCCTGCAT
CACATC-3′ and 5′-CGCAGTACACAGGCTCCAGA-3′, and GAPDH,
5′-TGGAAGGACTCATGACCACA-3′ and 5′-TCAGCTCAGGGATGACCTT-3′. The
transcriptional level was determined using a SYBR Premix EX Taq II
kit (Takara Bio, Inc., Otsu, Japan) and a CFX96 RT-qPCR detection
system (Applied Biosystems; Thermo Fisher Scientific, Inc.),
according to the manufacturer’s protocol. The thermocycling
conditions were as follows: 95°C for 30 sec, followed by 40 cycles
of 95°C for 5 sec and 60°C for 30 sec, then 95°C for 10 sec and
melting curve at 65–95°C with interval changes of 0.5°C every 5
sec. The 2−ΔΔCq method was used to analyze the
fold-change in gene expression relative to GAPDH (15).
Flow cytometry
Cells were digested with EDTA-free trypsin (Corning,
Inc.), harvested and washed twice with PBS. For apoptosis assays,
cells were treated with an Annexin V-FITC/PI kit (BestBio Ltd.,
Shanghai, China), according to the manufacturer’s protocols. A
single-cell suspension was established using 400 μl binding
buffer and cells were stained with 5 μl Annexin V-FITC for
30 min followed by staining with 7 μl 20 mg/ml propridium
iodide (PI) for 5 min at room temperature. To detect cytomembrane
DRs, cells were suspended in 50 μl PBS containing 1% goat
serum at room temperature for 30 min. Cells were washed with PBS 3
times and incubated with the primary antibodies presented in
Table I overnight at 4°C.
Subsequently, cells were washed 3 times with PBS and incubated with
the secondary antibodies presented in Table II at room temperature for 30 min.
Cells were then washed with PBS and suspended in 500 μl PBS.
Cells were also processed as described but without primary antibody
treatment, as a negative control. The samples were analyzed using a
flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). All
experiments were repeated 3 times.
 | Table IList of primary antibodies used. |
Table I
List of primary antibodies used.
| Antigen | Species | Method | Dilution | Supplier (cat.
no.) |
|---|
| β-catenin,
human | Rabbit,
polyclonal | WB | 1:1,000 | Abcam (6302) |
| GAPDH, human | Rabbit,
polyclonal | WB | 1:5,000 | Proteintech
(10494-1-AP) |
| BCL-2, human | Rabbit,
monoclonal | WB | 1:2,000 | Abcam (34124) |
| BAX, human | Rabbit,
monoclonal | WB | 1:1,000 | Proteintech
(50599-2-Ig) |
| Caspase-3,
human | Rabbit,
polyclonal | WB | 1:1,500 | Proteintech
(19677-1-AP) |
| Cleaved
caspase-3 | Rabbit,
polyclonal | WB | 1:1,000 | CST (9661) |
| Caspase-8,
human | Rabbit,
polyclonal | WB | 1:1,500 | Proteintech
(13423-1-AP) |
| Cleaved
caspase-8 | Rabbit,
monoclonal | WB | 1:1,000 | CST (9496) |
| FADD, human | Rabbit,
polyclonal | WB | 1:1,000 | Abcam (24533) |
| C-FLIP, human | Rabbit,
polyclonal | WB | 1:1,000 | Abcam (ab6144) |
| Mcl-1, human | Rabbit,
polyclonal | WB | 1:1,000 | Abcam
(ab32087) |
| DcR1, human | Rabbit,
polyclonal | WB | 1:1,000 | Abcam (ab2087) |
| DcR2, human | Rabbit,
polyclonal | WB | 1:1,000 | Abcam (ab2019) |
| DR4, human | Rabbit,
monoclonal | WB | 1:1,000 | CST (42533) |
| DR5, human | Rabbit,
monoclonal | WB | 1:1,000 | CST (8074) |
| DR4, human | Rabbit,
monoclonal | IF | 1:100 | CST (42533) |
| DR5, human | Rabbit,
monoclonal | IF | 1:50 | CST (8074) |
| DR4, human | Rabbit,
monoclonal | FC | 1:50 | CST (42533) |
| DR5, human | Rabbit,
monoclonal | FC | 1:20 | Bioss
(bs-7352R) |
| Caveolin-1 | Mouse,
polyclonal | IF | 1:100 | R&D
(MAB5736-SP) |
 | Table IISecondary antibodies and DAPI
stain. |
Table II
Secondary antibodies and DAPI
stain.
| Secondary detection
system antibody | Host | Method | Dilution | Supplier (cat.
no.) |
|---|
| Anti-Mouse-IgG
(H+L)-HRP | Goat | WB | 1:10,000 | Sungene
(LK2001) |
| Anti-Rabbit-IgG
(H+L)-HRP | Goat | WB | 1:10,000 | Sungene
(LK2003) |
| Hoechst 33342
nucleic acid staining (DAPI) | – | IF | 1 μg/ml | Sigma (D8417) |
| Anti-Mouse-IgG
(H+L)-Cy3 | Goat | IF, FC | 1:100 | Proteintech
(SA00009-1) |
| Anti-Mouse-IgG
(H+L)-FITC | Goat | IF, FC | 1:100 | Proteintech
(SA00003-11) |
| Anti-Rabbit-IgG
(H+L)-R-PE | Goat | IF, FC | 1:100 | Proteintech
(SA00008-2) |
Immunofluorescence
Cells were fixed with 4% paraformaldehyde (Sangon
Biotech Co., Ltd., Shanghai, China) for 30 min and washed 3 times
with PBS. Cells were blocked with normal goat serum (MultiSciences
Biotech Co., Ltd., Zhejiang, Hangzhou, China) for 30 min at room
temperature, and incubated with primary antibodies (Table I) at 4°C overnight. Following
incubation with a FITC/Cy3-conjugated secondary antibody (Table II) for 45 min at room temperature,
the cells were washed with PBS and the slides were stained with
DAPI (1 μg/ml; 100 μl; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) at room temperature for 3 min. Images were
captured with a confocal laser scanning microscope (Nikon
Corporation, Tokyo, Japan).
β-catenin-knockdown in H460 cells using
shRNA lentivirus
The sense sequence of shRNA1 was 5′-GTGCTATCTGTCTGCT
CTA-3′, and the sense sequence of the negative control (NC) shRNA
was 5′-TTCTCCGAACGTGTCACGT-3′. Lentiviral pGMLV was used to
construct shRNA1 (cat. no. 19761 pLKO.1.sh.β-catenin.1248). The
H460 cells (7×105 cells/well) were infected with shRNA1
or NC lentiviruses (8×105 particles/well), and screened
using 3 mg/ml puromycin (Sangon Biotech Co., Ltd.) for 2 weeks.
β-catenin levels were subsequently measured using RT-qPCR and
western blot analyses.
β-catenin-overexpression in H460-TR cells
using plasmid transfection
H460-TR cells were cultured in 6-well plates and
transfected with the pCMV-β-catenin plasmid. A total of 5 μl
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) and 10 μg plasmid were added to separate
500-μl aliquots of OptiMEM (Gibco; Thermo Fisher Scientific,
Inc.), and incubated at room temperature for 5 min. The diluted
plasmid was added to the diluted Lipofectamine® 2000 at
a 1:1 ratio, and the mixture was incubated at room temperature for
20 min. When the cells reached 70–90% confluence, they were
incubated with the plasmid mixture for 6 h. RT-qPCR and western
blot analyses were performed to measure the transfection
efficiency. For the negative control, a pCMV-C-Tag2C-flag plasmid
(pCMV) was transfected following the same protocol.
Western blot analysis
Whole cell lysates were extracted using
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Haimen, China) supplemented with protease inhibitor
cocktail (Sangon Biotech Co., Ltd.). Membrane DR4 and DR5 proteins
were extracted using ProteoExtract® Transmembrane
Protein Extraction kit (Merck KGaA, Darmstadt, Germany), according
to the manufacturer’s protocol. The protein content of the
supernatant was detected using a bicinchoninic acid assay (Pierce;
Thermo Fisher Scientific, Inc.) using bovine serum albumin as a
standard. A total of 40 μg protein/lane was separated by 10%
SDS-PAGE. The proteins were transferred to polyvinylidene fluoride
membranes, which were subsequently probed overnight at 4°C with the
primary antibodies listed in Table
I. The membranes were washed 3 times with Tris-buffered saline
with Tween and incubated with the appropriate secondary antibodies
(Table II) at room temperature
for 90 min. Membranes were visualized using an enhanced
chemiluminescence kit and exposed using a gel imaging analyzer
(both from Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
All experiments were repeated 3 times and the
results are presented as the mean ± standard deviation. 50%
effective concentration (EC50) values were analyzed
using GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA,
USA) with the following equation: Y = minimum dose + maximum
dose-minimum dose)/1+10LogEC50-X. Differences between 2
groups were analyzed using unpaired Student’s t-tests. P<0.05
was considered to indicate a statistically significant
difference.
Results
Establishment and identification of the
acquired TRAIL-resistant H460-TR cell line
To explore the molecular mechanism underlying
acquired TRAIL-resistance, TRAIL-resistant H460-TR cells were
established using H460 cells. No morphological differences were
observed between H460-TR and H460 cell lines; both were polygonal,
adherent and island-like. When exposed to 80 ng/ml TRAIL for 4 h,
the majority of H460 cells presented apoptotic features, while
H460-TR cells maintained a typical epithelioid monolayer (Fig. 1A). The toxicity of TRAIL to H460-TR
and H460 cells was assessed using CCK-8 assays and was demonstrated
to be dose-dependent. The EC50 value of TRAIL in H460-TR
cells was 201.4 ng/ml, compared with 65.47 ng/ml in H460 cells
(Fig. 1B). Furthermore, the number
of Annexin V and TUNEL-positive cells decreased in the H460-TR cell
line compared with the parental H460 cell line (Fig. 1C and D). These results indicate the
successful establishment of an acquired TRAIL-resistant cell
line.
The extrinsic and intrinsic apoptotic
signaling pathways were inhibited in acquired TRAIL-resistant NSCLC
cells
The effects of TRAIL on the extrinsic and intrinsic
apoptotic signaling pathways in the established TRAIL-resistant
H460-TR cells were investigated. No significant difference was
observed in the expression of FADD or caspase-8 protein between
H460 and H460-TR cells; however, the mRNA and protein expression
levels of c-FLIP were increased in the TRAIL-resistant cells
(Fig. 2A and B). The endogenous
anti-apoptotic protein, Mcl-1, was upregulated in H460-TR cells
(Fig. 2C). Furthermore, the
expression of cleaved caspase-3/8 was significantly decreased in
H460-TR cells following 80 ng/ml TRAIL treatment (Fig. 2D). These results indicate that the
extrinsic and intrinsic apoptotic signaling pathways are
downregulated in cells with acquired TRAIL-resistance compared with
cells without.
TRAIL-induced redistribution of DR4 and
DR5 to the cytomembrane is inhibited in H460-TR cells
TRAIL-induced apoptosis was reported to be initiated
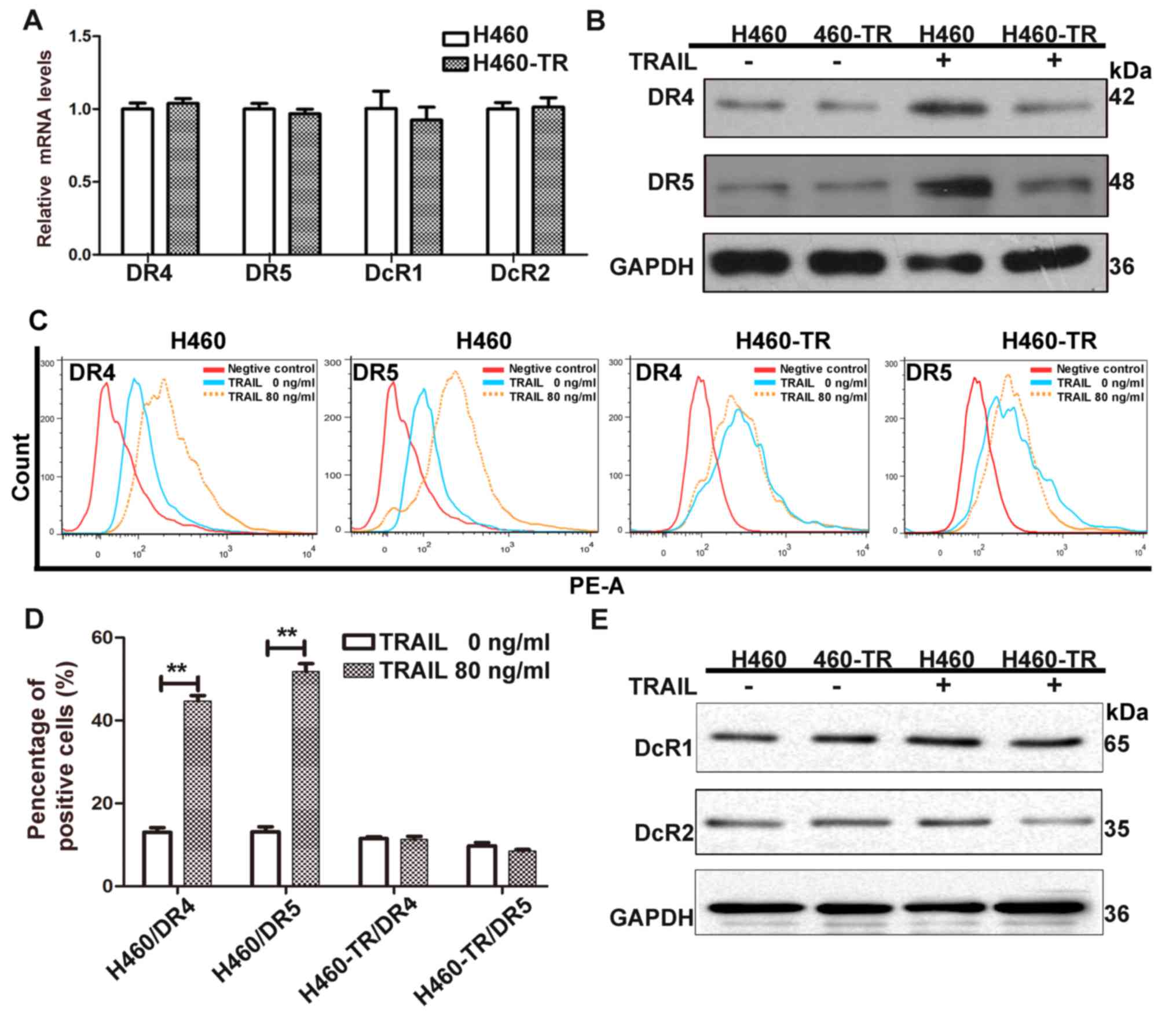
by TRAIL binding to DRs. The mRNA expression of TRAIL receptors was
comparable between TRAIL-sensitive and TRAIL-resistant cell lines
prior to TRAIL treatment (Fig.
3A). The levels of DR4 and DR5 expression in the cytomembrane
were increased in H460 cells following TRAIL treatment, whereas no
significant difference was observed in H460-TR cells following
treatment compared with pre-treatment levels (Fig. 3B and D). Furthermore, the
expression of DcR1 and DcR2 was not affected by TRAIL treatment in
H460 or H460-TR cells (Fig. 3E).
The lack of TRAIL-induced DR redistribution may be a key factor
affecting TRAIL sensitivity in H460-TR cells.
β-catenin is relevant to TRAIL
sensitivity
To evaluate whether β-catenin is involved in TRAIL
resistance, the expression of β-catenin was analysed using RT-qPCR
and western blot analysis. β-catenin was expressed in H460 cells at
approximately twice the level of that observed in H460-TR cells
(P<0.05; Fig. 4A and B). These
results indicate that β-catenin is positively associated with TRAIL
sensitivity.
Knockdown of β-catenin reduced drug
sensitivity in TRAIL-sensitive H460 cells
shRNA was used to suppress β-catenin transcription
in H460 cells and to investigate its effect on TRAIL sensitivity
and caspase activation. β-catenin mRNA expression was reduced
following infection with shRNA1 to a greater degree than with
shRNA2, therefore, shRNA1 was selected for use in further
experiments (Fig. 4C). Silencing
of β-catenin rendered H460 cells less sensitive to TRAIL, as
revealed by the results of a CCK-8 assay (Fig. 4D). Flow cytometry was performed to
analyze the apoptosis of H460 cells following treatment with or
without 80 ng/ml TRAIL for 8 h. The results further demonstrated
that β-catenin-silencing attenuates the cytotoxicity of TRAIL in
H460 cells (Fig. 4E and F). The
western blotting proteins bands for cleaved-caspase-3/8 were also
reduced following β-catenin-silencing (Fig. 4G). These results indicate that
β-catenin-silencing decreases TRAIL-sensitivity by inactivating
caspase proteins.
Overexpression of β-catenin enhances drug
sensitivity in TRAIL-resistant H460-TR cells
To further investigate the association between
β-catenin expression and TRAIL sensitivity, TRAIL-resistant H460-TR
cells were transfected with pCMV-β-catenin, which was successfully
constructed in our previous study (14). Following transfection and puromycin
screening, the expression of β-catenin protein was markedly
upregulated in H460-TR cells compared with untransfected cells
(Fig. 5A). The cells were next
treated with TRAIL for 24 h and the results demonstrated that
β-catenin overexpression rendered H460-TR cells more sensitive to
TRAIL compared with untransfected cells (Fig. 5B). Annexin V-FITC/PI staining also
indicated that β-catenin overexpression increased the apoptotic
rate of H460-TR cells treated with 80 ng/ml TRAIL for 8 h (Fig. 5C and D) compared with untreated
cells. As expected, the protein expression bands of
cleaved-caspase-3/8 were larger following β-catenin overexpression
(Fig. 5E). These results indicate
that β-catenin overexpression increases TRAIL sensitivity via
activating caspase.
β-catenin upregulates DR4 and DR5 in
NSCLC cells
To determine the effects of β-catenin on DR4 and DR5
in the context of altered TRAIL sensitivity, the expression of
these DRs on the cell membrane was assessed following
β-catenin-silencing or overexpression. The expression of DR4 and
DR5 mRNA and protein was reduced in TRAIL-sensitive H460 cells
following β-catenin downregulation (Fig. 6A and B). The expression of DR4 and
DR5 was increased in TRAIL-resistant H460-TR cells following
β-catenin overexpression (Fig. 6C
and D). Flow cytometry confirmed that β-catenin overexpression led
to an increase in DR expression in TRAIL-resistant H460-TR cells
compared with untransfected cells (Fig. 6E and F). Caveolin-1 is a marker of
lipid rafts. The colocalization of Caveolin-1 and DR4/5 was
assessed using immunofluorescence and confocal microscopy. The
results suggest that β-catenin enhanced the localization and
redistribution of DR4 and DR5 to lipid rafts (Fig. 6G). This indicates that β-catenin
promotes DR translocation to the cell membrane, allowing them to
combine more effectively with TRAIL and activate pro-apoptotic
caspase proteins, ultimately inducing apoptosis and reversing
TRAIL-resistance.
 | Figure 6β-catenin upregulates DR4 and DR5
expression in NSCLC cells. DR4 and DR5 (A) mRNA and (B) protein
expression in H460 cells infected with or without shRNA1 was
detected. The mRNA and protein expression levels were normalized to
GAPDH. DR4 and DR5 (C) mRNA and (D) protein expression levels in
H460-TR cells with or without pCMV-β-catenin. (E) Cytomembranal DR4
and DR5 in H460-TR cells transfected with or without pCMV-β-catenin
was assessed by flow cytometry, suggesting that β-catenin
upregulated the cytomembranal expression of DR4 and DR5. (F)
Quantified flow cytometry data. (G) Immunofluorescence microscopy
showing the localization and redistribution of DR4 and DR5 in lipid
rafts induced by β-catenin overexpression. Green, red and blue
areas indicate expression of DR, caveolin-1 and DAPI, respectively.
Yellow areas indicate the colocalization of DR and caveolin-1
expression. Magnification, ×800. Scale bar, 25 μm. All
assays were repeated 3 times and data are presented as the mean ±
standard deviation. *P<0.05. NC, negative control;
shRNA, short hairpin RNA; DR, death receptor. TRAIL, tumor necrosis
factor related apoptosis inducing ligand. |
Discussion
TRAIL is able to selectively target and kill tumor
cells without causing damage to normal cells (3,4). DRs
are often located on tumor cells and, upon activation by TRAIL, the
oligomerization of DRs recruits the linker molecule, Fas-associated
death domain (FADD), and pro-caspase-8, which together comprise the
death inducing signaling complex (DISC) (6). Activated caspase-8 directly induces
apoptosis via activating caspase-3, which cleaves a broad range of
apoptosis-associated protein substrates and executes the extrinsic
apoptosis pathway (16,17). In addition, activated caspase-8
truncates BH3 interacting domain death agonist along with the
pro-apoptotic proteins, BCL-2 associated X, apoptosis regulator and
BCL-2 antagonist/killer. However, the clinical application of TRAIL
is limited due to the prevalence of drug resistance (16,17).
TRAIL-resistance may be intrinsic, occurring at the first exposure
to TRAIL, or acquired resistance, developing during treatment
(18). At present, the mechanism
of acquired TRAIL resistance remains to be elucidated. TRAIL
resistance is caused by various factors, including endoplasmic
reticulum stress (19), protein
synthesis disorders (20),
decreased DRs expression (21) and
increased anti-apoptotic protein expression (22). Our results revealed that β-catenin
expression is positively associated with TRAIL sensitivity via
promoting the cytomembrane redistribution of DR4 and DR5.
c-FLIP, which is similar in structure to caspase-8,
and competitively binds FADD molecules, thus impeding the cleavage
of caspase-8 and subsequent signal transduction in the intrinsic
apoptotic signaling pathway (23).
It has previously been demonstrated that TRAIL and a DR5 agonist,
AD5-10, cleave c-FLIP in H460 cells (24). In human renal carcinoma Caki cells,
TRAIL has been reported to downregulate c-FLIP expression and
induce apoptosis (25). Mcl-1, an
anti-apoptotic protein that belongs to the BCL-2 family, has also
been reported to induce TRAIL resistance (26). TRAIL inhibits Mcl-1 expression via
activating the pro-apoptotic activity of p38 in the receptor
interacting serine/threonine kinase 1-dependent pathway in H460
cells (27). It has also been
demonstrated that YM155 sensitizes TRAIL-induced apoptosis via
cathepsin S-dependent downregulation of Mcl-1 expression, and
nuclear factor-κB-mediated downregulation of c-FLIP expression in
Caki cells (28). In the present
study, the expression of c-FLIP and Mcl-1 was higher in H460-TR
cells compared with H460 cells. These results indicate that the
DR-associated apoptotic pathway and the intrinsic apoptotic pathway
are responsible for changes in TRAIL-sensitivity.
The binding of TRAIL to its receptors is the first
step in TRAIL-induced apoptotic signaling. The cytomembrane
expression of DR4 and DR5, rather than the general expression of
these DRs, is the main determinant of TRAIL sensitivity (6). SW480 colon cancer cells are
characterized as TRAIL-resistant cells that express high levels of
DR4, even though cytomembranal DR4 is undetectable (29). The present study revealed that
baseline DR4 and DR5 expression was not affected by
TRAIL-sensitivity. Nevertheless, the redistribution of DR4 and DR5
to the membrane of TRAIL-sensitive H460 cells was increased
following TRAIL treatment, while the cytomembranal expression
levels of DR4 and DR5 in TRAIL-resistant H460-TR cells were not
significantly altered (Fig. 3B and
D). A previous study reported that TRAIL-induced DR transportation
into lipid rafts in TRAIL-sensitive cells, while redistribution was
not observed in TRAIL-resistant cells (30). As such, the cytomembranal
expression levels of DR4 and DR5 after TRAIL treatment is critical
for determining TRAIL sensitivity.
Lung cancer comprises a group of molecularly
heterogeneous diseases that are characterized by a range of genomic
and epigenomic alterations (31).
In ongoing experiments by our research group involving a gene
expression profiling chip in TRAIL-sensitive H460 cells, the
acquired TRAIL-resistant H460-TR cells and the primary resistant
A549 cells have demonstrated that multiple targets and genetic
alternations may be associated with the resistance process
(unpublished). In further experiments, it was demonstrated that
β-catenin was highly expressed in TRAIL-sensitive cells compared
with resistant cells. Further investigation of the association
between β-catenin and TRAIL sensitivity is required.
In the canonical Wnt pathway, β-catenin accumulates
in the cytoplasm upon Wnt stimulation and eventually translocates
to the nucleus to act as a transcriptional coactivator (32). The Wnt/β-catenin signaling pathway
participates in cell adhesion, embryonic development and
tumorigenesis (33). β-catenin and
DRs are co-expressed in colonic tumor tissues. DR expression
gradually increased during colon carcinoma tumorigenesis, possibly
due to upregulation of β-catenin expression (12). In TRAIL-resistant melanoma cells,
Wnt-3A was revealed to activate Wnt/β-catenin signaling, promote
the expression of the apoptotic molecules, BIM and PUMA, and reduce
the expression of the anti-apoptotic protein, Mcl-1, thus
increasing sensitivity to TRAIL (34). It was also demonstrated that Wnt-3A
upregulated β-catenin expression and activated caspase-3/8 in
H460-TR cells. β-catenin expression was markedly lower in cells
with acquired TRAIL resistance compared with TRAIL-sensitive cells,
and thus induced sensitivity to TRAIL.
While the present study demonstrated that β-catenin
is a critical determinant of acquired TRAIL resistance in NSCLC
cells, the exact molecular mechanisms responsible for TRAIL
resistance remain unclear. β-catenin-knockdown or overexpression
was used to determine whether β-catenin regulates TRAIL-sensitivity
via affecting DR4 and DR5 expression. When β-catenin expression was
downregulated in H460 cells, DR4 and DR5 expression levels were
reduced and resistance to TRAIL increased. These results were
consistent with a previous report that downregulation of β-catenin
expression in melanoma cells reduced their sensitivity to TRAIL
(34). Accordingly, β-catenin
overexpression in H460-TR cells significantly increased DR4 and DR5
expression levels and induced TRAIL-sensitivity. Upregulation of
β-catenin using lithium chloride has been reported to sensitize
A549 cells to TRAIL (35). A
previous study using APC-null colorectal cancer cells revealed that
β-catenin upregulated c-MYC expression, which subsequently
downregulated c-FLIP expression to promote TRAIL-induced apoptosis
(36). It has been reported that
Wnt/β-catenin signalling induces apoptosis via a caspase-dependent
apoptosis mechanism by downregulating expression of Mcl-1 (37). In the present study, it was
revealed that β-catenin expression is positively associated with
cytomembranal expression levels of DR4 and DR5, indicating that
β-catenin may promote TRAIL-sensitivity by inducing the
cytomembranal redistribution of DR4 and DR5.
β-catenin forms a complex with E-cadherin.
Interestingly, β-catenin, E-cadherin and DR expression has been
detected in the lipid rafts of the cell membrane (38,39).
Altering β-catenin and E-cadherin-mediated intercellular adhesion
has been reported to induce epithelial-mesenchymal-transition and
thus inhibit apoptosis in tumor cells (40). It was therefore speculated that the
E-cadherin/β-catenin complex enhanced cytomembranal translocation
of DR4 and DR5 in β-catenin-overexpressing H460 cells. As β-catenin
expression levels decline in TRAIL-resistant cells, the
E-cadherin/β-catenin complex may dissociate and the cytomembranal
expression levels of DRs and DISC may be attenuated. As such, low
β-catenin expression is associated with acquired TRAIL-resistance
in NSCLC cells.
In the present study, an acquired TRAIL-resistant
lung cancer cell line, H460-TR, was successfully constructed. It
was demonstrated that the expression levels of DRs induced by TRAIL
on the cell membrane is a key factor affecting acquired TRAIL
resistance. TO the best of our knowledge, the present study it the
first to indicate that β-catenin promotes DR-translocation to the
cell membrane, allowing them to combine with TRAIL more
effectively, to activate the downstream family of caspase
pro-apoptotic molecules, to induce apoptosis and reverse
TRAIL-resistance. In future studies, an in vivo nude mouse
recombinant TRAIL xenograft model should be used to validate our
in vitro results. The detailed molecular mechanisms by which
β-catenin enhances TRAIL sensitivity remain to be elucidated.
β-catenin overexpression or induction using Wnt-3A presents a
potential therapeutic strategy to enhance TRAIL sensitivity of
NSCLC cells.
Funding
The present study was funded by the Chinese National
Natural Science Foundation (grant nos. 81572967, 81372498 and
81773236), Hubei Natural Science Foundation (grant no. 2013CFA006),
Zhongnan Hospital of Wuhan University Science, Technology and
Innovation Seed Fund (grant nos. znpy2016050 and znpy2017049),
Wuhan City Huanghe Talents Plan and Chinese National Key Clinical
Speciality Construction Program (CX), the Zhongnan Hospital of
Wuhan University Science, Technology and Innovation Seed Fund
(grant no. znpy2017001) and the Fundamental Research Funds for the
Central Universities (grant no. 2042018kf0066, YG).
Availability of data and materials
All data generated or analyzed during this study
were included in this published article.
Authors’ contributions
CY, SHIMIN Z, YG and CX designed the present study.
CY, SHIMIN Z, YS and SHIYU Z acquired the data. CY, SHIMIN Z, GT
and FT analyzed the data. XL, YX and JZ interpreted the data. CY
and SHIMIN Z drafted the manuscript. CY, SHIMIN Z, YG and CX
provided critical revision. All authors approved the version to be
published.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Conflicts of interest
The authors declare that they have no conflicts of
interest.
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
c-FLIP
|
cellular FADD-like interleukin-1β
converting enzyme inhibitory protein
|
|
DR
|
death receptor
|
|
DcR
|
decoy receptor
|
|
DISC
|
death inducing signaling complex
|
|
EMT
|
epithelial-mesenchymal transition
|
|
FADD
|
Fas associated death domain
|
|
BAX
|
BCL-2 associated X protein
|
|
BAK
|
BCL-2-antagonist/killer
|
Acknowledgments
The authors would like to thank Xiaohua Leng for
excellent technical assistance.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cao C, D’Amico T, Demmy T, Dunning J,
Gossot D, Hansen H, He J, Jheon S, Petersen RH, Sihoe A, et al
International VATS Interest Group: Surgery versus SABR for
resectable non-small-cell lung cancer. Lancet Oncol. 16:e370–e371.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wiley SR, Schooley K, Smolak PJ, Din WS,
Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA,
et al: Identification and characterization of a new member of the
TNF family that induces apoptosis. Immunity. 3:673–682. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hao C, Song JH, Hsi B, Lewis J, Song DK,
Petruk KC, Tyrrell DL and Kneteman NM: TRAIL inhibits tumor growth
but is nontoxic to human hepatocytes in chimeric mice. Cancer Res.
64:8502–8506. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pitti RM, Marsters SA, Ruppert S, Donahue
CJ, Moore A and Ashkenazi A: Induction of apoptosis by Apo-2
ligand, a new member of the tumor necrosis factor cytokine family.
J Biol Chem. 271:12687–12690. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ashkenazi A: Directing cancer cells to
self-destruct with pro-apoptotic receptor agonists. Nat Rev Drug
Discov. 7:1001–1012. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hotte SJ, Hirte HW, Chen EX, Siu LL, Le
LH, Corey A, Iacobucci A, MacLean M, Lo L, Fox NL, et al: A phase 1
study of mapatumumab (fully human monoclonal antibody to TRAIL-R1)
in patients with advanced solid malignancies. Clin Cancer Res.
14:3450–3455. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Merchant MS, Geller JI, Baird K, Chou AJ,
Galli S, Charles A, Amaoko M, Rhee EH, Price A, Wexler LH, et al:
Phase I trial and pharmacokinetic study of lexatumumab in pediatric
patients with solid tumors. J Clin Oncol. 30:4141–4147. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Greco FA, Bonomi P, Crawford J, Kelly K,
Oh Y, Halpern W, Lo L, Gallant G and Klein J: Phase 2 study of
mapatumumab, a fully human agonistic monoclonal antibody which
targets and activates the TRAIL receptor-1, in patients with
advanced non-small cell lung cancer. Lung Cancer. 61:82–90. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ashkenazi A, Pai RC, Fong S, Leung S,
Lawrence DA, Marsters SA, Blackie C, Chang L, McMurtrey AE, Hebert
A, et al: Safty and antitumor activity of recombinant soluble Apo 2
ligand. J Clin Invest. 104:155–162. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hao C, Beguinot F, Condorelli G, Trencia
A, Van Meir EG, Yong VW, Parney IF, Roa WH and Petruk KC: Induction
and intracellular regulation of tumor necrosis factor-related
apoptosis-inducing ligand (TRAIL) mediated apotosis in human
malignant glioma cells. Cancer Res. 61:1162–1170. 2001.PubMed/NCBI
|
|
12
|
Jalving M, Heijink DM, Koornstra JJ,
Boersma-van Ek W, Zwart N, Wesseling J, Sluiter WJ, de Vries EG,
Kleibeuker JH and de Jong S: Regulation of TRAIL receptor
expression by β-catenin in colorectal tumours. Carcinogenesis.
35:1092–1099. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu M, Marsters S, Ye X, Luis E, Gonzalez L
and Ashkenazi A: E-cadherin couples death receptors to the
cytoskeleton to regulate apoptosis. Mol Cell. 54:987–998. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang B, Zhang S, Wang Z, Yang C, Ouyang W,
Zhou F, Zhou Y and Xie C: Deubiquitinase USP9X deubiquitinates
β-catenin and promotes high grade glioma cell growth. Oncotarget.
7:79515–79525. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
16
|
Rudner J, Jendrossek V, Lauber K, Daniel
PT, Wesselborg S and Belka C: Type I and type II reactions in
TRAIL-induced apoptosis - results from dose-response studies.
Oncogene. 24:130–140. 2005. View Article : Google Scholar
|
|
17
|
Ozören N and El-Deiry WS: Defining
characteristics of types I and II apoptotic cells in response to
TRAIL. Neoplasia. 4:551–557. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
de Miguel D, Lemke J, Anel A, Walczak H
and Martinez-Lostao L: Onto better TRAILs for cancer treatment.
Cell Death Differ. 23:733–747. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Teng Y, Gao M, Wang J, Kong Q, Hua H, Luo
T and Jiang Y: Inhibition of eIF2α dephosphorylation enhances
TRAIL-induced apoptosis in hepatoma cells. Cell Death Dis.
5:e10602014. View Article : Google Scholar
|
|
20
|
Fan S, Li Y, Yue P, Khuri FR and Sun SY:
The eIF4E/eIF4G interaction inhibitor 4EGI-1 augments
TRAIL-mediated apoptosis through c-FLIP down-regulation and DR5
induction independent of inhibition of cap-dependent protein
translation. Neoplasia. 12:346–356. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Haimovici A, Humbert M, Federzoni EA,
Shan-Krauer D, Brunner T, Frese S, Kaufmann T, Torbett BE and
Tschan MP: PU.1 supports TRAIL-induced cell death by inhibiting
NF-κB-mediated cell survival and inducing DR5 expression. Cell
Death Differ. 24:866–877. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mert U and Sanlioglu AD: Intracellular
localization of DR5 and related regulatory pathways as a mechanism
of resistance to TRAIL in cancer. Cell Mol Life Sci. 74:245–255.
2017. View Article : Google Scholar
|
|
23
|
Safa AR and Pollok KE: Targeting the
anti-apoptotic protein c-FLIP for cancer therapy. Cancers (Basel).
3:1639–1671. 2011. View Article : Google Scholar
|
|
24
|
Chen F, Guo J, Zhang Y, Zhao Y, Zhou N,
Liu S, Liu Y and Zheng D: Knockdown of c-FLIP(L) enhanced AD5-10
anti-death receptor 5 monoclonal antibody-induced apoptosis in
human lung cancer cells. Cancer Sci. 100:940–947. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jeon MY, Min KJ, Woo SM, Seo SU, Kim S,
Park JW and Kwon TK: Volasertib enhances sensitivity to TRAIL in
renal carcinoma Caki cells through downregulation of c-FLIP
expression. Int J Mol Sci. 18:1–12. 2017. View Article : Google Scholar
|
|
26
|
Murphy ÁC, Weyhenmeyer B, Noonan J,
Kilbride SM, Schimansky S, Loh KP, Kögel D, Letai AG, Prehn JH and
Murphy BM: Modulation of Mcl-1 sensitizes glioblastoma to
TRAIL-induced apoptosis. Apoptosis. 19:629–642. 2014. View Article : Google Scholar :
|
|
27
|
Azijli K1, Yuvaraj S, van Roosmalen I,
Flach K, Giovannetti E, Peters GJ, de Jong S and Kruyt FA: MAPK p38
and JNK have opposing activities on TRAIL-induced apoptosis
activation in NSCLC H460 cells that involves RIP1 and caspase-8 and
is mediated by Mcl-1. Apoptosis. 18:851–860. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Woo SM, Min KJ, Seo BR and Kwon TK: YM155
sensitizes TRAIL-induced apoptosis through cathepsin S-dependent
down-regulation of Mcl-1 and NF-κB-mediated down-regulation of
c-FLIP expression in human renal carcinoma Caki cells. Oncotarget.
7:61520–61532. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jin Z, McDonald ER III, Dicker DT and
El-Deiry WS: Deficient tumor necrosis factor-related
apoptosis-inducing ligand (TRAIL) death receptor transport to the
cell surface in human colon cancer cells selected for resistance to
TRAIL-induced apoptosis. J Biol Chem. 279:35829–35839. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ouyang W, Yang C, Zhang S, Liu Y, Yang B,
Zhang J, Zhou F, Zhou Y and Xie C: Absence of death receptor
translocation into lipid rafts in acquired TRAIL-resistant NSCLC
cells. Int J Oncol. 42:699–711. 2013. View Article : Google Scholar
|
|
31
|
Inamura K: Lung Cancer: Understanding its
molecular pathology and the 2015 WHO classification. Front Oncol.
7:1932017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Polakis P: Drugging Wnt signalling in
cancer. EMBO J. 31:2737–2746. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Valenta T, Hausmann G and Basler K: The
many faces and functions of β-catenin. EMBO J. 31:2714–2736. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zimmerman ZF, Kulikauskas RM, Bomsztyk K,
Moon RT and Chien AJ: Activation of Wnt/β-catenin signaling
increases apoptosis in melanoma cells treated with trail. PLoS One.
8:e695932013. View Article : Google Scholar
|
|
35
|
Lan Y, Liu X, Zhang R, Wang K, Wang Y and
Hua ZC: Lithium enhances TRAIL-induced apoptosis in human lung
carcinoma A549 cells. Biometals. 26:241–254. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang L, Ren X, Alt E, Bai X, Huang S, Xu
Z, Lynch PM, Moyer MP, Wen XF and Wu X: Chemoprevention of
colorectal cancer by targeting APC-deficient cells for apoptosis.
Nature. 464:1058–1061. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu X, Deng G, Hao X, Li Y, Zeng J, Ma C,
He Y, Liu X and Wang Y: A caspase-dependent pathway is involved in
Wnt/β-catenin signaling promoted apoptosis in Bacillus
Calmette-Guerin infected RAW264.7 macrophages. Int J Mol Sci.
15:5045–5062. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Galbiati F, Volonte D, Brown AM, Weinstein
DE, Ben-Ze’ev A, Pestell RG and Lisanti MP: Caveolin-1 expression
inhibits Wnt/beta-catenin/Lef-1 signaling by recruiting
beta-catenin to caveolae membrane domains. J Biol Chem.
275:23368–23377. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gajate C and Mollinedo F:
Cytoskeleton-mediated death receptor and ligand concentration in
lipid rafts forms apoptosis-promoting clusters in cancer
chemotherapy. J Biol Chem. 280:11641–11647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Brozovic A: The relationship between
platinum drug resistance and epithelial-mesenchymal transition.
Arch Toxicol. 91:605–619. 2017. View Article : Google Scholar
|