Introduction
Esophageal squamous cell carcinoma (ESCC) is the
most prevalent type of esophageal cancer and is the sixth leading
cause of cancer mortality worldwide (1). Due to ESCC's aggressive nature, the
prognosis of ESCC patients with local invasion and distant
metastasis at diagnosis is poor (2,3).
Surgical resection is recognized as the preferred treatment for
patients with newly diagnosed ESCC. However, high rates of tumor
recurrence are notable (4,5). Neoadjuvant chemotherapy or
chemoradiotherapy have been demonstrated to prolong overall
survival for patients with ESCC (6-8).
However, treatment options for recurrent cases are limited, and
recently approved targeted therapies have not observed effective
therapeutic effects (9,10). Therefore, ESCC patients with
recurrence and metastasis require novel and effective treatment
strategies.
The latest genomic analyses of ESCC cells have
exhibited epigenetic modifications, e.g., DNA methylation, histone
deacetylation, chromatin remodeling and non-coding RNA regulation
(11,12). In that regard, microRNAs (miRNAs or
miRs) consist of a class of small, well-conserved, non-coding RNAs
that regulate RNA transcripts in a sequence-dependent manner
(13). They participate in
physiological and pathological conditions, e.g., cell
differentiation, proliferation, motility and metabolism (14). A single miRNA can control a vast
number of RNA transcripts in normal and diseased cells (15). Therefore, aberrantly expressed
miRNAs may break down regulated RNA networks and contribute to
cancer cells' development, metastasis and drug resistance (16).
A large number of miRNAs exhibit differential
expression in ESCC, and they contribute to ESCC pathogenesis
through their activities as oncogenes or tumor suppressors
(11). Analyses of our original
miRNA expression signatures by RNA-sequencing revealed that both
strands of the miR-145 duplex (miR-145-5p, the guide
strand and miR-145-3p, the passenger strand) were
significantly downregulated in several types of cancers (17-20).
The traditional view of miRNA function has held that only one
strand of the miRNA duplex is incorporated into the RNA-induced
silencing complex (RISC), becoming the active strand (guide
strand). In contrast, the other strand (the passenger strand or
miRNA*) was thought to be degraded and to have no
function (21,22). However, recent studies of miRNA
biogenesis have demonstrated that certain miRNA passenger strands
are functional in plant and human cells (20,23).
Our recent studies have demonstrated that both
strands of the miR-145 duplex have antitumor roles in lung
cancer, bladder cancer, prostate cancer and head and neck cancer
(17-20). In ESCC cells, both strands of the
miR-150 duplex (miR-150-5p, the guide strand, and
miR-150-3p, the passenger strand) acted as antitumor miRNAs
through their targeting of SPOCK1 (24). A number of studies demonstrated
that miR-145-5p acted as a pivotal antitumor miRNA in human
cancers (25), including ESCC
(26). In contrast, the functional
significance and the targets of miR-145-3p are still obscure.
The aim of the present study was to demonstrate that
miR-145-3p possesses antitumor functions and to identify its
molecular targets, thereby elucidating ESCC pathogenesis. Thus, in
ESCC cells, it was demonstrated that ectopic expression of
miR-145-3p significantly blocked cancer cell proliferation,
migration and invasion, similar to the actions of
miR-145-5p. Furthermore, it was demonstrated that two genes,
dehydrogenase/reductase member 2 (DHRS2) and myosin IB
(MYO1B), were directly regulated by antitumor
miR-145-3p in ESCC cells. Involvement of miR-145-3p
(the passenger strand) is a novel concept in ESCC oncogenesis. The
present approach, based on the roles of antitumor miRNA and its
targets, will contribute to improved understanding of the molecular
pathogenesis of ESCC.
Materials and methods
Human ESCC clinical specimens and cell
lines
The present study was approved by the Bioethics
Committee of Kagoshima University (Kagoshima, Japan; approval no.
28-65). Written prior informed consent and approval were obtained
from all of the patients. All subjects in the patient cohort (n=29)
were diagnosed with ESCC based upon pathologic criteria. ESCC with
curative resection was included, salvage surgery was excluded. From
this group, 22 clinical specimens and 12 noncancerous esophageal
tissues were obtained. All samples were collected at the Kagoshima
University hospital from March 2010 to September 2014. Collection
of resected tissues occurred prior to preoperative therapy. The
clinicopathological features of the patients are presented in
Tables I and II. ESCC is more prevalent among males,
thus almost all cases recruited for the present study were
male.
 | Table IClinicopathological features of
esophageal squamous cell carcinoma patients. |
Table I
Clinicopathological features of
esophageal squamous cell carcinoma patients.
| No. | Age (years) | Sex |
Differentiation | T | N | M | Stage | ly | v | Recurrence |
|---|
| 1 | 52 | Male | Poor | 1b | 0 | 0 | IA | 1 | 1 | + |
| 2 | 72 | Male | Moderate | 1b | 0 | 0 | IA | 0 | 1 | − |
| 3 | 69 | Male | Moderate | 1b | 0 | 0 | IA | 0 | 0 | − |
| 4 | 56 | Male | Moderate | 2 | 0 | 0 | IB | 0 | 1 | − |
| 5 | 66 | Male | Moderate | 3 | 0 | 0 | IIA | 1 | 1 | − |
| 6 | 70 | Male | Moderate | 3 | 0 | 0 | IIA | 1 | 1 | + |
| 7 | 66 | Male | Moderate | 3 | 0 | 0 | IIA | 1 | 1 | − |
| 8 | 71 | Male | Well | 3 | 0 | 0 | IIA | 1 | 2 | − |
| 9 | 62 | Male | Well | 1a | 1 | 0 | IIB | 0 | 0 | − |
| 10 | 68 | Male | Moderate | 1b | 1 | 0 | IIB | 1 | 1 | − |
| 11 | 60 | Male | Moderate | 1b | 1 | 0 | IIB | 1 | 1 | − |
| 12 | 71 | Male | Moderate | 1b | 1 | 0 | IIB | 0 | 0 | − |
| 13 | 84 | Male | Well | 2 | 1 | 0 | IIB | 1 | 1 | − |
| 14 | 79 | Male | Moderate | 2 | 1 | 0 | IIB | 1 | 1 | − |
| 15 | 60 | Male | Moderate | 2 | 1 | 0 | IIB | 1 | 2 | − |
| 16 | 68 | Male | Poor | 1b | 2 | 0 | IIIA | 1 | 3 | + |
| 17 | 67 | Male | Well | 3 | 2 | 0 | IIIB | 2 | 2 | + |
| 18 | 55 | Male | Moderate | 3 | 2 | 0 | IIIB | 1 | 1 | + |
| 19 | 75 | Male | Moderate | 3 | 2 | 0 | IIIB | 1 | 1 | + |
| 20 | 74 | Male | Moderate | 2 | 3 | 0 | IIIC | 3 | 1 | + |
| 21 | 57 | Male | Poor | 3 | 3 | 0 | IIIC | 1 | 1 | + |
| 22 | 63 | Male | Well | 3 | 3 | 0 | IIIC | 2 | 1 | + |
 | Table IIFeatures of patients in noncancerous
esophageal tissues. |
Table II
Features of patients in noncancerous
esophageal tissues.
| No. | Age (years) | Sex |
|---|
| 1 | 66 | Male |
| 2 | 55 | Male |
| 3 | 52 | Male |
| 4 | 78 | Male |
| 5 | 75 | Male |
| 6 | 60 | Male |
| 7 | 71 | Male |
| 8 | 64 | Male |
| 9 | 79 | Female |
| 10 | 81 | Male |
| 11 | 69 | Male |
| 12 | 84 | Male |
In addition, 2 ESCC cell lines were used: TE-8,
moderately differentiated and TE-9, poorly differentiated (27) (RIKEN BioResource Center, Tsukuba,
Japan). Cell culture, extraction of total RNA and extraction of
protein were performed as described in previous reports (28,29).
Transfection of mimic and inhibitor
miRNA, small interfering (si)RNA into ESCC cells
In the present study, the following mimic and
inhibitor miRNAs or siRNAs were transfected: mimic miRNAs (Ambion
Pre-miR miRNA precursor; miR-145-5p:
5'-GUCCAGUUUUCCCAGGAAUCCCU-3', ID: PM11480; hsa-miR-145-3p:
5'-GGAUUCCUGGAAAUACUGUUCU-3', ID: PM13036; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), inhibitor miRNAs (Anti-miR
miRNA Inhibitor; has-miR-145-3p: 5'-GGAUUCCUGGAAAUACUG
UUCU-3', ID: AM13036; Applied Biosystems; Thermo Fisher Scientific,
Inc.) and siRNAs (Stealth Select RNAi siRNA; si-DHRS2, ID:
HSS145497 and HSS173461; si-MYO1B, ID: HSS106714 and
HSS106716; Invitrogen; Thermo Fisher Scientific, Inc.) and negative
control miRNA/siRNA (product ID: AM17111; Thermo Fisher Scientific,
Inc.). The transfection procedures were performed as previously
described (28-30).
Incorporation of miR-145-3p into the
RISC: Assessment by argonaute 2 (Ago2) immunoprecipitation
miRNAs were transfected into TE-8 cells and miRNAs
were isolated using an microRNA Isolation Kit, Human Ago2 (Wako
Pure Chemical Industries, Ltd., Osaka, Japan) as described
previously (28-30). The expression levels of
Ago2-conjugated miRNAs were assessed by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
miR-21 (assay ID; 000397; Applied Biosystems) was used as the
internal control.
RT-qPCR
Quantification of miRNAs and mRNAs was performed by
StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific, Inc.).
The procedure used for RT-qPCR has been described previously
(28,29). The expression levels of miRNAs were
analyzed using TaqMan RT-qPCR assays (miR-145-5p, assay ID:
002278; miR-145-3p assay ID: 002149; Applied Biosystems;
Thermo Fisher Scientific, Inc.). Data were normalized to
RNU48 (assay ID: 001006; Applied Biosystems). In addition,
the expression levels of DHRS2 and MYO1B were
assessed with the following TaqMan probes: DHRS2, assay ID:
Hs01061575_g1; MYO1B, assay ID: Hs00362654_m1; Applied
Biosystems; Thermo Fisher Scientific, Inc.), and normalized to
glucuronidase β (assay ID: Hs00939627_ml; Applied Biosystems;
Thermo Fisher Scientific, Inc.).
Cell proliferation, migration, invasion
and apoptosis assays
Protocols for determining cell proliferation (XTT
assays), migration and invasion were described previously (28,29).
For apoptosis assays, double staining with fluorescein
isothiocyanate (FITC)-Annexin V and propidium iodide was carried
out using a FITC Annexin V Apoptosis Detection kit (BD Biosciences,
Franklin Lakes, NJ, USA) according to the manufacturer's
recommendations. Stains were analyzed within 1 h using a flow
cytometer (CyAn ADP analyzer; Beckman Coulter, Inc., Brea, CA,
USA). Cells were identified as viable cells, dead cells, early
apoptotic cells and late apoptotic cells using Summit 4.3 software
(Beckman Coulter, Inc.). The percentages of early apoptotic and
late apoptotic cells from each experiment were then compared. As a
positive control, 1 µM gemcitabine hydrochloride (Tokyo Chemical
Industry Co., Ltd., Tokyo, Japan) was used.
Identification of putative target genes
regulated by miR-145-3p in ESCC cells
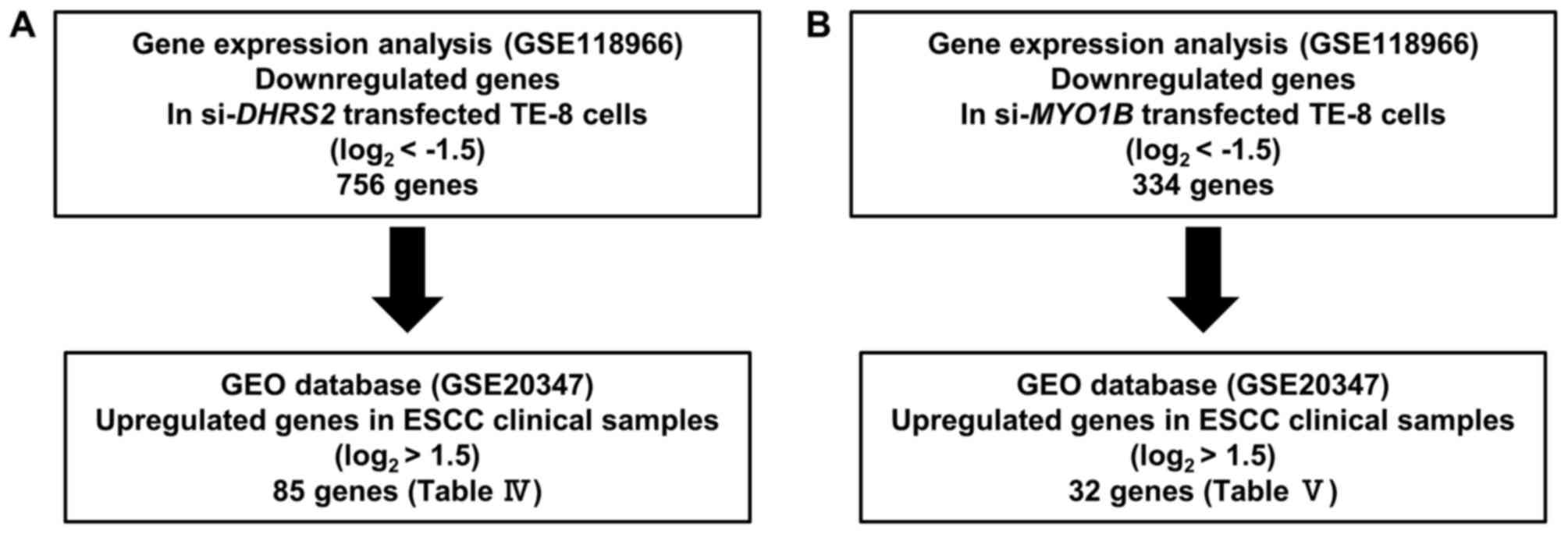
The present strategy for identification of
miR-145-3p target genes is outlined in Fig. 1. The microarray data were deposited
in the Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo/) under
accession number GSE107008. Putative target genes with a binding
site for miR-145-3p were detected by TargetScanHuman ver.7.1
(http://www.targetscan.org/vert_71/).
The GEO database (GSE20347) was used for assessment of the
association between target genes and ESCC.
Exploration of downstream targets
regulated by si-DHRS2 and si-MYO1B in ESCC
Genome-wide microarray analysis was used for
identification of DHRS2 and MYO1B downstream targets.
Expression data were deposited in a GEO database (GSE118966). A GEO
database (GSE20347) was used for assessment of the association
between target genes and ESCC. Our strategy for identification of
DHRS2 and MYO1B downstream targets is outlined in
Fig. 2. Expression analysis was
performed on microarray data using SurePrint G3 Human 8×60K v3
(Agilent Technologies, Inc., Santa Clara, CA, USA).
Western blot analysis
Anti-human DHRS2 rabbit polyclonal immunoglobulin
(Ig)G (1:1,000; HPA069551; Sigma-Aldrich; Merck KGaA) and
anti-human MYO1B rabbit polyclonal IgG (1:250; HPA013607;
Sigma-Aldrich; Merck KGaA) were used as primary antibodies.
Anti-human β-actin mouse monoclonal IgG (1:2,000; A1978;
Sigma-Aldrich; Merck KGaA) was used as an internal control. The
protocol for Western blot analysis was described previously
(29,30).
Immunohistochemistry
Tumor specimens were fixed, embedded and sectioned
as described previously (31).
Anti-human DHRS2 rabbit polyclonal IgG (1:250; HPA069551;
Sigma-Aldrich, St. Louis, MO, USA) and anti-human MYO1B rabbit
polyclonal IgG (1:300; HPA013607; Sigma-Aldrich; Merck KGaA were
used as primary antibodies. The protocol followed was described
previously (32).
Luciferase reporter assays
The following sequences were inserted into the
psiCHECk-2 vector (C8021; Promega Corporation, Madison, WI, USA):
The wild-type sequences of the 3'-untranslated regions (UTRs) of
DHRS2 and MYO1B, or the deletion-type, which lacks
the miR-145-3p target sites from DHRS2 (position 270-276) or
MYO1B (position 88-94 or position 1,117-1,123). The cloned
vectors were co-transfected into ESCC cells with mature
miR-145-3p. The procedures for transfection and
dual-luciferase reporter assays have been reported previously
(28,29).
Statistical analysis
Associations between groups were analyzed using the
Mann-Whitney U test or Tukey's multiple comparisons test following
one-way analysis of variance. The differences between survival
rates were analyzed by Kaplan-Meier survival curves and log-rank
statistics. Spearman's rank test was used to evaluate the
correlations between the expression levels of miR-145-3p,
miR-145-5p, DHRS2 and MYO1B. Data are
presented as the mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Expert StatView version 5.0 (SAS Institute, Inc., Cary, NC, USA)
and GraphPad Prism version 7.04 (GraphPad Software, Inc., La Jolla,
CA, USA) were used in these analyses.
Results
Expression levels of miR-145-5p and
miR-145-3p in ESCC clinical specimens
Expression levels of miR-145-5p and
miR-145-3p were significantly downregulated in cancer
tissues and ESCC cell lines relative to normal tissues (Fig. 3A). Spearman's rank test
demonstrated a positive correlation between the expression levels
of miR-145-5p and miR-145-3p (Fig. 3B).
It was demonstrated that 5-year survival was
significantly higher in patients who had high miR-145-3p
expression than in patients with low miR-145-3p expression
(Fig. 3C). There was no
significant association between the expression level of
miR-145-5p and patient survivals (Fig. 3C).
Ectopic expression of miR-145-5p and
miR-145-3p: Impact on ESCC cells
To assess the ectopic expression of
miR-145-5p and miR-145-3p, the mimic miRNA was
transfected to ESCC cell lines (Fig.
3D). XTT assays demonstrated significant inhibition of cell
proliferation in miR-145-5p and miR-145-3p
transfectants (Fig. 3E). Likewise,
cell migration and invasion were significantly inhibited following
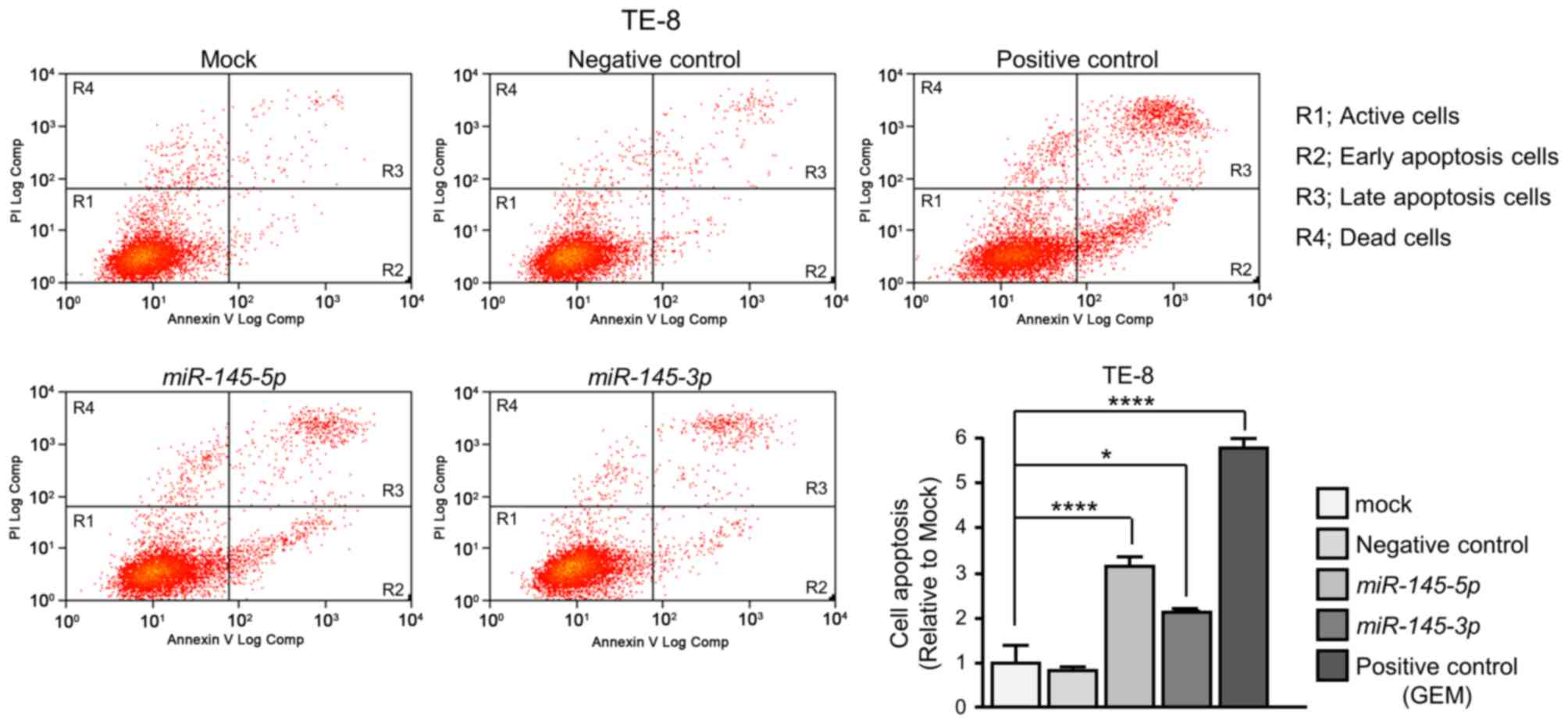
miR-145-5p or miR-145-3p transfection (Fig. 3F and G). The numbers of early
apoptotic and late apoptotic cells were significantly larger in
miR-145-5p or miR-145-3p transfectants than in mock
or negative control transfectants (Fig. 4).
Incorporation of miR-145-3p into the RISC
in ESCC cells
It was anticipated that the passenger strand of
miR-145-3p was incorporated into the RISC and served as a
tumor suppressor in ESCC cells. To verify that hypothesis, Ago2 was
immunoprecipitated in cells that had been transfected with either
miR-145-5p or miR-145-3p (Fig. 5A). Ago2 is an essential component
of the RISC (16). Isolated
Ago2-bound miRNAs were analyzed by RT-qPCR to confirm whether
miR-145-5p and miR-145-3p bound to Ago2. In TE-8
cells, miR-145-5p transfectants demonstrated higher
expression levels of miR-145-5p than mock transfectants,
miR-control or miR-145-3p transfectants. Similarly,
following miR-145-3p transfection, miR-145-3p was
detected by Ago2 immunoprecipitation (Fig. 5B). miR-145-5p and
miR-145-3p were demonstrated to bind to Ago2 separately and
were incorporated into RISC, thereby demonstrating miRNA
function.
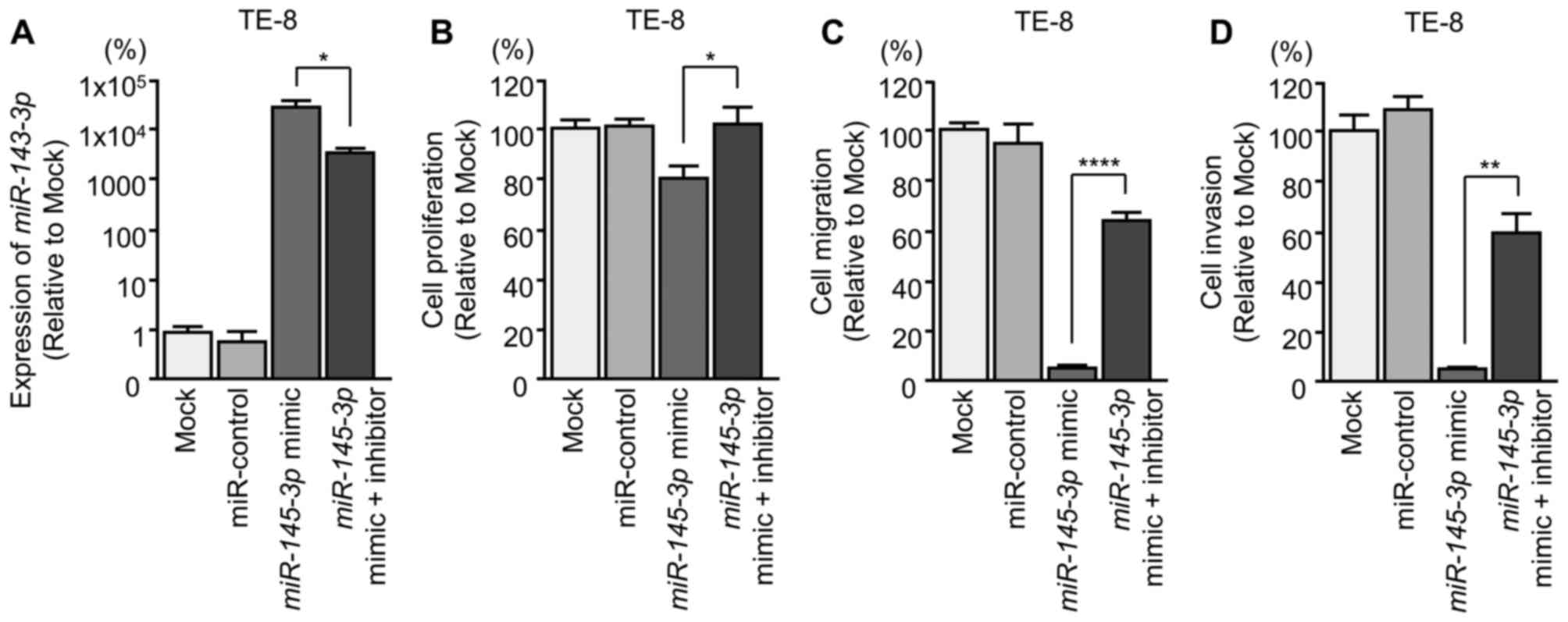
Effects of co-transfection of mimic and
inhibitor miR-145-3p into ESCC cells
To confirm the antitumor effects of
miR-145-3p, rescue experiments were performed using mimic
and inhibitor miR-145-3p with TE-8 cells (Fig. 6A). The rescue experiments indicated
that cancer cell proliferation, migration and invasion were rescued
in miR-145-3p inhibitor transfectants compared with restored
miR-145-3p mimic only (Fig.
6B-D).
Searching for putative targets regulated
by miR-145-3p in ESCC cells
The strategy to identify miR-145-3p target
genes is presented in Fig. 1. Gene
expression analyses demonstrated that 1,374 genes were
downregulated (log2 ratio <-1.0) in miR-145-3p
transfected TE-8 cells compared with control transfectants. The
present expression data were deposited in the GEO repository under
accession no. GSE107008. Among these downregulated genes, genes
that had putative miR-145-3p binding sites in their 3'-UTRs
were selected using information in the TargetScan database. A total
of 280 genes were identified. Then, 30 genes were selected by
restricting the identified genes to those strongly upregulated in
ESCC clinical specimens (log2 ratio >1.0; GEO
accession no. GSE20347; Table
III).
 | Table IIIPutative targets of miR-145-3p
regulation in ESCC cells. |
Table III
Putative targets of miR-145-3p
regulation in ESCC cells.
| Entrez gene ID | Gene symbol | Gene name | TE-8
miR-145-3p transfectant | ESCC GSE20347
fold-change | Target site
count | Prognosis P-value:
TCGA OncoLnc data
|
|---|
| ESCA | ESCC |
|---|
| 10202 | DHRS2 |
Dehydrogenase/reductase (SDR family)
member 2 | −2.69 | 2.02 | 1 | 0.047 | 0.708 |
| 1848 | DUSP6 | Dual specificity
phosphatase 6 | −2.61 | 1.00 | 1 | 0.456 | 0.469 |
| 55157 | DARS2 | Aspartyl-tRNA
synthetase 2, mitochondrial | −2.09 | 1.17 | 2 | 0.504 | 0.706 |
| 6646 | SOAT1 | Sterol
O-acyltransferase 1 | −2.06 | 1.81 | 1 | 0.732 | 0.667 |
| 4430 | MYO1B | Myosin IB | −1.98 | 1.61 | 2 | 0.372 | 0.856 |
| 2115 | ETV1 | Ets variant 1 | −1.84 | 1.10 | 1 | 0.142 | 0.119 |
| 983 | CDK1 | Cyclin-dependent
kinase 1 | −1.63 | 1.95 | 1 | 0.621 | 0.136 |
| 1719 | DHFR | Dihydrofolate
reductase | −1.48 | 1.14 | 1 | 0.199 | 0.465 |
| 51053 | GMNN | Geminin, DNA
replication inhibitor | −1.46 | 1.37 | 1 | 0.274 | 0.189 |
| 23321 | TRIM2 | Tripartite motif
containing 2 | −1.35 | 1.45 | 1 | <0.001a | 0.037a |
| 55697 | VAC14 | Vac14 homolog (S.
cerevisiae) | −1.34 | 1.53 | 1 | 0.052 | 0.095 |
| 79789 | CLMN | Calmin
(calponin-like, transmembrane) | −1.33 | 1.79 | 2 | 0.519 | 0.352 |
| 5654 | HTRA1 | HtrA serine
peptidase 1 | −1.30 | 1.44 | 1 | 0.863 | 0.252 |
| 54830 | NUP62CL | Nucleoporin 62 kDa
C-terminal like | −1.28 | 1.10 | 1 | 0.313 | 0.313 |
| 204 | AK2 | Adenylate kinase
2 | −1.24 | 1.21 | 2 | 0.691 | 0.972 |
| 126321 | MFSD12 | Major facilitator
superfamily domain containing 12 | −1.23 | 1.05 | 1 | 0.838 | 0.098 |
| 9532 | BAG2 | BCL2-associated
athanogene 2 | −1.20 | 1.80 | 2 | 0.109 | 0.563 |
| 51029 | DESI2 | Desumoylating
isopeptidase 2 | −1.18 | 1.25 | 2 | 0.261 | 0.19 |
| 6711 | SPTBN1 | Spectrin, β,
non-erythrocytic 1 | −1.18 | 1.21 | 1 | 0.515 | 0.504 |
| 1163 | CKS1B | CDC28 protein
kinase regulatory subunit 1B | −1.14 | 2.02 | 1 | 0.658 | 0.658 |
| 8534 | CHST1 | Carbohydrate
(keratan sulfate Gal-6) Sulfotransferase 1 | −1.14 | 1.52 | 1 | 0.943 | 0.983 |
| 5174 | PDZK1 | PDZ domain
containing 1 | −1.14 | 1.65 | 1 | 0.565 | 0.462 |
| 875 | CBS |
Cystathionine-β-synthase | −1.14 | 3.55 | 2 | 0.199 | 0.160 |
| 23516 | SLC39A14 | Solute carrier
family 39 (zinc transporter), member 14 | −1.12 | 2.68 | 1 | 0.322 | 0.101 |
| 6790 | AURKA | Aurora kinase
A | −1.07 | 2.17 | 1 | 0.322 | 0.051 |
| 79718 | TBL1XR1 | Transducin (β)-like
1 X-linked receptor 1 | −1.06 | 1.30 | 1 | 0.579 | 0.377 |
| 23141 | ANKLE2 | Ankyrin repeat and
LEM domain containing 2 | −1.05 | 1.16 | 1 | 0.418 | 0.585 |
| 23649 | POLA2 | Polymerase (DNA
directed), α 2, accessory subunit | −1.04 | 1.35 | 1 | 0.642 | 0.842 |
| 64151 | NCAPG | Non-SMC condensin I
complex, subunit G | −1.03 | 1.22 | 1 | 0.198 | 0.056 |
| 22848 | AAK1 | AP2 associated
kinase 1 | −1.03 | 1.27 | 3 | 0.634 | 0.346 |
In this fashion, DHRS2 was focused on, as its
expression was the most downregulated in miR-145-3p
transfectants and the most upregulated in ESCC clinical specimens.
Additionally, MYO1B was examined, as it was more highly
downregulated in miR-145-3p transfectants and was more
upregulated in ESCC clinical specimens. In addition, our previous
study had demonstrated that the activation of MYO1B was
associated with cancer cell aggressiveness (20).
Direct regulation of DHRS2 and MYO1B by
miR-145-3p in ESCC cells
The finding that DHRS2 and MYO1B were
downregulated by expression of miR-145-3p was further
investigated (Figs. 7 and 8). ESCC cells (TE-8) that had been
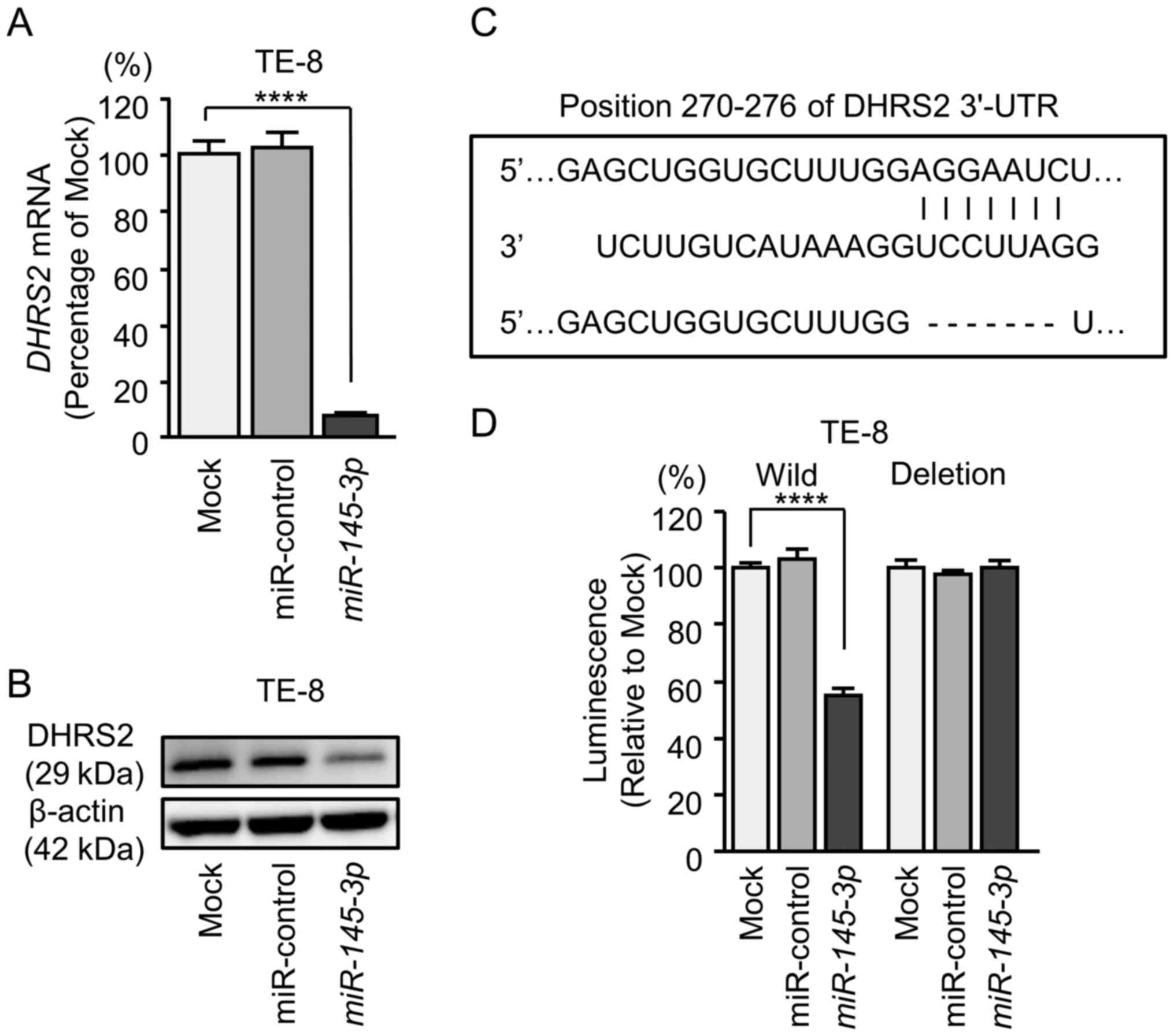
transfected with miR-145-3p were examined. Using RT-qPCR, it
was demonstrated that DHRS2 and MYO1B mRNA levels
were significantly reduced by miR-145-3p transfection
(Figs. 7A and 8A). Furthermore, western blot analysis
was performed to measure the expression levels of DHRS2 and MYO1B
proteins in the transfectants. Results demonstrated that the
proteins were also reduced by miR-145-3p transfection
(Figs. 7B and 8B).
Whether miR-145-3p directly regulated
DHRS2 and MYO1B genes in a sequence-dependent manner
was then evaluated.
The Human TargetScan database predicted that
DHRS2 had 1 binding site (positions 270-276) for
miR-145-3p in the 3'-UTR (Fig.
7C). Accordingly, luciferase reporter assays were carried out
with vectors that included either the wild-type or deletion-type
3'-UTR of DHRS2. Co-transfection with miR-145-3p and
vectors including the wild-type sequence significantly reduced
luciferase activity compared with those in mock and miR-control
transfectants in position 270-276 of the DHRS2 3'-UTR
(Fig. 7D).
MYO1B had 2 binding sites (positions 88-94
and 1,117-1,123) for miR-145-3p in the 3'-UTR (Fig. 8C). Luciferase activities were
significantly reduced in position 1,117-1,123 of the MYO1B
3'-UTR (Fig. 8D).
Effects of silencing DHRS2 and MYO1B in
ESCC cells
Subsequently, siRNAs were transfected into TE-8
cells to examine the function of DHRS2 and MYO1B in
ESCC cells (Figs. 9 and 10). The mRNA and protein expression
levels of DHRS2 and MYO1B were decreased by
si-DHRS2 and si-MYO1B, respectively (Figs. 9A and B, and 10A and B). Subsequently, the effects of
DHRS2 or MYO1B knockdown on ESCC cell proliferation,
migration and invasion were investigated.
Cancer cell proliferation was significantly
suppressed in si-DHRS2 or si-MYO1B transfectants
compared with mock and si-RNA-control transfectants. Additionally,
migration and invasion activities were significantly inhibited in
si-DHRS2 or si-MYO1B transfectants (Figs. 9C-E and 10C-E). In the apoptosis assays,
si-DHRS2_1/si-DHRS2_2 and
si-MYO1B_1/si-MYO1B_2 transfections significantly
increased apoptotic TE-8 cells (Figs.
9F and 10F).
Expression of DHRS2 and MYO1B in ESCC
clinical specimens
Based upon the findings above, it was of great
interest to use RT-qPCR to determine the expression levels of
DHRS2 and MYO1B in clinical specimens. DHRS2
and MYO1B expression levels were significantly upregulated
in ESCC tumor tissues (Fig. 11A).
Additionally, the 5-year survival rates of ESSC patients were
significantly shorter in those with elevated DHRS2
expression compared with those with low expression (Fig. 11B). There was no significant
association between the expression levels of MYO1B and the
survival rate (Fig. 11B).
Spearman's rank test demonstrated a negative
correlation between the expression of DHRS2 and
miR-145-3p, and MYO1B and miR-145-3p (Fig. 11C). Furthermore, the protein
expression levels of DHRS2 and MYO1B were examined in ESCC clinical
specimens by immunostaining. Both DHRS2 and MYO1B were strongly
expressed in cancer tissues, but not in noncancerous epithelia
(Fig. 12).
Exploration of downstream targets
regulated by si-DHRS2 and si-MYO1B in ESCC
The present strategy for selecting downstream genes
regulated by DHRS2 and MYO1B is demonstrated in
Fig. 2. A total of 756 genes were
commonly downregulated (log2 ratio <-1.5) in
si-DHRS2-transfected TE-8 cells. The upregulated genes in
ESCC tissues were also assessed by GEO database analyses (GEO
accession no. GSE20347). With that approach, 85 candidate genes
downstream from DHRS2 were identified (Table IV). Furthermore, a total of 334
genes were commonly downregulated (log2 ratio <−1.5)
in si-MYO1B-transfected TE-8 cells. In a similar approach,
32 candidate genes downstream from MYO1B were identified
(Table V).
 | Table IVPutative targets of si-DHRS2
regulation in ESCC cells. |
Table IV
Putative targets of si-DHRS2
regulation in ESCC cells.
| Entrez gene ID | Gene symbol | Gene name | ESCC GSE20347
fold-change | TE-8
si-DHRS2 transfectant |
|---|
| 1592 | CYP26A1 | Cytochrome P450,
family 26, subfamily A, polypeptide 1 | 1.86 | −3.78 |
| 10112 | KIF20A | Kinesin family
member 20A | 1.50 | −3.28 |
| 259266 | ASPM | Asp (abnormal
spindle) homolog, microcephaly associated (Drosophila) | 1.56 | −3.08 |
| 27074 | LAMP3 |
Lysosomal-associated membrane protein
3 | 1.56 | −3.03 |
| 11098 | PRSS23 | Protease, serine,
23 | 1.86 | −3.02 |
| 10202 | DHRS2 |
Dehydrogenase/reductase (SDR family)
member 2 | 2.02 | −2.98 |
| 4322 | MMP13 | Matrix
metallopeptidase 13 (collagenase 3) | 5.12 | −2.93 |
| 79075 | DSCC1 | DNA replication and
sister chromatid cohesion 1 | 1.95 | −2.80 |
| 983 | CDK1 | Cyclin-dependent
kinase 1 | 1.95 | −2.79 |
| 332 | BIRC5 | Baculoviral IAP
repeat containing 5 | 1.59 | −2.72 |
| 995 | CDC25C | Cell division cycle
25C | 1.51 | −2.71 |
| 11065 | UBE2C |
Ubiquitin-conjugating enzyme E2C | 1.68 | −2.70 |
| 4751 | NEK2 | NIMA-related kinase
2 | 1.66 | −2.70 |
| 1033 | CDKN3 | Cyclin-dependent
kinase inhibitor 3 | 1.94 | −2.70 |
| 11339 | OIP5 | Opa interacting
protein 5 | 1.63 | −2.68 |
| 2842 | GPR19 | G protein-coupled
receptor 19 | 2.12 | −2.64 |
| 10615 | SPAG5 | Sperm associated
antigen 5 | 1.59 | −2.60 |
| 9055 | PRC1 | Protein regulator
of cytokinesis 1 | 1.58 | −2.60 |
| 699 | BUB1 | BUB1 mitotic
checkpoint serine/threonine kinase | 2.04 | −2.59 |
| 991 | CDC20 | Cell division cycle
20 | 1.54 | −2.58 |
| 55355 | HJURP | Holliday junction
recognition protein | 1.79 | −2.51 |
| 7153 | TOP2A | Topoisomerase (DNA)
II alpha 170 kDa | 1.91 | −2.46 |
| 10403 | NDC80 | NDC80 kinetochore
complex component | 1.76 | −2.45 |
| 55388 | MCM10 | Minichromosome
maintenance complex component 10 | 1.90 | −2.45 |
| 2263 | FGFR2 | Fibroblast growth
factor receptor 2 | 1.65 | −2.44 |
| 3161 | HMMR | Hyaluronan-mediated
motility receptor (RHAMM) | 1.60 | −2.42 |
| 1063 | CENPF | Centromere protein
F, 350/400 kDa | 2.31 | −2.40 |
| 7272 | TTK | TTK protein
kinase | 1.58 | −2.36 |
| 9401 | RECQL4 | RecQ protein-like
4 | 1.92 | −2.35 |
| 9355 | LHX2 | LIM homeobox 2 | 2.63 | −2.33 |
| 22836 | RHOBTB3 | Rho-related BTB
domain containing 3 | 1.98 | −2.33 |
| 54478 | FAM64A | Family with
sequence similarity 64, member A | 1.60 | −2.33 |
| 9133 | CCNB2 | Cyclin B2 | 1.68 | −2.27 |
| 9787 | DLGAP5 | Discs, large
(Drosophila) homolog-associated protein 5 | 1.72 | −2.26 |
| 9156 | EXO1 | Exonuclease 1 | 1.89 | −2.26 |
| 3833 | KIFC1 | Kinesin family
member C1 | 2.15 | −2.19 |
| 347733 | TUBB2B | Tubulin, beta 2B
class IIb | 1.86 | −2.18 |
| 220134 | SKA1 | Spindle and
kinetochore associated complex subunit 1 | 1.73 | −2.18 |
| 4291 | MLF1 | Myeloid leukemia
factor 1 | 1.90 | −2.15 |
| 8438 | RAD54L | RAD54-like (S.
cerevisiae) | 2.31 | −2.14 |
| 79019 | CENPM | Centromere protein
M | 2.10 | −2.14 |
| 51514 | DTL | Denticleless E3
ubiquitin protein ligase homolog (Drosophila) | 1.62 | −2.12 |
| 55872 | PBK | PDZ binding
kinase | 1.70 | −2.11 |
| 3790 | KCNS3 | Potassium
voltage-gated channel, modifier subfamily S, member 3 | 2.32 | −2.11 |
| 51512 | GTSE1 | G-2 and S-phase
expressed 1 | 2.02 | −2.10 |
| 9493 | KIF23 | Kinesin family
member 23 | 1.96 | −2.10 |
| 5983 | RFC3 | Replication factor
C (activator 1) 3, 38 kDa | 1.74 | −2.09 |
| 81611 | ANP32E | Acidic
(leucine-rich) nuclear phosphoprotein 32 family, member E | 1.52 | −2.07 |
| 83461 | CDCA3 | Cell division cycle
associated 3 | 2.14 | −2.05 |
| 23350 | U2SURP | U2 snRNP-associated
SURP domain containing | 1.62 | −2.01 |
| 6790 | AURKA | Aurora kinase
A | 2.17 | −2.00 |
| 55165 | CEP55 | Centrosomal protein
55 kDa | 1.94 | −1.99 |
| 80178 | C16orf59 | Chromosome 16 open
reading frame 59 | 1.61 | −1.98 |
| 2305 | FOXM1 | Forkhead box
M1 | 2.16 | −1.96 |
| 24137 | KIF4A | Kinesin family
member 4A | 1.95 | −1.94 |
| 22974 | TPX2 | TPX2,
microtubule-associated | 1.65 | −1.94 |
| 55215 | FANCI | Fanconi anemia,
complementation group I | 1.70 | −1.91 |
| 10635 | RAD51AP1 | RAD51 associated
protein 1 | 2.20 | −1.88 |
| 993 | CDC25A | Cell division cycle
25A | 1.88 | −1.88 |
| 2175 | FANCA | Fanconi anemia,
complementation group A | 1.93 | −1.85 |
| 4171 | MCM2 | Minichromosome
maintenance complex component 2 | 2.55 | −1.82 |
| 2491 | CENPI | Centromere protein
I | 1.81 | −1.81 |
| 655 | BMP7 | Bone morphogenetic
protein 7 | 1.54 | −1.77 |
| 4998 | ORC1 | Origin recognition
complex, subunit 1 | 1.53 | −1.76 |
| 10036 | CHAF1A | Chromatin assembly
factor 1, subunit A (p150) | 1.75 | −1.76 |
| 4085 | MAD2L1 | MAD2 mitotic arrest
deficient-like 1 (yeast) | 1.67 | −1.76 |
| 3149 | HMGB3 | High mobility group
box 3 | 2.01 | −1.74 |
| 29028 | ATAD2 | ATPase family, AAA
domain containing 2 | 1.96 | −1.73 |
| 9837 | GINS1 | GINS complex
subunit 1 (Psf1 homolog) | 1.64 | −1.73 |
| 51659 | GINS2 | GINS complex
subunit 2 (Psf2 homolog) | 1.86 | −1.72 |
| 5984 | RFC4 | Replication factor
C (activator 1) 4, 37 kDa | 2.08 | −1.69 |
| 55839 | CENPN | Centromere protein
N | 1.72 | −1.66 |
| 7078 | TIMP3 | TIMP
metallopeptidase inhibitor 3 | 2.23 | −1.66 |
| 27346 | TMEM97 | Transmembrane
protein 97 | 1.67 | −1.65 |
| 9928 | KIF14 | Kinesin family
member 14 | 2.14 | −1.64 |
| 3625 | INHBB | Inhibin, β B | 1.75 | −1.63 |
| 10721 | POLQ | Polymerase (DNA
directed), θ | 1.51 - | 1.62 |
| 1663 | DDX11 | DEAD/H
(Asp-Glu-Ala-Asp/His) box helicase 11 | 2.08 | −1.60 |
| 9319 | TRIP13 | Thyroid hormone
receptor interactor 13 | 2.02 | −1.58 |
| 4605 | MYBL2 | V-myb avian
myeloblastosis viral oncogene | 3.08 | −1.56 |
| | homolog-like 2 | | |
| 51762 | RAB8B | RAB8B, member RAS
oncogene family | 1.76 | −1.54 |
| 91860 | CALML4 | Calmodulin-like
4 | 1.90 | −1.54 |
| 10293 | TRAIP | TRAF interacting
protein | 1.50 | −1.52 |
| 2237 | FEN1 | Flap
structure-specific endonuclease 1 | 1.54 | −1.51 |
| 55753 | OGDHL | Oxoglutarate
dehydrogenase-like | 2.37 | −1.51 |
 | Table VPutative targets of si-MYO1B
regulation in ESCC cells. |
Table V
Putative targets of si-MYO1B
regulation in ESCC cells.
| Entrez gene ID | Gene symbol | Gene name | ESCC GSE20347
fold-change | TE-8
si-MYO1B transfectant |
|---|
| 4430 | MYO1B | Myosin IB | 1.61 | −3.60 |
| 347733 | TUBB2B | Tubulin, β 2B class
IIb | 1.86 | −2.89 |
| 9837 | GINS1 | GINS complex
subunit 1 (Psf1 homolog) | 1.64 | −2.72 |
| 4322 | MMP13 | Matrix
metallopeptidase 13 (collagenase 3) | 5.12 | −2.66 |
| 79075 | DSCC1 | DNA replication and
sister chromatid cohesion 1 | 1.95 | −2.55 |
| 4312 | MMP1 | Matrix
metallopeptidase 1 (interstitial collagenase) | 6.54 | −2.40 |
| 51514 | DTL | Denticleless E3
ubiquitin protein ligase homolog (Drosophila) | 1.62 | −2.38 |
| 4319 | MMP10 | Matrix
metallopeptidase 10 (stromelysin 2) | 4.51 | −2.28 |
| 23657 | SLC7A11 | Solute carrier
family 7 (anionic amino acid transporter light chain, xc- system),
member 11 | 1.97 | −2.20 |
| 5983 | RFC3 | Replication factor
C (activator 1) 3, 38 kDa | 1.74 | −2.11 |
| 10202 | DHRS2 |
Dehydrogenase/reductase (SDR family)
member 2 | 2.02 | −2.04 |
| 2491 | CENPI | Centromere protein
I | 1.81 | −2.01 |
| 4085 | MAD2L1 | MAD2 mitotic arrest
deficient-like 1 (yeast) | 1.67 | −1.99 |
| 6574 | SLC20A1 | Solute carrier
family 20 (phosphate transporter), member 1 | 1.52 | −1.91 |
| 4998 | ORC1 | Origin recognition
complex, subunit 1 | 1.53 | −1.84 |
| 81611 | ANP32E | Acidic
(leucine-rich) nuclear | 1.52 | −1.83 |
| | phosphoprotein 32
family, member E | | |
| 10669 | CGREF1 | Cell growth
regulator with EF-hand domain 1 | 1.62 | −1.81 |
| 55388 | MCM10 | Minichromosome
maintenance complex component 10 | 1.90 | −1.79 |
| 2119 | ETV5 | Ets variant 5 | 2.05 | −1.79 |
| 11199 | ANXA10 | Annexin A10 | 2.06 | −1.76 |
| 655 | BMP7 | Bone morphogenetic
protein 7 | 1.54 | −1.73 |
| 983 | CDK1 | Cyclin-dependent
kinase 1 | 1.95 | −1.72 |
| 55872 | PBK | PDZ binding
kinase | 1.70 | −1.70 |
| 4072 | EPCAM | Epithelial cell
adhesion molecule | 2.56 | −1.68 |
| 8914 | TIMELESS | Timeless circadian
clock | 1.54 | −1.65 |
| 9518 | GDF15 | Growth
differentiation factor 15 | 1.58 | −1.65 |
| 55215 | FANCI | Fanconi anemia,
complementation group I | 1.70 | −1.62 |
| 11339 | OIP5 | Opa interacting
protein 5 | 1.63 | −1.61 |
| 10635 | RAD51AP1 | RAD51 associated
protein 1 | 2.20 | −1.59 |
| 332 | BIRC5 | Baculoviral IAP
repeat containing 5 | 1.59 | −1.58 |
| 995 | CDC25C | Cell division cycle
25C | 1.51 | −1.54 |
| 8438 | RAD54L | RAD54-like (S.
cerevisiae) | 2.31 | −1.52 |
Discussion
Downregulation of miR-145-5p has frequently
been observed in a wide range of cancers, including ESCC (32). A number of previous studies have
demonstrated that ectopic expression of miR-145-5p
suppressed cancer cell proliferation, migration, invasion and drug
resistance both in vitro and in vivo (25,26,33).
Notably, the promoter region of pre-miR-145 has a p53
response element and its expression is controlled by activation of
p53 under various conditions (34). Therefore, both strands of the
miR-145 duplex are pivotal tumor suppressor miRNAs
controlled by p53.
Analyses of the miRNA expression signatures
demonstrated that certain miRNA passenger strands were
downregulated and acted as antitumor miRNAs in several cancers,
e.g., miR-145-3p, miR-150-3p, miR-148a-5p and
miR-99a-3p (20,24,35,36).
Our previous studies demonstrated that antitumor miR-145-3p
directly targeted oncogenes, e.g., MTDH in lung
adenocarcinoma, UHRF1 in bladder cancer, MYO1B in
head and neck cancer and MELK, NCAPG, BUB1 and
CDK1 in prostate cancer (17-20).
Another group demonstrated that miR-145-3p inhibited cell
growth, motility and chemotaxis in non-small cell lung cancer by
targeting pyruvate dehydrogenase kinase 1 through suppressing the
mechanistic target of rapamycin pathway (37). These findings indicated that
antitumor miR-145-3p is associated with cancer pathogenesis.
Exploring the RNA network controlled by antitumor
miR-145-3p expands the understanding of the novel molecular
pathogenesis of ESCC. In the present study, 30 genes were
identified as putative oncogenes based on miR-145-3p
regulation in ESCC cells. Among these genes, the following 3 were
reported to be cancer-promoting genes in ESCC: cyclin dependent
kinase 1 (CDK1), aurora kinase A (AURKA) and
transducin β like 1 X-linked receptor 1 (TBL1XR1) (38-43).
Identification of the target genes controlled by miR-145-3p
is important for understanding the underlying molecular
pathogenesis of ESCC.
The present study demonstrated that both
DHRS2 and MYO1B were directly regulated by
miR-145-3p in ESCC cells. Overexpression of DHRS2 and
MYO1B was observed in ESCC clinical specimens, and
overexpression was associated with cancer cell aggressiveness.
DHRS2 was initially cloned from a HepG2 human
hepatocarcinoma cDNA library and named HEP27 (44). DHRS2 is a member of the
short-chain dehydrogenase/reductase (SDR) family that metabolizes
many different compounds (45).
HEP27 protein interacts with MDM2, which is a negative
regulator of p53, resulting in p53 stabilization and induction of
p53 transcriptional target genes (46). More recently, downregulation of
DHRS2 was reported in ESCC tissues and its downregulation
was associated with ESCC aggressiveness and clinical staging
(47). Thus, that report arrived
at the opposite conclusions from those in our study. Further
investigation of DHRS2 function in cancer cells will be
necessary.
MYO1B is a member of the membrane-associated class
I myosin family and it bridges membrane and actin cytoskeleton in
several cellular processes (48).
It was recently demonstrated that overexpression of MYO1B is
associated with head and neck cancer pathogenesis (20). Importantly, antitumor
miR-145-3p directly regulated expression of MYO1B in
head and neck cells (20). A
previous in vivo study demonstrated that downregulation of
MYO1B inhibited cervical lymph node metastasis in head and
neck cancer cells (49). These
findings indicate that aberrantly expressed MYO1B is
associated with cancer cell aggressiveness and metastasis.
MYO1B may be a novel diagnostic and therapeutic target for
patients with ESCC.
Downstream genes modulated by DHRS2 or
MYO1B in ESCC cells were investigated. Previous studies have
demonstrated that several aberrantly expressed oncogenes
(CDK1, BIRC5, BUB1, TOP2A,
CENPF, FOXM1 and AURKA) enhanced cancer cell
aggressiveness (38,50-54).
Notably, MMP13 may be controlled by DHRS2 and
MYO1B in ESCC cells. Our recent study demonstrated that
overexpression of MMP13 occurred in ESCC clinical specimens
and the expression of MMP13 promoted cancer cell
proliferation, migration and invasion (28). Identification of the downstream
genes regulated by the miR-145-3p/DHRS2 or
miR-145-3p/MYO1B axis may improve the understanding
of ESCC aggressiveness.
In conclusion, genes controlled by the antitumor
activity of miR-145-3p were closely associated with ESCC
pathogenesis. Association of the passenger strand of miRNA is a
novel concept of cancer research. DHRS2 and MYO1B
were directly regulated by miR-145-3p in ESCC cells.
Aberrantly expressed DHRS2 and MYO1B enhanced ESCC
cell aggressiveness. Elucidation of antitumor miRNAs controlling
RNA networks may provide novel prognostic markers and therapeutic
targets for this disease.
Funding
The present study was supported by KAKENHI grants
nos. 15K10801, 18K16322, 16K10508, 17K10706, 16K10510, 18K08687,
18K08626 and 17H04285.
Availability of data and materials
All data generated or analyzed during this study
are included in this published article.
Authors' contributions
MS and TI performed the majority of the study and
wrote the manuscript. NS and SN designed the study and wrote the
manuscript. YY, TAra, YK and HK performed the experiments and data
interpretation. TAri, KS, IO, YU and KM provided sample collection
and clinical support. All authors reviewed, edited and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Bioethics
Committee of Kagoshima University (Kagoshima, Japan; approval no.
28-65). Written prior informed consent and approval were obtained
from all patients.
Patient consent for publication
All patients had provided written informed consent
prior to surgery.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
2
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mariette C, Piessen G and Triboulet JP:
Therapeutic strategies in oesophageal carcinoma: Role of surgery
and other modalities. Lancet Oncol. 8:545–553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cai XW, Yu WW, Yu W, Zhang Q, Feng W, Liu
MN, Sun MH, Xiang JQ, Zhang YW and Fu XL: Tissue-based quantitative
proteomics to screen and identify the potential biomarkers for
early recurrence/metastasis of esophageal squamous cell carcinoma.
Cancer Med. 7:2504–2517. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cooper JS, Guo MD, Herskovic A, Macdonald
JS, Martenson JA Jr, Al-Sarraf M, Byhardt R, Russell AH, Beitler
JJ, Spencer S, et al Radiation Therapy Oncology Group:
Chemoradiotherapy of locally advanced esophageal cancer: Long-term
follow-up of a prospective randomized trial (RTOG 85-01). JAMA.
281:1623–1627. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Natsugoe S, Ikeda M, Baba M, Churei H,
Hiraki Y, Nakajo M and Aikou T; Kyushu Study Group for Adjuvant
Therapy of Esophageal Cancer: Long-term survivors of advanced
esophageal cancer without surgical treatment: A multicenter
questionnaire survey in Kyushu, Japan. Dis Esophagus. 16:239–242.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ando N, Kato H, Igaki H, Shinoda M, Ozawa
S, Shimizu H, Nakamura T, Yabusaki H, Aoyama N, Kurita A, et al: A
randomized trial comparing postoperative adjuvant chemotherapy with
cisplatin and 5-fluorouracil versus preoperative chemotherapy for
localized advanced squamous cell carcinoma of the thoracic
esophagus (JCOG9907). Ann Surg Oncol. 19:68–74. 2012. View Article : Google Scholar
|
|
9
|
Dutton SJ, Ferry DR, Blazeby JM, Abbas H,
Dahle-Smith A, Mansoor W, Thompson J, Harrison M, Chatterjee A,
Falk S, et al: Gefitinib for oesophageal cancer progressing after
chemotherapy (COG): A phase 3, multicentre, double-blind,
placebo-controlled randomised trial. Lancet Oncol. 15:894–904.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Suntharalingam M, Winter K, Ilson D,
Dicker AP, Kachnic L, Konski A, Chakravarthy AB, Anker CJ, Thakrar
H, Horiba N, et al: Effect of the addition of cetuximab to
paclitaxel, cisplatin, and radiation therapy for patients with
esophageal cancer: The NRG Oncology RTOG 0436 phase 3 randomized
clinical trial. JAMA Oncol. 3:1520–1528. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hemmatzadeh M, Mohammadi H, Karimi M,
Musavishenas MH and Baradaran B: Differential role of microRNAs in
the pathogenesis and treatment of Esophageal cancer. Biomed
Pharmacother. 82:509–519. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin DC, Wang MR and Koeffler HP: Genomic
and epigenomic aberrations in esophageal squamous cell carcinoma
and implications for patients. Gastroenterology. 154:374–389. 2018.
View Article : Google Scholar :
|
|
13
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar :
|
|
16
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mataki H, Seki N, Mizuno K, Nohata N,
Kamikawaji K, Kumamoto T, Koshizuka K, Goto Y and Inoue H:
Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and
miR-145-3p) coordinately targeted MTDH in lung squamous cell
carcinoma. Oncotarget. 7:72084–72098. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Matsushita R, Yoshino H, Enokida H, Goto
Y, Miyamoto K, Yonemori M, Inoguchi S, Nakagawa M and Seki N:
Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145
(miR-145-5p and miR-145-3p): Inhibition of bladder cancer cell
aggressiveness. Oncotarget. 7:28460–28487. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Goto Y, Kurozumi A, Arai T, Nohata N,
Kojima S, Okato A, Kato M, Yamazaki K, Ishida Y, Naya Y, et al:
Impact of novel miR-145-3p regulatory networks on survival in
patients with castration-resistant prostate cancer. Br J Cancer.
117:409–420. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamada Y, Koshizuka K, Hanazawa T, Kikkawa
N, Okato A, Idichi T, Arai T, Sugawara S, Katada K, Okamoto Y, et
al: Passenger strand of miR-145-3p acts as a tumor-suppressor by
targeting MYO1B in head and neck squamous cell carcinoma. Int J
Oncol. 52:166–178. 2018.
|
|
21
|
Schwarz DS, Hutvágner G, Du T, Xu Z,
Aronin N and Zamore PD: Asymmetry in the assembly of the RNAi
enzyme complex. Cell. 115:199–208. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mah SM, Buske C, Humphries RK and
Kuchenbauer F: miRNA*: A passenger stranded in
RNA-induced silencing complex? Crit Rev Eukaryot Gene Expr.
20:141–148. 2010. View Article : Google Scholar
|
|
23
|
Liu WW, Meng J, Cui J and Luan YS:
Characterization and Function of MicroRNA*s in Plants.
Front Plant Sci. 8:22002017. View Article : Google Scholar
|
|
24
|
Osako Y, Seki N, Koshizuka K, Okato A,
Idichi T, Arai T, Omoto I, Sasaki K, Uchikado Y, Kita Y, et al:
Regulation of SPOCK1 by dual strands of pre-miR-150 inhibit cancer
cell migration and invasion in esophageal squamous cell carcinoma.
J Hum Genet. 62:935–944. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sachdeva M and Mo YY: MicroRNA-145
suppresses cell invasion and metastasis by directly targeting mucin
1. Cancer Res. 70:378–387. 2010. View Article : Google Scholar :
|
|
26
|
Kano M, Seki N, Kikkawa N, Fujimura L,
Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M and
Matsubara H: miR-145, miR-133a and miR-133b: Tumor-suppressive
miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J
Cancer. 127:2804–2814. 2010. View Article : Google Scholar
|
|
27
|
Nishihira T, Hashimoto Y, Katayama M, Mori
S and Kuroki T: Molecular and cellular features of esophageal
cancer cells. J Cancer Res Clin Oncol. 119:441–449. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Osako Y, Seki N, Kita Y, Yonemori K,
Koshizuka K, Kurozumi A, Omoto I, Sasaki K, Uchikado Y, Kurahara H,
et al: Regulation of MMP13 by antitumor microRNA-375 markedly
inhibits cancer cell migration and invasion in esophageal squamous
cell carcinoma. Int J Oncol. 49:2255–2264. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yonemori K, Seki N, Kurahara H, Osako Y,
Idichi T, Arai T, Koshizuka K, Kita Y, Maemura K and Natsugoe S:
ZFP36L2 promotes cancer cell aggressiveness and is regulated by
antitumor microRNA-375 in pancreatic ductal adenocarcinoma. Cancer
Sci. 108:124–135. 2017. View Article : Google Scholar :
|
|
30
|
Yoshino H, Chiyomaru T, Enokida H,
Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N and Nakagawa
M: The tumour-suppressive function of miR-1 and miR-133a targeting
TAGLN2 in bladder cancer. Br J Cancer. 104:808–818. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Harada K, Baba Y, Ishimoto T, Kosumi K,
Tokunaga R, Izumi D, Ohuchi M, Nakamura K, Kiyozumi Y, Kurashige J,
et al: Suppressor microRNA-145 is epigenetically regulated by
promoter hypermethylation in esophageal squamous cell carcinoma.
Anticancer Res. 35:4617–4624. 2015.PubMed/NCBI
|
|
32
|
Kita Y, Nishizono Y, Okumura H, Uchikado
Y, Sasaki K, Matsumoto M, Setoyama T, Tanoue K, Omoto I, Mori S, et
al: Clinical and biological impact of cyclin-dependent kinase
subunit 2 in esophageal squamous cell carcinoma. Oncol Rep.
31:1986–1992. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zeng JF, Ma XQ, Wang LP and Wang W:
MicroRNA-145 exerts tumor-suppressive and chemo-resistance lowering
effects by targeting CD44 in gastric cancer. World J Gastroenterol.
23:2337–2345. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sachdeva M, Zhu S, Wu F, Wu H, Walia V,
Kumar S, Elble R, Watabe K and Mo YY: p53 represses c-Myc through
induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA.
106:3207–3212. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Arai T, Okato A, Yamada Y, Sugawara S,
Kurozumi A, Kojima S, Yamazaki K, Naya Y, Ichikawa T and Seki N:
Regulation of NCAPG by miR-99a-3p (passenger strand) inhibits
cancer cell aggressiveness and is involved in CRPC. Cancer Med.
7:1988–2002. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Idichi T, Seki N, Kurahara H, Fukuhisa H,
Toda H, Shimonosono M, Okato A, Arai T, Kita Y, Mataki Y, et al:
Molecular pathogenesis of pancreatic ductal adenocarcinoma: Impact
of passenger strand of pre-miR-148a on gene regulation. Cancer Sci.
109:2013–2026. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen GM, Zheng AJ, Cai J, Han P, Ji HB and
Wang LL: microRNA-145-3p inhibits non-small cell lung cancer cell
migration and invasion by targeting PDK1 via the mTOR signaling
pathway. J Cell Biochem. 119:885–895. 2018. View Article : Google Scholar
|
|
38
|
Dutertre S, Descamps S and Prigent C: On
the role of aurora-A in centrosome function. Oncogene.
21:6175–6183. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fung TK, Yam CH and Poon RY: The
N-terminal regulatory domain of cyclin A contains redundant
ubiquitination targeting sequences and acceptor sites. Cell Cycle.
4:1411–1420. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li JY, Daniels G, Wang J and Zhang X:
TBL1XR1 in physiological and pathological states. Am J Clin Exp
Urol. 3:13–23. 2015.PubMed/NCBI
|
|
41
|
Liu L, Lin C, Liang W, Wu S, Liu A, Wu J,
Zhang X, Ren P, Li M and Song L: TBL1XR1 promotes lymphangiogenesis
and lymphatic metastasis in esophageal squamous cell carcinoma.
Gut. 64:26–36. 2015. View Article : Google Scholar
|
|
42
|
Tamotsu K, Okumura H, Uchikado Y, Kita Y,
Sasaki K, Omoto I, Owaki T, Arigami T, Uenosono Y, Nakajo A, et al:
Correlation of Aurora-A expression with the effect of
chemoradiation therapy on esophageal squamous cell carcinoma. BMC
Cancer. 15:3232015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang HF, Alshareef A, Wu C, Jiao JW,
Sorensen PH, Lai R, Xu LY and Li EM: miR-200b induces cell cycle
arrest and represses cell growth in esophageal squamous cell
carcinoma. Carcinogenesis. 37:858–869. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gabrielli F, Donadel G, Bensi G, Heguy A
and Melli M: A nuclear protein, synthesized in growth-arrested
human hepa-toblastoma cells, is a novel member of the short-chain
alcohol dehydrogenase family. Eur J Biochem. 232:473–477. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gabrielli F and Tofanelli S: Molecular and
functional evolution of human DHRS2 and DHRS4 duplicated genes.
Gene. 511:461–469. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Deisenroth C, Thorner AR, Enomoto T, Perou
CM and Zhang Y: Mitochondrial Hep27 is a c-Myb target gene that
inhibits Mdm2 and stabilizes p53. Mol Cell Biol. 30:3981–3993.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou Y, Wang L, Ban X, Zeng T, Zhu Y, Li
M, Guan XY and Li Y: DHRS2 inhibits cell growth and motility in
esophageal squamous cell carcinoma. Oncogene. 37:1086–1094. 2018.
View Article : Google Scholar :
|
|
48
|
Wessels D, Murray J, Jung G, Hammer JA III
and Soll DR: Myosin IB null mutants of Dictyostelium exhibit
abnormalities in motility. Cell Motil Cytoskeleton. 20:301–315.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ohmura G, Tsujikawa T, Yaguchi T, Kawamura
N, Mikami S, Sugiyama J, Nakamura K, Kobayashi A, Iwata T, Nakano
H, et al: Aberrant Myosin 1b Expression Promotes Cell Migration and
Lymph Node Metastasis of HNSCC. Mol Cancer Res. 13:721–731. 2015.
View Article : Google Scholar
|
|
50
|
Elowe S: Bub1 and BubR1: At the interface
between chromosome attachment and the spindle checkpoint. Mol Cell
Biol. 31:3085–3093. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Athanasoula KC, Gogas H, Polonifi K,
Vaiopoulos AG, Polyzos A and Mantzourani M: Survivin beyond
physiology: Orchestration of multistep carcinogenesis and
therapeutic potentials. Cancer Lett. 347:175–182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Aytes A, Mitrofanova A, Lefebvre C,
Alvarez MJ, Castillo-Martin M, Zheng T, Eastham JA, Gopalan A,
Pienta KJ, Shen MM, et al: Cross-species regulatory network
analysis identifies a synergistic interaction between FOXM1 and
CENPF that drives prostate cancer malignancy. Cancer Cell.
25:638–651. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Asghar U, Witkiewicz AK, Turner NC and
Knudsen ES: The history and future of targeting cyclin-dependent
kinases in cancer therapy. Nat Rev Drug Discov. 14:130–146. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chen T, Sun Y, Ji P, Kopetz S and Zhang W:
Topoisomerase IIα in chromosome instability and personalized cancer
therapy. Oncogene. 34:4019–4031. 2015. View Article : Google Scholar
|