Introduction
Cystatin B (CSTB, stefin B) is an endogenous
inhibitor of intracellular cysteine proteases (1,2).
Previously, we reported that CSTB is a progression marker of human
epithelial ovarian cancer (OC) (3), a disease ranked as the eight most
common cancer worldwide in females (4) and gynecological cancer with the
highest rate of mortality in the United States of America (5). CSTB has been implicated in
inflammation (6), autosomal
recessive disorders (7), and
cancer (3,8-11);
however, the regulatory mechanism and the function of CSTB on cell
proliferation in epithelial OC (EOC) remains unknown.
MicroRNAs (miRNAs/miRs) are a class of small
noncoding RNA of ~22 nucleotides in length that can regulate a gene
by binding to its 3'-untranslated region (3'-UTR), triggering the
degradation of mRNA or the suppression of protein translation
(12,13). Accumulating evidence suggests that
numerous miRNAs of multi-gene regulatory capacity are dysregulated
in cancer (14,15), and have been associated the
regulation of biological processes, such as carcinogenesis,
metastasis, epithelial-mesenchymal transition (EMT) and
chemoresistance in EOC (16,17),
and are regulated by cytokines (18,19).
Transforming growth factor (TGF)-β belongs to a
superfamily of secreted cytokines and serves a key role in many
cellular processes. Members of the TGF-β superfamily transmit
signals via the Smad-dependent and -independent pathways (20,21).
The canonical pathway of TGF-β signaling occurs through the
activation of its corresponding membrane serine/threonine kinase
receptors (type I and type II receptors) to activate the
intracellular signaling transducer Smad2/3 proteins, which form a
complex with Smad4 (22). After
the Smad-complex translocates into the nucleus, it acts as a
transcription factor to regulate the expression of a target gene
(21). Smad-independent pathways,
such as the MAPK and PI3K signaling pathways, can also be induced
by TGF-β to initiate signal transduction and gene regulation
(23). The Smad-dependent and
-independent pathways have been demonstrated to contribute to the
pathogenesis of OC (24,25). Our previous study has revealed that
CSTB is mediated by the TGF-β signaling pathway (3); however, the regulatory mechanism by
which TGF-β1 regulates CSTB and its function in OC remain
unclear.
The present study aimed to explore the mechanism
underlying TGF-β1-mediated CSTB regulation via miR-143-3p, and
examine the function of CSTB on OC cell proliferation and
apoptosis.
Materials and methods
Bioinformatics analyses
To predict the miRNAs targeting CSTB, miRWalk
(version 1.0; http://zmf.mm.uni-heidelberg.de/apps/zmf/mirwalk/predictedmirnagene.html)
was applied. To analyze the expression of CSTB mRNA and miR-143-3p
between normal ovarian tissues and ovarian malignant tumors, three
gene expression datasets (GSE36668, GSE40595, GSE63885) for CSTB
and one microRNA expression dataset (GSE47841) for miR-143-3p were
downloaded from the public Gene Expression Omnibus (GEO) database
(http://www.ncbi.nlm.nih.gov/geo/). All
three CSTB mRNA expression profiles from 12 normal ovary samples
and 108 ovarian serous carcinoma samples were generated by the
Affymetrix Human Genome U133 Plus 2.0 Array and a miR-143-3p
expression profile from 9 normal ovary samples and 12 ovarian
serous carcinoma samples was generated by the Affymetrix
Multispecies miRNA-2 Array. These transcriptome microarray datasets
were integrated and analyzed by R software version 3.4.2
(https://www.r-project.org/).
Additionally, to analyze the differential expression of CSTB
between ovarian surface epithelium and ovarian serous carcinoma,
the online Oncomine tool (www.oncomine.org) was used. In this analysis, the
median levels of CSTB mRNA expression were calculated from two
datasets: Lu (26) and Bonome
(27). To analyze the overall
survival (OS) of patients with OC, the Kaplan-Meier Plotter
database (www.kmplot.com) was employed. Patients
with OC selected for OS analysis were divided into two groups
according to the cutoff value set as the median expression levels
of CSTB. The OS data were presented as a survival plot, and
analyzed with a log rank test.
Patients and ovarian tissue samples
A total of 35 fresh ovarian tissues samples were
obtained from patients (age range: 24-87 years) who underwent
surgery at Jinshan Hospital from January 2012 to December 2015 (11
normal ovarian samples from patients with non-ovarian tumor and 24
ovarian serous tumor samples, including 10 benign, 3 borderline,
and 11 malignant tumors). All tissue samples were immediately
snapped frozen in liquid nitrogen and stored at -80°C until use.
None of the patients had received chemotherapy or radiotherapy
prior to surgery. The present study was approved by the Ethics
Committee of Jinshan Hospital, Fudan University.
Cell culture and TGF-β treatment
The human OC cell lines OVCAR-3 and SK-OV-3 cells
were obtained from American Type Culture Collection (ATCC) and
maintained in RPMI-1640 and Dulbecco's modified Eagle's media
(Corning Inc.), respectively, with 10% (v/v) fetal bovine serum
(Invitrogen; Thermo Fisher Scientific, Inc.). Cells were cultured
as described previously (24). For
TGF-β1 treatment, after seeding cells for 24 h, 10 ng/ml
recombinant human TGF-β1 (cat. no. 240-B, R&D Systems, Inc.)
was added and incubated at 37°C for 3 h. Cells without any
treatment were used as a blank control. For blocking of the TGF-β1
signaling pathway, cells were pre-treated with 10 µM
SB431542, a TGF-β type I receptor inhibitor (Sigma-Aldrich; Merck
KGaA) for 30 min, followed by 10 ng/ml TGF-β1 treatment at 37°C for
3 h.
Total RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells or tissue samples
using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocols. Primary miRNA, mature miRNA and mRNA
were reversely transcribed using a Transcriptor First Strand cDNA
Synthesis Kit (Roche Applied Science) according to the
manufacturer's instructions. The primer sequences were listed in
Table S1. RT-qPCR was performed
at 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and
60°C for 30 sec using the FastStart Universal SYBR-Green Master
(Roche Applied Science) on an ABI PRISM 7300 Real Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The relative
expression of mRNA, primary miRNA or mature miRNA was evaluated
using the 2−∆∆Cq method (28) and all experiments were conducted in
triplicate. 18S and U6 was used as an internal control for mRNA and
miRNA, respectively.
Lentivirus-delivered short hairpin RNA
(shRNA) infection, siRNA and miRNA mimics/inhibitors
transfection
Lentiviral vectors containing shRNA for CSTB
(CSTB-shRNA, sh-CSTB) and negative control (NC-shRNA, sh-NC) were
obtained from Shanghai GenePharma Co., Ltd. Three specific
CSTB-siRNAs (5′-GUCCCAGCUUGAAGAGAAATT-3′ for
si-CSTB-1, 5′-GGACAAACUACUUCAUCAATT-3′ for si-CSTB-2,
and 5′-CCCUUGACCUUAUCUAACUTT-3′ for si-CSTB-3) and a
negative control (NC-siRNA, si-NC) were synthesized by Shanghai
GenePharma Co., Ltd. The miR-143-3p mimics, miR-negative control
(miR-NC), miR-143-3p inhibitor (anti-miR-143), and the negative
control of inhibitor (anti-miR-NC) was obtained from Guangzhou
RiboBio Co., Ltd. (Table S1).
After seeding at 2×105 cells/well in 6-well plates and
incubating for 24 h, cells were transfected with 50 nM si-CSTB or
si-NC, miR-143-3p mimics or miR-NC, 200 nM miR-143-3p inhibitors or
miR-NC using X-tremeGENE siRNA Transfection Reagent (Roche Applied
Science). Cyanine 3 (Cy3) dye-labeled miR-NC was used to evaluate
the transfection efficiency detected by fluorescence microscopy.
The sequence of si-CSTB-2 was used for subsequent experimentation.
Knockdown of CSTB was performed in OC cells by infecting cells with
CSTB-shRNA lentivirus according to the manufacturer's protocols.
Briefly, after cells were plated in 6-well plates and incubated at
37°C for 24 h, the amount of lentiviral particle solution
(multiplicity of infection=20) was added into the culture media
containing 8 µg/ml/well Polybrene for 24 h. The media was
replaced with fresh media containing 2 µg/ml puromycin for
selection every few days. Following puromycin selection for several
passages, the detection of protein knockdown or the phenotypic
assay were performed.
Western blotting
Cells were lysed in radioimmunoprecipitation assay
lysis buffer (Thermo Fisher Scientific, Inc.) with 1%
phenylmethanesulfonyl fluoride (Beyotime Institute of
Biotechnology) and 1% phosphatase inhibitor (Nanjing KeyGen Biotech
Co., Ltd.), followed by sonication. The protein was extracted from
the supernatant after 20 min of 18,000 × g centrifugation at 4°C.
Proteins were quantified via a BCA assay. Equal amount proteins
were separated on 15% SDS-PAGE and transferred to a polyvinylidene
difluoride membrane (EMD Millipore). After blocking with 5% non-fat
milk in Tris-buffered saline with Tween 20 at room temperature for
1 h, the membrane was incubated with a primary antibody at 4°C
overnight and subsequently incubated with horseradish
peroxidase-conjugated goat anti-rabbit or anti-mouse IgG (cat. nos.
7074 and 7076, respectively; 1:5,000 dilution; Cell Signaling
Technology, Inc.) for 1 h at room temperature. The following
primary antibodies were used: Rabbit anti-CSTB (1:5,000 dilution;
Abcam) and mouse anti-β-actin (1:5,000 dilution; ProteinTech
Group., Inc.). Signals were detected using Immobilon™ Western
Chemiluminescent HRP Substrate (EMD Millipore) and quantified using
Tanon-4500 Gel Imaging System with GIS ID Analysis Software v4.1.5
(Tanon Science and Technology Co., Ltd.).
Plasmid construction and dual-luciferase
reporter assay
The whole 3'-UTR of CSTB that contains the binding
site of miR-143-3p predicted by the miRWalk program (http://mirwalk.umm.uni-heidelberg.de/)
was amplified from genomic DNA using the Pfu Ultra II Fusion HS DNA
Polymerase (Stratagene; Agilent Technologies, Inc.) with
well-designed primers (Table S1).
After the dual-luciferase reporter vector pEZX-FR2 (GeneCopoeia,
Inc.) was linearized by EcoRI and XhoI restriction
enzymes, the PCR product was inserted into pEZX-FR2 using the
EasyGeno Assembly Cloning Kit (Tiangen Biotech Co., Ltd.) to
construct a wild-type clone named as CSTB-3UTR-wt. A mutation was
induced in the consensus sequence of the miR-143-3p binding site in
the 3'-UTR of CSTB using the QuikChange II Site-Directed
Mutagenesis Kit (Stratagene; Agilent Technologies, Inc.) to
construct a mutated clone named as CSTB-3UTR-mut. All clones were
verified by restriction enzymes EcoRI and XhoI
digestion and DNA sequencing with primer (5′-GATCCG
CGAGATCCTGAT-3′) by GENEWIZ, Inc. For the dual-luciferase reporter
assay, 293T cells (ATCC) were cultured in 24-well plates. Once
subconfluent, the cells were transfected with CSTB-3UTR-wt or
CSTB-3UTR-mut plasmid (0.5 µg), plus miR-143-3p mimics,
inhibitors or their negative controls using Roche X-tremeGENE siRNA
Transfection Reagent (2 µl). After transfection for 24 h,
the cells were lysed and luciferase activities were determined
using the Luc-Pair™ Duo-Luciferase Assay Kit (GeneCopoeia, Inc.)
according to the manufacturer's instructions. Renilla
luciferase activity was used for the normalization of Firefly
luciferase activity.
Cell proliferation assay and cell cycle
detection
For the cell proliferation assay, NC-shRNA- or
CSTB-shRNA-infected OVCAR-3 cells were plated in 96-well plates at
a density of 3×103 cells/well. Cell proliferation was
measured at 24, 48, and 72 h post-infection by the Cell
Proliferation Reagent (Cell Counting Kit-8; Dojindo Molecular
Technologies, Inc.) according to the manufacturer's protocols. The
signal in optical density was read by a microplate reader (BioTek
Instruments, Inc.) at 450 nm.
The cell cycle was detected by flow cytometry.
Briefly, CSTB-shRNA-infected OVCAR-3 cells were cultured in 6-well
plates for 48 h. The cells were harvested, washed twice with PBS,
and fixed in cold 70% ethanol for 2 h at -20°C. After washing with
PBS twice, the cells were resuspended in 500 µl of propidium
iodide (PI)/RNase Staining Buffer (BD Pharmingen) and incubated in
the dark for 15 min at room temperature. Cells (15,000) were then
detected by flow cytometry (Beckman Coulter, Inc.). The data were
analyzed using ModFit LT software v4.1.7 (Verity Software House,
Inc.), and presented as the percentage of the cell population at
the G0/G1, S and G2/M phases. Scramble-shRNA-infected OVCAR-3 cells
were used as a control.
Detection of apoptotic cells
Flow cytometry and western blotting were conducted
to detect apoptotic cells. Briefly, cells were cultured in 6-well
plates for 48 h. After washing with PBS, the cells were collected
and stained with 1 µl Annexin V conjugated with
allophycocyanin (BD Pharmingen) and 5 µl PI for 15 min at
room temperature. Apoptotic cells were detected by flow cytometry
with FlowJo X software v10.0.7r2 (BD Biosciences). The expression
of pro-apoptotic protein Bcl-2-associated X protein (Bax) was
analyzed by western blotting as aforementioned using an antibody
against Bax (1:2,000 dilution; Cell Signaling Technology,
Inc.).
Statistical analyses
All experiments were repeated at least three times
and all analyses were performed with SPSS 21 for Windows (IBM
Corp.). For comparisons between two groups in an experiment, a
Student's t-test was applied. For multiple comparisons, one-way
analysis of variance was used, followed by a Tukey's post-hoc test.
For the correlation analysis between CSTB and miR-143-3p,
nonparametric Spearman rank correlation was used. The results were
presented as the mean ± the standard error of the mean. P<0.05
was considered to indicate a statistically significant
difference.
Results
CSTB is negatively correlated with
miR-143-3p in human OC
miRNAs which can bind to the CSTB 3'-UTR were
screened using web-based programs in the miRWalk database. For
targeting CSTB, miR-143-3p was proposed as a potential candidate by
5 out of 10 available prediction programs (Fig. 1A). Additionally, the expression
data of transcripts were extracted from GSE36668, GSE40595 and
GSE63885 datasets to obtain the mRNA expression profile of CSTB,
and GSE47841 dataset for miR-143-3p in an online GEO database using
integrated bioinformatics analyses. Upregulated expression of CSTB
mRNA was detected in ovarian serous carcinoma (n=108) compared with
normal ovary controls (n=12) (Fig.
1B), while miR-143-3p expression was lower in ovarian serous
carcinoma (n=12) than normal ovary controls (n=9) (Fig. 1C). As the patients with CSTB mRNA
data available in the online datasets differed from patients with
miR-143-3p information, the expression of CSTB mRNA and miR-143-3p
was validated in the same freshly isolated ovarian tissue from the
same patient; correlation analysis of these transcripts was
performed.
 | Figure 1Detection of CSTB mRNA and miR-143-3p
expression in human ovarian tissues, and their correlation. (A)
Prediction of miR-143-3p binding to CSTB. miR-143-3p was proposed
to be a candidate from 5 out of 10 programs in the miRWalk
database. (B) Comparison of CSTB mRNA expression between normal
ovary controls and ovarian serous carcinoma. Data were obtained
from three gene expression datasets (GSE36668, GSE40595, GSE63885)
in the GEO database. (C) Comparison of miR-143-3p expression
between normal ovary controls and ovarian serous carcinoma. Data
were obtained from a miRNA expression dataset (GSE47841) in the GEO
database. (D and E) The expression of CSTB mRNA and miR-143-3p was
detected by reverse transcription-quantitative PCR in freshly
isolated ovarian tissues, including normal ovarian tissues (n=11),
Bn (n=10), Bd (n=3) and Mg (n=11). (F) The correlation between CSTB
mRNA and miR-143-3p in human ovarian tissues (total n=35). Data are
presented as the mean ± standard error of the mean.
*P<0.05; **P<0.01. Bn, benign tumor;
Bd, borderline tumor; CSTB, cystatin B; GEO, Gene Expression
Omnibus; hsa, homo sapiens; Mg, malignant tumor; miR,
microRNA. |
CSTB mRNA expression levels were significantly
increased in malignant tumors than in normal ovarian tissue, and
benign and borderline tumors (P<0.01; Fig. 1D), which supported the findings of
our previous study (3). On the
contrary, miR-143-3p was downregulated in malignant tumors compared
with normal ovarian tissues and benign tumors (P<0.05; Fig. 1E). Spearman correlation analysis
revealed a significant negative correlation between the expression
levels of miR-143-3p and CSTB mRNA (r=-0.362, P<0.05; Fig. 1F).
miR-143-3p directly binds to the 3′-UTR
of CSTB
To determine whether miR-143-3p directly binds to
the 3'-UTR of CSTB (Fig. 2A), we
cloned two CSTB 3'-UTR plasmids: CSTB-3UTR-wt and CSTB-3UTR-mut.
The predicted binding site and mutant site of 3'-UTR (Fig. 2B) were confirmed by sequencing
analysis. The luciferase activity in 293T cells was measured after
co-transfection of CSTB-3'UTR-containing plasmids with miR-143-3p
mimics or miR-NC by a dual-luciferase reporter assay. Transfection
of cells with CSTB 3'-UTR-wt and miR-143-3p revealed a significant
reduction in the luciferase activity compared with the
corresponding control (P<0.01). However, the cells expressing
the mutated CSTB 3'-UTR-mut demonstrated no marked alterations in
luciferase activity, suggesting that mutations in the 3'UTR of CSTB
abrogated the binding to the miR-143-3p (Fig. 2C). These findings indicate that
miR-143-3p can directly target CSTB.
CSTB is regulated by miR-143-3p in OC
cells
To further confirm that CSTB is a target of
miR-143-3p, gain-of-function and loss-of-function approaches were
applied. The transfection efficiency of miR-143-3p mimics and
inhibitors in OVCAR-3 cells was evaluated. A significant increase
and decrease in miR-143-3p expression were detected by RT-qPCR
after cells were transfected with miR-143-3p mimics of inhibitors
(anti-miR-143) for 12 h, respectively, compared with the
corresponding NC group (Fig. S1A and
B). The transfection efficiency of mimics was further detected
by fluorescence microscopy at 24 h post-transfection and ~90% of
cells were Cy3-mimic positive (Fig.
S1C). Additionally, the expression of CSTB mRNA and protein
were examined by RT-qPCR and western blot analyses of OVCAR-3 cells
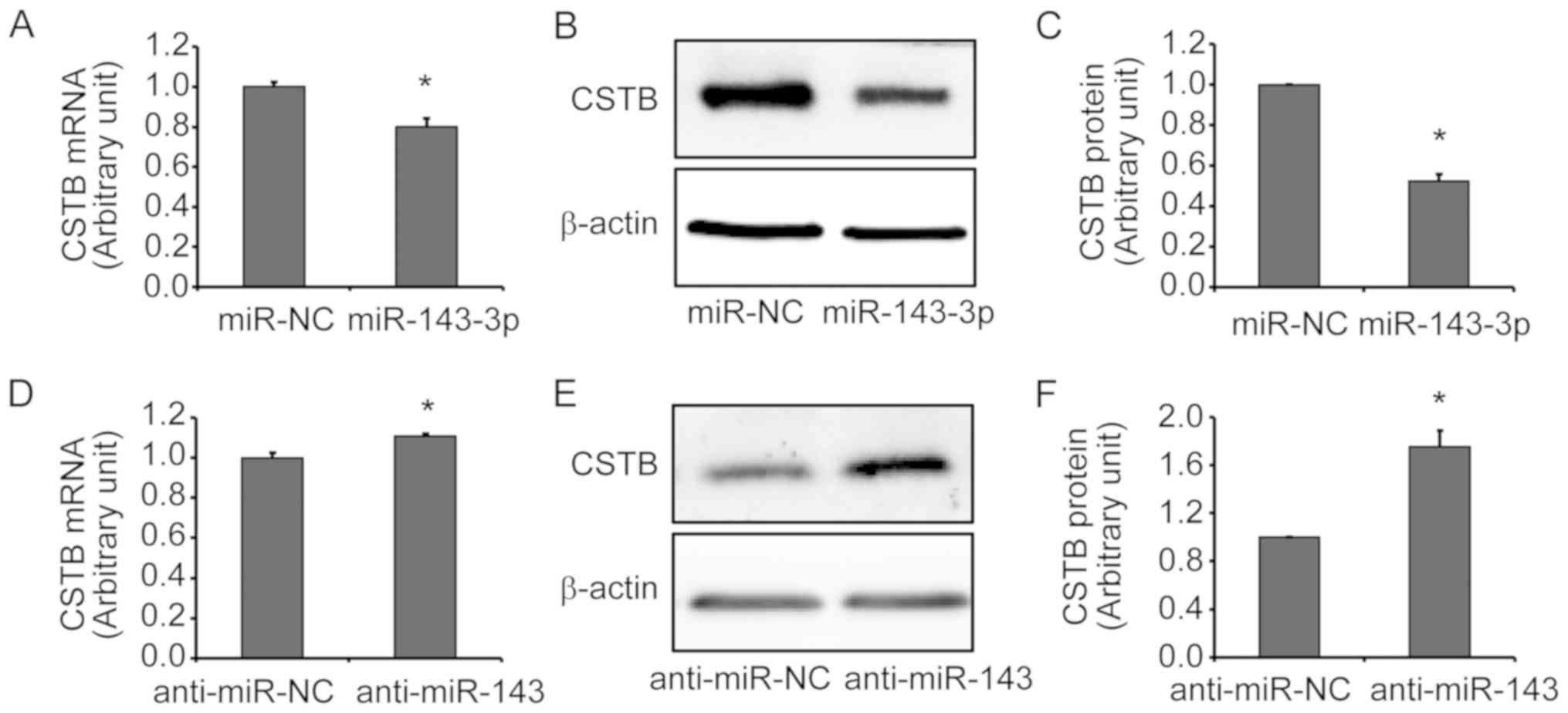
transfected with miR-143-3p mimics or inhibitors. CSTB expression
was significantly downregulated at the mRNA (Fig. 3A) and protein (Fig. 3B and C) levels after miR-143-3p
mimics transfection. Silencing of miR-143-3p by anti-miR-143
resulted in a significant increase in CSTB expression at the mRNA
(Fig. 3D) and protein (Fig. 3C and D) levels in OVCAR-3 cells.
These results indicated that miR-143-3p may negatively regulate
CSTB expression in OC cells.
miR-143-3p expression is regulated by the
TGF-β signaling pathway
As our previous findings indicated that CSTB is
mediated by the TGF-β signaling pathway in epithelial OC cells
(3) and the present study revealed
that miR-143-3p can negatively regulate CSTB expression, whether
the TGF-β signaling pathway affects miR-143-3p expression was
investigated. RT-qPCR revealed that primary miR-143-3p expression
was significantly increased after treatment with 10 ng/ml TGF-β1 in
OVCAR-3 cells compared with the control (P<0.01; Fig. 4A). The effects of TGF-β1 were
partially abolished by the blocking of the TGF-β type I receptor
using its inhibitor SB431542. In addition, an increase in the
levels of mature miR-143-3p was observed following TGF-β1
treatment, which was significantly abolished in the presence of
SB431542 (P<0.05; Fig. 4B).
High levels of CSTB expression are
associated with poor OS in patients with ovarian malignant
tumors
Overexpression of CSTB in OC was detected in two
microarray datasets from the Oncomine database. The median value of
CSTB mRNA expression was calculated from two datasets. CSTB mRNA
levels were higher in ovarian serous carcinoma than normal ovarian
surface epithelium in the Lu dataset (Reporter ID: 35816_at;
Fig. 5A) and in the Bonome dataset
(Reporter ID: 201201_at; Fig. 5B).
Furthermore, OS analysis was performed using the Kaplan-Meier
Plotter dataset. Increased CSTB mRNA expression levels were
associated with poor OS in patients with OC (Fig. 5C).
Knockdown of CSTB inhibits OC cell
proliferation
To determine the functional effects of CSTB on the
biological behaviors of OC cells, loss-of-function experiments were
applied. A total of three siRNAs specific to human CSTB
(CSTB-siRNA) were synthesized; transfection efficiency was
determined prior to subsequent experiments. Knockdown of CSTB
expression at the mRNA (Fig. S2A and
B) and protein (Fig. S2C and
D) levels was confirmed by RT-qPCR and western blotting in
OVCAR-3 (Fig. S2A and C) and
SK-OV-3 (Fig. S2B and D) cells,
respectively. Increased transfection efficiency was determined in
cells transfected with the second CSTB-siRNA (si-CSTB-2), which was
then used for the subsequent experiments and to generate shRNAs.
Additionally, a cell viability assay was performed after OVCAR-3
cells were transfected with si-CSTB-2. The results revealed that
knockdown of CSTB expression by CSTB-siRNA significantly decreased
cell proliferation at 48 and 72 h compared with the NC group
(Fig. S3A). The suppression of
CSTB by CSTB-siRNA appeared to have arrested the cell cycle in
OVCAR-3 cells at G2/M phase as detected by flow cytometry (Fig. S3B and C). Similar results were
observed in OVCAR-3 cells infected with CSTB-shRNA lentivirus.
Knockdown of CSTB protein was confirmed by western blotting
(Fig. 6A and B). Cell
proliferation was significantly reduced following lentiviral
transduction of CSTB-shRNA compared with the NC group at 72 h
(Fig. 6C). In addition, the
suppression of CSTB notably increased the number of OVCAR-3 cells
at G2/M phase compared with negative controls (Fig. 6D-F).
Knockdown of CSTB induces OC cell
apoptosis
Apoptotic cells were detected by flow cytometry
after infecting OVCAR-3 cells with CSTB-shRNA or sh-NC via
lentivirus for 48 h (Fig. 7A and
B). The cell population in the bottom right of quadrant graphs
represented the early apoptotic cells which were significantly
increased in the CSTB-shRNA group compared with the NC group
(Fig. 7C). Additionally, an
increase in the expression of pro-apoptotic protein Bax was
observed by western blotting in OVCAR-3 cells infected with
CSTB-shRNA lentivirus (Fig.
7D).
Discussion
CSTB is one of the endogenous inhibitors of
lysosomal cysteine proteases, and was reported to be dysregulated
in several types of cancer (3,8-11).
Our previous study showed that CSTB is overexpressed in human EOC
and is mediated by the TGF-β signaling pathway (3). To the best of our knowledge, the
present study is the first to demonstrate that CSTB could be
downregulated by miR-143-3p, whereas miR-143-3p was upregulated by
TGF-β1. Our findings suggested the existence of the
TGF-β/miR-143-3p/CSTB axis in OC cells. Furthermore, CSTB was
proposed to function as an oncogene, which affected OC cell
proliferation; the expression levels of CSTB were associated with
the OS of patients with OC.
Upregulated CSTB expression has also been detected
in ovarian clear cell carcinoma (29). Similarly, overexpression of CSTB
has been observed in other malignant tumors, such as
hepatocellular, bladder and colorectal cancers (9,30,31).
CSTB ablation was determined to retard breast tumor growth in a
mouse model (32). Our previous
study revealed that overexpression of CSTB is associated with the
clinicopathological features of EOC (3). In the present study, upregulated CSTB
expression was proposed to be associated with the OS of OC
patients. It has been shown that CSTB is a prognostic factor, while
high serum levels of CSTB were linkted to increased risk of
mortality in patients with colorectal cancer (31).
As serous carcinoma, arising from the ovarian
surface epithelium and/or fallopian tube epithelium, is the most
common type of OC (33), the
current study analyzed the expression of CSTB mRNA and miR-143-3p
between normal ovary controls and ovarian serous carcinoma from the
public GEO database. Our findings were verified in freshly isolated
human ovarian serous tumors from our hospital; functional assays
using the serous type of human OC cell lines, such as OVCAR-3 and
SK-OV-3, were also conducted. Using a loss-of-function approach to
knockdown CSTB expression in EOC cells, we found that downregulated
CSTB expression significantly inhibited cell proliferation.
CSTB-shRNA significantly increased the number of cells in early
apoptosis. Elevated pro-apoptotic protein Bax expression was also
observed in OVCAR-3 cells after CSTB downregulation. The effects of
CSTB downregulation on OC cell proliferation the cell cycle
appeared to be more pronounced than the effects of cell apoptosis.
These data indicated that CSTB serves a role in the behavior of OC
cells and suggest a potential role of CSTB in targeting OC.
Furthermore, a regulatory mechanism has been investigated; in the
majority of mammalian cells, miRNAs are major regulators of gene
expression, and can affect the expression of numerous oncogenes and
tumor suppressor genes (16,34,35).
Alterations in miRNA expression contributes to a substantial cell
re-organization, and thus, is involved in the pathogenesis of many
diseases (13,15).
Using the miRWalk database, miR-143-3p was
determined to be a candidate by 5 of 10 prediction programs. The
present study reported a low level of miR-143-3p expression in OC
patients; miR-143-3p negatively correlated with CSTB expression.
Treating OC cells with miR-143-3p mimics or inhibitors resulted in
a respective decrease and increase in CSTB expression within OC
cells. Our findings suggest that CSTB is a target of miR-143-3p.
miRNAs can function as suppressors of target mRNA expression by the
binding base pairing with a complementary sequence, leading to the
degradation of target mRNA or the suppression of mRNA translation,
resulting in reduced protein expression. An inhibitor of miRNA such
as anti-miR-143-3p has the opposite function of miR-143-3p mimics.
As it is an antisense RNA and complementary to sense miRNA, the
anti-miRNA can inhibit the endogenous miRNA, leading to reduced
inhibition of mRNAs mediated by miRs. Upregulation of CSTB protein
was detected after anti-miR143-3p treatment, indicating that the
suppression of miRNAs can lead to the overexpression of their
target transcripts.
Several studies have suggested that miR-143-3p
functions as a tumor suppressor. For instance, miR-143-3p inhibits
cell proliferation and induces cell apoptosis in esophageal
squamous cell carcinoma (36), and
suppresses EMT in breast cancer (37). In prostate and bladder cancers,
miR-143-3p inhibits cell proliferation and migration (38,39).
High-throughput miRNA profiling demonstrated that miR-143-3p is
significantly downregulated in OC tissues compared with normal
ovarian tissues, while elevated miR-143-3p expression in the OC
cell lines OVCAR-3, SK-OV-3 and ES2 significantly reduced their
proliferation, migration and invasion (40). Consistently with these studies, our
findings indicate that miR-143-3p is dysregulated in OC. Low levels
of miR-143-3p may lead to an increase in the expression of CSTB.
The present study provides notable evidence that CSTB could be
regulated by miR143-3p as suggested by gain-of-function and
loss-of-function approaches, indicating the association between
miR-143-3p and CSTB in OC.
Our previous study has shown that CSTB expression is
mediated by the TGF-β1 signaling pathway in OC cells (3); however, the molecular mechanism
underlying TGF-β-mediated CSTB expression remains unknown. The
current study demonstrated that TGF-β1 increased the expression of
primary and mature miR-143-3p. SB431542 (an inhibitor of TGF-β type
I receptor) markedly increased pri-miR-143-3p expression and
partially abolished the effects of TGF-β, whereas the
TGF-β1-induced upregulation of mature miR-143-3p was significantly
abolished following SB431542 exposure. A single primary miRNA may
contain one to six miRNA hairpin loop structures comprising ~70
nucleotides each, which can be cleaved by Drosha (ribonuclease III
enzyme) together with DiGeorge Syndrome Critical Region 8 to form
precursor miRNAs in the nucleus. Subsequently, precursor miRNAs are
transported from the nucleus into the cytoplasm by Exportin-5 and
cleaved by Dicer to generate double-stranded mature miRNA (12,14,16);
TGF-β may influence the processing of miRNAs (19). There is a CArG box and a Smad
binding element located in the 580-bp enhancer region of
miR-143-3p, which fully respond to TGF-β1 stimulation upon the
binding of the Smad3/Smad4 to the enhancer region (41). Similarly, in vascular smooth muscle
cells, TGF-β binds to miR-143 via the CArG box (42). These findings suggest the existence
of a TGF-β/miR-143-3p/CSTB axis in which the levels of miR-143-3p
are upregulated after TGF-β1 stimulation, and interacts with the
3'-UTR of CSTB mRNA, leading to the downregulation of CSTB
expression.
In conclusion, CSTB is overexpressed and negatively
correlated with miR-143-3p expression in human EOC. High levels of
CSTB expression were associated with the poor OS of patients with
OC. Suppression of CSTB was demonstrated to inhibit OC cell
proliferation and induce apoptosis. Mature miR-143-3p directly
bound the 3'UTR of CSTB, leading to a decrease in CSTB expression
in OC cells, which is regulated by TGF-β1. Our findings suggested
the therapeutic potential of targeting the TGF-β/miR-143-3p/CSTB
axis for treating patients with OC.
Supplementary Materials
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81872121),
the Natural Science Foundation of Shanghai (grant no. 17ZR1404100),
the Shanghai Municipal Commission of Health and Family Planning
(grant no. 201640287 to G.X.), and a grant from Jinshan District
Commission of Health and Family Planning (grant no.
JSKJ-KTMS-2016-01 to W.G.).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
WG performed the majority of experiments, analyzed
the data, generated figures and wrote the manuscript. XW conducted
the siRNA experiments. QL, JZ, WR were involved in the experiments
and performed bioinformatics analysis. GX made substantial
contributions to the design of the study, conducted data analysis
and figure generation, and wrote the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Jinshan Hospital (approval no. E-2013-018-01;
Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
3′-UTR
|
3′-untranslated region
|
|
CSTB
|
cystatin B
|
|
EMT
|
epithelial to mesenchymal
transition
|
|
miR
|
microRNA
|
|
TGF-β
|
transforming growth factor-β
|
|
OC
|
ovarian cancer
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
Acknowledgments
Not applicable.
References
|
1
|
Turk V, Stoka V and Turk D: Cystatins:
Biochemical and structural properties, and medical relevance. Front
Biosci. 13:5406–5420. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Žerovnik E: Putative alternative functions
of human stefin B (cystatin B): Binding to amyloid-beta, membranes,
and copper. J Mol Recognit. 30:302017. View
Article : Google Scholar
|
|
3
|
Wang X, Gui L, Zhang Y, Zhang J, Shi J and
Xu G: Cystatin B is a progression marker of human epithelial
ovarian tumors mediated by the TGF-β signaling pathway. Int J
Oncol. 44:1099–1106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maher K, Jerič Kokelj B, Butinar M,
Mikhaylov G, Manček-Keber M, Stoka V, Vasiljeva O, Turk B,
Grigoryev SA and Kopitar-Jerala N: A role for stefin B (cystatin B)
in inflammation and endotoxemia. J Biol Chem. 289:31736–31750.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lalioti MD, Scott HS, Buresi C, Rossier C,
Bottani A, Morris MA, Malafosse A and Antonarakis SE: Dodecamer
repeat expansion in cystatin B gene in progressive myoclonus
epilepsy. Nature. 386:847–851. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gashenko EA, Lebedeva VA, Brak IV,
Tsykalenko EA, Vinokurova GV and Korolenko TA: Evaluation of serum
proca-thepsin B, cystatin B and cystatin C as possible biomarkers
of ovarian cancer. Int J Circumpolar Health. 72:212152013.
View Article : Google Scholar
|
|
9
|
Feldman AS, Banyard J, Wu CL, McDougal WS
and Zetter BR: Cystatin B as a tissue and urinary biomarker of
bladder cancer recurrence and disease progression. Clin Cancer Res.
15:1024–1031. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee MJ, Yu GR, Park SH, Cho BH, Ahn JS,
Park HJ, Song EY and Kim DG: Identification of cystatin B as a
potential serum marker in hepatocellular carcinoma. Clin Cancer
Res. 14:1080–1089. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shiraishi T, Mori M, Tanaka S, Sugimachi K
and Akiyoshi T: Identification of cystatin B in human esophageal
carcinoma, using differential displays in which the gene expression
is related to lymph-node metastasis. Int J Cancer. 79:175–178.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Treiber T, Treiber N and Meister G:
Regulation of microRNA biogenesis and its crosstalk with other
cellular pathways. Nat Rev Mol Cell Biol. 20:5–20. 2019. View Article : Google Scholar
|
|
13
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
van Kouwenhove M, Kedde M and Agami R:
MicroRNA regulation by RNA-binding proteins and its implications
for cancer. Nat Rev Cancer. 11:644–656. 2011. View Article : Google Scholar
|
|
16
|
Zhang L, Nadeem L, Connor K and Xu G:
Mechanisms and therapeutic targets of microRNA-associated
chemoresistance in epithelial ovarian cancer. Curr Cancer Drug
Targets. 16:429–441. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Behbahani GD, Ghahhari NM, Javidi MA,
Molan AF, Feizi N and Babashah S: MicroRNA-mediated
post-transcriptional regulation of epithelial to mesenchymal
transition in cancer. Pathol Oncol Res. 23:1–12. 2017. View Article : Google Scholar
|
|
18
|
Schetter AJ, Heegaard NH and Harris CC:
Inflammation and cancer: Interweaving microRNA, free radical,
cytokine and p53 pathways. Carcinogenesis. 31:37–49. 2010.
View Article : Google Scholar
|
|
19
|
Ottley E and Gold E: microRNA and
non-canonical TGF-β signalling: Implications for prostate cancer
therapy. Crit Rev Oncol Hematol. 92:49–60. 2014. View Article : Google Scholar
|
|
20
|
Shi Y and Massagué J: Mechanisms of
TGF-beta signaling from cell membrane to the nucleus. Cell.
113:685–700. 2003. View Article : Google Scholar
|
|
21
|
Attisano L and Wrana JL: Signal
transduction by the TGF-beta superfamily. Science. 296:1646–1647.
2002. View Article : Google Scholar
|
|
22
|
Schmierer B and Hill CS: TGFbeta-SMAD
signal transduction: Molecular specificity and functional
flexibility. Nat Rev Mol Cell Biol. 8:970–982. 2007. View Article : Google Scholar
|
|
23
|
Derynck R and Zhang YE: Smad-dependent and
Smad-independent pathways in TGF-beta family signalling. Nature.
425:577–584. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun W, Gui L, Zuo X, Zhang L, Zhou D, Duan
X, Ren W and Xu G: Human epithelial-type ovarian tumour marker
beta-2-micro-globulin is regulated by the TGF-β signaling pathway.
J Transl Med. 14:752016. View Article : Google Scholar
|
|
25
|
Matsumoto T, Yokoi A, Hashimura M, Oguri
Y, Akiya M and Saegusa M: TGF-β-mediated LEFTY/Akt/GSK-3β/Snail
axis modulates epithelial-mesenchymal transition and cancer stem
cell properties in ovarian clear cell carcinomas. Mol Carcinog.
57:957–967. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu KH, Patterson AP, Wang L, Marquez RT,
Atkinson EN, Baggerly KA, Ramoth LR, Rosen DG, Liu J, Hellstrom I,
et al: Selection of potential markers for epithelial ovarian cancer
with gene expression arrays and recursive descent partition
analysis. Clin Cancer Res. 10:3291–3300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bonome T, Levine DA, Shih J, Randonovich
M, Pise-Masison CA, Bogomolniy F, Ozbun L, Brady J, Barrett JC,
Boyd J, et al: A gene signature predicting for survival in
suboptimally debulked patients with ovarian cancer. Cancer Res.
68:5478–5486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) μethod. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
29
|
Takaya A, Peng WX, Ishino K, Kudo M,
Yamamoto T, Wada R, Takeshita T and Naito Z: Cystatin B as a
potential diagnostic biomarker in ovarian clear cell carcinoma. Int
J Oncol. 46:1573–1581. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin YY, Chen ZW, Lin ZP, Lin LB, Yang XM,
Xu LY and Xie Q: Tissue Levels of Stefin A and Stefin B in
Hepatocellular Carcinoma. Anat Rec (Hoboken). 299:428–438. 2016.
View Article : Google Scholar
|
|
31
|
Kos J, Krasovec M, Cimerman N, Nielsen HJ,
Christensen IJ and Brünner N: Cysteine proteinase inhibitors stefin
A, stefin B, and cystatin C in sera from patients with colorectal
cancer: Relation to prognosis. Clin Cancer Res. 6:505–511.
2000.PubMed/NCBI
|
|
32
|
Butinar M, Prebanda MT, Rajkovic J, Jerič
B, Stoka V, Peters C, Reinheckel T, Krüger A, Turk V, Turk B, et
al: Stefin B deficiency reduces tumor growth via sensitization of
tumor cells to oxidative stress in a breast cancer model. Oncogene.
33:3392–3400. 2014. View Article : Google Scholar
|
|
33
|
Jelovac D and Armstrong DK: Recent
progress in the diagnosis and treatment of ovarian cancer. CA
Cancer J Clin. 61:183–203. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cheng CJ, Bahal R, Babar IA, Pincus Z,
Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM,
et al: MicroRNA silencing for cancer therapy targeted to the tumour
microenvironment. Nature. 518:107–110. 2015. View Article : Google Scholar
|
|
35
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
He Z, Yi J, Liu X, Chen J, Han S, Jin L,
Chen L and Song H: MiR-143-3p functions as a tumor suppressor by
regulating cell proliferation, invasion and epithelial-mesenchymal
transition by targeting QKI-5 in esophageal squamous cell
carcinoma. Mol Cancer. 15:512016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhai L, Ma C, Li W, Yang S and Liu Z:
miR-143 suppresses epithelial-mesenchymal transition and inhibits
tumor growth of breast cancer through down-regulation of ERK5. Mol
Carcinog. 55:1990–2000. 2016. View Article : Google Scholar
|
|
38
|
Xu B, Niu X, Zhang X, Tao J, Wu D, Wang Z,
Li P, Zhang W, Wu H, Feng N, et al: miR-143 decreases prostate
cancer cells proliferation and migration and enhances their
sensitivity to docetaxel through suppression of KRAS. Mol Cell
Biochem. 350:207–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Song T, Zhang X, Wang C, Wu Y, Dong J, Gao
J, Cai W and Hong B: Expression of miR-143 reduces growth and
migration of human bladder carcinoma cells by targeting
cyclooxygenase-2. Asian Pac J Cancer Prev. 12:929–933. 2011.
|
|
40
|
Shi H, Shen H, Xu J, Zhao S, Yao S and
Jiang N: MiR-143-3p suppresses the progression of ovarian cancer.
Am J Transl Res. 10:866–874. 2018.
|
|
41
|
Long X and Miano JM: Transforming growth
factor-beta1 (TGF-beta1) utilizes distinct pathways for the
transcriptional activation of microRNA 143/145 in human coronary
artery smooth muscle cells. J Biol Chem. 286:30119–30129. 2011.
View Article : Google Scholar
|
|
42
|
Davis-Dusenbery BN, Chan MC, Reno KE,
Weisman AS, Layne MD, Lagna G and Hata A: down-regulation of
Kruppel-like factor-4 (KLF4) by microRNA-143/145 is critical for
modulation of vascular smooth muscle cell phenotype by transforming
growth factor-beta and bone morphogenetic protein 4. J Biol Chem.
286:28097–28110. 2011. View Article : Google Scholar : PubMed/NCBI
|