Introduction
Oral cancer is the 11th most prevalent cancer
worldwide and the 3rd most common cancer in developing nations
(1,2). Squamous cell carcinoma represents
~90% of oral cancer cases. The standard treatment strategies for
patients with resectable advanced-stage oral squamous cell
carcinoma (OSCC) are surgery following adjuvant radiotherapy or
concurrent chemoradiotherapy (3).
Although significant technological advances in the diagnosis and
treatment of patients with OSCC have been made in last few decades,
prognosis remains poor, with a 5-year overall survival rate of
50-60%, becoming even lower in advanced stages (2). OSCC frequently leads to lymphatic
metastasis, which is associated with the depth of invasion of the
primary tumor. This represents the most important predictor of
survival in patients with OSCC and has led to a major change in the
T stage classification described in the 8th edition of the American
Joint Committee on Cancer (4,5). The
revised staging system now reflects not only tumor size, but also
the depth of invasion (4).
Therefore, in order to provide more effective treatment for
patients with OSCC and to improve their clinical outcomes, an
increased understanding of the molecular and biological mechanisms
of tumor invasion and metastasis in OSCC is required.
The receptor tyrosine kinase, recepteur d'origine
nantais (RON), also known as macrophage-stimulating 1 receptor
(MST1R), is a member of the MET proto-oncogene family that includes
c-Met (6). RON is activated by the
ligand macrophage-stimulation protein (MSP) (7), and it has essential functional roles
during embryonic development and organogenesis (8,9). As
with other proteins involved in embryonic development, RON is
associated with carcinogenesis, cancer progression and metastasis.
RON is overexpressed in various cancers and its overexpression is
associated with poor prognosis in several types of cancer (10-18).
Thus, RON has received much interest in the fields of cancer
biology and cancer therapy targets over the past 20 years. Agents
targeting RON and/or c-Met for cancer therapy, such as low
molecular weight kinase inhibitors or monoclonal antibodies that
neutralize the receptors or ligands, are in various phases of
clinical trials and/or preclinical testing (19-21).
Nevertheless, little is known about the role of RON in human
OSCC.
The present study assessed whether RON affects
aggressive cancer cell behaviors of human OSCC cells, such as
invasion, migration and apoptosis inhibition and the oncogenic
signaling pathways involved in these changes, including
epithelial-mesenchymal transition and mitogen-activated protein
kinase (MAPK) signaling pathways. The expression of RON in human
OSCC tissues was analyzed by immunohistochemistry and reverse
transcription-quantitative PCR (RT-qPCR). The potential association
between RON expression and various clinicopathological variables,
including survival, was evaluated in a well-defined series of OSCCs
with complete long-term follow-up data. This study may provide an
important basis for the application of new RON-targeting therapies
for the treatment of OSCC.
Materials and methods
Cell culture and transfection
The human OSCC cell line, SCC25, was purchased from
the American Type Culture Collection and the human OSCC cell line,
PCI50, was provided by Dr Sung (Seoul National University, Seoul,
South Korea). SCC25 cells were cultured in DMEM/F12 medium (Gibco;
Thermo Fisher Scientific, Inc.) and PCI50 cells were cultured in
DMEM medium (Gibco; Thermo Fisher Scientific, Inc.), supplemented
with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) in a
humidified atmosphere of 5% CO2 at 37°C. Images of the
cells were captured using the EVOS FL microscope (EVOS® FL; Thermo
Fisher Scientific, Inc.). Small interfering RNAs (siRNAs) were used
to knockdown the endogenous gene expression of RON or
snail family transcriptional repressor 2 (SLUG) in OSCC
cells. OSCC cells were seeded in 6-well plates at a density of
2×105 cells/well and transfected with a 100 pmol
RON-specific (cat. no. sc-36434; Santa Cruz Biotechnology,
Inc), 100 pmol SLUG-specific (cat. no. 6591; Bioneer
Corporation) or 100 pmol negative control siRNA (cat. no. 1027281;
Qiagen, Inc.), using Lipofectamine RNAiMAX (Invitrogen; Thermo
Fisher Scientific, Inc.) for 48 h at 37°C, and then after 24-48 h,
the subsequent experiments were performed.
Protein isolation and western blotting
analysis
Cells were lysed in RIPA assay buffer (Biosesang,
Inc.) and protein concentrations were determined using a
bicinchoninic acid assay. Protein lysates (20-30 μg) were
separated by SDS-PAGE (10-12% gels) and then electrophoretically
transferred onto PVDF membranes. Membranes were incubated for 1 h
with 5% BSA (Bioshop Canada, Inc.) in 0.1% TBS-Tween-20 at room
temperature and then washed four times, for 15 min each, with 0.1%
TBS-Tween-20. Specific proteins were sequentially detected with
primary antibodies against RON (cat. no. sc-25781) and GAPDH (cat.
no. sc-25778) from Santa Cruz Biotechnology, Inc., and cleaved
caspase-3 (cat. no. 9664), cleaved caspase-7 (cat. no. 9491),
cleaved caspase-9 (cat. no. 9501), cleaved
poly(ADP-ribose)polymerase (PARP; cat. no. 5625), SLUG (cat. no.
9585), phospho-ERK1/2 (cat. no. 4370), phospho-c-JNK (cat. no.
4511), phospho-p38 (cat. no. 4511), total ERK (cat. no. 4695),
total JNK (cat. no. 9258) and total p-38 (cat. no. 9212) obtained
from Cell Signaling Technology, Inc. Primary antibodies were
diluted 1:1,000 and incubated with membranes for 24 h at 4°C.
Anti-rabbit (cat. no. 7074; Cell Signaling Technology, Inc.) or
anti-mouse (cat. no. 7076; Cell Signaling Technology, Inc.)
horseradish peroxidase (HRP)-conjugated secondary antibodies were
diluted 1:2,000 and incubated with membranes at room temperature
for 2 h. Immunoreactive proteins were visualized using an enhanced
chemiluminescence detection system for HRP (EMD Millipore), and
images were showed using an LAS-4000 luminescent image analyzer
(Image Leader LAS 4000IR, Fujifilm). Band densitometry was analyzed
using an Multi gauge V3.2 (Fujifilm).
RNA isolation, reverse transcription
polymerase chain reaction (RT-PCR) and RT-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. RT was performed using 1 μg of
total RNA, M-MLV reverse transcriptase (Invitrogen; Thermo Fisher
Scientific, Inc.), 1 μl 10 mM dNTP mix (Enzynomics Co.,
Ltd.), 1 μl oligo dT (500 μg/ml; Promega
Corporation), 2 μl 0.1 M dithiothreitol (Invitrogen; Thermo
Fisher Scientific, Inc.), 4 μl 5X first-strand buffer
(Invitrogen; Thermo Fisher Scientific, Inc.) and 1 μl RNase
inhibitor (Promega Corporation). For the first step, dNTP and oligo
dT were added at 65°C for 10 min, and the second step was performed
using RNA, dithiothreitol, 5X first-strand buffer, RNase inhibitor,
M-MLV reverse transcriptase at 37°C for 60 min, and then 70°C for
10 min, according to the manufacturer's protocol. cDNA was then
amplified using primers specific for RON, SLUG and
GAPDH (Bioneer Corporation), as previously described
(22). PCR was performed using
GoTaq DNA Polymerase and 5X Green GoTaq reaction buffer (Promega
Corporation). The primer sequences were as follows: RON forward,
5′-TCGCCTCGATGGAGCTCCTC-3′; RON reverse,
5′-CATGTGTGCCACTGTGACGT-3′; SLUG forward,
5′-CGAACTGGACACACATACAGTG-3′; SLUG reverse,
5′-CTGAGGATCTCTGGTTGTGGT-3′; GAPDH forward,
5′-ACCACAGTCCATGCCATCAC-3′; and GAPDH reverse,
5′-TCCACCCTGTTGCTGTA-3′. PCR products were separated by
electrophoresis on a 1% agarose gel containing ethidium
bromide.
qPCR analysis was performed using the QuantiSpeed
SYBR Green kit (cat. no. 105-02; Philekorea, Korea) with the
Rotor-Gene 6000 real-time rotary analyzer (Corbett Life Science),
and confirmed using melting curve analysis. Amplification plots
were used to evaluate the quantification cycle (Cq). The sequences
of the qPCR primers used were as follows: RON forward,
5′-CTTTGACGTGAAGTACGTGGT-3′; RON reverse,
5′-CGTATGGCTACAAACACAGCAC-3′; SLUG forward,
5′-CGAACTGGACACACATACAGTG-3′; SLUG reverse,
5′-CTGAGGATCTCTGGTTGTGGT-3′; 18S rRNA forward,
5′-GTAACCCGTTGAACCCCATT-3′; 18S rRNA reverse
5′-CCATCCAATCGGTAGTAGCG-3. qPCR was performed at 95°C for 2 min for
1 cycle, and then 95°C for 5 sec, 60°C for 20 sec for 40
cycles.
Cell invasion assay
Cell invasion ability was determined by the number
of cells that invaded through a Transwell invasion apparatus with
8.0-μm pores (Costar; Corning, Inc.). At 48 h after
transfection, cells transfected with RON or SLUG
siRNA or a negative control siRNA were seeded at 3×105
cells in 120 μl medium containing 0.2% BSA (BioShop, Inc.)
suspension in the upper chamber. The lower chamber was filled with
400 μl medium containing 0.2% BSA containing 100 ng/ml MSP
(cat. no. 352-MS; R&D Systems, Inc.) into the lower chamber as
the chemoattractant. After incubation for 24 h, cells that had
moved to the bottom Transwell surface were stained with Diff-Quik
solution (Sysmex Corporation) and were counted in five random
squares in the microscopic field of view. Results are presented as
the mean ± SE of the number of cells/field from three individual
experiments.
Cell migration assay (wound healing
assay)
At 24 h after transfection, cells transfected with a
RON-specific, SLUG-specific or negative control siRNA were seeded
in each well of Culture-Inserts (Ibidi GmbH) at 1.5×105
cells/well. After incubation for 24 h, each insert was detached and
the progression of cell migration was ascertained by imaging at 0,
8, 12 and 24 h, using an inverted microscope. Distances between
gaps were normalized to 1 cm after capture of three random
sites.
Apoptosis assay
Apoptosis was determined using an Annexin V-FITC
assay (cat. no. 556547; BD Biosciences). At 48 h after
transfection, cells transfected with a RON-specific siRNA or
a negative control siRNA were collected using trypsin, washed twice
in PBS, and re-suspended in binding buffer (BD Biosciences).
Annexin V-FITC and 7-amino-actinomycin D (BD Biosciences) were
added to the cells, which were incubated in the dark for 15 min and
then re-suspended in 400 ml binding buffer. Cells were analyzed
using a FACSCalibur flow cytometer (BD Biosciences; Becton,
Dickinson and Company) and BD CellQuest version 3.3 software
(Becton, Dickinson and Company). Data analysis was performed using
WinMDI version 2.9 (The Scripps Research Institute).
Patients and tumor specimens
To evaluate RON protein expression,
paraffin-embedded tissue sections were collected from 99 patients
who had undergone a diagnostic biopsy or definitive surgery for
OSCC at Chonnam National University Hwasun Hospital (Jeonnam,
Korea) between May 2004 and August 2013. None of the collected
tissues were obtained after radiotherapy or chemotherapy. Of these,
10 patients were excluded because of follow-up loss or palliative
treatment intent. A total of 67 men and 22 women participated in
the present study with a mean age of 62.7±12.8 years (range, 26-87
years). Of the 89 patients, 86 were treated with definitive
surgery, with or without adjuvant radiotherapy, or with
cisplatin-based concurrent chemoradiotherapy (CRT). Three patients
that refused surgery were treated with induction chemotherapy,
followed by cisplatin-based concurrent CRT, with curative intent.
The incidence of recurrence included locoregional recurrence and/or
distant metastasis after primary treatment with curative intent.
Patients with locoregional recurrence underwent salvage surgery or
CRT. Treatment failure was defined as disease with inoperable
locoregional progression or distant metastasis, even with salvage
treatment. Patients' clinicopathologic characteristics and complete
medical history, were reviewed in hospital records. Tumors were
staged according to the 7th edition of the American Joint Committee
on Cancer staging system (23).
Survival duration (months) was measured from the date of starting
treatment to the date of death or the date last seen. Disease-free
survival (DFS) was measured to the first observed date of any
recurrence or death and disease-specific survival (DSS) to the date
of OSCC-related death.
To evaluate RON mRNA expression, fresh OSCC
tissues and paired normal oral mucosa were collected from 20
patients that underwent diagnostic biopsy or definitive surgery of
OSCC at CNUHH. A total of 12 men and 8 women participated in the
present study with a mean age of 66.2±15.4 years (range, 23-86
years). RON mRNA expression was analyzed by RT-PCR and RT-qPCR,
performed as described above. Due to the small amount of fresh
tissue obtained during diagnostic biopsy, western blotting could
not be performed to measure RON protein expression.
Patient dataset from The Cancer Genome
Atlas (TCGA)
The TCGA (portal.gdc.cancer.gov/) head and neck cancer dataset
included 530 samples across all anatomical sites of the head and
neck. For this study, only OSCC patients with DFS status
information were selected, eliminated duplicate samples of the same
patient in the dataset, and finally included 226 patients in this
analysis. RNA-seq data was analyzed as expected maximization (RSEM)
values of MST1R expression, which were log2 transformed (RSEM+1).
The mean of MST1R expression levels was used as a cutoff value for
classification into low or high RON expression groups in survival
analyses.
Immunohistochemistry
Tissue processing and immunohisto-chemical analysis
were performed as previously described (22). Tissue sections were incubated with
primary antibodies against RON (cat. no. sc-25781; Santa Cruz
Biotechnology). Two independent observers interpreted RON staining
with no knowledge of the associated clinical information. Staining
intensity was scored as follows: 0, no staining of tumor cells; 1+,
weak-to-comparable staining in the cytoplasm and/or nucleus,
compared to non-tumor cell staining; 2+, readily appreciable or
dark brown staining distinctly marking the tumor cell cytoplasm
and/or nucleus. Percentages of stained cells were scored as
follows: 0, 0%; 1, 1-25%; 2, 26-50%; 3, 51-75%; and 4, >75%.
Final staining scores were the product of the intensity and
percentage scores, with ≤4 defined as low RON expression and >4
defined as high RON expression.
Statistical analysis
The significance of experimental differences was
assessed using Student's t-test. Association between RON expression
and various clinicopathological parameters were compared using the
χ2 test or Fisher's exact test. Survival curves were
calculated by the Kaplan-Meier method and were compared using the
log-rank test. Analyses were performed using SPSS version 21.0
(IBM, Corp.). P<0.05 was considered to indicate a statistically
significant difference.
Results
Knockdown of RON suppresses tumor cell
invasion in human OSCC cells
To examine the role of RON in tumor
progression, the present study used siRNA to inhibit the endogenous
expression of RON in the human OSCC cells, PCI50 and SCC25.
The mRNA and protein levels of RON were lower in the
RON-specific siRNA-treated PCI50 and SCC25 cells than in the
negative control siRNA-treated cells (Fig. 1A).
The number of invading RON-knockdown PCI50
cells was 109.0±6.4, compared with 406.3±11.7 invading negative
control PCI50 cells (Fig. 1B). The
number of invading RON-knockdown SCC25 cells was 99.5±5.3, compared
with 396.5±27.1 invading negative control SCC25 cells (Fig. 1B). The difference between the two
groups was statistically significant (P<0.05).
Knockdown of RON suppresses tumor cell
migration and RON activation by MSP enhances cell migration in
human OSCC cells
With time, the artificial wound gap became
significantly narrower in plates of negative control PCI50 and
SCC25 cells at 8, 12 and 24 h, compared with RON-knockdown cells
(P<0.05; Fig. 2A and C). Wound
gaps were not filled, even after 24 h, in plates of negative
control PCI50 and SCC25 cells. Therefore, in order to achieve
greater activation of RON signaling, 100 ng/ml of the RON ligand,
MSP, was applied to the plates of PCI50 and SCC25 cells. Migration
of PCI50 and SCC25 cells increased after incubation with MSP and
the artificial wound gap in plates of negative control cells became
significantly narrower than the wound gap in RON-knockdown cells at
8, 12, and 24 h (P<0.05; Fig. 2B
and C). This result demonstrated that ligand-mediated
activation of RON enhanced cell migration.
Knockdown of RON enhances tumor cell
apoptosis in human OSCC cells
To investigate the effect of RON on apoptosis, we
used an Annexin V apoptosis assay. Flow cytometric analysis
indicated that RON knockdown significantly increased the
proportion of apoptotic cells (Fig.
3A). The proportion of early and late apoptotic cells induced
by transfection of RON-specific siRNA were greater than in
the cells transfected with negative control siRNA (58.1 vs. 7.0%
and 14.2 vs. 4.0%, respectively) in PCI50 and SCC25 cells. To
confirm this effect of RON knockdown on tumor cell
apoptosis, apoptosis regulatory proteins were evaluated after siRNA
treatment. Levels of cleaved caspase-3, -7 and -9 and PARP were
higher in RON-knockdown PCI50 and SCC25 cells, compared with
negative control cells (Fig. 3B).
These results indicate that RON knockdown induces tumor cell
apoptosis by modulating apoptosis regulatory proteins, such as
caspases-3, -7 and -9 and PARP in human OSCC cells.
 | Figure 3Effect of RON knockdown on cell
apoptosis in human oral squamous cell carcinoma cells. (A)
Representative dot plots and histogram showed that RON knockdown
PCI50 and SCC25 cells displayed more apoptosis than in control
cells. (B) Levels of cleaved caspases-3, -7, and -9, and cleaved
PARP were greater in RON knockdown PCI50 and SCC25 cells than in
control cells (*P<0.05 vs. C). siRNA, small
interfering RNA; 7-AAD, 7-amino-actinomycin D; C, negative control
siRNA transfected cells; R, RON-specific siRNA transfected cells;
M1, normal cells; M2, apoptotic cells; RON, recepteur d'origine
nantais; PARP, poly(ADP-ribose)polymerase. |
Knockdown of RON decreases the expression
of epithelial-mesenchymal transition (EMT)-related transcription
factor, SLUG, and the phosphorylation of MAPK signaling proteins in
human OSCC cells
To examine the potential mechanisms involved in the
effects of RON knockdown on aggressive tumor cell behavior,
we investigated the impact of RON knockdown on the
EMT-related protein, SLUG and the MAPK signaling proteins. SLUG is
known to be involved in cell invasion and metastasis, and the MAPK
signaling pathways are essential for cell growth and survival.
RON knockdown reduced the expression of SLUG mRNA and
protein in PCI50 and SCC25 cells (Fig.
4A). In addition, the phosphorylation levels of ERK1/2 and p38
were decreased significantly by RON knockdown in PCI50 and
SCC25 cells. RON knockdown also decreased the levels of
phospho-JNK in PCI50 cells, but not in SCC25 cells (Fig. 4B).
Knockdown of SLUG eliminates the effect
of RON activation by MSP to aggressive cell behavior in human OSCC
cells
The mRNA and protein levels of SLUG were lower in
SLUG-specific siRNA-transfected PCI50 and SCC25 cells
compared with those in the negative control siRNA-transfected cells
(Fig. 5A).
 | Figure 5Effect of SLUG knockdown on cell
invasion and migration under RON activation by MSP in human oral
squamous cell carcinoma cells. (A) SLUG mRNA and protein were
reduced by SLUG siRNA in PCI50 and SCC25 cells compared with
negative control siRNA as shown by RT-PCR (upper left panel) and
RT-qPCR (upper right panel), as well as by western blotting (lower
panel). (B) In cell invasion assay, significantly fewer SLUG
knockdown PCI50 and SCC25 cells invaded than did negative control
cells. Stained invading cells were counted (bar graph; mean ± SE,
experiments were run in triplicate; *P<0.05). (C)
Cell migration with MSP, was significantly less in SLUG knockdown
PCI50 and SCC25 cells than in negative control cells. Cell
migration is displayed as relative healing distances measured in
three random sites. Values indicate mean ± SE for three independent
experiments (*P<0.05). RON, recepteur d'origine
nantais; siRNA, small interfering RNA; C, negative control siRNA
transfected cells; S, SLUG-specific siRNA transfected cells; SLUG,
snail family transcriptional repressor 2; MSP,
macrophage-stimulation protein. |
To verify the relationship between RON and SLUG,
cell invasion and migration were compared in negative control OSCC
cells and SLUG-knockdown OSCC cells after RON activation by
treatment with 200 ng/ml MSP. The number of invading
SLUG-knockdown PCI50 cells was 1.2±0.7, compared with
153.3±14.1 invading negative control PCI50 cells (Fig. 5B). The number of invading
SLUG-knockdown SCC25 cells was 5.5±1.4, compared with
170.1±14.3 invading negative control SCC25 cells (Fig. 5B). The difference between the two
groups was statistically significant (P<0.05). With time, the
artificial wound gap became significantly narrower in plates of
negative control PCI50 and SCC25 cells at 4, 8 and 12 h, compared
with SLUG-knockdown cells after MSP-induced RON activation
(P<0.05; Fig. 5C). This result
demonstrated that SLUG knockdown blocks the enhancement of
cell migration induced by RON activation in OSCC cells.
RON activation by its ligand, MSP,
induces morphological changes in human OSCC cells
To demonstrate the role of RON in EMT, morphological
changes in PCI50 and SCC25 cells treated with 500 ng/ml MSP for 48
and 72 h were analyzed. Light microscopy showed a scattering
effect, indicating cell-cell dissociation (Fig. 6). The individual cell morphology
also was changed to an elongated and spindle-like shape. These
findings suggested that MSP-induced RON activation is involved in
the process of EMT at the cellular level.
Expression of RON is high in human OSCC
tissues
The patients in this study included 67 men and 22
women. The mean age was 62.7±12.8 years (mean ± SD), with a range
of 26-87 years. The mean patient follow-up period was 59.5±38.1
months, with a range of 3.6-156.3 months. RON protein expression
was investigated by immunohistochemical staining of formalin-fixed,
paraffin-embedded biopsy tissue obtained from 89 patients with
OSCC. Immunostaining patterns were heterogenous, with predominantly
nuclear and/or cytoplasmic staining in tumor cells, but weak or no
staining in the normal adjacent tissues (Fig. 7). Based on the grading criteria,
high RON expression was observed in 43.8% of patients (39/89) and
low RON expression was observed in 56.2% of patients (50/89).
Patient data and the correlation between RON
expression and various OSCC clinicopathological variables are shown
in Table I. RON expression in OSCC
was not associated with age, sex, tumor location, tumor stage, T
stage, N stage or chemoradiotherapy treatment (P>0.05). Patients
with high RON expression tended to have a second primary malignancy
more often than those with low RON expression [24.0% (12/50) in the
low-expression group vs. 41.0% (16/39) in the high-expression
group]. This may be weak evidence that RON is involved in the
carcinogenesis of human cancers. Patients with high RON expression
had marginally higher rates of recurrence and treatment failure
rate than those with low RON expression, but this was not a
significant difference (P>0.05; Table II).
 | Table IAssociation between RON expression
and clinico-pathological parameters in patients with oral squamous
cell carcinoma. |
Table I
Association between RON expression
and clinico-pathological parameters in patients with oral squamous
cell carcinoma.
| Parameters | Total (n=89) | RON expression
| P-value |
|---|
| Low (n=50) | High (n=39) |
|---|
| Age (years) | | | | 0.21 |
| <62.7 | 41 | 20 | 21 | |
| ≥62.7 | 48 | 30 | 18 | |
| Sex | | | | 0.32 |
| Male | 67 | 40 | 27 | |
| Female | 22 | 10 | 12 | |
| Location | | | | 0.11 |
| Oral tongue | 61 | 38 | 23 | |
| FOM, BM, RMT | 28 | 12 | 16 | |
| Stage | | | | 1.00 |
| I, II | 59 | 33 | 26 | |
| III, IV | 30 | 17 | 13 | |
| T stage | | | | 1.00 |
| T1, T2 | 79 | 44 | 35 | |
| T3, T4 | 10 | 6 | 4 | |
| N stage | | | | 0.82 |
| N0 | 63 | 36 | 27 | |
| N1, N2 | 26 | 14 | 12 | |
| Chemotherapy and/or
radiotherapy | | | | 1.00 |
| No | 36 | 20 | 16 | |
| Yes | 53 | 30 | 12 | |
| Second primary
malignancy | | | | 0.11 |
| No | 61 | 38 | 23 | |
| Yes | 28 | 12 | 16 | |
 | Table IIAssociation between RON expression
and recurrence/treatment failure in patients with oral squamous
cell carcinoma. |
Table II
Association between RON expression
and recurrence/treatment failure in patients with oral squamous
cell carcinoma.
| Parameters | Total (n=89) | RON expression | P-value |
|---|
| Low (n=50) | High (n=39) |
|---|
| Recurrence | | | | 0.39 |
| No | 53 (59.6%) | 32 (64%) | 21 (53.8%) | |
| Yes | 36 (40.4%) | 18 (36%) | 18 (46.2%) | |
| Treatment
failure | | | | 0.50 |
| No | 59 (66.3%) | 35 (70%) | 24 (61.5%) | |
| Yes | 30 (33.7%) | 15 (30%) | 15 (38.5%) | |
For the 89 patients with OSCC enrolled in this
study, the 3/5-year DSS was 68/67% and the 3/5-year DFS was 64/58%.
The 3-year DSS/DFS rates in patients with high RON expression were
lower than those with low RON expression (72/70% in the
low-expression group vs. 64/56% in the high-expression group);
however, analysis of Kaplan-Meier curves for DSS/DFS using the
log-rank test, did not demonstrate a significant difference between
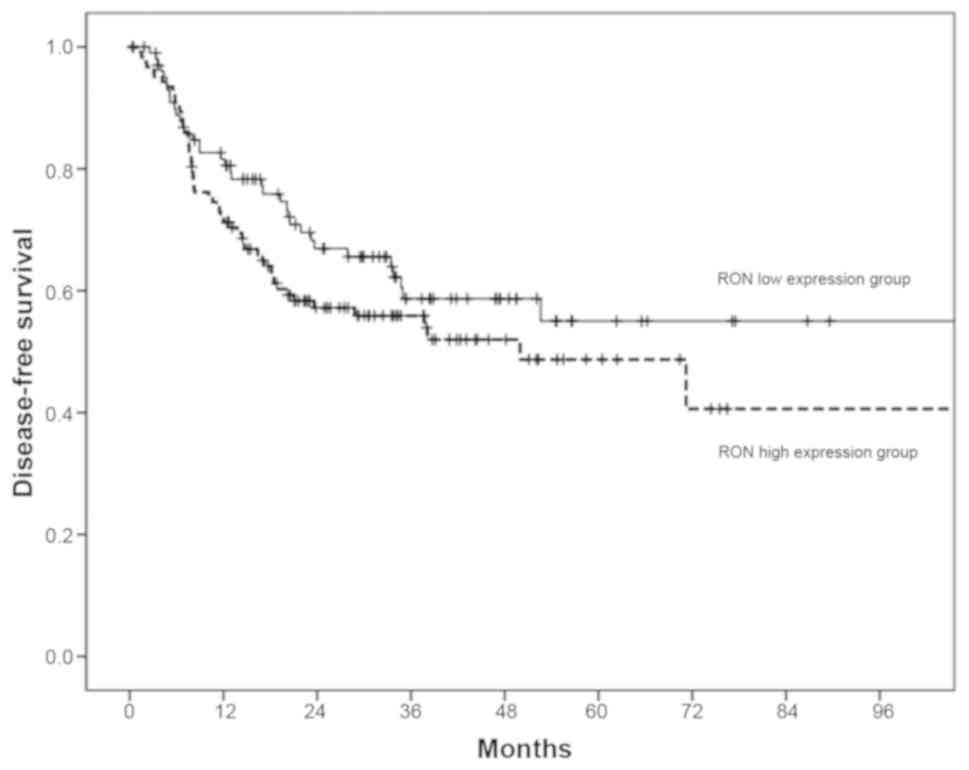
these two groups of patients (P>0.05; Fig. 8). For the 226 patients with OSCC in
the TCGA dataset, the 3-year DFS rates in patients with high RON
expression were lower than in those with low RON expression (60% in
the low-expression group vs. 51% in the high-expression group).
Similar to the previous results of patients from Chonnam National
University Hwasun Hospital, the TCGA data showed no statistically
significant association was found between RON expression and DFS
(P>0.05; Fig. 9).
To provide further support for the
immunohistochemical data on RON protein expression, RON mRNA
expression was analyzed using using RT-PCR and RT-qPCR in 20 fresh
human OSCC tissue samples and paired normal oral mucosa samples.
RON mRNA expression was significantly higher in OSCC tissue
compared with paired normal oral tissue (P<0.01; Fig. 10).
Discussion
OSCC typically has locoregional progression, and as
it grows, it invades adjacent tissues and undergoes regional
lymphatic metastasis, but rarely develops distant metastases
(5). The depth of tumor invasion
in OSCC is an important parameter for predicting lymphatic
metastasis and patient survival (5). The depth of tumor invasion affects
adjuvant treatment decisions, including the option of elective
lymph node dissection. These are characteristics specific for OSCC,
which differ from other types of head and neck cancers. Thus, the
discovery of molecular targets involved in tumor invasion in OSCC
patients is highly important.
RON is an attractive molecular target for OSCC
therapy. In this study, RON knockdown reduced tumor cell invasion
and migration. Conversely, RON activation by MSP, enhanced cell
migration in human OSCC cells. Several previous studies support
these findings. Maggiora et al (11) reported that overexpression of RON
increased cell invasion and migration, and that MSP caused a
12-fold increase in migration of human breast carcinoma cells. In
human colorectal cancer cells and hepatocellular cancer cells,
RON knockdown suppresses tumor cell invasion and migration
(24,25). These findings suggest that
RON-targeting therapy may control the invasiveness of cancer cells
in OSCC.
RON has an important role in EMT. EMT has been
highlighted as a key regulator of metastasis, by promoting tumor
cell invasion. The EMT process is tightly controlled through a
complex system of regulation of EMT-related transcription factors,
such as Snail1, SLUG, Zeb1, Zeb2 and Twist1, which are responsible
for inducing and sustaining the mesenchymal phenotype (26). Preliminary experiments were
performed to determine the association between RON and various
EMT-related transcription factors. Unfortunately, EMT-related
factors, other than SLUG, showed variable results that were
difficult to interpret among different cell line types, cell
conditions and cell preparation timings. Additionally, it was
difficult to obtain reproducibility in repeated experiments, even
under the same conditions. Therefore, the experiments were focused
on the association between RON and SLUG. RON activation by MSP
induced cell morphological changes to spindle shape and RON
knockdown suppressed SLUG expression. In addition, the promotion of
cell invasion and migration by MSP-mediated RON activation was
suppressed by SLUG knockdown. These findings suggest that
increased tumor cell invasion and migration by RON activation may
be mediated by SLUG. The role of RON in EMT further supports the
contribution of RON to tumor invasion and metastasis in OSCC.
The MAPK pathway, which includes ERK1/2, JNK and
p38, plays a key role in various steps of cancer development
(27). These proteins are involved
in controlling cell proliferation, differentiation, migration and
apoptosis in response to a variety of stresses, including cancers.
In this study, RON knockdown suppressed the activation of
ERK1/2, JNK and p38 in OSCC cells. In addition, RON
knockdown was associated with the reversal of aggressive tumor cell
behavior, such as apoptosis inhibition, cell invasion and cell
migration in OSCC cells. Although additional evaluation may be
required to clarify this mechanism, these findings suggested that
RON knockdown inhibits aggressive tumor cell behavior by
regulating the MAPK pathway in OSCC.
RON is involved in EMT through the MAPK pathway.
Several studies have reported that RON activation induces
spindle-shaped cell morphology and that RON is essential for the
expression of a specialized mesenchymal marker (28-31).
In addition, a MAPK inhibitor restored the original morphology of
the epithelial cells or blocked the expression of EMT-related
factors (28-30). These findings support the
hypothesis that RON is associated with aggressive tumor cell
behavior and is involved in EMT through the MAPK pathway in
OSCC.
The abundant expression of a targeted molecule is
the first requirement for an effective molecular target therapy.
Elevated RON expression has been found to be associated with poor
prognosis in multiple types of cancer, such as breast, stomach,
colon, lung, bladder, ovary and cholangiocarcinoma (14,15,32-36).
It was also demonstrated that RON is overexpressed in both fresh
OSCC tissue and paraffin-embedded tissue from human OSCC. However,
contrary to expectations, there was no significant association
between RON expression and prognostic indicators, including
recurrence, treatment failure and survival in human OSCC. Although
this is a limitation of our study, this is possible because the
treatment outcomes of patients with cancer are affected by many
factors, such as multiple genetic mutations, various other
signaling pathways and specific microenvironments that contribute
to cancer progression.
Several different splice variant isoforms of RON
have been identified in several cancers (37-40).
They are involved in RON activation in tumor cells and contribute
to oncogenic activities (37).
More importantly, efforts to block the RON activity, using low
molecular weight kinase inhibitors or monoclonal antibodies, can be
disrupted by unknown splice variants, because, despite having
similar sequences, they exhibit different localizations and have
varied functions (38). Thus, the
influence of the RON splice variants needs to be considered when
developing RON-targeting anticancer therapies.
RON is a promising target in various types of
cancer. Many studies and clinical trials have been conducted to
overcome cancer by inhibiting RON. The phase I/II clinical trials
of the tyrosine kinase inhibitor, foretinib, multi-targeting c-Met,
vascular endothelial growth factor 2 and RON, and anti-RON
monoclonal antibody, narnatumab, have reported for advanced solid
tumors, although the clinical trials were abandoned (21,41-43).
Several studies recently reported that the anti-RON antibody
Zt/g4-drug conjugate (ADC) is effective in targeted inhibition of
colorectal cancer, bladder cancer, triple-negative breast cancer
and pancreatic cancer (44-47).
In addition, Ruiz-Torres et al (48) demonstrated that the RON signaling
pathway is associated with the immunosuppressive microenvironment
in breast cancer. Ekiz et al (49) described that inhibition of RON
kinase potentiates checkpoint immunotherapy in breast cancer. It is
expected that the effectiveness of RON targeting anticancer therapy
can be improved through the ADC technique and/or the combination
therapy with immunotherapy.
In summary, RON knockdown suppressed cell
invasion, migration and inhibition of apoptosis mediated by the
MAPK pathway and the EMT-related factor, SLUG, in human OSCC cells.
RON was overexpressed in human OSCC tissue. The results of the
present study provide a theoretical basis for the application of
RON-targeting agents, under active investigation, for the treatment
of OSCC.
Acknowledgments
Not applicable.
Funding
This study was supported by a research grant from
the Research Institute of Medical Sciences, Chonnam National
University (2013-CURIMS-DR008).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Author's contributions
TMY and SAK analyzed the data and drafted
manuscript. SAK performed the experimental study. DHL and JKL
collected the clinical data. KHL analyzed the pathological data.
TMY and YEJ participated in the design of the study. SCL, YEJ, IJC
and MGN contributed to the interpretation of data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the ethics committee of
the Institutional Review Board of Chonnam National University
Hwasun Hospital (2011-027). Patients provided written informed
consent for the use of resected tissue specimens.
Patient consent for publication
Not applicable
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fitzmaurice C, Dicker D, Pain A, Hamavid
H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R,
Wolfe C, et al Global Burden of Disease Cancer Collaboration: The
global burden of cancer 2013. JAMA Oncol. 1:505–527. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pfister DG, Spencer S, Adelstein D, Adkins
D, Brizel DM, Burtness B, Busse PM, Caudell JJ, Cmelak AJ, Colevas
AD, et al: National Comprehensive Cancer Network: NCCN Clinical
Practice Guidelines in Oncology (NCCN Guidelines) Head and Neck
Cancers (Version 1). 2018, http://www.nccn.org. Accessed
February 15, 2018.
|
|
4
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Wincheser DP: American Joint Committee on Cancer-Cancer Staging
Manual. 8th edition. Springer; New York, NY: 2018
|
|
5
|
Pentenero M, Gandolfo S and Carrozzo M:
Importance of tumor thickness and depth of invasion in nodal
involvement and prognosis of oral squamous cell carcinoma: A review
of the literature. Head Neck. 27:1080–1091. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang MH, Wang D and Chen YQ: Oncogenic and
invasive potentials of human macrophage-stimulating protein
receptor, the RON receptor tyrosine kinase. Carcinogenesis.
24:1291–1300. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gaudino G, Follenzi A, Naldini L, Collesi
C, Santoro M, Gallo KA, Godowski PJ and Comoglio PM: RON is a
heterodimeric tyrosine kinase receptor activated by the HGF
homologue MSP. EMBO J. 13:3524–3532. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Muraoka RS, Sun WY, Colbert MC, Waltz SE,
Witte DP, Degen JL and Friezner Degen SJ: The Ron/STK receptor
tyrosine kinase is essential for peri-implantation development in
the mouse. J Clin Invest. 103:1277–1285. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Waltz SE, Eaton L, Toney-Earley K, Hess
KA, Peace BE, Ihlendorf JR, Wang MH, Kaestner KH and Degen SJ:
Ron-mediated cytoplasmic signaling is dispensable for viability but
is required to limit inflammatory responses. J Clin Invest.
108:567–576. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen Q, Seol DW, Carr B and Zarnegar R:
Co-expression and regulation of Met and Ron proto-oncogenes in
human hepato-cellular carcinoma tissues and cell lines. Hepatology.
26:59–66. 1997.PubMed/NCBI
|
|
11
|
Maggiora P, Marchio S, Stella MC, Giai M,
Belfiore A, De Bortoli M, Di Renzo MF, Costantino A, Sismondi P and
Comoglio PM: Overexpression of the RON gene in human breast
carcinoma. Oncogene. 16:2927–2933. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin HS, Berry GJ, Fee WE Jr, Terris DJ and
Sun Z: Identification of tyrosine kinases overexpressed in head and
neck cancer. Arch Otolaryngol Head Neck Surg. 130:311–316. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Camp ER, Liu W, Fan F, Yang A, Somcio R
and Ellis LM: RON, a tyrosine kinase receptor involved in tumor
progression and metastasis. Ann Surg Oncol. 12:273–281. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee WY, Chen HH, Chow NH, Su WC, Lin PW
and Guo HR: Prognostic significance of co-expression of RON and MET
receptors in node-negative breast cancer patients. Clin Cancer Res.
11:2222–2228. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheng HL, Liu HS, Lin YJ, Chen HH, Hsu PY,
Chang TY, Ho CL, Tzai TS and Chow NH: Co-expression of RON and MET
is a prognostic indicator for patients with transitional-cell
carcinoma of the bladder. Br J Cancer. 92:1906–1914. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Camp ER, Yang A, Gray MJ, Fan F, Hamilton
SR, Evans DB, Hooper AT, Pereira DS, Hicklin DJ and Ellis LM:
Tyrosine kinase receptor RON in human pancreatic cancer:
Expression, function, and validation as a target. Cancer.
109:1030–1039. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Welm AL, Sneddon JB, Taylor C, Nuyten DS,
van de Vijver MJ, Hasegawa BH and Bishop JM: The
macrophage-stimulating protein pathway promotes metastasis in a
mouse model for breast cancer and predicts poor prognosis in
humans. Proc Natl Acad Sci USA. 104:7570–7575. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Catenacci DV, Cervantes G, Yala S, Nelson
EA, El-Hashani E, Kanteti R, El Dinali M, Hasina R, Brägelmann J,
Seiwert T, et al: RON (MST1R) is a novel prognostic marker and
therapeut-ictarget for gastroesophageal adenocarcinoma. Cancer Biol
Ther. 12:9–46. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bieniasz M, Radhakrishnan P, Faham N, De
La OJP and Welm AL: Preclinical efficacy of Ron kinase inhibitors
alone and in combination with PI3K inhibitors for treatment of
sfRon-expressing breast cancer patient-derived xenografts. Clin
Cancer Res. 21:5588–5600. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yao HP, Zhou YQ, Ma Q, Guin S, Padhye SS,
Zhang RW and Wang MH: The monoclonal antibody Zt/f2 targeting RON
receptor tyrosine kinase as potential therapeutics against tumor
growth-mediated by colon cancer cells. Mol Cancer. 10:822011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shah MA, Wainberg ZA, Catenacci DV,
Hochster HS, Ford J, Kunz P, Lee FC, Kallender H, Cecchi F, Rabe
DC, et al: Phase II study evaluating 2 dosing schedules of oral
foretinib (GSK1363089), cMET/VEGFR2 inhibitor, in patients with
metastatic gastric cancer. PLoS One. 8:e540142013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yoon TM, Kim SA, Park YL, Lee KH, Sung MW,
Lee JK, Lim SC, Chung IJ and Joo YE: Expression of the receptor
tyrosine kinase recepteur d'origine nantais and its association
with tumor progression in hypopharyngeal cancer. Head Neck.
35:1106–1113. 2013. View Article : Google Scholar
|
|
23
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: American Joint Committee on Cancer-Cancer
Staging Manual. 7th edition. Springer; New York, NY: 2010
|
|
24
|
Xu XM, Wang D, Shen Q, Chen YQ and Wang
MH: RNA-mediated gene silencing of the RON receptor tyrosine kinase
alters oncogenic phenotypes of human colorectal carcinoma cells.
Oncogene. 23:8464–8474. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cho SB, Park YL, Song YA, Kim KY, Lee GH,
Cho DH, Myung DS, Park KJ, Lee WS, Chung IJ, et al: Small
interfering RNA-directed targeting of RON alters invasive and
oncogenic phenotypes of human hepatocellular carcinoma cells. Oncol
Rep. 26:1581–1586. 2011.PubMed/NCBI
|
|
26
|
Santamaria PG, Moreno-Bueno G, Portillo F
and Cano A: EMT: Present and future in clinical oncology. Mol
Oncol. 11:718–738. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wagner EF and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang D, Shen Q, Chen YQ and Wang MH:
Collaborative activities of macrophage-stimulating protein and
transforming growth factor-beta1 in induction of epithelial to
mesen-chymal transition: Roles of the RON receptor tyrosine kinase.
Oncogene. 23:1668–1680. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xiangming X, Yun Q, Guoliang Z, Jianjiang
L and Lisong T: Mechanisms of RON-mediated epithelial-mesenchymal
transition in MDCK cells through the MAPK pathway. Braz J Med Biol
Res. 44:634–641. 2011.PubMed/NCBI
|
|
30
|
Wang D, Shen Q, Xu XM, Chen YQ and Wang
MH: Activation of the RON receptor tyrosine kinase attenuates
transforming growth factor-beta1-induced apoptotic death and
promotes phenotypic changes in mouse intestinal epithelial cells.
Carcinogenesis. 26:27–36. 2005. View Article : Google Scholar
|
|
31
|
Côté M, Miller AD and Liu SL: Human RON
receptor tyrosine kinase induces complete epithelial-to-mesenchymal
transition but causes cellular senescence. Biochem Biophys Res
Commun. 360:219–225. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee CT, Chow NH, Su PF, Lin SC, Lin PC and
Lee JC: The prognostic significance of RON and MET receptor
coexpression in patients with colorectal cancer. Dis Colon Rectum.
51:1268–1274. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song YA, Park YL, Kim KY, Myung E, Chung
CY, Cho SB, Lee WS, Jung YD, Kweon SS and Joo YE: RON is associated
with tumor progression via the inhibition of apoptosis and cell
cycle arrest in human gastric cancer. Pathol Int. 62:127–136. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Han WL, Li WD, Hu J, Rusidanmu A, Chen LF,
Shen L and Zheng SS: Expression of the recepteur d'originenantais
receptor tyrosine kinase in non-small cell lung cancer and its
clinical significance. Chin Med J (Engl). 125:1110–1114. 2012.
|
|
35
|
Ferrandina G, Martinelli E, Petrillo M,
Prisco MG, Zucconi A, Santaguida S, Zannoni G, Scambia G and
Ferlini C: Prognostic role of the recepteur d'origine nantais (RON)
expression in ovarian cancer patients. Gynecol Oncol. 111:237–243.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cheng CT, Chen YY, Wu RC, Tsai CY, Chiang
KC, Yeh TS, Chen MH and Yeh CN: MET-RON dual inhibitor, BMS-777607,
suppresses cholangiocarcinoma cell growth, and MET-RON upregulation
indicates worse prognosis for intra-hepatic cholan-giocarcinoma
patients. Oncol Rep. 40:1411–1421. 2018.PubMed/NCBI
|
|
37
|
Ling Y, Kuang Y, Chen LL, Lao WF, Zhu YR,
Wang LQ and Wang D: A novel RON splice variant lacking exon 2
activates the PI3K/AKT pathway via PTEN phosphorylation in
colorectal carcinoma cells. Oncotarget. 8:39101–39116. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Krishnaswamy S, Mohammed AK, Tripathi G,
Alokail MS and Al-Daghri NM: Splice variants of the extracellular
region of RON receptor tyrosine kinase in lung cancer cell lines
identified by PCR and sequencing. BMC Cancer. 17:7382017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chakedis J, French R, Babicky M, Jaquish
D, Howard H, Mose E, Lam R, Holman P, Miyamoto J, Walterscheid Z,
et al: A novel protein isoform of the RON tyrosine kinase receptor
transforms human pancreatic duct epithelial cells. Oncogene.
35:3249–3259. 2016. View Article : Google Scholar :
|
|
40
|
Krishnaswamy S, Bukhari I, Mohammed AK,
Amer OE, Tripathi G, Alokail MS and Al-Daghri NM: Identification of
the splice variants of Recepteur d'Origine nantais (RON) in lung
cancer cell lines. Gene. 679:335–340. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Leighl NB, Tsao MS, Liu G, Tu D, Ho C,
Shepherd FA, Murray N, Goffin JR, Nicholas G, Sakashita S, et al: A
phase I study of foretinib plus erlotinib in patients with
previously treated advanced non-small cell lung cancer: Canadian
cancer trials group IND.196. Oncotarget. 8:69651–69662. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yau TCC, Lencioni R, Sukeepaisarnjaroen W,
Chao Y, Yen CJ, Lausoontornsiri W, Chen PJ, Sanpajit T, Camp A, Cox
DS, et al: A phase I/II multicenter study of single-agent foretinib
as first-line therapy in patients with advanced hepatocellular
carcinoma. Clin Cancer Res. 23:2405–2413. 2017. View Article : Google Scholar :
|
|
43
|
LoRusso PM, Gounder M, Jalal SI, André V,
Kambhampati SRP, Loizos N, Hall J, Holzer TR, Nasir A, Cosaert J,
et al: Phase 1 study of narnatumab, an anti-RON receptor monoclonal
antibody, in patients with advanced solid tumors. Invest New Drugs.
35:442–450. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Feng L, Yao HP, Wang W, Zhou YQ, Zhou J,
Zhang R and Wang MH: Efficacy of anti-RON antibody Zt/g4-drug
maytan-sinoid conjugation (anti-RON ADC) as a novel therapeutics
for targeted colorectal cancer therapy. Clin Cancer Res.
20:6045–6058. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen JF, Yu BX, Yu R, Ma L, Lv XY, Cheng Y
and Ma Q: Monoclonal antibody Zt/g4 targeting RON receptor tyrosine
kinase enhances chemosensitivity of bladder cancer cells to
Epirubicin by promoting G1/S arrest and apoptosis. Oncol Rep.
37:721–728. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Suthe SR, Yao HP, Weng TH, Hu CY, Feng L,
Wu ZG and Wang MH: RON receptor tyrosine kinase as a therapeutic
target for eradication of triple-negative breast cancer: Efficacy
of anti-RON ADC Zt/g4-MMAE. Mol Cancer Ther. 17:2654–2664. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yao HP, Feng L, Weng TH, Hu CY, Suthe SR,
Mostofa AGM, Chen LH, Wu ZG, Wang WL and Wang MH: Preclinical
efficacy of anti-RON antibody-drug conjugate Zt/g4-MMAE for
targeted therapy of pancreatic cancer overexpressing RON receptor
tyrosine kinase. Mol Pharm. 15:3260–3271. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ruiz-Torres SJ, Benight NM, Karns RA,
Lower EE, Guan JL and Waltz SE: HGFL-mediated RON signaling
supports breast cancer stem cell phenotypes via activation of
non-canonical β-catenin signaling. Oncotarget. 8:58918–58933. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ekiz HA, Lai SA, Gundlapalli H, Haroun F,
Williams MA and Welm AL: Inhibition of RON kinase potentiates
anti-CTLA-4 immunotherapy to shrink breast tumors and prevent
metastatic outgrowth. Oncoimmunology. 7:e14802862018. View Article : Google Scholar : PubMed/NCBI
|