Introduction
Breast cancer (BC) is the most common type of
malignant tumor in females and is composed of numerous subtypes
with a high heterogeneity (1). In
total, ~60-70% of human BC cases are associated with an
overexpression of estrogen receptor α (ER) and are sensitive to
endocrine therapy (2,3). Compared with ER-positive patients,
ER-negative patients exhibit a more aggressive phenotype,
metastasis and a poor prognosis (4,5).
There is a marked difference in the gene expression profiles of
ER-negative and ER-positive BC (6-8).
However, few specific factors associated with ER-negative BC have
been identified. Therefore, it remains a major challenge to
identify novel molecular targets for the treatment of ER-negative
BC, which may prevent progression.
Chemokine (C-X-C motif) ligand 1 (CXCL1) belongs to
the CXC chemokine family, a family composed of small peptides, and
was originally identified in melanoma tumors (9,10).
CXCL1 binds specifically to the G protein-coupled receptor
chemokine (C-X-C motif) receptor 2 (CXCR2), which is a member of
the CXC chemokine receptor family (11). Aberrant expression of CXCL1 has
been identified in numerous types of malignancy, and has been
associated with oncogenesis, metastasis, angiogenesis and
chemoresistance (12-14). Acharyya et al (12) reported that CXCL1, as an important
molecule, was involved in the endothelial-cancer-marrow signaling
network, and linked tumor metastasis and drug resistance. Wang
et al (15) also identified
that CXCL1 secreted by lymphatic endothelial cells promoted gastric
cancer progression via integrin subunit β1/focal adhesion
kinase/AKT signaling. These findings indicated that CXCL1 may act
as a pro-tumorigenic molecule in a paracrine manner following its
secretion by non-tumor cells. Previously, the overexpression of
CXCL1 in tumor cells has been reported in various types of cancer,
including prostate cancer, hepatocellular carcinoma and gastric
carcinoma (16-18). Previous studies have also
demonstrated that CXCL1 is upregulated in the plasma and stroma of
patients with BC (19,20); however, whether there is a
difference in CXCL1 expression depending on the expression levels
of ER in BC remains unclear and requires further investigation.
The present study analyzed CXCL1 expression in
breast tumor tissues by reverse transcription-quantitative PCR
(RT-qPCR) and immunohistochemistry (IHC), which revealed that CXCL1
was highly expressed in ER-negative BC tissues compared with
ER-positive BC tissues. In addition, the present study further
investigated the expression of CXCL1 in BC cell lines. Furthermore,
it was revealed that CXCL1 secreted by tumor cells may promote
ER-negative BC cell metastasis via the ERK/matrix metalloproteinase
(MMP)2/9 signaling pathway in a CXCR2-dependent manner. IHC assays
also suggested that phosphorylated (p)-ERK1/2 was positively
associated with CXCL1 protein in BC tissues.
Materials and methods
Cell culture and reagents
The ER-negative BC cell lines BT-549, MDA-MB-231
MDA-MB-468, and HS578t, and the ER-positive BC cell lines T47D,
MCF-7 and ZR-75-1 were purchased from the American Type Culture
Collection and maintained in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) or Dulbecco's modified Eagle's medium/F12 with
10% fetal bovine serum (both from Gibco; Thermo Fisher Scientific,
Inc.), 100 µg/ml streptomycin and 100 U/ml penicillin
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in a 5%
CO2 standard humidified incubator. For time- and
dose-dependence experiments, MDA-MB-231 and BT-549 BC cells were
treated with recombinant human (rh)CXCL1 at concentrations of 0,
0.1, 1 or 10 ng/ml for 1 h at 37°C, and treated with 1 ng/ml
rhCXCL1 for 0, 10, 30, 60 min at 37°C in a 5% CO2
standard humidified incubator. MDA-MB-231 and BT-549 BC cells were
treated the combinations of treatments (1 ng/ml rhCXCL1 + 200 nM
SB225002, 1 ng/ml rhCXCL1 + 5 µM U0126, 1 ng/ml rhCXCL1 +
200 nM SB225002 + 5 µM U0126) at 37°C in a 5% CO2
standard humidified incubator. rhCXCL1 (cat. no. 275-GR) was
obtained from R&D Systems, Inc. The CXCR2 inhibitor SB225002
(cat. no. S7651) and the MEK1/2 inhibitor U0126 (cat. no. S1102)
were purchased from Selleck Chemicals.
Cell migration and invasion assays
For the cell migration assay, 3×105
MDA-MB-231 or BT-549 cells in 200 µl serum-free medium were
seeded into the upper chamber of a Transwell plate (EMD Millipore),
and complete medium with or without various concentrations (0.1,
1.0 and 10 ng/ml) of CXCL1, U0126 (5 µM) and SB225002 (200
nM) was added to the lower compartment. Wells without CXCL1 served
as controls. The cells were incubated for 12 h at 37°C, then the
Transwell inserted were removed and washed, and cells were fixed
with 4% paraformaldehyde for 15 min at room temperature and stained
with 0.5% crystal violet for 5 min at room temperature. The numbers
of migratory cells in five randomly selected fields were counted
under an inverted light microscope (magnification, ×200; TE2000-U;
Nikon Corporation).
For the cell invasion assay the upper chamber was
coated with Matrigel (EMD Millipore) as described previously
(21). The remaining steps were
the same as the migration assay. After 24 h of incubation at 37°C,
the numbers of invaded cells in five randomly selected fields were
counted (magnification, ×200).
Knockdown of CXCL1
For knockdown of CXCL1 in MDA-MB-231 and BT-549
cells, lentiviral expression vectors containing CXCL1 short hairpin
RNA (shRNA) or control shRNA were obtained from Shanghai GenePharma
Co., Ltd. The sequence of CXCL1 shRNA was 5′-GCACATCTGTTTT
GTAACT-3′, and the control shRNA sequence was 5′-TTC
TCCGAACGTGTCACGT-3′. Cells at a density of 30-50% in 6-well plates
were transfected with sh-CXCL1 or sh-Ctrl lentivirus
(1×108 TU/ml). After 8-12 h of incubation, the medium
was replaced with complete medium containing FBS and puromycin.
Further experiments were performed after ≥2 weeks.
RNA isolation and RT-qPCR
Total RNA was extracted from human tissue specimens
and cells using TRIzol (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. RNA was reverse
transcribed using the PrimeScript RT Master Mix kit (Takara
Biotechnology, Co., Ltd.), according to the manufacturer's
protocol. RT was conducted as follows: 15 min at 37°C for three
times, followed by inactivation at 85°C for 5 sec. qPCR was
performed with SYBR Pre-mix Ex Taq™ II (Takara Biotechnology, Co.,
Ltd.) according to the manufacturer's protocol. qPCR was conducted
as follows: 2 min at 95°C, followed by 39 cycles at 95°C for 30
sec, 30 sec at 58°C and 20 sec at 72°C. The sequences of the
primers for CXCL1, GAPDH, MMP2 and MMP9 are listed in Table I. Relative gene expression was
normal ized to GAPDH and calculated using the 2−ΔΔCq
method (22). The experiment was
independently repeated in triplicate.
 | Table IPrimers used for reverse
transcription-quantitative PCR analysis. |
Table I
Primers used for reverse
transcription-quantitative PCR analysis.
| Gene | Forward | Reverse |
|---|
| CXCL1 |
5′-TCCTGCATCCCCCATAGTTA-3′ |
5′-CTTCAGGAACAGCCACCAGT-3′ |
| GAPDH |
5′-CTCTGCTCCTCCTGTTCGAC-3′ |
5′-GCGCCCAATACGACCAAATC-3′ |
| MMP2 |
5′-TTGATGGCATCGCTCAGATC-3′ |
5′-TGTCACGTGGCGTCACAGT-3′ |
| MMP9 |
5′-GGTTCAGGCGAGGACCATAGAG-3′ |
5′-TTTGACAGCGACAAGAAGTGG-3′ |
Western blot analysis
Total proteins were extracted using RIPA lysis
buffer with PMSF (both from Beyotime Institute of Biotechnology).
Protein concentrations were assessed using a BCA Protein Assay kit
(Beyotime Institute of Biotechnology). A total of 40 µg
protein was separated by 8-10% SDS-PAGE and transferred onto PVDF
membranes. After blocking with 5% skim milk for 1 h at room
temperature, the membranes were incubated at 4°C overnight with the
following primary antibodies: p-ERK1/2 (1:1,000; cat. no. AF1891;
Beyotime Institute of Biotechnology); ERK1/2 (1:5,000; cat. no.
ab184699); p-AKT (1:1,000; cat. no. ab38449) (both from Abcam); AKT
(1:1,000; cat. no. 9272S); STAT3 (1:1,000; cat. no. 12640S) (both
from Cell Signaling Technology, Inc.); p-STAT3 (1:5,000; cat. no.
ab76315); p-ribosomal S6 kinase P90 (p-RSK1P90; 1:5,000; cat. no.
ab32203); RSK1P90 (1:5,000; cat. no. ab32114); MMP9 (1:5,000; cat.
no. ab76003); MMP2 (1:2,000; cat. no. ab92536) (all from Abcam);
and GAPDH (1:1,000; cat. no. 5174S; Cell Signaling Technology,
Inc.). Subsequently, appropriate horseradish peroxidase
(HRP)-conjugated secondary antibodies (1:1,000; cat. nos. 7074S and
7076S; Cell Signaling Technology, Inc.) were applied for 1 h at
37°C. The immunoreactive bands were detected using an enhanced
chemiluminescence reagent (EMD Millipore).
Immunofluorescence (IF)
An IF assay was performed as described previously
(21). Cells were grown on glass
coverslips for 24 h, fixed with 4% paraformaldehyde for 20 min at
room temperature, permeabilized with 0.1% Triton X-100 for 15 min
and then blocked with 10% normal goat serum (cat. no. C0265;
Beyotime Institute of Biotechnology) for 30 min at room
temperature. The cells were incubated overnight at 4°C with
specific primary antibodies against CXCL1 (1:200; cat. no. ab89318;
Abcam). After washing three times with PBS, the cells were stained
with FITC-conjugated goat anti-rabbit secondary antibody (1:200;
cat. no. TA130022; OriGene Technologies, Inc.) for 1 h at room
temperature. The cell nucleus was stained with DAPI for 5 min at
room temperature. IF images were obtained with a Nikon Eclipse 80i
microscope (magnification, ×400; Nikon Corporation)
Patients and samples
A total of 87 paired human breast tissue specimens,
including tumor and adjacent non-tumor tissue, were obtained from
the First Affiliated Hospital of Chongqing Medical University. All
patients (20-72 years old) underwent surgery for BC at the First
Affiliated Hospital of Chongqing Medical University between
November 2015 and June 2016. All patients had their primary site in
the breast and were diagnosed specifically with BC for the first
time by the Clinical Diagnostic Pathology Center of Chongqing
Medical University. The ER status of the patient was determined
according to the results of immunohistochemistry by the Clinical
Diagnostic Pathology Center of Chongqing Medical University. The
study was approved by the Ethics Committee of Chongqing Medical
University. Written informed consent was obtained from all
patients.
IHC
IHC staining was performed as described previously
(21). The human tissues were
fixed with 4% formaldehyde buffer for 12-24 h at room temperature.
Deparaffinized specimens were then sectioned (4-µm thick
slices). The slices were autoclaved at 115°C for 5 min for antigen
retrieval in citric acid buffer (pH 6.0), quenched for endogenous
peroxidase activity with 0.3% H2O2 solution
for 10-15 min, blocked for non-specific binding with 10% normal
goat serum for 10-15 min at room temperature, and incubated with
specific rabbit primary antibodies against CXCL1 (1:400; cat. no.
ab89318; Abcam) and p-ERK1/2 (1:200; cat. no. AF1891; Abcam)
overnight at 4°C. Subsequently, the sections were treated with
HRP-conjugated goat anti-rabbit IgG secondary antibody (1:200; cat.
no. TA140003; OriGene Technologies, Inc.) for 30 min at room
temperature. After staining with diaminobenzidine (OriGene
Technologies, Inc.) and hematoxylin for 5 sec at room temperature,
images were captured using a Nikon Eclipse 80i microscope
(magnification, ×200; Nikon Corporation). CXCL1 and p-ERK1/2
staining intensities (I) were scored as: 0, 1, 2, 3. The percentage
of the stained area (A) was scored as: 1 (0-25%), 2 (26-50%), 3
(51-75%) and 4 (76-100%). The sum of the intensity and percentage
scores (I + A) was used as the final IHC score. Expression was
analyzed using Image-Pro Plus 6.0 software (Media Cybernetics,
Inc.).
Enzyme-linked immunosorbent assay
(ELISA)
BC cells were seeded in a 6-well cultured plate at a
density of 5×105 cells. Following culture for 12 h, the
suspension was replaced with 1 ml serum-free media. After the cells
were starved for 24 h, the supernatants were harvested and
centrifuged in 1,000 × g for 10 min at room temperature.
Concentrations of secreted CXCL1 in the supernatants were
determined using a human CXCL1/GROα Quantikine ELISA kit (cat. no.
DGR00B; R&D Systems, Inc.) according to the manufacturer's
protocol.
Oncomine database analysis
Oncomine, a cancer microarray database, was screened
for breast cancer datasets where ER status was determined
(wwww.oncomine.org) (23). A total of 4 independent
microarrays, including Bittner (GSE2109), The Cancer Genome Atlas
database, Sorlie (24) and Desmedt
(25) were obtained from the
Oncomine database. CXCL1 expression was analyzed in ER-negative and
ER-positive BC with the R (version 3.5.1) package ggstatspot
(indrajeet-patil.github.io/ggstatsplot).
Statistical analysis
SPSS 20.0 software (IBM Corp.) was used for all
statistical analysis. Data of three independent experiments are
presented as the mean ± standard deviation. One-way ANOVA followed
by Dunnett's multiple comparisons tests was used to evaluate the
significant differences among multiple groups. Fisher's exact test
was used to evaluate associations between the detected protein
expression levels of CXCL1 and p-ERK1/2. P<0.05 was considered
to indicate a statistically significant difference.
Results
Increased expression of CXCL1 mRNA in
ER-negative BC tissues
To analyze the expression of CXCL1 in human BC
tissues, the relative mRNA expression levels of CXCL1 in all 87
samples were examined. The clinical parameters of the patients with
BC are presented in Table II. The
CXCL1 mRNA levels in ER-negative BC tissues (n=55) were
significantly upregulated compared with the ER-positive BC tissues
(n=32; Fig. 1A). In addition, four
independent microarrays obtained from the Oncomine public database
were analyzed. The mRNA expression levels of CXCL1 were
significantly upregulated in the ER-negative BC cases compared with
the ER-positive BC cases in the Bittner, Sorlie and Desmedt breast
databases and The Cancer Genome Atlas database (Fig. 1B-E). In summary, these results
suggest that there is high expression of CXCL1 mRNA in ER-negative
breast tumors.
 | Table IIClinicopathological characteristics
of breast tumors (n=87). |
Table II
Clinicopathological characteristics
of breast tumors (n=87).
|
Characteristics | Number (%) |
|---|
| Age (years) | |
| <45 | 30 (34.5) |
| ≥45 | 57 (65.5) |
| Lymph node
metastasis | |
| Negative | 50 (57.5) |
| Positive | 37 (42.5) |
| Tumor size
(cm) | |
| <2 | 20 (23.0) |
| ≥2 to <5 | 64 (73.6) |
| ≥5 | 3 (3.4) |
| Histological grade
(54) | |
| I | 1 (1.1) |
| II | 57 (65.6) |
| III | 18 (20.7) |
| Unknown | 11 (12.6) |
| ER status | |
| Negative | 54 (62.1) |
| Positive | 33 (37.9) |
| PR status | |
| Negative | 55 (63.2) |
| Positive | 32 (36.8) |
| HER2 status | |
| Negative | 47 (54.0) |
| Positive | 38 (43.7) |
| Unknown | 2 (2.3) |
| Ki 67 (%) | |
| <14 | 26 (29.9) |
| ≥14 | 61 (70.1) |
| p53 | |
| Negative | 23 (26.4) |
| Positive | 64 (73.6) |
| Chemotherapy | |
| Yes | 20 (23.0) |
| No | 67 (77.0) |
CXCL1 is upregulated in ER-negative BC
cells
To further verify the association between CXCL1
expression and ER-negative BC, four ER-negative BC cell lines
(BT-549, MDA-MB-231, MDA-MB-468 and HS578t) and three ER-positive
BC cell lines (T47D, MCF-7, ZR-75-1) were analyzed. The mRNA and
protein CXCL1 expression levels in these cells were detected by
RT-qPCR and ELISA. The levels of CXCL1 mRNA (Fig. 2A) and protein (Fig. 2B) were markedly upregulated in the
ER-negative BC cells compared with the ER-positive cells. CXCL1 was
predominantly located in the cell cytoplasm, as determined via IF
assays (Fig. 2C). These data
demonstrated that CXCL1 exhibits increased expression in
ER-negative BC cells compared with ER-positive BC cells.
 | Figure 2Upregulation of CXCL1 in ER-negative
cancer cells, and the CXCL1/CXCR2-induced migration and invasion of
ER-negative cancer cells. (A) Quantification of CXCL1 mRNA was
performed via reverse transcription-quantitative PCR analysis in
four ER-negative cell lines (BT-549, MDA-MB-231, MDA-MB-468,
HS578t) and three ER-positive cell lines (T47D, MCF-7, ZR-75-1).
Data are presented as the mean ± standard deviation from three
independent experiments. (B) Secreted CXCL1 protein in the
supernatant from BC cells was collected and measured by ELISA. Data
are presented as the mean ± standard deviation from three
independent experiments. (C) Expression and localization of CXCL1
in BC cells as determined by immunofluorescence staining. Scale
bars, 100 µm. Magnification, ×400. (D) Migratory and (E)
invasive abilities of MDA-MB-231 and BT-549 cells incubated
with/without rhCXCL1 (0.1, 1.0 and 10 ng/ml) were evaluated by
Transwell assays. Magnification, ×200. **P<0.001. (F)
Migration and (G) invasion of MDA-MB-231 and BT-549 cells were
evaluated using Transwell assays following treatment with/without
rhCXCL1 (10 ng/ml) or the CXCR2 antagonist SB225002 (200 nM).
**P<0.001. BC, breast cancer; CXCL1, chemokine (C-X-C
motif) ligand 1; ER, estrogen receptor; rh, recombinant human. |
CXCL1 promotes ER-negative BC cell
migration and invasion in a CXCR2-dependent manner
Based on the aforementioned findings, it was
hypothesized that CXCL1 overexpression in ER-negative BC may be
associated with the aggressive nature of ER-negative BC. To
investigate the effect of CXCL1 on the invasion of ER-negative BCs,
MDA-MB-231 and BT-549 cells were treated with/without rhCXCL1 (0.1,
1.0 and 10 ng/ml). A Transwell assay revealed that CXCL1
significantly increased the migration (Fig. 2D) and invasion (Fig. 2E) of MDA-MB-231 and BT-549 cells in
a dose-dependent manner, compared with control treatment.
Subsequently, SB225002, a specific CXCR2 antagonist, was used to
determine whether the effects of CXCL1 on the migration and
invasion of ER-negative cells were associated with CXCR2. The
CXCL1-induced increases in cell migration (Fig. 2F) and invasion (Fig. 2G) were significantly attenuated by
treatment with SB225002. In summary, these data suggested that
enhanced CXCL1 in ER-negative BC promotes cell migration and
invasion in a CXCR2-dependent manner.
CXCL1/CXCR2 induces ER-negative BC cell
invasion and migration via the ERK1/2 pathway
Previous studies have reported that chemokines can
bind to their receptors to induce cancer progression by stimulating
a series of downstream signaling pathways, including the PI3K/AKT,
Janus kinase (JAK)/STAT3 and ERK1/2 pathways (26-29).
Therefore, possible signaling mechanisms associated with the
CXCL1/CXCR2-induced promotion of ER-negative BC cell migration and
invasion were examined by western blot analysis. It was identified
that only p-ERK1/2 was activated by rhCXCL1 in MDA-MB-231 and
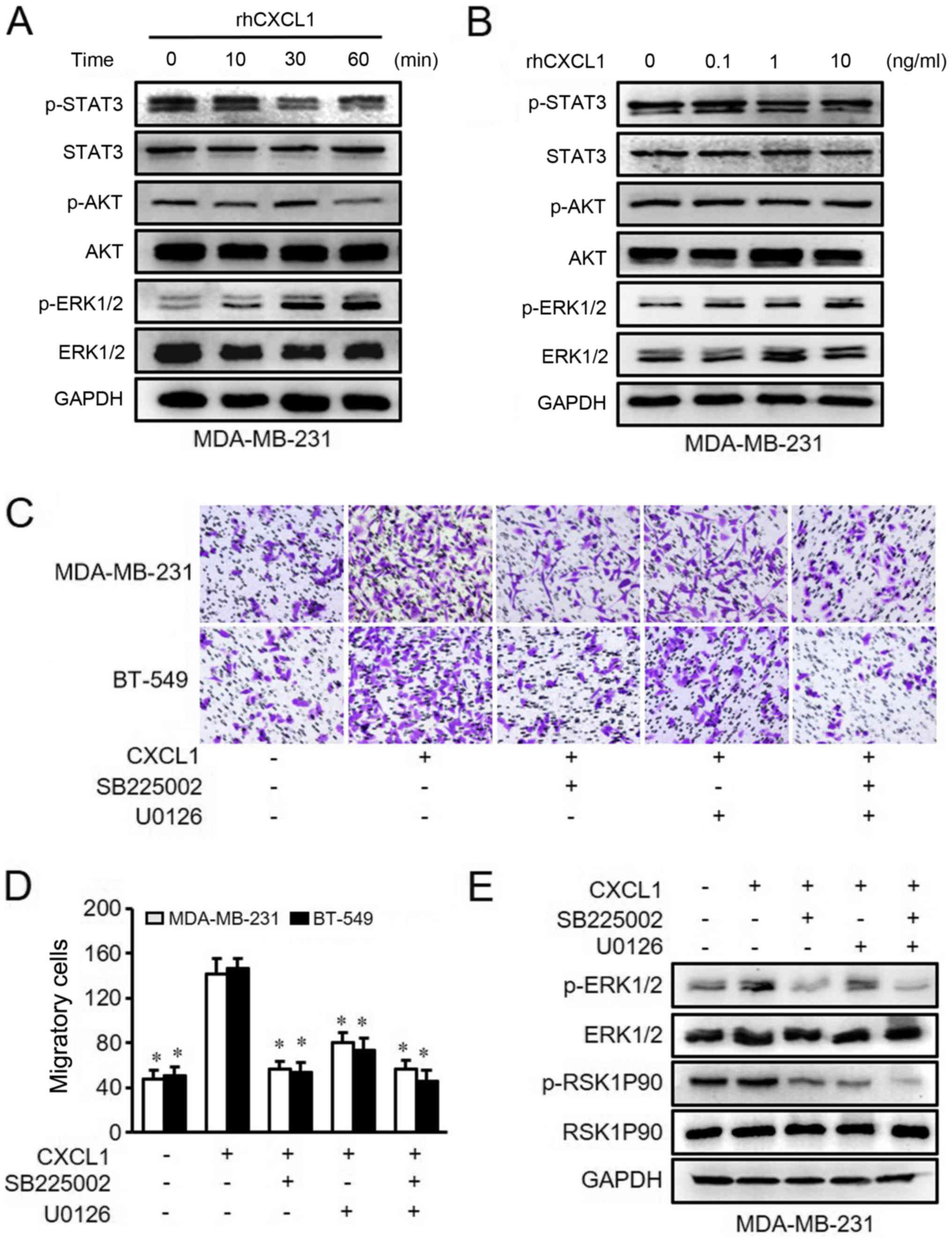
BT-549 cells in a time- and dose-dependent manner (Figs. 3A and B, and 4A and B).
Next, the present study used inhibitors
of CXCR2 and MEK to treat MDA-MB-231ß and BT-549 cells
The results demonstrated that CXCL1-mediated cell
migration and invasion were significantly inhibited by either
SB225002 or U0126 compared with rhCXCL1 treatment alone (Figs. 3C and D, and 4C and D). Similarly, the activated ERK1/2
and RSK1P90 proteins in the ERK pathway that were stimulated by
CXCL1 were inhibited following treatment with SB225002 and U0126
(Figs. 3E and 4E). These findings suggested that CXCL1
regulates the migration and invasion of ER-negative cells via ERK
signaling in a CXCR2-dependent manner.
Knockdown of CXCL1 reduces ER-negative BC
cell migration and invasion via the ERK1/2 pathway
To further determine the role of CXCL1 in the
invasion of ER-negative BC cells, the lentivirus-mediated shCXCL1
and control vector were stably transduced into ER-negative
MDA-MB-231 and BT-549 cells. The efficiency of knockdown was
verified via RT-qPCR analysis and ELISAs (Fig. 5A-D). As hypothesized, reduced CXCL1
significantly attenuated the migratory abilities of MDA-MB-231 and
BT-549 cells (Fig. 6A and B).
Similar results were observed in the cell invasion assay (Fig. 6C and D). Subsequently, the levels
and phosphorylation of ERK and RSK1P90, key proteins associated
with ERK signaling activation, were detected via western blot
analysis. It was identified that knockdown of CXCL1 in MDA-MB-231
and BT-549 cells inhibited ERK1/2 pathway activation (Fig. 6E and F). These data demonstrated
that silencing CXCL1 in ER-negative cells prevents cell migration
and invasion due to inhibition of the ERK1/2 pathway.
Effects of CXCL1 stimulation on MMP2/9
expression by ERK1/2 activation
It has been reported that MMP2 and MMP9 are strongly
associated with tumor metastasis (30-33).
Thus, it was hypothesized that activated ERK1/2 signaling may
contribute to CXCL1-mediated MMP2/9 expression in ER-negative
cells. To verify this hypothesis, the MDA-MB-231 and BT-549 cells
pretreated with SB225002, U0126 and/or rhCXCL1 were evaluated for
their mRNA and protein expression levels of MMP2/9 via RT-qPCR and
western blot analyses. As presented in Fig. 7A and C, rhCXCL1 treatment
significantly increased the mRNA and protein levels of MMP2/9;
however, the effects of CXCL1 on the activation of MMP2/9 in
MDA-MB-231 cells were reversed by pretreatment with SB225002 or
U0126. Similar results were observed in BT-549 cells (Fig. 7B and D). Furthermore, it was
determined that knockdown of CXCL1 in MDA-MB-231 and BT-549 cells
by shCXCL1 inhibited ERK//MMP2/9 signaling, and this inhibitory
effect could be reversed by the treatment of these cells with
rhCXCL1 (Fig. 7E and F). In
summary, these data suggested that CXCL1 can stimulate MMP2/9
expression in ER-negative cells via ERK1/2 activation in a
CXCR2-dependent manner.
CXCL1 protein is highly expressed in
ER-negative BC tissues and positively associated with p-ERK1/2 in
BC tissues
The protein expression levels of CXCL1 and p-ERK1/2
were detected in 88 BC tissue samples via IHC. CXCL1 and p-ERK1/2
were expressed in 62.5% (55/88) and 52.3% (46/88) of these tumor
cases, respectively. Representative images are presented in
Fig. 8A, and quantitative analysis
revealed that CXCL1 expression was significantly increased in
ER-negative BC tissues compared with ER-positive tissues
(P<0.05; Fig. 8B). Furthermore,
a significant association between CXCL1 and p-ERK1/2 expression was
observed via IHC; p-ERK1/2 expression was observed in 61.8% (34.55)
of CXCL1-positive tissues, but only 36.4% (12/33) of CXCL1-negative
tissues (P<0.05; Fig. 8C).
These data suggested an enhanced CXCL1 protein expression in
ER-negative BC, that is associated with the expression of p-ERK1/2
protein.
Discussion
Chemokine systems, including chemokines and their
receptors, serve important roles in cancer biology by inducing
tumor cell growth, migration, invasion, chemoresistance and
angiogenesis (11,34). Chemokines can interact with cancer
cells via two pathways; the autocrine pathway and the paracrine
pathway (35). There is extensive
evidence that CXCL1 is produced by immune cells and stromal cells,
and acts in a paracrine manner in the tumor microenvironment during
carcinogenesis (14,36). However, tumor-derived CXCL1 has
rarely been reported to promote cell metastasis in an autocrine
manner in human BC. In the present study, CXCL1 mRNA levels and
CXCL1 secretion levels in the supernatant were determined to be
upregulated in ER-negative cells. Similar results have been
previously reported for another chemokine, IL-8 (37). The present study further revealed
that CXCL1 could increase the metastatic potential of MDA-MB-231
and BT-549 cells in a dose-dependent manner in vitro. These
results indicated that tumor-derived CXCL1 may be associated with
the invasive ability of ER-negative BC cells.
Certain studies have suggested that patients with
pancreatic, gastric or hepatocellular cancer exhibit increased
levels of CXCL1 in cancer tissues (38-40).
By contrast, other studies have demonstrated that CXCL1 mRNA
expression levels in hepatic tumors were similar between cancerous
and non-cancerous tissues (41).
Notably, in the present study, no difference in the mRNA expression
level of CXCL1 was identified between the adjacent non-tumor and
tumor tissues for all patients with BC (data not shown). However,
it was revealed that CXCL1 mRNA was upregulated in patients with
ER-negative BC compared with ER-positive BC. In addition, a marked
difference was observed in the CXCL1 protein levels between
ER-negative and ER-positive BC tumor tissues via IHC staining.
These findings indicated that CXCL1 may be a biomarker for
ER-negative BC.
Chemokines can bind to specific G-protein coupled
receptors to activate multiple downstream signaling pathways in
cancer. In addition to JAK/STAT3 and PI3K/AKT signaling, the
MAPK/ERK signaling pathway is one of these targeted pathways
(42-45). However, in the present study, it
was identified that only ERK signaling was stimulated by rhCXCL1 in
ER-negative cells in a dose- and time-dependent manner; knockdown
of CXCL1 in MDA-MB-231 and BT-549 cells inhibited the activation of
the ERK pathway. Furthermore, the present results demonstrated that
CXCL1-mediated ER-negative BC cell migration and invasion could be
significantly suppressed following inhibition of the ERK1/2 pathway
using U0126. ERK1/2 phosphorylation stimulated by CXCL1 has been
reported in other types of cell, including endothelial cells,
muscle cells and astrocytes (46-48).
Furthermore, cellular migration and invasion stimulated by the MAPK
pathway has been well reported (45). However, to the best of our
knowledge, no previous study has reported that the ERK pathway may
serve a key role in the CXCL1-induced metastasis of ER-negative
BC.
Activation of the ERK/MMP2/9 pathway axis regulated
by CXCL1 may serve a crucial role in ER-negative cell metastasis.
MMP2 and MMP9, members of the MMP family, have been reported to
drive metastasis in various cancer types, including pancreatic,
hepatocellular and lung cancers (49). The upregulation of MMP2 and MMP9 is
associated with poor prognosis in patients with ovarian and breast
cancers (50,51). Furthermore, it has been reported
that MMP2 and MMP9 promote the migration and invasion of cancer
cells via regulation of the ERK signaling pathway (52,53).
The present study demonstrated that CXCL1 could upregulate the
expression of MMP2/9 in ER-negative cells, which could be reversed
by treatment with the ERK inhibitor U0126. Additionally, knockdown
of CXCL1 in ER-negative cells downregulated MMP2/9 expression, and
this effect was significantly reversed by addition of rhCXCL1.
Although MMP2/9 upregulation induced by CXCL1 derived from
lymphatic endothelial cells has previously been reported in gastric
cancer (14), this study did not
report that the CXCL1-induced upregulation of MMP2/9 expression is
dependent on ERK1/2 signaling, as was indicated in the present
study for ER-negative BC.
In summary, the present findings revealed that the
expression levels of CXCL1 were upregulated in ER-negative BC. It
was demonstrated that CXCL1 can stimulate tumor cell invasion via
the ERK1/2/MMP2/9 pathway axis. Therefore, CXCL1 may serve as a
potential therapeutic target in ER-negative BC.
Funding
This work was supported in part by National Natural
Science Foundation of China (grant nos. NSFC 81472658 and NSFC
81772979) and Chongqing Natural Science Foundation (grant no.
cstc2015shmszx0269) for Shengchun Liu; and partly supported by
National Natural Science Foundation of China (grant nos. NSFC
81472476 and NSFC 31671481) for Manran Liu.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
SL, ML, CY and HY designed the study. CY and HY
performed the majority of the experiments and were major
contributors in writing the manuscript. XL, LJ and KT participated
in the collection of clinical samples and prepared experimental
materials. RC and MP conducted the statistical analysis of clinical
data and analyzed a substantial quantity of experimental data. All
authors have read and approved the final submitted manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics committee of
Chongqing Medical University. Written informed consent was obtained
from all patients.
Patient consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Polyak K: Heterogeneity in breast cancer.
J Clin Invest. 121:3786–3788. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carey LA, Perou CM, Livasy CA, Dressler
LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S,
et al: Race, breast cancer subtypes, and survival in the Carolina
Breast Cancer Study. JAMA. 295:2492–2502. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Litzenburger BC and Brown PH: Advances in
Preventive Therapy for Estrogen-Receptor-Negative Breast Cancer.
Curr Breast Cancer Rep. 6:96–109. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Barcellos-Hoff MH: Does microenvironment
contribute to the etiology of estrogen receptor-negative breast
cancer? Clin Cancer Res. 19:541–548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen JQ and Russo J: ERalpha-negative and
triple negative breast cancer: Molecular features and potential
therapeutic approaches. Biochim Biophys Acta. 1796:162–175.
2009.PubMed/NCBI
|
|
6
|
Shen K, Rice SD, Gingrich DA, Wang D, Mi
Z, Tian C, Ding Z, Brower SL, Ervin PR Jr, Gabrin MJ, et al:
Distinct genes related to drug response identified in ER positive
and ER negative breast cancer cell lines. PLoS One. 7:e409002012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bianchini G, Qi Y, Alvarez RH, Iwamoto T,
Coutant C, Ibrahim NK, Valero V, Cristofanilli M, Green MC,
Radvanyi L, et al: Molecular anatomy of breast cancer stroma and
its prognostic value in estrogen receptor-positive and -negative
cancers. J Clin Oncol. 28:4316–4323. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Milne RL, Kuchenbaecker KB, Michailidou K,
Beesley J, Kar S, Lindström S, Hui S, Lemaçon A, Soucy P, Dennis J,
et al: ABCTB Investigators; EMBRACE; GEMO Study Collaborators;
HEBON; kConFab/AOCS Investigators; NBSC Collaborators:
Identification of ten variants associated with risk of
estrogen-receptor-negative breast cancer. Nat Genet. 49:1767–1778.
2017. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Silva RL, Lopes AH, Guimarães RM and Cunha
TM: CXCL1/CXCR2 signaling in pathological pain: Role in peripheral
and central sensitization. Neurobiol Dis. 105:109–116. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang D, Yang W, Du J, Devalaraja MN, Liang
P, Matsumoto K, Tsubakimoto K, Endo T and Richmond A:
MGSA/GRO-mediated melanocyte transformation involves induction of
Ras expression. Oncogene. 19:4647–4659. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Balkwill FR: The chemokine system and
cancer. J Pathol. 226:148–157. 2012. View Article : Google Scholar
|
|
12
|
Acharyya S, Oskarsson T, Vanharanta S,
Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N,
Seshan VE, et al: A CXCL1 paracrine network links cancer
chemoresistance and metastasis. Cell. 150:165–178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Miyake M, Hori S, Morizawa Y, Tatsumi Y,
Nakai Y, Anai S, Torimoto K, Aoki K, Tanaka N, Shimada K, et al:
CXCL1-mediated interaction of cancer cells with tumor-associated
macrophages and cancer-associated fibroblasts promotes tumor
progression in human bladder cancer. Neoplasia. 18:636–646. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu J, Zhang C, He Y, Wu H, Wang Z, Song W,
Li W, He W, Cai S and Zhan W: Lymphatic endothelial cell-secreted
CXCL1 stimulates lymphangiogenesis and metastasis of gastric
cancer. Int J Cancer. 130:787–797. 2012. View Article : Google Scholar
|
|
15
|
Wang Z, Wang Z, Li G, Wu H, Sun K, Chen J,
Feng Y, Chen C, Cai S, Xu J, et al: CXCL1 from tumor-associated
lymphatic endothelial cells drives gastric cancer cell into
lymphatic system via activating integrin β1/FAK/AKT signaling.
Cancer Lett. 385:28–38. 2017. View Article : Google Scholar
|
|
16
|
Kuo PL, Shen KH, Hung SH and Hsu YL:
CXCL1/GROα increases cell migration and invasion of prostate cancer
by decreasing fibulin-1 expression through NF-κB/HDAC1 epigenetic
regulation. Carcinogenesis. 33:2477–2487. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han KQ, He XQ, Ma MY, Guo XD, Zhang XM,
Chen J, Han H, Zhang WW, Zhu QG and Zhao WZ: Targeted silencing of
CXCL1 by siRNA inhibits tumor growth and apoptosis in
hepatocellular carcinoma. Int J Oncol. 47:2131–2140. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang L, Zhang C, Xu J, Wu H, Peng J, Cai S
and He Y: CXCL1 gene silencing inhibits HGC803 cell migration and
invasion and acts as an independent prognostic factor for poor
survival in gastric cancer. Mol Med Rep. 14:4673–4679. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Divella R, Daniele A, Savino E, Palma F,
Bellizzi A, Giotta F, Simone G, Lioce M, Quaranta M, Paradiso A, et
al: Circulating levels of transforming growth factor-βeta (TGF-β)
and chemokine (C-X-C motif) ligand-1 (CXCL1) as predictors of
distant seeding of circulating tumor cells in patients with
metastatic breast cancer. Anticancer Res. 33:1491–1497.
2013.PubMed/NCBI
|
|
20
|
Zou A, Lambert D, Yeh H, Yasukawa K,
Behbod F, Fan F and Cheng N: Elevated CXCL1 expression in breast
cancer stroma predicts poor prognosis and is inversely associated
with expression of TGF-β signaling proteins. BMC Cancer.
14:7812014. View Article : Google Scholar
|
|
21
|
Wang L, Hou Y, Sun Y, Zhao L, Tang X, Hu
P, Yang J, Zeng Z, Yang G, Cui X, et al: c-Ski activates
cancer-associated fibroblasts to regulate breast cancer cell
invasion. Mol Oncol. 7:1116–1128. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-ΔΔC(T) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
23
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sorlie T, Tibshirani R, Parker J, Hastie
T, Marron JS, Nobel A, Deng S, Johnesn H, Pesich R, Geisler S, et
al: Repeated observation of breast tumor subtypes in independent
gene expression data sets. Proc Natl Acad Sci USA. 100:8418–8423.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Desmedt C, Piette F, Loi S, Wang Y,
Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, ZhangY
d'Assignies MS, et al: Strong time dependence of the 76-gene
prognostic signature for node-negative breast cancer patients in
the TRANSBIG multicenter independent validation series. Clin Cancer
Res. 13:3207–3214. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao J, Ou B, Han D, Wang P, Zong Y, Zhu
C, Liu D, Zheng M, Sun J, Feng H, et al: Tumor-derived CXCL5
promotes human colorectal cancer metastasis through activation of
the ERK/Elk-1/Snail and AKT/GSK3β/β-catenin pathways. Mol Cancer.
16:702017. View Article : Google Scholar
|
|
27
|
Fu XT, Dai Z, Song K, Zhang ZJ, Zhou ZJ,
Zhou SL, Zhao YM, Xiao YS, Sun QM, Ding ZB, et al:
Macrophage-secreted IL-8 induces epithelial-mesenchymal transition
in hepatocellular carcinoma cells by activating the
JAK2/STAT3/Snail pathway. Int J Oncol. 46:587–596. 2015. View Article : Google Scholar
|
|
28
|
Li S, Lu J, Chen Y, Xiong N, Li L, Zhang
J, Yang H, Wu C, Zeng H and Liu Y: MCP-1-induced ERK/GSK-3β/Snail
signaling facilitates the epithelial-mesenchymal transition and
promotes the migration of MCF-7 human breast carcinoma cells. Cell
Mol Immunol. 14:621–630. 2017. View Article : Google Scholar
|
|
29
|
Ou B, Zhao J, Guan S, Feng H, Wangpu X,
Zhu C, Zong Y, Ma J, Sun J, Shen X, et al: CCR4 promotes metastasis
via ERK/NF-κB/MMP13 pathway and acts downstream of TNF-α in
colorectal cancer. Oncotarget. 7:47637–47649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jabłońska-Trypuć A, Matejczyk M and
Rosochacki S: Matrix metalloproteinases (MMPs), the main
extracellular matrix (ECM) enzymes in collagen degradation, as a
target for anticancer drugs. J Enzyme Inhib Med Chem. 31(Suppl 1):
177–183. 2016. View Article : Google Scholar
|
|
31
|
Nishio K, Motozawa K, Omagari D, Gojoubori
T, Ikeda T, Asano M and Gionhaku N: Comparison of MMP2 and MMP9
expression levels between primary and metastatic regions of oral
squamous cell carcinoma. J Oral Sci. 58:59–65. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng Y, Miu Y, Yang X, Yang X and Zhu M:
CCR7 mediates TGF-β1-induced human malignant glioma invasion,
migration, and epithelial-mesenchymal transition by activating
MMP2/9 through the nuclear factor kappaB signaling pathway. DNA
Cell Biol. 36:853–861. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen SX, Yin JF, Lin BC, Su HF, Zheng Z,
Xie CY and Fei ZH: Upregulated expression of long noncoding RNA
SNHG15 promotes cell proliferation and invasion through regulates
MMP2/MMP9 in patients with GC. Tumour Biol. 37:6801–6812. 2016.
View Article : Google Scholar
|
|
34
|
Balkwill F: Cancer and the chemokine
network. Nat Rev Cancer. 4:540–550. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mantovani A, Savino B, Locati M, Zammataro
L, Allavena P and Bonecchi R: The chemokine system in cancer
biology and therapy. Cytokine Growth Factor Rev. 21:27–39. 2010.
View Article : Google Scholar
|
|
36
|
Zhang T, Tseng C, Zhang Y, Sirin O, Corn
PG, Li-Ning-Tapia EM, Troncoso P, Davis J, Pettaway C, Ward J, et
al: CXCL1 mediates obesity-associated adipose stromal cell
trafficking and function in the tumour microenvironment. Nat
Commun. 7:116742016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Freund A, Chauveau C, Brouillet JP, Lucas
A, Lacroix M, Licznar A, Vignon F and Lazennec G: IL-8 expression
and its possible relationship with estrogen-receptor-negative
status of breast cancer cells. Oncogene. 22:256–265. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lian S, Zhai X, Wang X, Zhu H, Zhang S,
Wang W, Wang Z and Huang J: Elevated expression of growth-regulated
oncogene-alpha in tumor and stromal cells predicts unfavorable
prognosis in pancreatic cancer. Medicine (Baltimore). 95:e43282016.
View Article : Google Scholar
|
|
39
|
Han KQ, Han H, He XQ, Wang L, Guo XD,
Zhang XM, Chen J, Zhu QG, Nian H, Zhai XF, et al: Chemokine CXCL1
may serve as a potential molecular target for hepatocellular
carcinoma. Cancer Med. 5:2861–2871. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xiang Z, Jiang DP, Xia GG, Wei ZW, Chen W,
He Y and Zhang CH: CXCL1 expression is correlated with Snail
expression and affects the prognosis of patients with gastric
cancer. Oncol Lett. 10:2458–2464. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cui X, Li Z, Gao J, Gao PJ, Ni YB and Zhu
JY: Elevated CXCL1 increases hepatocellular carcinoma
aggressiveness and is inhibited by miRNA-200a. Oncotarget.
7:65052–65066. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou J, Yi L, Ouyang Q, Xu L, Cui H and Xu
M: Neurotensin signaling regulates stem-like traits of glioblastoma
stem cells through activation of IL-8/CXCR1/STAT3 pathway. Cell
Signal. 26:2896–2902. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou B, Sun C, Li N, Shan W, Lu H, Guo L,
Guo E, Xia M, Weng D, Meng L, et al: Cisplatin-induced CCL5
secretion from CAFs promotes cisplatin-resistance in ovarian cancer
via regulation of the STAT3 and PI3K/Akt signaling pathways. Int J
Oncol. 48:2087–2097. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lin HY, Sun SM, Lu XF, Chen PY, Chen CF,
Liang WQ and Peng CY: CCR10 activation stimulates the invasion and
migration of breast cancer cells through the ERK1/2/MMP-7 signaling
pathway. Int Immunopharmacol. 51:124–130. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xiong Y, Huang F, Li X, Chen Z, Feng D,
Jiang H, Chen W and Zhang X: CCL21/CCR7 interaction promotes
cellular migration and invasion via modulation of the MEK/ERK1/2
signaling pathway and correlates with lymphatic metastatic spread
and poor prognosis in urinary bladder cancer. Int J Oncol.
51:75–90. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Miyake M, Goodison S, Urquidi V, Gomes
Giacoia E and Rosser CJ: Expression of CXCL1 in human endothelial
cells induces angiogenesis through the CXCR2 receptor and the
ERK1/2 and EGF pathways. Lab Invest. 93:768–778. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Al-Alwan LA, Chang Y, Rousseau S, Martin
JG, Eidelman DH and Hamid Q: CXCL1 inhibits airway smooth muscle
cell migration through the decoy receptor Duffy antigen receptor
for chemokines. J Immunol. 193:1416–1426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Filipovic R and Zecevic N: The effect of
CXCL1 on human fetal oligodendrocyte progenitor cells. Glia.
56:1–15. 2008. View Article : Google Scholar
|
|
49
|
Alaseem A, Alhazzani K, Dondapati P,
Alobid S, Bishayee A and Rathinavelu A: Matrix Metalloproteinases:
A challenging paradigm of cancer management. Semin Cancer Biol.
56:100–115. 2019. View Article : Google Scholar
|
|
50
|
Zeng L, Qian J, Zhu F, Wu F, Zhao H and
Zhu H: The prognostic values of matrix metalloproteinases in
ovarian cancer. J Int Med Res. May 17–2019.Epub ahead of print.
View Article : Google Scholar
|
|
51
|
Ren F, Tang R, Zhang X, Madushi WM, Luo D,
Dang Y, Li Z, Wei K and Chen G: Overexpression of MMP family
members functions as prognostic biomarker for breast cancer
patients: A systematic review and meta-snalysis. PLoS One.
10:e01355442015. View Article : Google Scholar
|
|
52
|
Bai L, Lin G, Sun L, Liu Y, Huang X, Cao
C, Guo Y and Xie C: Upregulation of SIRT6 predicts poor prognosis
and promotes metastasis of non-small cell lung cancer via the
ERK1/2/MMP9 pathway. Oncotarget. 7:40377–40386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang Y, Wu N, Pang B, Tong D, Sun D, Sun
H, Zhang C, Sun W, Meng X, Bai J, et al: TRIB1 promotes colorectal
cancer cell migration and invasion through activation MMP-2 via
FAK/Src and ERK pathways. Oncotarget. 8:47931–47942.
2017.PubMed/NCBI
|
|
54
|
Robbins P, Pinder S, de Klerk N, Dawkins
H, Harvey J, Sterrett G, Ellis I and Elston C: Histological grading
of breast carcinomas: A study of interobserver agreement. Hum
Pathol. 26:873–879. 1995. View Article : Google Scholar : PubMed/NCBI
|