Introduction
Breast cancer is one of the most common malignant
cancers and the leading cause of cancer-related mortality in women
globally (1). Breast cancer is a
heterogeneous disease generally divided into four major molecular
subtypes: Luminal A and luminal B [which are mostly positive for
estrogen receptor (ER) and progesterone receptor (PR) expression],
triple negative breast cancer (TNBC)/basal-like and erb-b2 receptor
tyrosine kinase 2 (HER2)-positive (2). Despite major developments in the
diagnostic and therapeutic strategies, the prognosis of women with
advanced breast cancer requires improvement (3). As a result, there is an urgent need
to further elucidate the underlying molecular mechanisms of breast
cancer progression.
Long non-coding RNAs (lncRNAs) are RNA molecules
>200 nucleotides in length that do not have significant
protein-coding potential (4).
Recent studies have revealed that lncRNAs are involved in
regulating multiple biological processes, including development,
differentiation and carcinogenesis (5,6).
Various lncRNAs have been reported to participate in the
progression of breast cancer. For example, X inactive specific
transcript suppresses breast cancer cell growth, migration, and
invasion via the microRNA (miR)-155/caudal type homeobox 1 axis
(7). HOX transcript antisense RNA
increases ligand-independent ER activities and contributes to
tamoxifen resistance in breast cancer (8). H19 imprinted maternally expressed
transcript, let-7 and RNA-binding protein LIN28 form a
double-negative feedback loop and have an important role in the
maintenance of breast cancer stem cells (9). Nuclear enriched abundant transcript 1
(NEAT1), also known as MENε/β, is a novel lncRNA localized to
nuclear paraspeckles. It is critical for the maintenance of
paraspeckles and associated with the development of several types
of cancer (10). A previous study
demonstrated that NEAT1 promotes breast cancer progression by
targeting the miR-448/zinc finger E-box binding homeobox 1 axis
(11); however, the precise
mechanisms by which NEAT1 promotes breast cancer progression remain
largely unknown.
miRNAs are small, non-coding RNA molecules that
regulate many cellular activities by binding the 3'-untranslated
region (3'-UTR) of corresponding mRNAs (12). Increasing evidence has indicated
that dysregulation of miRNAs has an important role in cancer
development (13,14). Previous studies have demonstrated
that miR-124 exerts a tumor suppressive role in various
malignancies, including gastric cancer (15), hepatocellular carcinoma (16), bladder cancer (17) and non-small cell lung cancer
(18). Notably, miR-124
overexpression inhibits cell growth, migration, invasion and
chemoresistance in breast cancer (19-21).
Furthermore, miR-124 overexpression enhances the sensitivity of
HER2-positive breast cancer cells to irradiation by directly
targeting STAT3 (22). However,
the association between NEAT1, miR-124 and STAT3 in breast cancer
is not fully understood.
The current study aimed to investigate the
interaction between NEAT1, miR-124 and STAT3 in breast cancer. It
was demonstrated that NEAT1 acts as a competing endogenous lncRNA
(ceRNA) to positively regulate STAT3 by sponging miR-124.
Furthermore, NEAT1 and STAT3 functioned coordinately to promote
breast cancer progression by forming a positive feedback loop.
Materials and methods
Cell culture
To gain comprehensive data of gene expression in
breast cancer cell lines, four cell lines were used in the present
study to cover three common breast cancer subtypes: the
ER/PR+ cell lines MCF-7 and T47D; the HER2+
cell line SKBR3; and the TNBC cell line MDA-MB-231 (23). The breast cancer cell lines and the
normal breast cell line MCF-10A were obtained from the American
Type Culture Collection. The breast cancer cells were cultured in
DMEM (Hyclone; GE Healthcare Life Sciences) supplemented with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) in a 5% CO2
environment at 37°C. The MCF-10A cell line was cultured in DMEM/F12
(1:1; Hyclone; GE Healthcare Life Sciences) supplemented with 5%
FBS, 10 µg/ml insulin, 20 ng/ml epidermal growth factor
(EGF), 100 ng/ml cholera toxin and 0.5 mg/ml hydrocortisone at 37°C
with 5% CO2. MCF-7 and MDA-MB-231 cells were used as
representative for ER+ and ER- breast cancer,
respectively, for subsequent functional assays.
Cell transfection
miR-124 mimics, miR-124 inhibitors, NEAT1 small
interfering RNA (siRNA), STAT3 siRNA and the corresponding negative
controls were synthesized by GenePharma Co., Ltd (the sequences of
the oligonucleotides are provided in Table I). The full-length sequences of
NEAT1 or STAT3 were amplified by PCR and subcloned into the
pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.) to
generate the pcDNA-NEAT1 (NEAT1) or pcDNA-STAT3 (STAT3)
overexpression plasmids, respectively. NEAT1, STAT3 and miR-124
levels were overexpressed by transfection with the
NEAT1-overexpressing vector, STAT3-overexpressing vector and
miR-124 mimics (50 nM). NEAT1, STAT3 and miR-124 levels were
depleted by transfection with NEAT1 siRNA, STAT3 siRNA and miR-124
inhibitors (100 nM). All transfections were performed using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Total RNA and protein were extracted at 48 or 72 h
post-transfection, respectively.
 | Table ISequences of oligonucleotides used in
this study. |
Table I
Sequences of oligonucleotides used in
this study.
| Oligo | Sequence
(5'-3') |
|---|
| miR-124 mimics
(sense) |
UAAGGCACGCGGUGAAUGCC |
| miR-124 mimics
(antisense) |
CAUUCACCGCGUGCCUUAUU |
| miR-124
inhibitor |
GGCAUUCACCGCGUGCCUUA |
| NEAT1 siRNA
(sense) |
GUGAGAAGUUGCUUAGAAACUUUCC |
| NEAT1 siRNA
(antisense) |
GGAAAGUUUCUAAGCAACUUCUCAC |
| STAT3 siRNA
(sense) |
GAAGGAGGCGUCACUUUCA |
| STAT3 siRNA
(antisense) |
UGAAAGUGACGCCUCCUUC |
Clinical samples
All clinical samples (31 pairs of matched breast
cancer and normal breast tissue samples; age, 29-65) were obtained
from the First Affiliated Hospital of Xi'an Jiaotong University
(Xi'an, China) between November 2016 and December 2017. The study
was approved by the Ethics Committee of Xi'an Jiaotong University
First Affiliated Hospital and each patient provided written
informed consent. The specimens were resected and frozen in liquid
nitrogen immediately after surgery. None of the patients had
received any preoperative local or systemic treatment.
Reverse transcription-quantitative PCR
(RT-qPCR). Total RNA was extracted from surgical specimens and
cultured cells using RNAiso Plus (Takara Biotechnology Co., Ltd.)
according to the manufacture's protocol. Reverse transcription was
performed using PrimeScript™ RT Reagent kit (Takara Biotechnology
Co., Ltd.) as previously described (24). PCR was conducted using
SYBR® Premix Ex Taq™ II (Tli RNaseH Plus2X; Takara
Biotechnology Co., Ltd.) on a CFX96TM Real-Time PCR Detection
System (Bio-Rad Laboratories, Inc.). The thermocycling conditions
were as follows: 30 sec at 95°C, followed by 40 cycles of 5 sec at
95°C and 30 sec at 60°C. β-actin was used as internal control for
NEAT1 and STAT3. U6 was used as internal control for miR-124. The
relative expression levels were calculated using the
2−ΔΔCq method (25).
Five normal breast tissue samples derived from breast cancer
patients were used as the normal control. The PCR primers used in
this study are provided in Table
II.
 | Table IIPrimers used for reverse
transcription-quantitative PCR analysis. |
Table II
Primers used for reverse
transcription-quantitative PCR analysis.
| Primer | Sequence
(5'-3') |
|---|
| NEAT1, F |
TGGCTAGCTCAGGGCTTCAG |
| NEAT1, R |
TCTCCTTGCCAAGCTTCCTTC |
| STAT3, F |
ATCACGCCTTCTACAGACTGC |
| STAT3, R |
CATCCTGGAGATTCTCTACCACT |
| ACTB, F |
CCTTCTACAATGAGCTGCGT |
| ACTB, R |
CCTGGATAGCAACGTACATG |
| miR-124, F |
GCGGCCGTGTTCACAGCGGACC |
| miR-124, R |
GTGCAGGGTCCGAGGT |
| U6, F |
GCTTCGGCAGCACATATACTAAAAT |
| U6, R |
CGCTTCACGAATTTGCGTGTCAT |
Western blot analysis
Total protein was extracted from cells by using a
Total Protein Extraction kit (Nanjing KeyGen Biotech Co., Ltd.),
according to the manufacturer's instructions. Measurement of
protein concentration was conducted with a protein bicinchoninic
acid assay kit (Thermo Fisher Scientific, Inc.). The total protein
extracts (20 µg) were separated by 10% SDS-PAGE and
transferred onto nitrocellulose membranes (Bio-Rad Laboratories,
Inc.). Non-specific binding sites were blocked by incubation in 5%
non-fat milk for 2 h at room temperature. Subsequently, the
membranes were incubated with anti-STAT3 antibody (1:1,000; cat.
no. ab68153; Abcam) at 4°C overnight. The membranes were then
incubated with horseradish peroxidase-conjugated secondary antibody
(1:5,000; cat. no. sc2004; Santa Cruz Biotechnology, Inc.). An
anti-β-actin antibody (1:5,000; cat. no. A5441; Sigma-Aldrich;
Merck KGaA) was used as the loading control. Protein signals were
visualized using an enhanced chemiluminescence kit (EMD
Millipore).
Luciferase activity assay
The putative binding sites for miR-124 were
predicted using Targetscan (targetscan.org/mamm_31/), StarBase
(starbase.sysu.edu.cn/) and miRcode (mircode.org/). The fragments
containing the wild-type or mutant miR-124-binding sites of NEAT1
and STAT3 3'-UTRs were cloned into the pGL3-control vector (Promega
Corporation) at the NheI and XhoI restriction sites
to construct the luciferase reporter vectors. The putative
STAT3-binding sites in the NEAT1 promoter were identified by using
the University of California Santa Cruz (UCSC) Genome Browser
(genome-asia.ucsc.edu/index.html) and the JASPAR
database (jaspardev.genereg.net/). The NEAT1 promoter fragments
containing putative STAT3-binding sites were inserted into the
pGL3-basic vector (Promega Corporation). MDA-MB-231 cells were
seeded in 24-well plates and cotransfected with luciferase reporter
vectors, miR-124 mimics, STAT3 siRNA or negative control, and
pRL-TK Renilla vector (Promega Corporation), using
Lipofectamine® 2000, according to the manufacturer's
instructions. At 48 h post-transfection, the cells were lysed and
the firefly and Renilla luciferase activities were detected
using a Dual-Luciferase Reporter Assay System (Promega
Corporation).
Cell viability assay
An MTT assay was used to measure the cell viability
of MCF-7 and MDA-MB-231 lines. At 24 h post-transfection, cells
were seeded in 96-well plates at a density of 4×103
cells/well and cultured for 24, 48 and 72 h. Then, the MTT assay
was conducted as previously described (26).
Colony formation assay
At 24 h post-transfection, MCF-7 and MDA-MB-231
cells were seeded in 6-well plates at a density of 500 cells/well
and cultured for 2 weeks with medium replacement every 3 days. Then
the cells were fixed and stained with 0.1% crystal violet at room
temperature. The colonies containing >50 cells were manually
counted and imaged under a microscope.
Cell cycle analysis
MCF-7 and MDA-MB-231 cells were seeded in 6-well
plates and transfected with oligonucleotides as indicated. At 48 h
post-transfection, the cells were trypsinized and fixed in 70%
ethanol at 4°C overnight. Then the cells were treated with RNase A
and stained with propidium iodide (PI; Sigma-Aldrich; Merck KGaA)
for 30 min. Flow cytometry was conducted on a BD FACSCalibur flow
cytometer (BD Biosciences). The results were analyzed using the
ModiFit LT V3.3.11 software (Verity Software House).
Statistical analysis
Statistical analyses were performed using SPSS
software (version 20.0; IBM Corp). Data are presented as the mean ±
standard deviation of at least three independent experiments.
Comparisons between two groups were conducted using the Student's
t-test (two-tailed). Multiple group comparisons were conducted
using one-way ANOVA followed by Dunnett's or least significant
difference post hoc tests. Correlation analyses between gene
expression were performed with Pearson's correlation test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of NEAT1 and miR-124 in breast
cancer
The expression levels of NEAT1 and miR-124 in breast
cancer samples and cell lines were examined by RT-qPCR. NEAT1
levels in breast cancer tissues and cell lines were significantly
elevated compared with normal breast tissues and MCF-10A cells
(Fig. 1A and B). By contrast,
miR-124 levels in breast cancer tissues and cell lines were
significantly reduced compared with normal breast tissues and
MCF-10A cells (Fig. 1C and D).
These results suggested that NEAT1 overexpression and miR-124
downregulation may be associated with breast carcinogenesis.
NEAT1 promotes cell proliferation and
cell cycle progression in breast cancer cells
To explore the biological function of NEAT1 in
breast cancer cells, MCF-7 and MDA-MB-231 cells were transfected
with the pcDNA-NEAT1 plasmid or NEAT1 siRNA, respectively. The
transfection efficiency was determined by RT-qPCR analysis
(Fig. 2A). MTT and colony
formation assays demonstrated that the proliferation of MCF-7 cells
was promoted by NEAT1 overexpression, whereas proliferation of
MDA-MB-231 cells was inhibited by NEAT1 silencing (Fig. 2B and C). In addition, NEAT1
overexpression accelerated MCF-7 cell cycle progression, whereas
G0/G1 cell cycle arrest was observed in the NEAT1-silenced
MDA-MB-231 cells (Fig. 2D). These
results suggested that NEAT1 promoted cell proliferation and cell
cycle progression in breast cancer cells.
NEAT1 acts as a sponge of miR-124
Bioinformatics analysis based on the online database
tools StarBase and miRcode indicated that NEAT1 contains a putative
binding site for miR-124. The complementary binding region between
miR-124 and NEAT1 is shown in Fig.
3A. To validate whether NEAT1 is a direct target of miR-124, a
luciferase activity assay was performed in MDA-MB-231 cells.
Luciferase activity of wild-type NEAT1 constructs (NEAT1-wt) was
significantly reduced when cotransfected with miR-124 mimics. By
contrast, the luciferase activity of the mutated NEAT1 construct
(NEAT1-mut) was not affected by miR-124 mimics transfection,
confirming the functionality of the miR-124 binding site (Fig. 3B). Furthermore, miR-124 levels were
increased following NEAT1 knockdown in MCF-7 and MDA-MB-231 cells
(Fig. 3C). In addition, there was
a negative correlation between NEAT1 and miR-124 levels in breast
cancer tissues (Fig. 3D). These
results suggested that NEAT1 may function as a miR-124 sponge in
breast cancer cells.
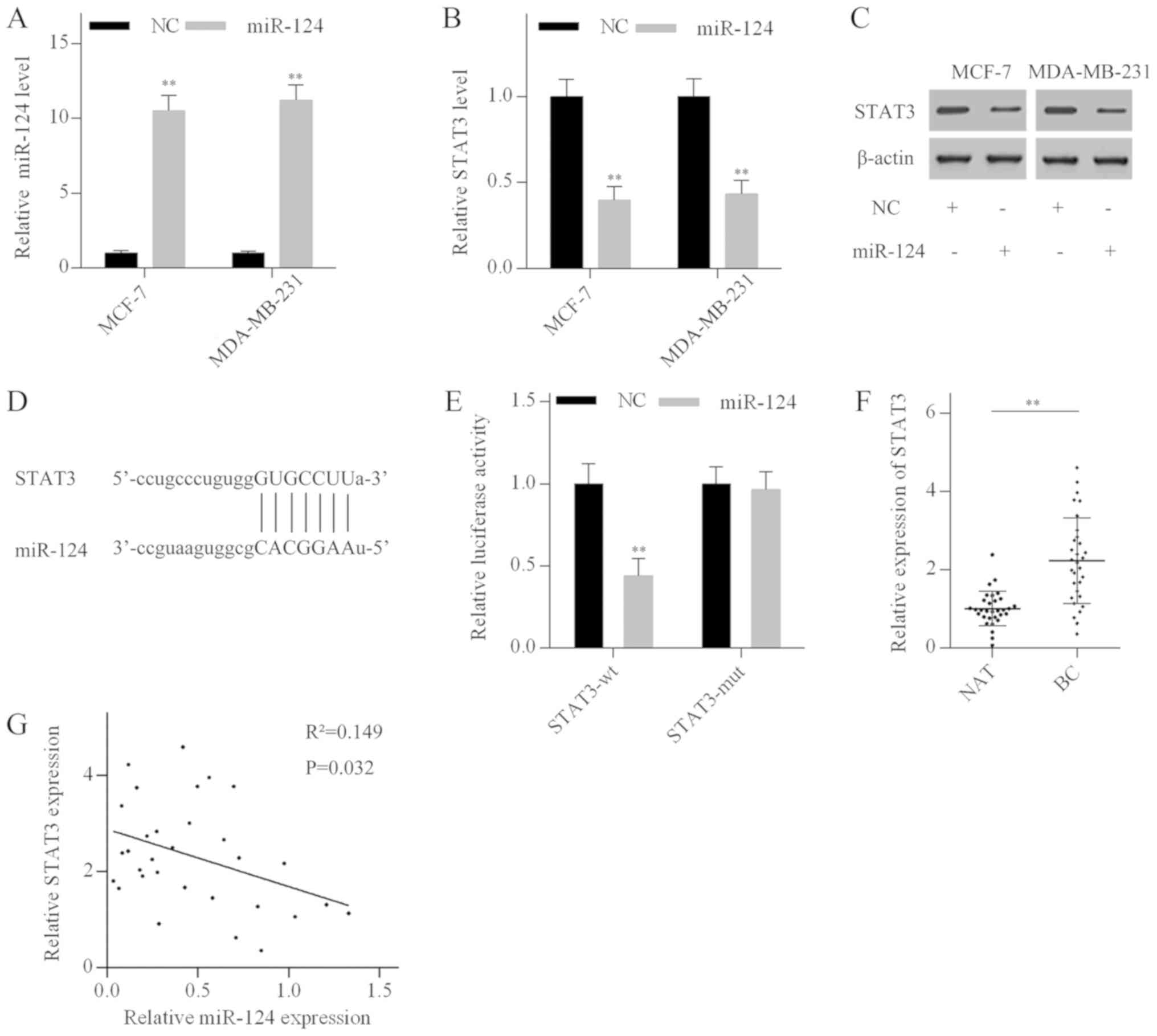
STAT3 is a direct target of miR-124
To investigate the role of miR-124 in breast cancer,
MCF-7 and MDA-MB-231 cells were transfected with miR-124 mimics.
RT-qPCR analysis revealed that miR-124 mimics significantly
elevated miR-124 levels (Fig. 4A).
Additionally, miR-124 overexpression suppressed STAT3 expression in
both the mRNA and protein levels (Fig.
4B and C). Bioinformatics analysis using Targetscan identified
a putative miR-124 binding site in the 3'-UTR of the STAT3 mRNA
(Fig. 4D). To investigate whether
miR-124 directly targeted STAT3, luciferase reporter vectors
containing a wild-type 3'-UTR fragment of STAT3 (STAT3-wt) or a
mutant 3'-UTR fragment of STAT3 (STAT3-mut) were generated. The
luciferase activity assay indicated that miR-124 overexpression
dramatically reduced the luciferase activity of the STAT3-wt
vector, but not the STAT3-mut vector (Fig. 4E). In addition, RT-qPCR analysis
revealed that STAT3 mRNA expression levels were significantly
elevated in breast cancer tissues compared with normal breast
tissues (Fig. 4F), and negatively
correlated with miR-124 levels in breast cancer tissues (Fig. 4G). These results suggested that
STAT3 was a direct target of miR-124 in breast cancer.
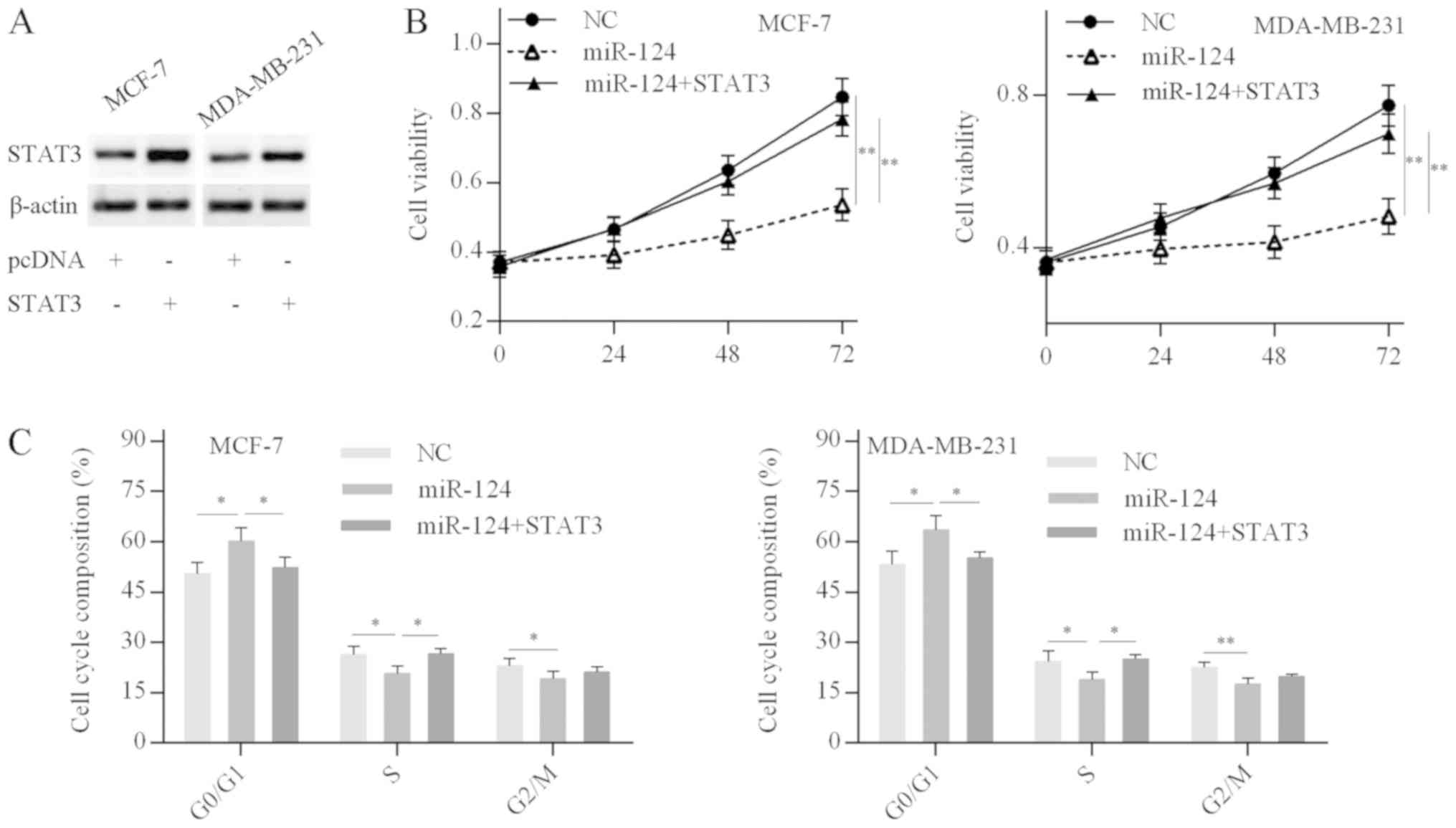
miR-124 inhibits cell proliferation and
induces cell cycle arrest in breast cancer cells
First, the efficiency of STAT3 overexpression was
confirmed by western blot analysis; STAT3 protein expression levels
were significantly elevated in cells transfected with the
pcDNA-STAT3 plasmid compared with the empty vector (Fig. 5A). MTT and cell cycle analysis
revealed that miR-124 overexpression in breast cancer cells
decreased the cell proliferation rate and resulted in a G0/G1 phase
cell cycle arrest, while these inhibitory effects were abolished by
STAT3 overexpression in miR-124-over-expressing breast cancer cells
(Fig. 5B and C). The results
suggested that miR-124 inhibits breast cancer cell proliferation
and cell cycle progression by targeting STAT3.
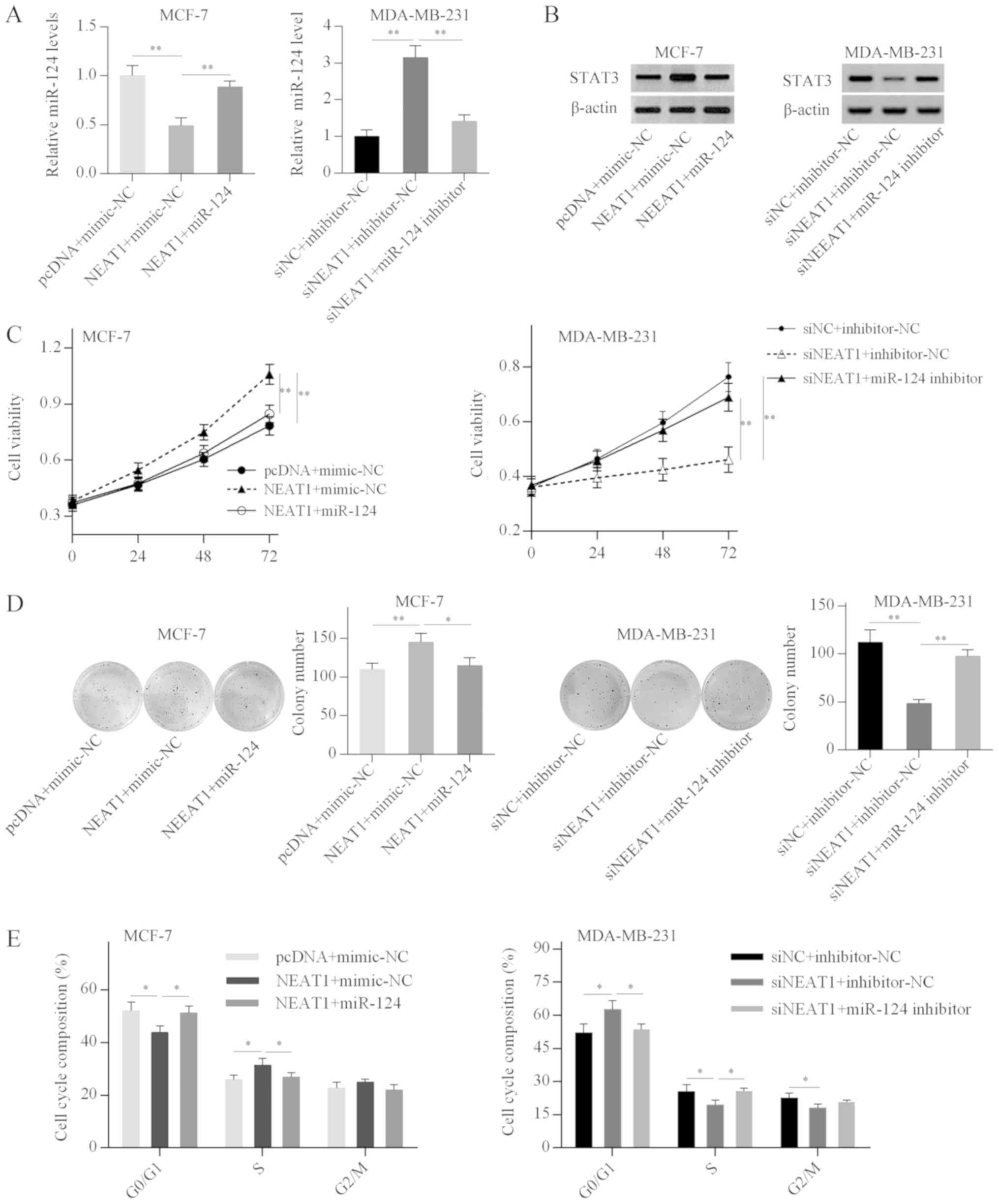
NEAT1 promotes breast cancer cell growth
via targeting the miR-124/STAT3 axis
Next, the present study investigated whether NEAT1
acts as a ceRNA to increase STAT3 expression in breast cancer
cells. RT-qPCR analysis demonstrated that miR-124 overexpression
reversed the NEAT1-mediated inhibitory effect on miR-124 expression
in MCF-7 cells, and miR-124 depletion weakened the NEAT1
silencing-induced miR-124 upregulation in MDA-MB-231 cells
(Fig. 6A). Western blot analysis
further revealed that NEAT1 overexpression increased STAT3 protein
expression in MCF-7 cells, while this effect was weakened by
miR-124 overexpression (Fig. 6B).
By contrast, STAT3 expression was decreased in NEAT1-silenced
MDA-MB-231 cells, and miR-124 knockdown alleviated the inhibitory
effect of NEAT1 silencing on STAT3 expression (Fig. 6B). These results suggested that
NEAT1 positively regulated STAT3 expression by acting as a ceRNA
that sponges miR-124 in breast cancer cell lines.
Subsequently, the present study investigated whether
NEAT1 promoted the proliferation and cell cycle progression of
breast cancer cells by targeting the miR-124/STAT3 axis. Functional
analysis revealed that miR-124 overexpression effectively reduced
NEAT1-induced cell proliferation and cell cycle progression in
MCF-7 cells (Fig. 6C-E).
Conversely, the inhibitory effects of NEAT1 silencing on cell
proliferation and cell cycle were abrogated by miR-124 depletion in
MDA-MB-231 cells (Fig. 6C-E).
These results suggested that NEAT1 promoted breast cancer cell
growth by targeting the miR-124/STAT3 axis.
NEAT1 is inhibited by STAT3
silencing
To explore the crosstalk between NEAT1 and STAT3,
their correlation in breast cancer specimens was evaluated. NEAT1
levels were positively correlated with STAT3 mRNA expression levels
(Fig. 7A). Then, STAT3 expression
was silenced in MCF-7 and MDA-MB-231 cells using specific siRNA
(Fig. 7B). RT-qPCR analysis
demonstrated that NEAT1 expression levels were decreased following
STAT3 silencing (Fig. 7C).
Furthermore, multiple putative STAT3 binding sites were identified
in the NEAT1 promoter by bioinformatics analysis using the UCSC
(genome-asia.ucsc.edu/index.html) and JASPAR
(jaspardev.genereg.net/) web-based tools.
Two binding sites (S1 and S2) with prediction scores >10 are
shown in Fig. 7D. Subsequently,
two fragments containing S1 or S2 were cloned into luciferase
reporter vectors. The vectors were cotransfected into MDA-MB-231
cells with STAT3 siRNA or negative control siRNA. Luciferase
activity analysis revealed that STAT3 silencing significantly
reduced the luciferase activity of fragment S1, whereas the
luciferase activity of fragment S2 was not affected (Fig. 7E). These results demonstrated that
STAT3 positively regulated NEAT1 transcription, and thus formed a
positive feedback loop in breast cancer cells.
Discussion
Emerging evidence has revealed that lncRNAs have a
pivotal role in the regulation of physiological and pathological
processes, including in breast cancer (6). According to bioinformatics analysis,
in the present study, miR-124 was predicted to directly target both
NEAT1 and STAT3. The association between NEAT1 and miR-124 in
breast cancer has not been elucidated in previous studies. As a
result, the present study focused on miR-124 as the target gene of
NEAT1. The results demonstrated that NEAT1 and STAT3 expression
levels were elevated in breast cancer, whereas miR-124 was
significantly reduced. Thus, it was speculated that NEAT1 may be
involved in breast cancer progression by modulating the
miR-124/STAT3 axis.
Accumulating evidence has suggested that NEAT1 has a
critical role in carcinogenesis (10). For example, NEAT1 is highly
expressed in hepatocellular carcinoma (HCC) and promotes the
proliferation of HCC cells by regulating the miR-129/valosin
containing protein/inhibitor of κB kinase axis (27). NEAT1 has been identified to be an
indicator of diagnosis and prognosis in colorectal cancer (28). Furthermore, high NEAT1 expression
was reported to be associated with TNM stage and overall survival
in breast cancer (29,30). In the present study, it was
demonstrated that NEAT1 expression was elevated in breast cancer
tissues and cell lines, as previously described (10,29,30).
In addition, NEAT1 overexpression promoted cell proliferation and
cell cycle progression in breast cancer cells, whereas NEAT1
silencing led to a reduced proliferation rate and cell cycle arrest
at G0/G1 phase.
NEAT1 has been reported to regulate gene expression
by a variety of mechanisms (10).
NEAT1 forms a complex with forkhead box protein N3 and paired
amphipathic helix protein SIN3A, and thus represses transacting
T-cell-specific transcription factor GATA3 expression, which is
involved in epithelial-mesenchymal transition (31). Additionally, NEAT1 epigenetically
represses E-cadherin expression through interaction with
histone-lysine N-methyltransferase EHMT2/ DNA
(cytosine-5)-methyltransferase 1/Snail complex (32). Previous studies have identified a
new regulatory mechanism in which lncRNAs act as endogenous sponges
of miRNAs (33,34). NEAT1 acts as a ceRNA to positively
regulate histone-lysine N-methyltransferase EZH2 expression by
sponging miR-101 in breast cancer cells (35). In the current study, miR-124
expression was shown to be negatively correlated with NEAT1 and
STAT3 expression in breast cancer tissues, suggesting potential
crosstalk between miR-124 and the other two RNAs. Furthermore,
bioinformatics prediction analysis indicated that NEAT1 and STAT3
might be potential direct targets of miR-124. The luciferase
activity assay and RT-qPCR analysis demonstrated that miR-124
directly targeted NEAT1 and STAT3 in breast cancer cells. However,
no significant correlation between NEAT1, miR-124 and STAT3
expression in breast cancer cell lines was identified (data not
shown). This may be attributed to the limited number of breast
cancer cell lines used in the present study.
In HCC, NEAT1 acts as a ceRNA to increase STAT3
expression by sponging miR-485, resulting in enhanced cancer
progression (36). Additionally,
NEAT1 promotes gastric cancer development by targeting
miR-506/STAT3 axis (37). In the
present study, it was confirmed that STAT3 protein levels were
elevated following NEAT1 overexpression, and partially attenuated
by miR-124 overexpression in NEAT1-overexpressing breast cancer
cells. The results suggested that NEAT1 acted as a ceRNA to
increase STAT3 expression by sponging miR-124 in breast cancer.
Subsequently, the findings of the present study demonstrated that
the effects of NEAT1 on the proliferation and cell cycle of breast
cancer cells were attenuated by miR-124 overexpression. Moreover,
miR-124 overexpression inhibited the growth of breast cancer cells
by targeting STAT3. Taken together, these findings indicated that
NEAT1 may promote the growth of breast cancer cells by targeting
the miR-124/STAT3 axis.
Previous studies have demonstrated that STAT3 is
constitutively activated in various types of cancer, including
breast cancer (38).
Phosphorylated STAT3 is translocated into the nucleus and binds to
the consensus promoter sequence of target genes to initiate
transcription (39). Recent
studies have demonstrated that lncRNAs are regulated by the
interleukin-6 (IL6)/STAT3 pathway in cancers. For example, a set of
lncRNAs were induced by IL-6-activated STAT3 in multiple myeloma
cells (40). In glioma cells,
activation of the EGF receptor pathway positively regulated NEAT1
expression via STAT3 and NF-κB (p65) (41). In addition, IL-6 promoted NEAT1
transcription via STAT3 and histone 3 lysine 4 trimethylation in
HCC cells (42). The current study
identified a positive correlation between NEAT1 and STAT3
expression levels in breast cancer tissues. Furthermore, it was
demonstrated that STAT3 silencing reduced NEAT1 expression levels
by inhibiting the promoter activity in breast cancer cells.
Therefore, NEAT1 and STAT3 formed a positive feedback loop mediated
by miR-124.
In conclusion, the findings of the present study
demonstrated that NEAT1 positively regulated STAT3 expression by
sponging miR-124 in breast cancer cells. Furthermore, a positive
feedback loop between NEAT1 and STAT3 that contributed to breast
cancer cell growth was identified. These results increase the
understanding of the molecular mechanisms underlying breast cancer
development and indicate the possibility of developing NEAT1 as a
potential target in breast cancer treatment.
Funding
No funding was received.
Availability of data and materials
All data and materials involved in this study are
available from the corresponding author on reasonable request.
Authors' contributions
GY, YP and JL were responsible for the study
conception and design. JW, XL, CW and MW performed the majority of
the experiments. YP and JL drafted the manuscript. All the authors
read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Xi'an Jiaotong University First Affiliated Hospital and each
patient provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Siegel RL: Cancer statistics, 2016. CA
Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer Genome Atlas N; Cancer Genome Atlas
Network: Comprehensive molecular portraits of human breast tumours.
Nature. 490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and trends - an update.
Cancer Epidemiol Biomarkers Prev. 25:16–27. 2016. View Article : Google Scholar
|
|
4
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar
|
|
6
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng R, Lin S, Guan L, Yuan H, Liu K, Liu
C, Ye W, Liao Y, Jia J and Zhang R: Long non-coding RNA XIST
inhibited breast cancer cell growth, migration, and invasion via
miR-155/CDX1 axis. Biochem Biophys Res Commun. 498:1002–1008. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xue X, Yang YA, Zhang A, Fong KW, Kim J,
Song B, Li S, Zhao JC and Yu J: LncRNA HOTAIR enhances ER signaling
and confers tamoxifen resistance in breast cancer. Oncogene.
35:2746–2755. 2016. View Article : Google Scholar :
|
|
9
|
Peng F, Li TT, Wang KL, Xiao GQ, Wang JH,
Zhao HD, Kang ZJ, Fan WJ, Zhu LL, Li M, et al: H19/let-7/LIN28
reciprocal negative regulatory circuit promotes breast cancer stem
cell maintenance. Cell Death Dis. 8:e25692017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu X, Li Z, Zheng H, Chan MT and Wu WK:
NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif.
50:502017.
|
|
11
|
Jiang X, Zhou Y, Sun AJ and Xue JL: NEAT1
contributes to breast cancer progression through modulating miR-448
and ZEB1. J Cell Physiol. 233:8558–8566. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar :
|
|
14
|
Pang Y, Liu J, Li X, Xiao G, Wang H, Yang
G, Li Y, Tang SC, Qin S, Du N, et al: MYC and DNMT3A-mediated DNA
methylation represses microRNA-200b in triple negative breast
cancer. J Cell Mol Med. 22:6262–6274. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang L, Lin T, Xu C, Hu S, Pan Y and Jin
R: miR-124 interacts with the Notch1 signalling pathway and has
therapeutic potential against gastric cancer. J Cell Mol Med.
20:313–322. 2016. View Article : Google Scholar
|
|
16
|
Lu Y, Yue X, Cui Y, Zhang J and Wang K:
MicroRNA-124 suppresses growth of human hepatocellular carcinoma by
targeting STAT3. Biochem Biophys Res Commun. 441:873–879. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu DH, Liang H and Lu SN: miR-124
suppresses pancreatic ductal adenocarcinoma growth by regulating
monocarboxylate transporter 1-mediated cancer lactate metabolism.
Cell Physiol Biochem. 50:924–935. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang M, Meng B and Liu Y, Yu J, Chen Q and
Liu Y: miR-124 inhibits growth and enhances radiation-induced
apoptosis in non-small cell lung cancer by inhibiting STAT3. Cell
Physiol Biochem. 44:2017–2028. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang YJ, Wang QY, Zhou CX, Yin QQ, He M,
Yu XT, Cao DX, Chen GQ, He JR and Zhao Q: miR-124 targets Slug to
regulate epithelial-mesenchymal transition and metastasis of breast
cancer. Carcinogenesis. 34:713–722. 2013. View Article : Google Scholar
|
|
20
|
Cai WL, Huang WD, Li B, Chen TR, Li ZX,
Zhao CL, Li HY, Wu YM, Yan WJ and Xiao JR: microRNA-124 inhibits
bone metastasis of breast cancer by repressing Interleukin-11. Mol
Cancer. 17:92018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen SM, Chou WC, Hu LY, Hsiung CN, Chu
HW, Huang YL, Hsu HM, Yu JC and Shen CY: The effect of microRNA-124
overexpression on anti-tumor drug sensitivity. PLoS One.
10:e01284722015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fu Y and Xiong J: MicroRNA-124 enhances
response to radiotherapy in human epidermal growth factor receptor
2-positive breast cancer cells by targeting signal transducer and
activator of transcription 3. Croat Med J. 57:457–464. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Holliday DL and Speirs V: Choosing the
right cell line for breast cancer research. Breast Cancer Res.
13:2152011. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pang Y, Liu J, Li X, Zhang Y, Zhang B,
Zhang J, Du N, Xu C, Liang R, Ren H, et al: Nano Let-7b
sensitization of eliminating esophageal cancer stem-like cells is
dependent on blockade of Wnt activation of symmetric division. Int
J Oncol. 51:1077–1088. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Liu J, Li X, Wang M, Xiao G, Yang G, Wang
H, Li Y, Sun X, Qin S, Du N, et al: A miR-26a/E2F7 feedback loop
contributes to tamoxifen resistance in ER-positive breast cancer.
Int J Oncol. 53:1601–1612. 2018.PubMed/NCBI
|
|
27
|
Fang L, Sun J, Pan Z, Song Y, Zhong L,
Zhang Y, Liu Y, Zheng X and Huang P: Long non-coding RNA NEAT1
promotes hepatocellular carcinoma cell proliferation through the
regulation of miR-129-5p-VCP-IκB. Am J Physiol Gastrointest Liver
Physiol. 313:G150–G156. 2017. View Article : Google Scholar
|
|
28
|
Li Y, Li Y, Chen W, He F, Tan Z, Zheng J,
Wang W, Zhao Q and Li J: NEAT expression is associated with tumor
recurrence and unfavorable prognosis in colorectal cancer.
Oncotarget. 6:27641–27650. 2015.PubMed/NCBI
|
|
29
|
Zhao D, Zhang Y, Wang N and Yu N: NEAT1
negatively regulates miR-218 expression and promotes breast cancer
progression. Cancer Biomark. 20:247–254. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li X, Wang S, Li Z, Long X, Guo Z, Zhang
G, Zu J, Chen Y and Wen L: The lncRNA NEAT1 facilitates cell growth
and invasion via the miR-211/HMGA2 axis in breast cancer. Int J
Biol Macromol. 105:346–353. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li W, Zhang Z, Liu X, Cheng X, Zhang Y,
Han X, Zhang Y, Liu S, Yang J, Xu B, et al: The FOXN3-NEAT1-SIN3A
repressor complex promotes progression of hormonally responsive
breast cancer. J Clin Invest. 127:3421–3440. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y and Cheng C: Long noncoding RNA NEAT1
promotes the metastasis of osteosarcoma via interaction with the
G9a DNMT1-Snail complex. Am J Cancer Res. 8:81–90. 2018.
|
|
33
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bayoumi AS, Sayed A, Broskova Z, Teoh JP,
Wilson J, Su H, Tang YL and Kim IM: Crosstalk between long
noncoding RNAs and microRNAs in health and disease. Int J Mol Sci.
17:3562016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Qian K, Liu G, Tang Z, Hu Y, Fang Y, Chen
Z and Xu X: The long non-coding RNA NEAT1 interacted with miR-101
modulates breast cancer growth by targeting EZH2. Arch Biochem
Biophys. 615:1–9. 2017. View Article : Google Scholar
|
|
36
|
Zhang XN, Zhou J and Lu XJ: The long
noncoding RNA NEAT1 contributes to hepatocellular carcinoma
development by sponging miR-485 and enhancing the expression of the
STAT3. J Cell Physiol. 233:6733–6741. 2018. View Article : Google Scholar
|
|
37
|
Tan HY, Wang C, Liu G and Zhou X: Long
noncoding RNA NEAT1-modulated miR-506 regulates gastric cancer
development through targeting STAT3. J Cell Biochem. 120:4827–4836.
2019. View Article : Google Scholar
|
|
38
|
Banerjee K and Resat H: Constitutive
activation of STAT3 in breast cancer cells: A review. Int J Cancer.
138:2570–2578. 2016. View Article : Google Scholar :
|
|
39
|
Srivastava J and DiGiovanni J:
Non-canonical Stat3 signaling in cancer. Mol Carcinog. 5:1889–1898.
2016. View Article : Google Scholar
|
|
40
|
Binder S, Hösler N, Riedel D, Zipfel I,
Buschmann T, Kämpf C, Reiche K, Burger R, Gramatzki M, Hackermüller
J, et al: STAT3-induced long noncoding RNAs in multiple myeloma
cells display different properties in cancer. Sci Rep. 7:79762017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang
J, Zhpu J, Kang C, Li M and Jiang C: Long noncoding RNA NEAT1,
regulated by the EGFR pathway, contributes to glioblastoma
progression through the WNT/beta-catenin pathway by scaffolding
EZH2. Clin Cancer Res. 24:684–695. 2018. View Article : Google Scholar
|
|
42
|
Wang S, Zhang Q, Wang Q, Shen Q, Chen X,
Li Z, Zhou Y, Hou J, Xu B, Li N, et al: NEAT1 paraspeckle promotes
human hepatocellular carcinoma progression by strengthening
IL-6/STAT3 signaling. OncoImmunology. 7:e15039132018. View Article : Google Scholar : PubMed/NCBI
|