1. Introduction
Hepatocellular carcinoma (HCC), a leading cause of
cancer-related mortality worldwide, accounts for 70-85% of all
liver cancer cases (1).
Epidemiological data reveal that hepatitis B virus (HBV)-related
HCC accounts for a large proportion of liver cancer cases,
particularly in developing countries (2). Although the management of HBV-related
HCC has improved in recent decades, its prognosis remains poor
(3). Therefore, exploring the
underlying mechanism is crucial for improving the prevention,
diagnosis and treatment of HBV-related HCC. Although HBV-related
HCC is associated with HBV genotype, mutation status, integration
and dysregulation of signalling pathways, its detailed mechanism
remains elusive (4,5).
According to the central dogma, genes exert their
effects by encoding proteins. Most previous research has focused on
protein-coding genes. In recent years, with the development of
high-resolution microarrays and massive parallel sequencing,
~70–90% of the human genome is confirmed to be actively transcribed
into RNA, although only a minority of the transcripts encode
proteins (6). Long non-coding RNAs
(lncRNAs), which are non-coding RNA molecules >200 nucleotides
(nt) in length, are usually transcribed by RNA polymerase II and
may be polyadenylated. Compared with mRNAs, lncRNAs usually have
fewer exons and are expressed at lower levels (7,8).
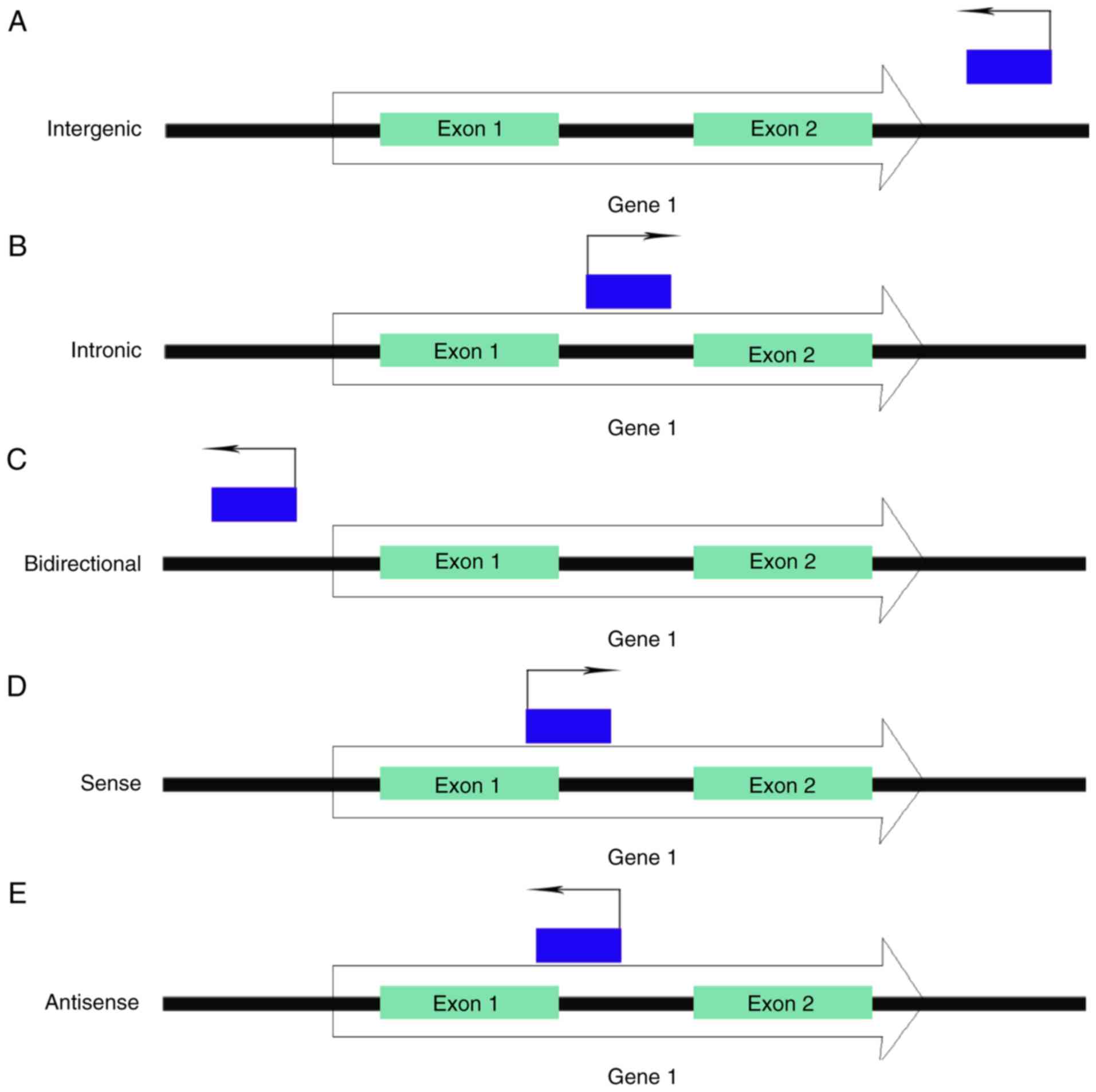
Based on genomic position and strand orientation, lncRNAs are
classified into five categories, including intergenic, intronic,
bidirectional, sense and antisense lncRNAs (Fig. 1) (9). Although considered to be
transcriptional noise in the past, lncRNAs are involved in the
tumourigenesis of several cancers (10). By functioning as signals, decoys,
guides and scaffolds, lncRNAs regulate invasion, tumourigenicity
and metastasis of HCC (11). In
recent years, several lncRNAs have been found to be aberrantly
expressed in HBV-related HCC and to affect the risk and prognosis
of HBV-related HCC. Further research has screened out circulating
lncRNAs and genetic polymorphisms in lncRNAs as risk factors for
the occurrence and prognosis of HBV-related HCC.
The aim of the present review was to briefly outline
the main mechanisms underlying the involvement of lncRNAs in
HBV-related HCC and introduce current findings on lncRNAs in
HBV-related HCC. Mounting evidence suggests that lncRNAs are
crucial regulators of HBV-related HCC, and investigation of lncRNAs
may help to further elucidate the molecular mechanisms implicated
in the development of HBV-related HCC.
2. Main mechanisms of action of lncRNAs in
HBV-related HCC
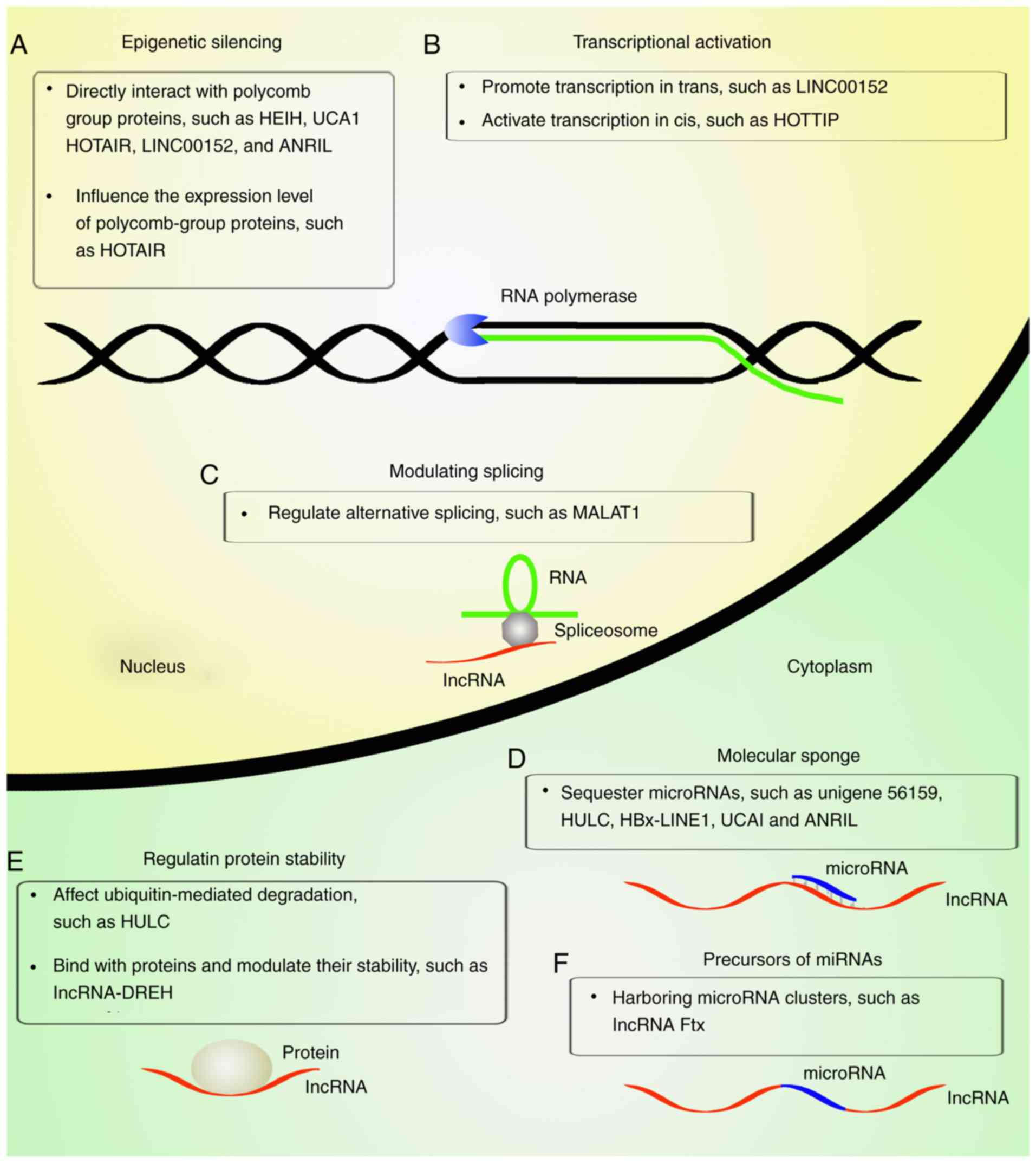
A number of lncRNAs are dysregulated in HBV-related
HCC. As illustrated in Fig. 2,
these lncRNAs affect the risk and prognosis of HBV-related HCC
through diverse mechanisms, such as epigenetic silencing,
transcriptional activation, alternative splicing regulation,
molecular sponging, modulating protein stability, and by serving as
precursors for miRNAs (Fig. 2).
These mechanisms will be briefly delineated below.
Epigenetic silencing
Epigenetic silencing, which decreases the expression
of target genes without changing DNA sequences, is a well-known
mechanism through which lncRNAs regulate the expression of target
genes. LncRNAs participate in epigenetic silencing in two different
ways: First, lncRNAs interact with polycomb-group proteins
directly, and then promote the epigenetic silencing of target
genes. Several lncRNAs, such as high expression in HCC (HEIH)
(12), urothelial carcinoma
associated 1 (UCA1) (13), HOX
transcript anti-sense RNA (HOTAIR) (14) and long intergenic ncRNA 152
(LINC00152) (15), repress gene
expression by interacting with enhancer of Zeste homolog 2 (EZH2),
a core component of polycomb repressive complex 2 (PRC2). CDKN2B
antisense RNA 1 (ANRIL), through binding with PRC2, can repress the
transcription of Kruppel-like factor 2 (KLF2) (16). Second, lncRNAs participate in
epigenetic silencing by altering the expression level of
polycomb-group proteins. For example, HOTAIR affects epigenetic
reprogramming by enhancing Plk1-dependent proteasomal degradation
of suppressor of Zeste 12 homolog (SUZ12), a key subunit of PRC2,
in HBV-related HCC (17,18).
Transcriptional activation
Several lncRNAs are implicated in the occurrence of
HBV-related HCC by activating the transcription of target genes in
cis and in trans. HOXA transcript at the distal tip
(HOTTIP), which was originally found to activate the transcription
of distal HOXA genes through interaction with the WDR5/MLL complex
during embryonic development (19), also promotes expression of
neighbouring homeobox A genes (HOAX genes) in HCC (20-22).
Epithelial cell adhesion molecule (EpCAM) is a mammalian target of
rapamycin (mTOR)-related oncogene near LINC00152. In HCC patients,
LINC00152 activates the transcription of EpCAM by binding to its
promoter (23). Although the
detailed mechanism remains elusive, metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1) is able to increase the
expression of latent transforming growth factor β-binding protein 3
(LTBP3) (24).
Regulation of alternative splicing
Through alternative splicing, a single pre-mRNA
produces several different mRNAs. Certain lncRNAs have been found
to participate in alternative splicing. MALAT1 is a
nuclear-retained lncRNA that regulates alternative splicing in HCC.
By interacting with the serine/arginine-rich family of nuclear
phosphoproteins (SR proteins), MALAT1 affects the distribution of
splicing factors in nuclear speckle domains, changes the cellular
levels of the phosphorylated forms of SR proteins, and modulates
alternative splicing (25,26).
Competitive endogenous RNA
The competitive endogenous RNA (ceRNA) hypothesis,
originally proposed by Salmena et al in 2011 (27), states that lncRNAs exert their
effects by acting as molecular sponges for miRNAs. A general model
for this process is that lncRNAs sequester miRNAs and then
de-repress the expression of miRNA target genes. As proposed by
Thomson and Dinger, instead of being a general mechanism for
predicting the function of individual lncRNAs, the ceRNA hypothesis
is of great value in exploring the mechanisms of lncRNAs in cancer
(28). Several lncRNAs, such as
Unigene56159 (29), highly
upregulated in liver cancer (HULC) (30-35),
HBx-LINE1 (36), UCA1 (37) and ANRIL (38), are involved in the development of
HBV-related HCC by titrating miRNAs away from their targets.
Several limitations of current research, such as the lack of a
physiological expression system and overdependence on the
application of miRNA-target prediction algorithms, should be taken
into consideration in future research.
Modulating protein stability
LncRNAs alter the stability of proteins via
different mechanisms. By upregulating ubiquitin-specific peptidase
22 (USP22), HULC decreases ubiquitin-mediated degradation of
cyclooxygenase-2 (COX2) and silent information regulator 1, and
then stabilizes these two proteins in HCC (34,39).
Moreover, lncRNAs modulate the stability of proteins by direct
binding. Vimentin is a type III intermediate filament (IF) and the
major cytoskeletal component of mesenchymal cells. Its filament
structure in liver cells can be altered through binding with lncRNA
downregulated expression by HBx (lncRNA-Dreh) (40).
Precursors of miRNAs
LncRNAs may be precursors of miRNAs. The Ftx
transcript, a conserved lncRNA in the X-inactivation centre,
encodes two clusters of miRNAs in its introns. miR-374a and
miR-545, located in one of these two clusters, are induced by HBV X
protein (HBx) and are upregulated in HBV-related HCC. These two
miRNAs are correlated with poor prognosis of HCC patients and
promote the proliferation, migration and invasion of HCC cells
(41).
3. LncRNAs dysregulated in HBV-related
HCC
As mentioned above, lncRNAs may affect the risk of
HBV-related HCC through diverse mechanisms. Thus, investigating
lncRNAs aberrantly expressed in HBV-related HCC is an effective way
for exploring the molecular mechanism of HBV-related HCC. In this
review, well-studied lncRNAs that affect the occurrence and
prognosis of HBV-related HCC are summarized in Table I and are discussed in detail
below.
 | Table IDysregulated lncRNAs in HBV-related
HCC. |
Table I
Dysregulated lncRNAs in HBV-related
HCC.
| LncRNA | Chromosome
location | Classification | Subcellular
location | Expression | Molecular
mechanism | Biological
function | Clinical
impact | (Refs.) |
|---|
| HULC | Chr6:8652209
-8653846 | Intergenic | Cytoplasm | Upregulated | Sequesters a series
of microRNAs and decreases the expression of p18 | Promotes
proliferation of hepatoma cells and tumour angiogenesis in
vitro and in vivo | A potential
therapeutic target | (30 −34,42,44,45) |
| HOTAIR | Chr12:53962308
−53974956 | Antisense | Nucleus and
cytoplasm | Upregulated | Participates in
epigenetic silencing in hepatoma cells | Promotes migration
and invasion of HCC cells | Affects symptoms
and prognosis of HCC patients | (14,46,48−55,57,58) |
| MALAT1 | Chr11: 65497688
-65506431 | Intergenic | Nucleus | Upregulated | Activates the
transcription of LTBP3 in HBV-related HCC | Promotes migration
and invasion of HCC cells in vitro and tumour growth in
vivo | Affects the
prognosis of HCC patients | (24,61-65) |
| UCA1 | Chr19:15828206
-15836136 | Intergenic | Nucleus and
cytoplasm | Upregulated | Recruits EZH2 to
the promoter of p27 in the nucleus and sequesters miR-216b and
miR-203 in the cytoplasm | Promotes growth,
metastasis and EMT of HCC cell lines | Correlates with
tumour size, vascular invasion, metastasis, postoperative survival
and disease stage of HCC patients | (13,37,70) |
| HEIH | Chr5:180829954
-180831605 | Intergenic | Nucleus and
cytoplasm | Upregulated | Binds with EZH2 and
participates in repression of EZH2 targets | Promotes cell
proliferation and tumour growth, modulates cell cycle | Correlates with the
prognosis of HBV-related HCC | (12,72) |
| LINC00152 | Chr2:87455368
-87584075 | Intergenic | Nucleus | Upregulated | Inhibits the
expression of E-cadherin and activates the mTOR pathway | Promotes the
proliferation and EMT of HCC cell lines and tumourigenesis | Correlates with
tumour size, HBV infection, tumour number and HBx | (15,23) |
| HOTTIP | Chr7:27201844
-27207259 | Intergenic | Nucleus | Upregulated | Promotes the
expression of HOXA genes | Inhibits
proliferation and migration of HCC cells in vitro and
reduces tumourigenesis and pulmonary metastasis in vivo | Correlates with
increased metastasis formation and decreased OS | (20 −22,77,78) |
| HBx-LINE1 | Chr8p11 | Not known | Nucleus | Upregulated | Activates
Wnt/β-catenin signalling pathway and sequesters miR-122 | Promotes cell
motility through EMT and causes mouse liver injury | Correlates with
poorer patient survival | (36,80) |
| ANRIL | Chr9:22113678
-22121097 | Antisense | Nucleus | Upregulated | Represses the
transcription of KLF2 and sequesters miR-122-5p | Inhibits apoptosis
of HCC cells in vitro and promotes proliferation, invasion and
migration of HCC cells in vitro | Correlates with
tumour size, BCLC stage, OS, histological grade and TNM stage of
HCC patients | (16,8,4) |
| Unigene56159 | Chr3 | Intronic | Not known | Upregulated | Sequesters
miR-140-5p | Promotes EMT,
migration and invasion of hepatoma cells | Highly expressed in
HBV-related HCC tissues | (29) |
| BAIAP2-AS1 | Chr17:81029130
-81034573 | Antisense | Cytoplasm | Upregulated | May function as a
ceRNA | May be an oncogene
for HCC | Highly expressed in
HBV-related HCC tissues | (91) |
| WEE2-AS1 | Chr7:141704338
-141738230 | Antisense | Not known | Upregulated | Up-regulate FERMT3
and activate PI3K/AKT/GSK3β signal pathway | Promotes the
proliferation, migration and invasion of HCC in vitro
and | Correlates with
hepatic vascular invasion, poor tumour differentiation and HBV
infection | (94) |
| DBH-AS1 | Chr9:133654586
-133657313 | Antisense | Nucleus | Uncertain | Activates MAPK
signal pathway | Inhibits serum
starvation- induced apoptosis of HCC cells, promotes tumour growth,
proliferation and cell-cycle progression of HCC cell | Correlated with
tumour size and seropositivity of HBsAg | (73,6) |
| DREH | Chr5:109213218
-109213911 | Sense | Not known | Downregulated | Inhibits the
expression of vimentin and alters its filament structure | Inhibits
proliferation of HCC cells in vitro and tumour growth in
vivo | Associates with RFS
and OS of HCC patients and correlates with tumour size and HBsAg in
HCC patients | (40,73,87) |
| uc.306 | Chr10 | Intronic | Not known | Downregulated | May participate in
the Wnt pathway | Not known | Affects the
prognosis and correlates with OS of HCC patients | (90) |
| n346077 | Chr11 | Antisense | Not known | Downregulated | Not known | Suppresses HCC cell
migration and invasion in vitro | Not known | (89) |
| lncRNA-6195 | Chr3:125647611
-125650486 | Intergenic | Not known | Downregulated | Binds with ENO1,
inhibits its enzymatic activity | Repressed energy
metabolism in HCC cells, reduces proliferation of HCC cells in
vitro and suppresses tumorigenesis in vivo | Lower expression
level correlates with high Edmondson-Steiner grade of the tumours
and shorter OS | (93) |
HULC
HULC, an oncogenic lncRNA of ~500 nt on chromosome
6p24.3, contains one intron, one canonical and two non-canonical
polyadenylation signals. Through microarray analysis and qPCR, HULC
was the first lncRNA identified to be specifically upregulated in
HCC (42,43). Further research has demonstrated
that cAMP response element-binding protein (CREB) increases the
expression of HULC by binding to its promoter (30).
As an lncRNA mainly localized in the cytoplasm, HULC
acts as a competitive endogenous RNA for miR-372 (30), miR-186 (31), miR-488 (32), miR-200a-5p (33), miR-6825-5p, miR-6845-5p and
miR-6886-3p (34), in HCC tissues
and cell lines. By titrating these miRNAs away from their target
mRNAs, HULC augments the expression of PRKACB, HMGA2, ADAM9, ZEB1
and USP22, and promotes HCC development. Moreover, via
deubiquitination mediated by USP22, HULC increases the stability of
COX2 and promotes the growth of HCC cell lines (39).
The function of HULC in HBV-related HCC has been
investigated in recent years. By interacting with CREB, HBx
increases the expression of HULC in HBV-related HCC tissues and
cell lines. The expression of p18, a tumour suppressor gene located
near HULC, is decreased by HULC and promotes the proliferation of
hepatoma cells both in vitro and in vivo (44). Moreover, HULC also acts as a
molecular sponge for miR-107 in HBV-related HCC. By sequestering
miR-107, HULC upregulates E2F1 and then activates transcription of
SPHK1 in HBV-related HCC tissues and cell lines. This process
promotes tumour angiogenesis in vitro and in vivo
(35). In 2017, Jiang et al
revealed that metformin is able to inhibit the expression of HULC
and suppress the progression of HBV-related HCC (45). Although it must be further
investigated, this finding suggests that HULC is a potential
therapeutic target for HBV-related HCC.
Taken together, these findings demonstrate that HULC
is an important oncogene in HBV-related HCC.
HOTAIR
The HOTAIR gene is ~12.6 kb in length and has six
exons. LncRNA HOTAIR, a 2,158-nt transcript of the HOTAIR gene, was
originally discovered by Rinn et al using tiling microarray
analysis in 2007 (46). Further
analyses revealed that HOTAIR interacts with PRC2 and enhances
repression of the HOXD locus by PRC2 (46). Growing evidence has demonstrated
that HOTAIR contributes to the risk of several cancers, including
HCC (47).
In 2011, the expression of HOTAIR was reported to be
increased in HCC tissues and cell lines (48). This finding was confirmed by a
series of studies (14,49-55).
Subsequent research has suggested that elevated expression of
HOTAIR promotes migration and invasion of HCC cells by inhibiting
RNA-binding motif protein 38 (49), and affects the symptoms and
prognosis of HCC patients (48,51,55).
Furthermore, overexpression of HOTAIR promotes autophagy by
elevating the expression of ATG3 and ATG7 (52), and promotes glycolysis by
activating glucose transporter isoform 1 and mTOR signalling
(53). Inhibition of HOTAIR
suppresses tumourigenesis, proliferation, viability, migration and
invasion of HCC cells (14,48,54,56).
Functional analyses have indicated that HOTAIR promotes HCC through
repression of miR-1, and is involved in the recruitment of
macrophages and myeloid-derived suppressor cells to the tumour
micro-environment (57). Moreover,
knockdown of HOTAIR was shown to increase the chemosensitivity of
HCC cells to cisplatin (58).
HOTAIR is also involved in the occurrence of
HBV-related HCC. qPCR has demonstrated that the expression of
HOTAIR is markedly increased in liver tumour tissues from X/c-myc
bitransgenic mice and HBV-infected patients (17). In HBV replicating cells, HOTAIR is
regulated by DEAD box protein 5 and participates in proteasomal
degradation of SUZ12 (18). In
addition, HOTAIR reduces the stability of SUZ12 and ZNF198 by
enhancing Plk1-dependent ubiquitination of these two proteins, and
it may affect epigenetic reprogramming involved in oncogenic
transformation (17). Taken
together, this evidence indicates that HOTAIR promotes the
occurrence of HCC and contributes to the risk of HBV-related
HCC.
MALAT1
MALAT1 (also known as nuclear-enriched abundant
transcript 2) is ~8,000 nt in length and localized in the nucleus
(25). MALAT1 tends to be a
nucleus-retained lncRNA and affects alternative splicing and gene
expression (25,26). It was one of the first lncRNAs that
was identified to be associated with a disease. In 2003, Ji et
al demonstrated that MALAT1 predicts prognosis of early-stage
lung cancer (59). MALAT1 is also
an important oncogene in several other cancers, including
esophageal squamous cell carcinoma, gastric and colorectal cancer
(60).
MALAT1 is downregulated in HCC tissues and is
associated with the overall survival (OS) of HCC (43). Higher expression of MALAT1
contributes to the risk of HCC recurrence after liver
transplantation (61).
Loss-of-function experiments have revealed that inhibition of
MALAT1 in HepG2 cells reduces cell viability, motility and
invasiveness and increases sensitivity to apoptosis (61). With the help of bioinformatics,
MALAT1 was found to promote HCC progression, metastasis and
multidrug resistance by sequestering a number of miRNAs, including
miR-216b (62), miR-143-3p
(63) and miR-146-5p (64). However, none of these studies
investigated the subcellular localization or cytoplasmic abundance
of MALAT1 (62-64). As an lncRNA mainly located in
nuclear speckles (25), it does
not appear likely that MALAT1 can sponge miRNAs in the
cytoplasm.
qPCR has revealed that Sp1, Sp3 and MALAT1 are
upregulated in HBV-related HCC tissues and cell lines. Further
investigation has indicated that Sp1 and Sp3 bind to the proximal
promoter region of MALAT1 and enhance its transcriptional activity
(65). The expression of MALAT1 is
elevated by HBx in HCC tissues and cell lines (24,66).
Loss- and gain-of-function experiments have indicated that MALAT1
regulates HBx-induced cancer stem cell properties, promotes
migration and invasion of HCC cells in vitro and tumour
growth in vivo. Further research demonstrated that MALAT1
promotes tumour growth and metastasis by upregu-lating LTBP3. These
findings indicate that MALAT1 mediates the oncogenic effect of HBx
by increasing the expression of LTBP3 (24).
Although the detailed mechanism requires further
investigation, MALAT1, a key oncogene for several cancers,
contributes to the risk of HBV-related HCC.
UCA1
The UCA1 gene is ~7.3 kb in length and contains
three exons (67). It is mapped to
chromosome 19p13.12 and has three transcriptional isoforms. UCA1, a
transcription isoform of the UCA1 gene that is ~1,400 nt in length,
is the most abundant isoform of UCA1 in various cancers (68). It was originally identified in the
bladder cancer cell line BLZ-211 and is a sensitive and specific
marker for bladder cancer (67).
UCA1 is also an oncogene for a number of other cancers, including
HCC (68). UCA1 is also involved
in anticancer drug resistance (69).
To determine whether UCA1 is involved in HBV-related
HCC, the expression of UCA1 was detected in HCC cell lines and
several groups of HCC patients mainly infected with HBV. Their
results demonstrated that UCA1 was upregulated in cancerous tissues
and HCC cell lines, and higher UCA1 expression in HCC was
positively associated with tumour size, vascular invasion,
metastasis, postoperative survival and disease stage (13,37,70).
These findings indicate the important role of UCA1 in HBV-related
HCC.
As an lncRNA localized in both the nucleus and
cytoplasm (13,71), UCA1 has diverse functions. In
hepatocytes, UCA1 is upregulated by HBx. Then, UCA1 recruits EZH2
to the promoter of p27 in the nucleus, suppresses the expression of
p27 and activates CDK2. This process contributes to G1/S transition
and promotes the growth of hepatic and hepatoma cells. These
findings unveil the crucial role of the HBx/UCA1/EZH2/p27
signalling pathway in HCC (13).
In the cytoplasm, UCA1 acts as a molecular sponge for miR-216b and
miR-203. By sequestering miR-216b, UCA1 increases the expression of
fibroblast growth factor receptor 1, activates the extracellular
signal-regulated kinase signalling pathway and promotes the growth
and metastasis of HCC cell lines (70). In addition, sponging miR-203
promotes epithelial-to-mesen-chymal transition (EMT) in HCC cells
via the upregulation of Snail2 (37). In conclusion, UCA1, a crucial
oncogene in several cancers, is regulated by HBx and contributes to
the risk of HBV-related HCC.
HEIH
The lncRNA HEIH is ~1,600 nt in length and maps to
chromosome 5. It is located in the nucleus and cytoplasm of HCC
cells. With the use of microarray analysis and qPCR, HEIH was found
to be upregulated in HBV-related HCC tumour tissues (12). In addition to being upregulated in
HCC cell lines, further investigation revealed that HEIH is
significantly increased only in HBV-related HCC tumour tissues
(72,73). Logistic multivariate regression has
demonstrated that the expression of HEIH is associated with disease
recurrence and OS in HCC patients. These findings suggest that HEIH
is an independent prognostic factor for OS in HBV-related HCC
(12).
Rather than DNA amplification, DNA methylation or
histone acetylation, upregulation of HEIH is induced by the
transcription factor Sp1. Loss- and gain-of-function experiments
were performed to evaluate the effect of HEIH on cell biological
behaviour. In vitro experiments demonstrated that HEIH
promotes cell proliferation by upregulating proliferating cell
nuclear antigen and modulating the cell cycle by decreasing the
expression of p16, p21 and p27 in HCC cell lines. In vivo
studies revealed that HEIH promotes tumour growth in nude mice. RNA
immunoprecipitation and pulldown revealed that HEIH is
physiologically associated with EZH2 and is required for repression
of EZH2 target genes (12). These
findings indicate that, by participating in epigenetic silencing,
HEIH contributes to the risk of HBV-related HCC.
LINC00152
LINC00152, an lncRNA located on chromosome 2p11.2,
contains four exons and encodes an 828-bp transcript. It is mainly
localized in the nucleus of HCC cells (23) and plays a vital role in
carcinogenesis of several cancers (74).
In a group of HCC patients with different
aetiologies, LINC00152 was found to be hypomethylated during
hepatocarcinogenesis (75). This
finding was confirmed in a group of HCC patients mainly infected
with HBV (23). The expression of
LINC00152 was found to be significantly increased in HCC patients
and cell lines (15,23). These results suggest that LINC00152
is involved in HCC. The expression of LINC00152 is correlated with
tumour size, HBV infection, tumour number and HBx expression
(15,23). Kaplan-Meier analysis has revealed
that higher expression of LINC00152 results in significantly
shorter OS time (15).
LINC00152 is activated by HBx and promotes
proliferation and EMT of HCC cell lines in vitro and
tumouri-genesis in vivo (15,23).
With the use of microarray analysis, the Gal4-λN/BoxB reporter
system and antisense oligonucle-otide technology, LINC00152 has
been shown to activate the mTOR pathway by binding to the promoter
of EpCAM in a cis-regulation pattern (23). In addition, LINC00152 promotes cell
EMT by decreasing the binding of EZH2 to the promoter of E-cadherin
and inhibiting expression of E-cadherin in HCC cell lines (15). These findings indicate that
LINC00152 contributes to the risk of HBV-related HCC.
HOTTIP
LncRNA HOTTIP is mapped to the HOXA locus and
encodes a 3,764-nt transcript. It was originally identified in
anatomically distal human fibroblasts. HOTTIP activates and then
promotes the development of several cancers by recruiting
histone-modifying enzymes to HOX genes and silencing tumour
suppressor genes (76).
The expression of HOTTIP is increased in HCC tumour
tissues and cell lines (20,21,77,78).
By analysing the expression of lncRNAs in HCC patients infected
with different hepatitis viruses, HOTTIP has been shown to be
significantly upregu-lated in HCV- and HBV-related HCC patients
(73). Higher expression levels of
HOTTIP are correlated with increased metastasis and decreased OS
(21). These findings show that
HOTTIP may be a prognostic indicator for HCC and should prompt
further investigation of its function.
Loss-of-function experiments have revealed that
knockdown of HOTTIP inhibits the proliferation and migration of HCC
cells in vitro and reduces tumourigenesis and pulmonary
metastasis in vivo (20).
In vitro and in vivo investigations have also
revealed that the expression of HOTTIP is suppressed by miR-192,
miR-204 and miR-125b (20,78), and it promotes the expression of a
panel of HOXA genes, including HOXA 10, 11 and 13, in HCC (20-22).
In addition to being inhibited by miRNAs, the expression of HOTTIP
is also repressed by HOXA13. This suggests the existence of a
bidirectional regulatory loop between HOTTIP and HOXA13 (21). These findings suggest that HOTTIP
is a crucial oncogene in HBV-related HCC.
HBx-LINE1
As 85-90% of HBV-related HCC tumours harbour at
least one HBV insertion site (79), it was reasonable to hypothesize
that integration of HBV into the host genome may play an important
role in the development of HBV-related HCC. To investigate the
effect of such integration on genome disruption, transcriptome
sequencing was performed to analyse six HBV-positive HCC cell lines
for viral integration. With the use of ViralFusionSeq, a specific
algorithm for accurate and unbiased detection of HBV integration, a
virus-human chimeric transcript named HBx-LINE1 was detected in
HKCI-4. Hemi-nested reverse transcription-PCR revealed that
HBx-LINE1 was expressed in 21 of 90 HBV-related HCC tumours.
Subsequent research demonstrated that HBx-LINE1 is an lncRNA that
promotes cell motility by activating the Wnt/β-catenin signalling
pathway (80).
HBx-LINE1 was detected in 17 of 40 HCC tissues by
qPCR. Subsequent investigations have revealed that the expression
of HBx-LINE1 is inversely correlated with miR-122 in HBV-related
HCC tissues and HuH7 cells. Functional analysis demonstrated that
HBx-LINE1 sponges miR-122 in HuH7 cells and mouse liver cells. By
sequestering miR-122, HBx-LINE1 activates the β-catenin signalling
pathway, promoting hepatic cell migration and causing mouse liver
injury (36).
However, these findings were not consistent with
those of other studies. In 2015, Ding et al reported that
HBx-LINE1 was not detected in 30 HCC tissues. In addition,
HBx-LINE1 integration was not detected using primers designed to
detect the HBx-LINE1 virus-host junction sequences at the DNA level
(81).
There are several possible reasons for this
discrepancy. HBV integration is a complex progress, and its
mechanism remains unclear. Cellular and viral factors affecting HBV
integration also remain unclear. Combined with the fact that these
studies were conducted in different areas, the small sample size
may have caused the discrepancy. More research with larger samples
may help further elucidate the role of HBx-LINE1 in the occurrence
and prognosis of HBV-related HCC.
ANRIL
ANRIL, originally identified in familial melanoma
patients, is transcribed as a 3,800-nt lncRNA in the anti-sense
direction of the INK4BARF-INK4A gene cluster (82). Accumulating evidence indicates that
ANRIL is upregulated in several cancers and acts as an oncogene
(83).
In HCC patients and cell lines, the expression of
ANRIL was found to be increased by qPCR (16,38,84).
A recent study revealed that ANRIL was upregulated in HCV- and
HBV-related HCC patients (73).
The expression of ANRIL was associated with tumour size, Barcelona
Clinic Liver Cancer (BCLC) stage, histological grade, TNM stage and
OS in HCC patients (16,38). These findings suggest that ANRIL
contributes to the risk of HCC, particularly HBV-related HCC.
Loss-of-function experiments have been conducted to
elucidate the effect of ANRIL on HCC cell behaviour. Knockdown of
ANRIL expression induced apoptosis and suppressed proliferation,
invasion and migration of HCC cells in vitro (16,38,84).
Inhibition of ANRIL led to slower tumour growth in vivo
(16,38). Functional analyses revealed that
ANRIL is activated by the transcription factor Sp1 and represses
the transcription of KLF2 through binding with PRC2 (16). ANRIL also exerts its effects by
sponging miR-122-5p (38). These
findings indicate that ANRIL is a crucial oncogene and contributes
to the risk of HBV-related HCC.
DBH-AS1
DBH-AS1, an lncRNA of ~2 kb residing at chr9q34, is
downregulated by quercetin in HepG2 cells (85). The expression of DBH-AS1, which is
induced by HBx and repressed by p53, is upregulated in HCC cell
lines and tumour tissues. The expression of DBH-As1 is positively
correlated with tumour size and hepatitis B surface antigen
(HBsAg). In vitro and in vivo experiments have
demonstrated that DBH-AS1 inhibits serum-starvation-induced
apoptosis of HCC cells, promotes proliferation and cell cycle
progression of HCC cells, and contributes to tumour growth. DBH-AS1
activates the mitogen-activated protein kinase (MAPK) signalling
pathway. Through activation of MAPK signalling, DBH-AS1 contributes
to the risk of HBV-related HCC (86).
However, a recent study by Zhang et al
demonstrated opposite results regarding the expression pattern of
DBH-AS1. In 11 HBV-related HCC patients, the expression of DBH-AS1
was significantly decreased in tumour tissues compared with
peritumoural tissues (73). There
are several possible reasons for this discrepancy. First, the
sample size of these two studies was small. Second, in the former
study, HCC patients with different viral aetiologies were included
in the HCC group. As the expression of lncRNAs in HCC patients with
different aetiologies may be different, this may have led to the
different results. To elucidate the expression pattern and
functional role of DBH-AS1 in HBV-related HCC, more studies on
HBV-related HCC with larger sample sizes are required.
DREH
DREH is ~700 nt in length and located on chromosome
5. By analysing the expression profile of lncRNAs induced by HBx
with the use of microarrays and qPCR, lncRNA-Dreh, the mouse
ortholog of DREH, was found to be downregulated in HBx transgenic
mice and mouse liver cells expressing HBx. By binding to vimentin,
lncRNA-Dreh inhibits its expression and alters its filament
structure. This process represses tumour growth and inhibits tumour
metastasis in vivo. Loss-of-function experiments have
demonstrated that inhibition of Dreh promotes proliferation and
migration of mouse live cell lines (40). These findings suggest that
lncRNA-Dreh acts as a tumour suppressor in HBV-related HCC.
The importance of lncRNA-Dreh led researchers to
investigate the function of its human orthologue in HBV-related
HCC. Although DREH is upregulated in HCV-related HCC tissues, qPCR
revealed that DREH is significantly downregulated in HBV-related
HCC tissues (40,73,87).
The expression of DREH was found to be inversely correlated with
HBx in HCC tissues and downregulated by HBx in HCC cell lines.
Inhibition of DREH promotes proliferation of HCC cells in
vitro and tumour growth in vivo (87). Survival and correlation analyses
have demonstrated that lower expression of DREH is closely
associated with the recurrence-free survival (RFS) and OS of HCC
patients (40) and is correlated
with tumour size and HBsAg in HCC patients (87). Although its detailed mechanism of
action in HCC patients remains elusive, DREH acts as a tumour
suppressor in HBV-related HCC.
Other lncRNAs in HBV-related HCC
In recent years, a growing number of lncRNAs are
found to be aberrantly expressed in HBV-related HCC via microarray
analyses and high-throughput sequencing. For example, the
expression of XLOC_007433 and AC144449.1 was found to be increased
and decreased, respectively, in male patients with HBV-related HCC
(88). In HBV-related HCC tissues
and cell lines, n346077 (89) and
uc.306 (90) are downregulated,
while BAIAP2-AS1, PRC1-AS1, LINC00665 and AC092171.4 are
upregulated (91,92). n346077 is associated with invasion
and migration of HCC cells (89),
and uc.306 is negatively correlated with the OS of HBV-related HCC
patients (90). LncRNA-6195, a
tumour repressor for HBV-related HCC, suppresses HCC cell
proliferation both in vitro and in vivo through
binding with α-enolase and then inhibiting its enzymatic activity
(93). The expression level of
Unigene56159, a 2653-nt-long lncRNA located in the second intron of
ROBO1, is significantly higher in HBV-positive compared with
HBV-negative HCC tissues and cell lines. Subsequent investigation
revealed that Unigene56159 is induced by HBV and promotes EMT,
migration and invasion of hepatoma cells via sequestering
miR-140-5p and increasing the expression of Slug (29). Upregulated by HBx, WEE2-AS1 is able
to accelerate the proliferation, migration, invasion and cell cycle
progression of HCC cells through increasing the expression of
Fermitin family member 3 (94).
Additionally, recent studies have shown that the expression levels
of lncRNAs are regulated by HBx (95-99).
Mining gene expression databases is also an
effective method for identifying lncRNAs that are involved in
HBV-related HCC. Through analysing data from the Gene Expression
Omnibus database and the Cancer Genome Atlas, a
LINC00346-miR-10a-5p-CDK1 axis was found to affect the progression
of HBV-related HCC (100).
Another research using bioinformatics identified MSC-AS1, POLR2J4,
EIF3J-AS1, SERHL, RMST and PVT1 as risk factors for RFS of
HBV-related HCC (101).
4. Circulating lncRNAs serve as novel
biomarkers for HBV-related HCC
Despite the marked advances in diagnostic methods
and surgical techniques, the prognosis of HBV-related HCC remains
poor. A number of serum markers, such as α-fetoprotein (AFP), lens
culinaris agglutinin-reactive fraction of AFP and des-γ-carboxy
prothrombin, are used for diagnosis and outcome prediction in HCC
(102,103). However, the specificity and
sensitivity of these tumour markers are not sufficient. This has
encouraged researchers to search for novel serum markers of HCC. As
an increasing number of lncRNAs have been demonstrated to affect
the risk of HBV-related HCC, researchers have tried to determine
whether these lncRNAs may be used as serum biomarkers for
HBV-related HCC.
As the first lncRNA confirmed to be upregulated in
HCC, the expression of HULC in the peripheral blood cells of HCC
patients was first found to be markedly increased in 3 of 4 HCC
patients in a pilot experiment in 2007 (42). Later research revealed that the
plasma HULC-positive rate was higher in HBV-positive HCC patients
compared with that in HBV-negative HCC patients, and that the level
of HULC was correlated with Edmondson grade (104). By screening in the training set
and validation in the validation set, plasma HULC was confirmed to
be increased in the HCC group and exhibited adequate diagnostic
accuracy for HCC (105). These
results indicate that plasma HULC is a useful biomarker for
HBV-related HCC.
In addition to HULC, a number of other circulating
lncRNAs, such as MALAT1 (106),
LINC00152 (105,107), RP11-160H22.5 (107), XLOC014172 (107), PVT1 (108) and uc002mbe.2 (108), are upregulated in HCC patients.
The serum levels of lncRNA uc003wbd (109), lncRNA-AF085935 (109), uc001ncr (110) and AX800134 (110) are elevated in HBV-related HCC
patients. These results indicate that circulating lncRNAs may be
useful biomarkers for predicting the risk and prognosis of
HBV-related HCC.
However, there are several limitations to these
studies. First, several studies recruited HCC patients with
different aetiologies. Although the detailed mechanism remains
elusive, Zhang et al observed that HCC patients with
different viral aetiologies had different dysregulated lncRNAs
(73). Assigning patients with
different aetiologies to one group may cause bias. Second, as
several studies included small samples, the results may not be as
reliable. Future research with larger samples and recruiting of
only HBV-related HCC patients will validate the findings and yield
more reliable results. Third, a number of these circulating lncRNAs
identified to be dysregulated in HBV-related HCC are also
aberrantly expressed in other tumours. Future research in patients
with different tumours using high-throughput technology will help
us identify circulating lncRNAs specific for HBV-related HCC.
However, despite these limitations, the research
mentioned above provides valuable clues for further screening of
circulating lncRNAs that may serve as biomarkers for HBV-related
HCC.
5. Genetic polymorphisms in lncRNAs and
HBV-related HCC
Genetic polymorphisms are the most abundant genetic
markers in the human genome. Association studies are widely used to
identify genetic variations that contribute to the occurrence and
prognosis of several cancers, including HBV-related HCC. Since
genome-wide association studies have discovered that the majority
of single-nucleotide polymorphisms (SNPs) associated with complex
disease are located in genomic regions not coding for proteins
(111,112), researchers started to explore the
associations between several SNPs in lncRNAs and cancer (113). The effect of SNPs in lncRNAs on
HBV-related HCC has also been investigated. In 2012, the variant
genotype of rs7763881 in HULC was shown to decrease the risk of
HBV-related HCC in a group of unrelated ethnic Han Chinese patients
(114). As shown in Table II, a number of SNPs in lncRNAs
affect the occurrence and prognosis of HBV-related HCC (Table II).
 | Table IIGenetic mutations in lncRNAs and
HBV-related HCC. |
Table II
Genetic mutations in lncRNAs and
HBV-related HCC.
| Polymorphism
ID | Substitutes | lncRNA | SNP location | Clinical
impact | Biological
function | (Refs.) |
|---|
| rs7763881 | A>C | HULC | Chr6:8653014 | Decreases the risk
of HCC | Not known | (114) |
| rs920778 | C>T | HOTAIR | Chr12:53966448 | Increases the risk
of HCC | Increases the
expression of HOTAIR and promotes the proliferation of HCC
cells | (115) |
| rs145204276 | AGGCA>- | GAS5 | Chr1:173868254 | Increases the risk
of HCC | Regulates the
expression of GAS5 via an epigenetic mechanism | (118) |
| rs10680577 | TACT>- | RERT-lncRNA | Chr19:40798691 | Increases the risk
of HCC | Increases
expression of RERT-lncRNA and EGLN2, may change structure of
RERT-lncRNA | (119) |
| rs35622507 | GAGT repeats | KCNQ1OT1 | Not known | Decreases the risk
of HCC | Alters the
structure of KCNQ1OT1 and modulates expression of KCNQ1OT1 and
CDKN1C | (120) |
| rs7248320 | A>G | AC008392.1 | Chr19:48256972 | Increases the risk
of HCC | eQTLs for
CARD8 | (123) |
| rs3757328, | G>A | ZNRD1-AS1 | Chr6:30060575 | Increases the risk
of HCC | eQTLs for
ZNRD1 | (124) |
| rs6940552 | G>A | ZNRD1-AS1 | Chr6:30044563 | Increases the risk
of HCC | eQTLs for
ZNRD1 | (124,125) |
| rs9261204 | A>G | ZNRD1-AS1 | Chr6:30037466 | Increases the risk
of HCC | eQTLs for
ZNRD1 | (124,125) |
| rs11489585 | A>G | APTR | Chr7:77685535 | Increases the risk
of HCC | eQTLs for
PTPN12 | (126) |
| rs79037040 | G>T | RP11-1149O23.3 | Chr8:23225458 | Decreases the risk
of HCC | eQTLs for
TNFRSF10A | (127) |
| rs2055822 | A>G | RP11-459E5.1 | Chr8:22779707 | Increases the risk
of HCC | eQTLs for
TNFRSF10B | (127) |
| rs1110839 | T>G | AC016683.6 | Chr2:113236840 | Is associated with
better prognosis of HCC | eQTLs for PAX8 | (128) |
| rs4848320 | C>T | AC016683.6 | Chr2:113253214 | Is associated with
better prognosis of HCC | eQTLs for PAX8 | (128) |
As mentioned above, HOTAIR contributes to the
susceptibility to HBV-related HCC. Recently, a case-control study
was conducted to evaluate the association between SNPs in HOTAIR
and the risk of HBV-related HCC. The minor allele of rs920778 was
found to be a risk factor for HBV-related HCC. With the use of
qPCR, functional investigation using luciferase activity and CCK-8
assays revealed that the minor allele of rs920778 increased the
expression of HOTAIR and was associated with a higher proliferation
rate of HCC cells. These findings suggest that the minor allele of
rs920778 promotes the occurrence of HBV-related HCC by upregulating
HOTAIR and increasing proliferation of HCC cells (115).
Growth arrest special 5 (GAS5), an lncRNA
identified in a mouse thymoma cell line, has been found to be
down-regulated in most HCC patients (116,117). rs145204276 is a 5-bp indel
polymorphism in the promoter region of GAS5. Association studies
have demonstrated that the deletion allele of rs145204276
significantly increases the risk of HBV-related HCC. Subsequent
analyses have revealed that rs145204276 increases the expression of
GAS5 in HCC tissues and cell lines by altering the methylation
status of the GAS5 promoter region (118).
A 4-bp deletion allele of rs10680577 in
RERT-lncRNA, an lncRNA whose sequence overlaps with Ras-related
GTP-binding protein 4b and prolyl hydroxylase 1 (EGLN2), promotes
the occurrence of HBV-related HCC. This allele leads to increased
expression of RERT-lncRNA and EGLN2 in HCC tissues and cell lines.
Bioinformatics analyses have demonstrated that this allele may
alter the structure of RERT-lncRNA. Thus, rs10680577 may affect the
occurrence of HBV-related HCC by regulating the expression of
RERT-lncRNA and EGLN2 and altering the structure of RERT-lncRNA
(119).
rs35622507, a novel short tandem repeat (STR)
polymorphism in the coding region of KCNQ1-overlapping transcript 1
(KCNQ1OT1), affects the occurrence of HBV-related HCC. In a group
of unrelated Han Chinese subjects, the homozygous 10-10 genotype
was shown to increase the risk of HCC. qPCR indicated that HCC cell
lines with the homozygous 10-10 genotype exhibited significantly
lower expression of KCNQ1OT1 and higher expression of
cyclin-dependent kinase inhibitor 1C (CDKN1C) compared with HCC
cell lines with other genotypes. Bioinformatics prediction
indicated that this STR polymorphism may alter the structure of
KCNQ1OT1. Thus, rs35622507 may affect the risk of HBV-related HCC
by altering the structure of KCNQ1OT1 and modulating the expression
of KCNQ1OT1 and CDKN1C (120).
Recently, a genome-wide association study explored
risk loci for familial HBV-related HCC. Although the detailed
molecular mechanism remained unclear, a cluster of SNPs overlapping
with LINC00272 was associated with increased risk of HBV-related
HCC (121).
Expression quantitative trait loci (eQTLs) are
genetic variants that may affect the expression of a specific gene
and contribute to the risk of complex diseases (122). They are significantly enriched
for disease-associated polymorphisms. Several SNPs in lncRNAs, such
as rs7248320 (123), rs3757328
(124), rs6940552 (124,125), rs9261204 (124,125), rs11489585 (126), rs79037040 (127) and rs2055822 (127), are eQTLs for a series of genes
and may contribute to the risk of HBV-related HCC. In addition to
affecting the risk of HBV-related HCC, some eQTLs, such as
rs1110839 (128) and rs4848320
(128), may affect the prognosis
of HBV-related HCC.
However, all the abovementioned studies were
conducted in Chinese populations. Future research in other
populations will help to better understand the effect of SNPs in
lncRNAs on HBV-related HCC. Taken together, although preliminary,
the evidence suggests that genetic polymorphisms in lncRNAs may
affect the occurrence and prognosis of HBV-related HCC.
6. Conclusions
During recent years, with the use of various
approaches, including loss- and gain-of-function experiments and
in vitro and in vivo analyses, the important role of
lncRNAs in the occurrence and prognosis of HBV-related HCC has
started to emerge. Several circulating lncRNAs and genetic
polymorphisms in lncRNAs are also screened out to affect the risk
and prognosis of HBV-related HCC. These studies will hopefully
elucidate the mechanism of action of lncRNAs and help in the
prevention, diagnosis and treatment of HBV-related HCC.
However, although the findings of these studies are
valuable, the functions of a large proportion of lncRNAs
dysregulated in HBV-related HCC remain elusive. Although Zhang
et al found that HCC patients infected with different
hepatitis viruses had different dysregulated lncRNAs (73), their mechanism of action remains
unclear. Moreover, a recent study indicated that an lncRNA was able
to exert its function by encoding a small polypeptide (129). Whether any lncRNAs affect the
risk of HBV-related HCC in this manner is yet to be investigated.
Future research in these areas will help unravel the function of
lncRNAs in HBV-related HCC.
In conclusion, although the functions of lncRNAs
have yet to be fully elucidated, recent studies indicate that
lncRNAs play key roles in HBV-related HCC, and investigation of
lncRNAs will pave the way for fully understanding the mechanism
underlying the development of HBV-related HCC.
Acknowledgments
The authors would like to thank professor Yunqiang
Liu of Sichuan University for his kind suggestions on the
manuscript.
Funding
This study was not supported by any specific grant
from any funding agency in the public, commercial, or
not-for-profit sectors.
Availability of data and materials
Not applicable.
Authors' contributions
ZH, CXB and LSL were involved in the conception and
design of this study. ZH summarized relevant literature and wrote
the manuscript; CXB revised the manuscript; ZJ, WXW, CHJ and LL
collected and evaluated relevant literature; LSL revised the
manuscript according to the reviewers' comments and was responsible
for further support. All authors have read and approved the final
version.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that there are no potential
conflicts of interest and no financial support related to this
study.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM: The global health burden of
infection-associated cancers in the year 2002. Int J Cancer.
118:3030–3044. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Giannelli G and Antonaci S: Novel concepts
in hepatocellular carcinoma: From molecular research to clinical
practice. J Clin Gastroenterol. 40:842–846. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Han YF, Zhao J, Ma LY, Yin JH, Chang WJ,
Zhang HW and Cao GW: Factors predicting occurrence and prognosis of
hepatitis-B-virus-related hepatocellular carcinoma. World J
Gastroenterol. 17:4258–4270. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tarocchi M, Polvani S, Marroncini G and
Galli A: Molecular mechanism of hepatitis B virus-induced
hepatocarcinogenesis. World J Gastroenterol. 20:11630–11640. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Costa FF: Non-coding RNAs: Could they be
the answer? Brief Funct Genomics. 10:316–319. 2011. View Article : Google Scholar
|
|
7
|
Jia H, Osak M, Bogu GK, Stanton LW,
Johnson R and Lipovich L: Genome-wide computational identification
and manual annotation of human long noncoding RNA genes. RNA.
16:1478–1487. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv
XW and Li J: Long noncoding RNAs: Novel insights into hepatocelluar
carcinoma. Cancer Lett. 344:20–27. 2014. View Article : Google Scholar
|
|
12
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long noncoding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu JJ, Song W, Zhang SD, Shen XH, Qiu XM,
Wu HZ, Gong PH, Lu S, Zhao ZJ, He ML and Fan H: HBx-upregulated
lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting
EZH2 and repressing p27Kip1/CDK2 signaling. Sci Rep. 6:235212016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang
H, Liang WC, Wang SS, Ko CH, Waye MM, et al: Hotair mediates
hepatocarcinogenesis through suppressing miRNA-218 expression and
activating P14 and P16 signaling. J Hepatol. 63:886–895. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Deng X, Zhao XF, Liang XQ, Chen R, Pan YF
and Liang J: Linc00152 promotes cancer progression in hepatitis B
virus-associated hepatocellular carcinoma. Biomed Pharmacother.
90:100–108. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang MD, Chen WM, Qi FZ, Xia R, Sun M, Xu
TP, Yin L, Zhang EB, De W and Shu YQ: Long non-coding RNA ANRIL is
upregulated in hepatocellular carcinoma and regulates cell
apoptosis by epigenetic silencing of KLF2. J Hematol Oncol.
8:502015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang H, Diab A, Fan H, Mani SK, Hullinger
R, Merle P and Andrisani O: PLK1 and HOTAIR accelerate proteasomal
degradation of SUZ12 and ZNF198 during Hepatitis B virus-induced
liver carcinogenesis. Cancer Res. 75:2363–2374. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang H, Xing Z, Mani SK, Bancel B,
Durantel D, Zoulim F, Tran EJ, Merle P and Andrisani O: RNA
helicase DEAD box protein 5 regulates Polycomb repressive complex
2/Hox transcript antisense intergenic RNA function in hepatitis B
virus infection and hepatocarcinogenesis. Hepatology. 64:1033–1048.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang KC, Yang YW, Liu B, Sanyal A,
Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta
RA, et al: A long noncoding RNA maintains active chromatin to
coordinate homeotic gene expression. Nature. 472:120–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tsang FH, Au SL, Wei L, Fan DN, Lee JM,
Wong CC, Ng IO and Wong CM: Long non-coding RNA HOTTIP is
frequently up-regulated in hepatocellular carcinoma and is targeted
by tumour suppressive miR-125b. Liver Int. 35:1597–606. 2015.
View Article : Google Scholar
|
|
21
|
Quagliata L, Matter MS, Piscuoglio S,
Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z,
Boldanova T, et al: Long noncoding RNA HOTTIP/HOXA13 expression is
associated with disease progression and predicts outcome in
hepatocellular carcinoma patients. Hepatology. 59:911–923. 2014.
View Article : Google Scholar :
|
|
22
|
Quagliata L, Quintavalle C, Lanzafame M,
Matter MS, Novello C, di Tommaso L, Pressiani T, Rimassa L,
Tornillo L, Roncalli M, et al: High expression of HOXA13 correlates
with poorly differentiated hepatocellular carcinomas and modulates
sorafenib response in in vitro models. Lab Invest. 98:95–105. 2018.
View Article : Google Scholar
|
|
23
|
Ji J, Tang J, Deng L, Xie Y, Jiang R, Li G
and Sun B: LINC00152 promotes proliferation in hepatocellular
carcinoma by targeting EpCAM via the mTOR signaling pathway.
Oncotarget. 6:42813–42824. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hou Z, Xu X, Fu X, Tao S, Zhou J, Liu S
and Tan D: HBx-related long non-coding RNA MALAT1 promotes cell
metastasis via up-regulating LTBP3 in hepatocellular carcinoma. Am
J Cancer Res. 7:845–856. 2017.PubMed/NCBI
|
|
25
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miyagawa R, Tano K, Mizuno R, Nakamura Y,
Ijiri K, Rakwal R, Shibato J, Masuo Y, Mayeda A, Hirose T and
Akimitsu N: Identification of cis- and trans-acting factors
involved in the localization of MALAT-1 noncoding RNA to nuclear
speckles. RNA. 18:738–751. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lv J, Fan HX, Zhao XP, Lv P, Fan JY, Zhang
Y, Liu M and Tang H: Long non-coding RNA Unigene56159 promotes
epithelial-mesenchymal transition by acting as a ceRNA of
miR-140-5p in hepatocellular carcinoma cells. Cancer Lett.
382:166–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y,
Chen N, Sun F and Fan Q: CREB up-regulates long non-coding RNA,
HULC expression through interaction with microRNA-372 in liver
cancer. Nucleic Acids Res. 38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Y, Chen F, Zhao M, Yang Z, Li J,
Zhang S, Zhang W, Ye L and Zhang X: The long noncoding RNA HULC
promotes liver cancer by increasing the expression of the HMGA2
oncogene via sequestration of the microRNA-186. J Biol Chem.
292:15395–15407. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hu D, Shen D, Zhang M, Jiang N, Sun F,
Yuan S and Wan K: MiR-488 suppresses cell proliferation and
invasion by targeting ADAM9 and lncRNA HULC in hepatocellular
carcinoma. Am J Cancer Res. 7:2070–2080. 2017.PubMed/NCBI
|
|
33
|
Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ,
Wang CY, Zhang HM, Zhang RX, Zhang JJ, et al: LncRNA HULC enhances
epithelial-mesenchymal transition to promote tumorigenesis and
metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1
signaling pathway. Oncotarget. 7:42431–42446. 2016.PubMed/NCBI
|
|
34
|
Xiong H, Ni Z, He J, Jiang S, Li X, He J,
Gong W, Zheng L, Chen S, Li B, et al: LncRNA HULC triggers
autophagy via stabilizing Sirt1 and attenuates the chemosensitivity
of HCC cells. Oncogene. 36:3528–3540. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lu Z, Xiao Z, Liu F, Cui M, Li W, Yang Z,
Li J, Ye L and Zhang X: Long non-coding RNA HULC promotes tumor
angiogenesis in liver cancer by up-regulating sphingosine kinase 1
(SPHK1). Oncotarget. 7:241–254. 2016.
|
|
36
|
Liang HW, Wang N, Wang Y, Wang F, Fu Z,
Yan X, Zhu H, Diao W, Ding Y, Chen X, et al: Hepatitis B
virus-human chimeric transcript HBx-LINE1 promotes hepatic injury
via sequestering cellular microRNA-122. J Hepatol. 64:278–291.
2016. View Article : Google Scholar
|
|
37
|
Xiao JN, Yan TH, Yu RM, Gao Y, Zeng WL, Lu
SW, Que HX, Liu ZP and Jiang JH: Long non-coding RNA UCA1 regulates
the expression of Snail2 by miR-203 to promote hepatocellular
carcinoma progression. J Cancer Res Clin Oncol. 143:981–990. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ma J, Li T, Han X and Yuan H: Knockdown of
LncRNA ANRIL suppresses cell proliferation, metastasis, and
invasion via regulating miR-122-5p expression in hepatocellular
carcinoma. J Cancer Res Clin Oncol. 144:205–214. 2018. View Article : Google Scholar
|
|
39
|
Xiong H, Li B, He J, Zeng Y, Zhang Y and
He F: lncRNA HULC promotes the growth of hepatocellular carcinoma
cells via stabilizing COX-2 protein. Biochem Biophys Res Commun.
490:693–699. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang
Y, Tang GN, Zhou WP and Sun SH: Hepatitis B virus X protein
(HBx)-related long noncoding RNA (lncRNA) down-regulated expression
by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by
targeting the intermediate filament protein vimentin. Hepatology.
57:1882–1892. 2013. View Article : Google Scholar
|
|
41
|
Zhao Q, Li T, Qi J, Liu J and Qin C: The
miR-545/374a cluster encoded in the Ftx lncRNA is overexpressed in
HBV-related hepatocellular carcinoma and promotes tumorigenesis and
tumor progression. PLoS One. 9:e1097822014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M and Zatloukal K: Characterization of HULC, a novel
gene with striking up-regulation in hepatocellular carcinoma, as
noncoding RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sonohara F, Inokawa Y, Hayashi M, Yamada
S, Sugimoto H, Fujii T, Kodera Y and Nomoto S: Prognostic value of
long non-coding RNA HULC and MALAT1 following the curative
resection of hepatocellular carcinoma. Sci Rep. 7:161422017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Du Y, Kong G, You X, Zhang S, Zhang T, Gao
Y, Ye L and Zhang X: Elevation of highly up-regulated in liver
cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell
proliferation via down-regulating p18. J Biol Chem.
287:26302–26311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jiang Z and Liu H: Metformin inhibits
tumorigenesis in HBV-induced hepatocellular carcinoma by
suppressing HULC overexpression caused by HBX. J Cell Biochem.
119:4482–4495. 2018. View Article : Google Scholar
|
|
46
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1123.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang J, Zhang P, Wang L, Piao HL and Ma
L: Long non-coding RNA HOTAIR in carcinogenesis and metastasis.
Acta Biochim Biophys Sin (Shanghai). 46:1–5. 2014. View Article : Google Scholar
|
|
48
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ding C, Cheng S, Yang Z, Lv Z, Xiao H, Du
C, Peng C, Xie H, Zhou L, Wu J and Zheng S: Long non-coding RNA
HOTAIR promotes cell migration and invasion via down-regulation of
RNA binding motif protein 38 in hepatocellular carcinoma cells. Int
J Mol Sci. 15:4060–4076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Su DN, Wu SP, Chen HT and He JH: HOTAIR, a
long non-coding RNA driver of malignancy whose expression is
activated by FOXC1, negatively regulates miRNA-1 in hepatocellular
carcinoma. Oncol Lett. 12:4061–4067. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gao JZ, Li J, Du JL and Li XL: Long
non-coding RNA HOTAIR is a marker for hepatocellular carcinoma
progression and tumor recurrence. Oncol Lett. 11:1791–1798. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yang L, Zhang X, Li H and Liu J: The long
noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and
ATG7 in hepatocellular carcinoma. Mol Biosyst. 12:2605–2612. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wei S, Fan Q, Yang L, Zhang X, Ma Y, Zong
Z, Hua X, Su D, Sun H, Li H and Liu Z: Promotion of glycolysis by
HOTAIR through GLUT1 upregulation via mTOR signaling. Oncol Rep.
38:1902–1908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Geng YJ, Xie SL, Li Q, Ma J and Wang GY:
Large intervening non-coding RNA HOTAIR is associated with
hepatocellular carcinoma progression. J Int Med Res. 39:2119–2128.
2011. View Article : Google Scholar
|
|
55
|
Ishibashi M, Kogo R, Shibata K, Sawada G,
Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et
al: Clinical significance of the expression of long non-coding RNA
HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 29:946–950.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wu Y, Xiong Q, Li S, Yang X and Ge F:
Integrated proteomic and transcriptomic analysis reveals long
noncoding RNA HOX transcript antisense intergenic RNA (HOTAIR)
promotes hepatocellular carcinoma cell proliferation by regulating
opioid growth factor receptor (OGFr). Mol Cell Proteomics.
17:146–159. 2018. View Article : Google Scholar :
|
|
57
|
Fujisaka Y, Iwata T, Tamai K, Nakamura M,
Mochizuki M, Shibuya R, Yamaguchi K, Shimosegawa T and Satoh K:
Long non-coding RNA HOTAIR up-regulates chemokine (C-C motif)
ligand 2 and promotes proliferation of macrophages and
myeloid-derived suppressor cells in hepatocellular carcinoma cell
lines. Oncol Lett. 15:509–514. 2018.PubMed/NCBI
|
|
58
|
Zhou JJ, Cheng D, He XY, Meng Z, Ye HL and
Chen RF: Knockdown of long non-coding RNA HOTAIR sensitizes
hepatocellular carcinoma cell to cisplatin by suppressing the
STAT3/ABCB1 signaling pathway. Oncol Lett. 14:7986–7992.
2017.PubMed/NCBI
|
|
59
|
Ji P, Diederichs S, Wang W, Boing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Tian X and Xu G: Clinical value of lncRNA
MALAT1 as a prognostic marker in human cancer: Systematic review
and meta-analysis. BMJ Open. 5:e0086532015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
over-expression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar
|
|
62
|
Yuan P, Cao W, Zang Q, Li G, Guo X and Fan
J: The HIF-2a-MALAT1-miR-216b axis regulates multi-drug resistance
of hepatocellular carcinoma cells via modulating autophagy. Biochem
Biophys Res Commun. 478:1067–1073. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chen L, Yao H, Wang K and Liu X: Long
non-coding RNA MALAT1 regulates ZEB1 expression by sponging
miR-143-3p and promotes hepatocellular carcinoma progression. J
Cell Biochem. 118:4836–4843. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li C, Miao R, Liu S, Wan Y, Zhang S, Deng
Y, Bi J, Qu K, Zhang J and Liu C: Down-regulation of miR-146b-5p by
long noncoding RNA MALAT1 in hepatocellular carcinoma promotes
cancer growth and metastasis. Oncotarget. 8:28683–28695.
2017.PubMed/NCBI
|
|
65
|
Huang Z, Huang L, Shen S, Li J, Lu H, Mo
W, Dang Y, Luo D, Chen G and Feng Z: Sp1 cooperates with Sp3 to
upregulate MALAT1 expression in human hepatocellular carcinoma.
Oncol Rep. 34:2403–2412. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
He B, Peng F, Li W and Jiang Y:
Interaction of lncRNA-MALAT1 and miR-124 regulates HBx-induced
cancer stem cell properties in HepG2 through PI3K/Akt signaling. J
Cell Biochem. 120:2908–2918. 2019. View Article : Google Scholar
|
|
67
|
Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW,
Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, et al: Rapid identification
of UCA1 as a very sensitive and specific unique marker for human
bladder carcinoma. Clin Cancer Res. 12:4851–4858. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Xue M, Chen W and Li X: Urothelial cancer
associated 1: A long noncoding RNA with a crucial role in cancer. J
Cancer Res Clin Oncol. 142:1407–1419. 2016. View Article : Google Scholar
|
|
69
|
Wang H, Guan Z, He K, Qian J, Cao J and
Teng L: LncRNA UCA1 in anti-cancer drug resistance. Oncotarget.
8:64638–64650. 2017.PubMed/NCBI
|
|
70
|
Wang F, Ying HQ, He BS, Pan YQ, Deng QW,
Sun HL, Chen J, Liu X and Wang SK: Upregulated lncRNA-UCA1
contributes to progression of hepatocellular carcinoma through
inhibition of miR-216b and activation of FGFR1/ERK signaling
pathway. Oncotarget. 6:7899–7917. 2015.PubMed/NCBI
|
|
71
|
Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu
M and Mo YY: Long non-coding RNA UCA1 promotes breast tumor growth
by suppression of p27 (Kip1). Cell Death Dis. 5:e10082014.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang Y, Li Z, Zhang Y, Zhong Q, Chen Q
and Zhang L: Molecular mechanism of HEIH and HULC in the
proliferation and invasion of hepatoma cells. Int J Clin Exp Med.
8:12956–12962. 2015.PubMed/NCBI
|
|
73
|
Zhang Q, Matsuura K, Kleiner DE, Zamboni
F, Alter HJ and Farci P: Analysis of long noncoding RNA expression
in hepatocellular carcinoma of different viral etiology. J Transl
Med. 14:3282016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yu Y, Yang J, Li Q, Xu B, Lian Y and Miao
L: LINC00152: A pivotal oncogenic long non-coding RNA in human
cancers. Cell Prolif. 50:2017. View Article : Google Scholar
|
|
75
|
Neumann O, Kesselmeier M, Geffers R,
Pellegrino R, Radlwimmer B, Hoffmann K, Ehemann V, Schemmer P,
Schirmacher P, Lorenzo Bermejo J and Longerich T: Methylome
analysis and integrative profiling of human HCCs identify novel
protumorigenic factors. Hepatology. 56:1817–1827. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lian Y, Cai Z, Gong H, Xue S, Wu D and
Wang K: HOTTIP: A critical oncogenic long non-coding RNA in human
cancers. Mol Biosyst. 12:3247–3253. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zhang Y, Huang JC, Cai KT, Yu XB, Chen YR,
Pan WY, He ZL, Lv J, Feng ZB and Chen G: Long noncoding RNA HOTTIP
promotes hepatocellular carcinoma tumorigenesis and development: A
comprehensive investigation based on bioinformatics, qRTPCR and
metaanalysis of 393 cases. Int J Oncol. 51:1705–1721. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ge Y, Yan X, Jin Y, Yang X, Yu X, Zhou L,
Han S, Yuan Q and Yang M: MiRNA-192 [corrected] and miRNA-204
directly suppress lncRNA HOTTIP and Interrupt GLS1-mediated
glutaminolysis in hepatocellular carcinoma. PLoS Genet.
11:e10057262015. View Article : Google Scholar
|
|
79
|
Brechot C, Gozuacik D, Murakami Y and
Paterlini-Brechot P: Molecular bases for the development of
hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC).
Semin Cancer Biol. 10:211–231. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Lau CC, Sun T, Ching AK, He M, Li JW, Wong
AM, Co NN, Chan AW, Li PS, Lung RW, et al: Viral-human chimeric
transcript predisposes risk to liver cancer development and
progression. Cancer Cell. 25:335–349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ding SL, Yang ZW, Wang J, Zhang XL, Chen
XM and Lu FM: Integrative analysis of aberrant Wnt signaling in
hepatitis B virus-related hepatocellular carcinoma. World J
Gastroenterol. 21:6317–6328. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Pasmant E, Laurendeau I, Hèron D, Vidaud
M, Vidaud D and Bièche I: Characterization of a germ-line deletion,
including the entire INK4/ARF locus, in a melanoma-neural system
tumor family: Identification of ANRIL, an antisense noncoding RNA
whose expression coclusters with ARF. Cancer Res. 67:3963–3969.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Li Z, Yu X and Shen J: ANRIL: A pivotal
tumor suppressor long non-coding RNA in human cancers. Tumour Biol.
37:5657–5661. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Hua L, Wang CY, Yao KH, Chen JT, Zhang JJ
and Ma WL: High expression of long non-coding RNA ANRIL is
associated with poor prognosis in hepatocellular carcinoma. Int J
Clin Exp Pathol. 8:3076–3082. 2015.PubMed/NCBI
|
|
85
|
Magkoufopoulou C, Claessen SM, Jennen DG,
Kleinjans JC and van Delft JH: Comparison of phenotypic and
transcriptomic effects of false-positive genotoxins, true
genotoxins and non-genotoxins using HepG2 cells. Mutagenesis.
26:593–604. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Huang JL, Ren TY, Cao SW, Zheng SH, Hu XM,
Hu YW, Lin L, Chen J, Zheng L and Wang Q: HBx-related long
non-coding RNA DBH-AS1 promotes cell proliferation and survival by
activating MAPK signaling in hepatocellular carcinoma. Oncotarget.
6:33791–33804. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Lv D, Wang Y, Zhang Y, Cui P and Xu Y:
Downregulated long non-coding RNA DREH promotes cell proliferation
in hepatitis B virus-associated hepatocellular carcinoma. Oncol
Lett. 14:2025–2032. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Niu J, Lin Y, Liu P, Yu Y, Su C and Wang
X: Microarray analysis on the lncRNA expression profile in male
hepatocelluar carcinoma patients with chronic hepatitis B virus
infection. Oncotarget. 7:76169–76180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Fan H, Zhang Q, Zhao X, Lv P, Liu M and
Tang H: Transcriptomic profiling of long non-coding RNAs in
hepatitis B virus-related hepatocellular carcinoma. Oncotarget.
8:65421–65434. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Luo HL, Chen J, Luo T, Wu FX, Liu JJ, Wang
HF, Chen M, Li LQ and Li H: Downregulation of macrophage-derived
T-UCR uc.306 associates with poor prognosis in hepatocellular
carcinoma. Cell Physiol Biochem. 42:1526–1539. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Gong X, Wei W, Chen L, Xia Z and Yu C:
Comprehensive analysis of long non-coding RNA expression profiles
in hepatitis B virus-related hepatocellular carcinoma. Oncotarget.
7:42422–42430. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Xia J, Inagaki Y, Sawakami T, Song P, Cai
Y, Hasegawa K, Sakamoto Y, Akimitsu N, Tang W and Kokudo N:
Preliminary investigation of five novel long non-coding RNAs in
hepatocel-lular carcinoma cell lines. Biosci Trends. 10:315–319.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Yu S, Li N, Huang Z, Chen R, Yi P, Kang R,
Tang D, Hu X and Fan X: A novel lncRNA, TCONS_00006195, represses
hepatocellular carcinoma progression by inhibiting enzymatic
activity of ENO1. Cell Death Dis. 9:11842018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Hu Z, Huang P, Yan Y, Zhou Z, Wang J and
Wu G: Hepatitis B virus X protein related lncRNA WEE2-AS1 promotes
hepatocellular carcinoma proliferation and invasion. Biochem
Biophys Res Commun. 508:79–86. 2019. View Article : Google Scholar
|
|
95
|
Huang JF, Wang Y, Liu F, Liu Y, Zhao CX,
Guo YJ and Sun SH: EVI1 promotes cell proliferation in HBx-induced
hepatocarcinogenesis as a critical transcription factor regulating
lncRNAs. Oncotarget. 7:21887–21899. 2016.PubMed/NCBI
|
|
96
|
Yu TT, Xu XM, Hu Y, Deng JJ, Ge W, Han NN
and Zhang MX: Long noncoding RNAs in hepatitis B virus-related
hepatocellular carcinoma. World J Gastroenterol. 21:7208–7217.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Pan YF, Qin T, Feng L and Yu ZJ:
Expression profile of altered long non-coding RNAs in patients with
HBV-associated hepatocellular carcinoma. J Huazhong Univ Sci
Technolog Med Sci. 33:96–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Kuang XY, Li N, Fu YM, Li J and Fan XG:
Expression profiles of long non-coding RNAs in human liver cell
line LO2 with stable expression of hepatitis B x gene. Zhonghua Gan
Zang Bing Za Zhi. 24:417–421. 2016.In Chinese. PubMed/NCBI
|
|
99
|
Jin Y, Wu D, Yang W, Weng M, Li Y, Wang X,
Zhang X, Jin X and Wang T: Hepatitis B virus x protein induces
epithelial-mesenchymal transition of hepatocellular carcinoma cells
by regulating long non-coding RNA. Virol J. 14:2382017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Li H, Zhao X, Li C, Sheng C and Bai Z:
Integrated analysis of lncRNA-associated ceRNA network reveals
potential biomarkers for the prognosis of hepatitis B virus-related
hepato-cellular carcinoma. Cancer Manag Res. 11:877–897. 2019.
View Article : Google Scholar :
|
|
101
|
Gu JX, Zhang X, Miao RC, Xiang XH, Fu YN,
Zhang JY, Liu C and Qu K: Six-long non-coding RNA signature
predicts recurrence-free survival in hepatocellular carcinoma.
World J Gastroenterol. 25:220–232. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Toyoda H, Kumada T, Tada T, Sone Y,
Kaneoka Y and Maeda A: Tumor markers for hepatocellular carcinoma:
Simple and significant predictors of outcome in patients with HCC.
Liver Cancer. 4:126–136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zhou L, Liu J and Luo F: Serum tumor
markers for detection of hepatocellular carcinoma. World J
Gastroenterol. 12:1175–1181. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Xie H, Ma H and Zhou D: Plasma HULC as a
promising novel biomarker for the detection of hepatocellular
carcinoma. Biomed Res Int. 2013:1361062013. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Li J, Wang X, Tang J, Jiang R, Zhang W, Ji
J and Sun B: HULC and Linc00152 Act as novel biomarkers in
predicting diagnosis of hepatocellular carcinoma. Cell Physiol
Biochem. 37:687–696. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Konishi H, Ichikawa D, Yamamoto Y, Arita
T, Shoda K, Hiramoto H, Hamada J, Itoh H, Fujita Y, Komatsu S, et
al: Plasma level of metastasis-associated lung adenocarcinoma
transcript 1 is associated with liver damage and predicts
development of hepatocellular carcinoma. Cancer Sci. 107:149–154.
2016. View Article : Google Scholar :
|
|
107
|
Yuan W, Sun Y, Liu L, Zhou B, Wang S and
Gu D: Circulating LncRNAs serve as diagnostic markers for
hepatocellular carcinoma. Cell Physiol Biochem. 44:125–132. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Yu J, Han J, Zhang J, Li G, Liu H, Cui X,
Xu Y, Li T, Liu J and Wang C: The long noncoding RNAs PVT1 and
uc002mbe.2 in sera provide a new supplementary method for
hepatocellular carcinoma diagnosis. Medicine (Baltimore).
95:e44362016. View Article : Google Scholar
|
|
109
|
Lu J, Xie F, Geng L, Shen W, Sui C and
Yang J: Investigation of serum lncRNA-uc003wbd and lncRNA-AF085935
expression profile in patients with hepatocellular carcinoma and
HBV. Tumour Biol. 36:3231–3236. 2015. View Article : Google Scholar
|
|
110
|
Wang K, Guo WX, Li N, Gao CF, Shi J, Tang
YF, Shen F, Wu MC, Liu SR and Cheng SQ: Serum LncRNAs profiles
serve as novel potential biomarkers for the diagnosis of
HBV-positive hepatocellular carcinoma. PLoS One. 10:e01449342015.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Freedman ML, Monteiro AN, Gayther SA,
Coetzee GA, Risch A, Plass C, Casey G, De Biasi M, Carlson C,
Duggan D, et al: Principles for the post-GWAS functional
characterization of cancer risk loci. Nat Genet. 43:513–518. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Hindorff LA, Sethupathy P, Junkins HA,
Ramos EM, Mehta JP, Collins FS and Manolio TA: Potential etiologic
and functional implications of genome-wide association loci for
human diseases and traits. Proc Natl Acad Sci USA. 106:9362–9367.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Rao A, Rajkumar T and Mani S: Perspectives
of long non-coding RNAs in cancer. Mol Biol Rep. 44:203–218. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J,
Zhang Y, Chen J, Shen H and Hu Z: A genetic variant in long
non-coding RNA HULC contributes to risk of HBV-related
hepatocellular carcinoma in a Chinese population. PLoS One.
7:e351452012. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Li H, Tang XM, Liu Y, Li W, Chen Q and Pan
Y: Association of functional genetic variants of HOTAIR with
hepatocellular carcinoma (HCC) susceptibility in a Chinese
population. Cell Physiol Biochem. 44:447–454. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Chang L, Li C, Lan T, Wu L, Yuan Y, Liu Q
and Liu Z: Decreased expression of long non-coding RNA GAS5
indicates a poor prognosis and promotes cell proliferation and
invasion in hepatocellular carcinoma by regulating vimentin. Mol
Med Rep. 13:1541–1550. 2016. View Article : Google Scholar :
|
|
117
|
Tu ZQ, Li RJ, Mei JZ and Li XH:
Down-regulation of long non-coding RNA GAS5 is associated with the
prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol.
7:4303–4309. 2014.PubMed/NCBI
|
|
118
|
Tao R, Hu S, Wang S, Zhou X, Zhang Q, Wang
C, Zhao X, Zhou W, Zhang S, Li C, et al: Association between indel
polymorphism in the promoter region of lncRNA GAS5 and the risk of
hepatocellular carcinoma. Carcinogenesis. 36:1136–1143. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Zhu Z, Gao X, He Y, Zhao H, Yu Q, Jiang D,
Zhang P, Ma X, Huang H, Dong D, et al: An insertion/deletion
polymorphism within RERT-lncRNA modulates hepatocellular carcinoma
risk. Cancer Res. 72:6163–6172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Wan J, Huang M, Zhao H, Wang C, Zhao X,
Jiang X, Bian S, He Y and Gao Y: A novel tetranucleotide repeat
polymorphism within KCNQ1OT1 confers risk for hepatocellular
carcinoma. DNA Cell Biol. 32:628–634. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Lin YY, Yu MW, Lin SM, Lee SD, Chen CL,
Chen DS and Chen PJ: Genome-wide association analysis identifies a
GLUL haplotype for familial hepatitis B virus-related
hepatocellular carcinoma. Cancer. 123:3966–3976. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Goring HH, Curran JE, Johnson MP, Dyer TD,
Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL,
Comuzzie AG, et al: Discovery of expression QTLs using large-scale
transcriptional profiling in human lymphocytes. Nat Genet.
39:1208–1216. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
123
|
Yin J, Wen J, Hang D, Han J, Jiang J, Song
C, Liu Y, Liu J, Liu L, Zhu L, et al: Expression quantitative trait
loci for CARD8 contributes to risk of two infection-related
cancers-hepatocellular carcinoma and cervical cancer. PLoS One.
10:e01323522015. View Article : Google Scholar
|
|
124
|
Wen J, Liu Y, Liu J, Liu L, Song C, Han J,
Zhu L, Wang C, Chen J, Zhai X, et al: Expression quantitative trait
loci in long non-coding RNA ZNRD1-AS1 influence both HBV infection
and hepatocellular carcinoma development. Mol Carcinog.
54:1275–1282. 2015. View Article : Google Scholar
|
|
125
|
Liu Z, Song C, Wen J, Xu L, Liu Y, Zhu J,
Zhu L, Hu Z, Ma H and Liu L: Hepatitis B virus genotypes,
expression quantitative trait loci for ZNRD1-AS1 and their
interactions in hepatocellular carcinoma. Oncotarget.
7:44076–44083. 2016.PubMed/NCBI
|
|
126
|
Song C, Liu Y, Xu L, Wen J, Jiang D, Chen
J, Zhai X, Hu Z, Liu L and Liu J: Hepatitis B virus mutations,
expression quantitative trait loci for PTPN12, and their
interactions in hepatocellular carcinoma. Cancer Med. 5:1687–1693.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Wen J, Song C, Liu J, Chen J, Zhai X and
Hu Z: Expression quantitative trait loci for TNFRSF10 influence
both HBV infection and hepatocellular carcinoma development. J Med
Virol. 88:474–480. 2016. View Article : Google Scholar
|
|
128
|
Ma S, Yang J, Song C, Ge Z, Zhou J, Zhang
G and Hu Z: Expression quantitative trait loci for PAX8 contributes
to the prognosis of hepatocellular carcinoma. PLoS One.
12:e01737002017. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Rion N and Rüegg MA: LncRNA-encoded
peptides: More than translational noise? Cell Res. 27:604–605.
2017. View Article : Google Scholar : PubMed/NCBI
|