Introduction
Bladder cancer is one of the most common types of
urinary cancer. The incidence of bladder cancer has been steadily
increasing and there was an estimated 81,190 new bladder cancer
cases diagnosed in the USA in 2018 (1). Furthermore, the risk of recurrence
and progression remains high (2).
Therefore, there is an urgent need to find stable new biomarkers
and to identify the mechanisms of bladder cancer progression in
order to support targeted bladder tumour therapy (3).
Cells can communicate with adjacent cells or distant
cells by secreting extracellular vesicles (EVs), which exist in a
variety of sizes ranging between 100 and 1,000 nm in diameter.
Exosomes are produced in EVs and secreted when the vesicles fuse
with the cell membrane. Exosomes are carriers with diameters of
<150 nm and are rich in proteins, lipids, RNA and other
components (4). As exosomes carry
surface molecules, they can induce signal transduction through
receptor-ligand interactions; alternatively, they can be
internalized by endocytosis and/or phagocytosis and can even fuse
with target cell membranes to deliver their contents into the
cytoplasm (5). Therefore, exosomes
derived from donor cells are capable of altering the physiological
states of recipient cells. In particular, exosomes derived from
non-tumour cells can promote adaptation of disseminated cancer
cells to the external environment, thereby reflecting the
occurrence of the dynamic changes in the metastatic
microenvironment (6,7).
Macrophages are one of the most abundant types of
stromal cells found in the tumour microenvironment (TME).
Macrophages can be induced to exhibit an immunological M1 phenotype
that inhibits tumours or an M2 phenotype that promotes tumour
inflammation/immunosuppression (8). Tumour-associated macrophages (TAMs),
which are considered to be similar to M2 macrophages, promote
tumour metastasis by enhancing tumour cell motility, promoting
angiogenesis and degrading the extracellular basal layer (9). In recent years, TAMs have been
extensively studied and are now considered to be important factors
in tumour progression (10,11).
Several methods are used to study macrophages in vitro,
among which evaluation of primary peripheral blood mononuclear
cells and assessment of variations in differentiation among
monocyte lines are the most common methods (12). TAM populations are commonly used to
mimic the functions of macrophage populations because primary
tissue-derived macrophages are difficult to grow and expand in
vitro (12,13). Currently, the driving force for the
differentiation of TAMs remains largely unknown, and it is unclear
whether tumour-derived exosomes are functionally essential for
changes in the TME. The present study identified a
microenvironmental mechanism that forms TAMs through
exosome-mediated communication between cancer cells and immune
cells.
Materials and methods
Cell culture
The human bladder cancer cell line T24 was a gift
from the Molecular Tumour and Epigenetic Experiment of Chongqing
Medical University (Chonqing, China). THP-1 human acute monocytic
leukaemia cells were kindly provided by the Stem Cell Bank, Chinese
Academy of Sciences. All cell lines were maintained in RPMI-1640
medium (cat. no. 10-040-CVR; Corning, Inc.) supplemented with 10%
EV-depleted foetal bovine serum (FBS; cat. no. SA102.02; CellMax),
100 U/ml penicillin and 100 µg/ml streptomycin (cat. no.
C0222; Beyotime Institute of Biotechnology), and bovine EV-depleted
medium was obtained by overnight ultracentrifugation at 100,000 × g
at 4°C (14). The cells were
cultured at 37°C in a standard humidified incubator with 5%
CO2. All experiments were performed when the cells
reached 70-80% confluency.
Macrophage differentiation
To generate M0 macrophages, monocytic THP-1 cells
were cultured in complete medium (RPMI-1640 with 10% EV-depleted
FBS) at a relatively low cell density (2.5×105 cells/ml)
with 100 ng/ml phorbol 12-myristate 13-acetate (PMA; cat. no.
P1585; Sigma-Aldrich; Merck KGaA) for 24 h for differentiation into
resting (M0) macrophages (15).
Flow cytometry
Single-cell suspensions were stained with FITC
anti-human CD11b and CD14 antibodies (1:20; cat. nos. 301329 and
301803, respectively; BioLegend, Inc.) for 30 min at 37°C. FITC
mouse IgG1 and IgG2a antibodies (1:20; cat. nos. 400109 and 400207;
BioLegend, Inc.) were used to stain single cell suspensions for 30
min at 37°C. These control antibodies were the isotype controls for
CD11b and CD14, respectively, and were used as a reference in each
experiment. In total, ~1×105 events were collected for
each sample with a Becton Dickinson FACS Calibur system (BD
Biosciences) and FlowJo software 7.6.1 (FlowJo, LLC) was used for
analysis.
Exosome isolation
According to a previously published protocol
(16), exosomes were collected by
differential ultracen-trifugation under appropriately optimized
conditions. Briefly, T24 cells were cultured for 48 h in complete
RPMI-1640 medium containing 10% EV-depleted FBS. The conditioned
medium was collected, centrifuged at 300 × g for 10 min at 4°C to
remove free cells and then centrifuged at 2,000 × g for 10 min at
4°C to remove dead cells. Next, centrifugation was performed at
10,000 × g for 30 min at 4°C to remove cell debris. The supernatant
was collected and filtered through a 0.22-µm filter (cat.
no. SLGP033RB; EMD Millipore) to remove contaminating apoptotic
bodies and microvesicles. Then, the clarified supernatant was
ultracentrifuged at 100,000 × g for 70 min at 4°C to precipitate
the exosomes. The supernatant was carefully removed and stored at
-20°C as the non-exosome-conditioned medium (non-exo-CM), and the
crude precipitate containing the exosomes was washed with 1 ml PBS.
Ultracentrifugation was performed again at 100,000 × g for 70 min
at 4°C. The supernatant was aspirated, and the exosome pellets were
resuspended in 200 µl PBS and stored at −20°C.
Transmission electron microscopy
(TEM)
For TEM, freshly isolated exosomes were used; these
were diluted with nuclease-free water to maximize image quality. A
10-µl suspension with exosomes was adsorbed onto a
carbon-coated 200-mesh nickel grid for 10 min at room temperature,
and then the excess was blotted with filter paper. The sample was
negatively stained with 10 µl 10% phosphotungstic acid (cat.
no. P4006; Sigma-Aldrich; Merck KGaA) for 10 sec at room
temperature, and excess stain was removed with filter paper. The
mesh was thoroughly dried before viewing. TEM sample preparation
and imaging were performed at the Electron Microscopy Laboratory of
Chongqing Medical University.
Nanoparticle tracking analysis (NTA)
NTA assays were performed using a ZetaView PMX 110
(Particle Metrix) and its corresponding software (ZetaView 8.02.28;
Particle Metrix) (17,18). For NTA, exosomes were diluted in 1x
PBS, and the resuspension volumes and dilution factors were used to
calculate the total number of isolated exosomes. A 10-µl
sample was transferred to 40 ml distilled water and resuspended.
The suspension was filtered through a 450-nm filter to separate the
exosomes from the larger particles, and the resulting filtered
suspension was used for particle measurement. Samples were taken at
11 different locations, and two cycles of readings were performed
at each location. After automatically analysing all locations and
removing any anomalous locations, the mean, median, diameter size
and sample concentration were calculated using optimized machine
software. The data obtained with the ZetaView instrument were
analysed using the corresponding software ZetaView 8.02.28.
Exosome labelling and tracking
For the exosome uptake experiment, exosomes were
labelled with a PKH67 Fluorescent Cell Ligation kit (Sigma-Aldrich;
Merck KGaA) according to the manufacturer's instructions. The
exosomes were then washed with 0.5% BSA/PBS (cat. no. P0007;
Beyotime Institute of Biotechnology) to remove excess dye. The
labelled exosomes were again ultracentrifuged at 100,000 x g for 1
h at 4°C to remove residual dye; then, the exosomes (at a
concentration of 10 µg/ml) were incubated with macrophages
at 37°C for 8 h in the dark. The cell nuclei and cytoskeleton were
separately stained with DAPI (cat. no. C1005; Beyotime Institute of
Biotechnology) for 5 min at room temperature and F-actin (cat. no.
KA4118; AmyJet Scientific) for 30 min at room temperature and then
examined by laser scanning confocal microscopy (magnification,
×800).
ELISA
Conditioned medium was separately collected from the
control (macrophages cultured in the normal medium),
non-exo-CM-treated (macrophages cultured in the
non-exosome-conditioned medium) and exo-treated (macrophages
treated with 10 µg/ml exosomes) groups, centrifuged at 1,000
× g for 5 min at 4°C, and stored at −20°C until further analysis.
Subsequently, IL-10 and IL-12 (p70) expression levels in the
conditioned medium were quantified using ELISA kits (cat. nos.
EK0416 and EK0421, respectively; Wuhan Boster Biological
Technology, Ltd.) according to the manufacturer's instructions.
MicroRNA (miRNA or miR) transfection
Macrophages were transfected with miR-21-5p mimics,
miR-21-5p inhibitor, mimic negative control (NC) or inhibitor NC
(Shanghai GenePharma Co. Ltd.) using Lipofectamine® 2000
(cat. no. 11668019; Invitrogen: Thermo Fisher Scientific, Inc.) at
a final concentration of 100 nM, and the same conditions were
applied for each transfection experiment. The efficiency of cell
transfection was assessed by reverse transcription-quantitative PCR
(RT-qPCR) after 48 h. Treatment with SF1670 (cat. no. S7310;
Selleck Chemicals; 500 or 1 µM) for 24 h at 37°C was
employed to inhibit phosphatase and tensin homolog (PTEN) after
transfecting cells with miR-21-5p inhibitor. The sequences of
mimic, inhibitor and negative controls were as follows: miR-21-5p
mimic, 5'-UAG CUU AUC AGA CUG AUG UUG A-3'; miR-21-5p inhibitor,
5'-CAG UAC UUU UGU GUA GUA CAA-3'; mimic negative control, 5'-UUC
UCC GAA CGU GUC ACG UTT-3'; and inhibitor negative control, 5'-CAG
UAC UUU UGU GUA GUA CAA-3'.
Co-culture
For experiments involving co-culture of T24 cells
and M0 macrophages, T24 cells were seeded onto 0.4 µm pore
size Transwell culture inserts (cat. no. 3450; Corning, Inc.) and
transfected with NC or miR-21-5p inhibitor. After 24 h, the inserts
were transferred to dishes seeded with M0 macrophages and
co-cultured for 24 h.
Luciferase reporter assay
The miR-21-5p target gene, PTEN, was predicted using
TargetScan software (version 7.2; http://www.targetscan.org/). These sequence of the
wild-type miR-21 binding site on PTEN and a binding site mutant
sequence were inserted into pmirGLO vectors (Shanghai GenePharma
Co., Ltd.). Lipofectamine® 2000 was then used for
transfection of the cells, which were seeded in 6-well plates for
24 h. According to the manufacturer's instructions, luciferase
activity was measured using a dual luciferase reporter gene assay
kit (cat. no. E1910; Promega Corporation). Renilla
luciferase activity was used to normalize to firefly luciferase
activity.
Migration and invasion assays
Cell migration and invasion assays were performed on
24-well Transwell cell culture chambers with 8-µm wells
pre-coated or not pre-coated with Matrigel basement membrane gel
(cat. no. 354234; Corning, Inc.). T24 cells (8×104
cells) in RPMI-1640 medium were plated into the upper chambers with
or without Matrigel (40 µl/well). Each different cell
supernatant (from T24 cells, untreated macrophages,
non-exo-CM-treated macrophages, exo-treated macrophages, negative
control macrophages, miR-21-5p mimic-transfected macrophages or
miR-21-5p inhibitor-transfected macrophages) was collected, diluted
with fresh FBS media (RPMI-1640 with 10% FBS) (1:1), and plated
into each lower chamber. For migration assays, T24 cells incubated
at 37°C for 8 h. For invasion assays, the incubation time was 24 h.
The membranes were collected and stained with 0.5% crystal violet
(cat. no. C0121; Beyotime Institute of Biotechnology) for 15 min at
room temperature. Migrating and invading cells were observed under
an optical microscope (magnification, ×100). Five fields of view
were randomly selected to calculate the number of cells that
migrated and invaded.
RT-qPCR
Total RNA was isolated from cells and exosomes using
TRIzol (cat. no. 15596026; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. RNA was quantified using
a Nanodrop ND-1000 (Thermo Fisher Scientific, Inc.). RT was carried
out using a PrimeScript™ RT Reagent kit with gDNA Eraser (Perfect
Real Time; cat. no. RR047A; Takara Bio, Inc.). qPCR was performed
on a CFX96 Real-Time PCR Detection system (Bio-Rad Laboratories,
Inc.) using TB Green Premix Ex Taq II (cat. no. RR820A; Takara Bio,
Inc.), and the expression of mRNA or miRNA was normalized to
β-actin or U6, respectively. The thermocycling parameters were as
follows: i) initial denaturation at 95°C for 30 sec; ii) 40 cycles
of denaturation at 95°C for 5 sec and 60°C for 30 sec; and
(3) annealing/extension at 65°C
for 5 sec with a 0.5°C increase for each repetition (60 cycles to
95°C). The results were analysed using the 2−ΔΔCq method
(19). All primers are listed in
Table I.
 | Table IPrimers used for reverse
transcription-quantitative PCR. |
Table I
Primers used for reverse
transcription-quantitative PCR.
| Gene | Primer
sequence |
|---|
| miR-21-5p | Forward:
5′-ACACTCCAGCTGGGTAGCTTATCAGACTGA-3′ |
| Reverse:
5′-TGGTGTCGTGGAGTCG-3′ |
| PTEN | Forward:
5′-TGGATTCGACTTAGACTTGACCT-3′ |
| Reverse:
5′-GGTGGGTTATGGTCTTCAAAAGG-3′ |
| IL-10 | Forward:
5′-GACTTTAAGGGTTACCTGGGTTG-3′ |
| Reverse:
5′-TCACATGCGCCTTGATGTCTG-3′ |
| TGF-β | Forward:
5′-GGTACCTGAACCCGTGTTGCT-3′ |
| Reverse:
5′-TGTTGCTGTATTTCTGGTAACAGCTC-3′ |
| β-actin | Forward:
5′-GTCTTCCCCTCCATCGTG-3′ |
| Reverse:
5′-AGGGTGAGGATGCCTCTCTT-3′ |
| U6 | Forward:
5′-CTCGCTTCGGCAGCACA-3′ |
| Reverse:
5′-AACGCTTCACGAATTTGCGT-3′ |
Western blot analysis
Cells and exosomes were lysed with RIPA buffer (cat.
no. P0013B; Beyotime Institute of Biotechnology). A BCA protein
assay kit (cat. no. P0010; Beyotime Institute of Biotechnology) was
used to detect protein concentrations, and a 12% sodium dodecyl
sulfate-polyacrylamide gel was applied for total protein (30
µg) separation. The proteins in the gel were transferred to
a 0.45-µM pore size PVDF membrane (cat. no. IPVH00010; EMD
Millipore) via wet electrophoretic transfer. The western blots were
blocked with skim milk powder for 1 h at room temperature and
incubated overnight at 4°C with anti-CD9 (cat. no. ab92726; Abcam),
anti-CD63 (cat. no. ab134045; Abcam), anti-tumour susceptibility
gene 101 (TSG101; cat. no. ab125011; Abcam), anti-PI3 kinase p85
(cat. no. 4257T; Cell Signalling Technology, Inc.), anti-PTEN (cat.
no. 9559T; Cell Signalling Technology, Inc.), anti-AKT (cat. no.
4691T; Cell Signalling Technology, Inc.), anti-phosphorylated
(p)-AKT (cat. no. 2965T; Cell Signalling Technology, Inc.),
anti-STAT3 (cat. no. 4904T; Cell Signalling Technology, Inc.) and
anti-p-STAT3 (cat. no. 9145T; Cell Signalling Technology, Inc.)
antibodies, all diluted to 1:1,000. An anti-b-actin antibody (cat.
no. 4970S; 1:2,000; Cell Signalling Technology, Inc.) and
anti-GAPDH (cat. no. 10494-1-AP; 1:20,000; ProteinTech Group, Inc.)
were used as internal loading controls. Following antibody
incubation, the protein bands were washed three times with
TBS-Tween 20 on an orbital shaker in 10 min intervals.
Subsequently, the membranes were incubated with a horseradish
peroxidase-conjugated secondary antibody (cat. no. A0208; 1:3,000;
Beyotime Institute of Biotechnology) for 1 h at room temperature
and then developed using an electrochemiluminescence western blot
detection reagent (cat. no. WBKLS0010; EMD Millipore). Fusion FX
Spectra software (version 7; Vilber) was used to detect the bands.
The protein expression levels in the various groups were compared
with those in the control group based on the relative intensities
of the bands.
Statistical analysis
Three independent experiments were performed and the
experimental results are shown as the mean ± standard deviation.
The data were compared using Student's t-test for two groups. The
differences among multiple groups were assessed using one-way ANOVA
followed by a post-hoc Bonferroni test. P<0.05 was considered to
indicate a statistically significant difference. All statistical
analyses were performed with SPSS statistical software 19.0 (IBM
Corp.).
Results
Morphological characteristics of THP-1
cells after differentiation
To obtain M0 macrophages, PMA was used to induce
polarization of THP-1 cells as described previously (15,20).
Macrophage-like morphological changes were observed in THP-1 cells
after stimulation with PMA for 24 h (Fig. 1A). The cytoplasmic volume was
increased in human monocyte-derived macrophages compared with
monocytes. Compared with no treatment, PMA treatment of THP-1 cells
results in a more mature phenotype with lower proliferation rates,
higher adherence levels and increased expression of cell surface
markers (13). The present study
analysed the expression levels of CD11b and CD14, as CD11b and CD14
are reported to be common markers for the differentiation of
monocytes into macrophages (15,20).
In the absence of any stimulation, 19.44 and 69.46% of cultured
THP-1 cells were positive for CD11b and CD14, respectively. After
treatment with PMA, marked increases in the numbers of CD11b- and
CD14-positive cells (65.46 and 90.72%) were observed (Fig. 1B). These results showed that M0
macrophages were successfully obtained from THP-1 cells and could
be used in subsequent experiments, which was consistent with other
studies (21,22).
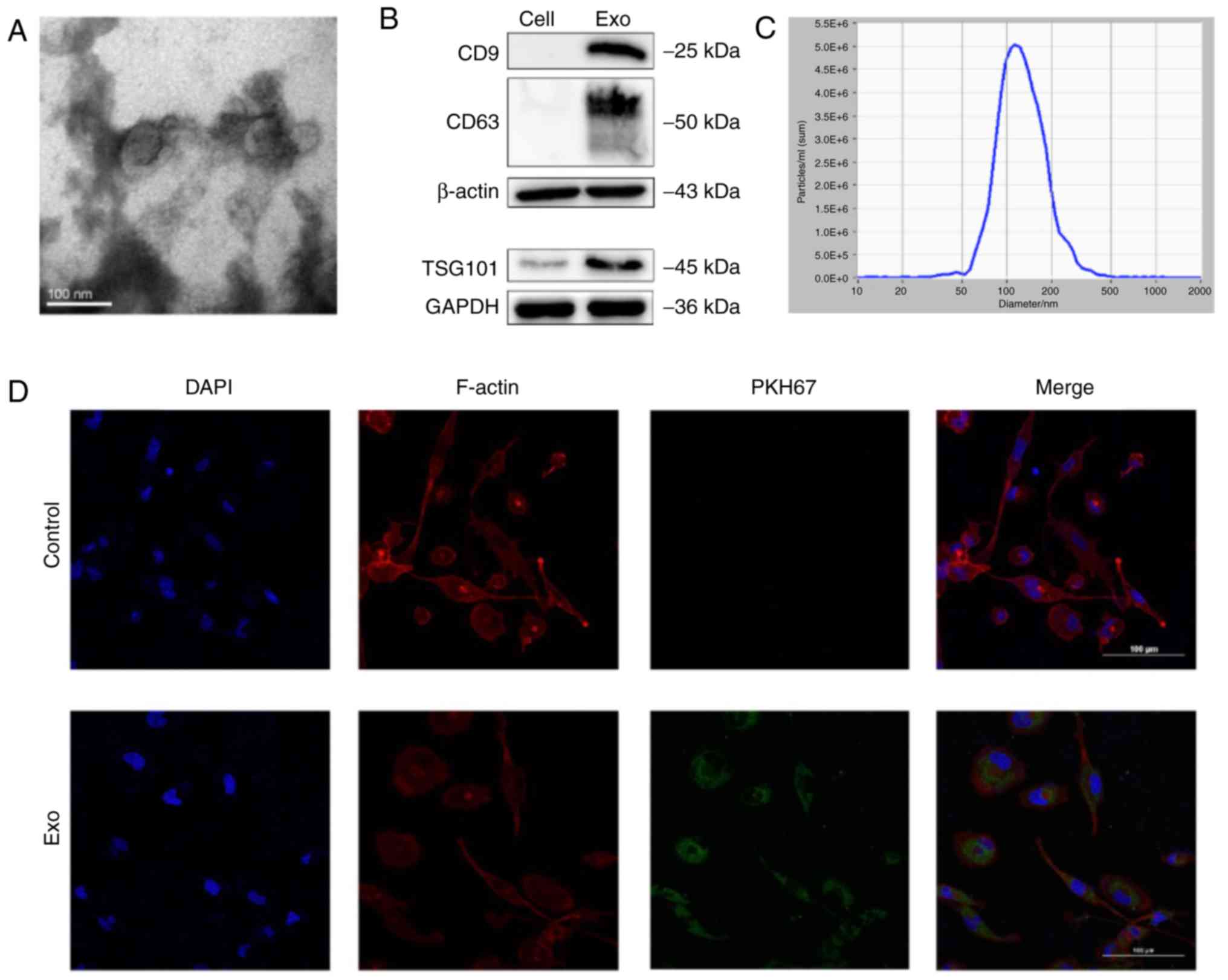
Identification of exosomes derived from
T24 cells and uptake of the exosomes in macrophages
T24 is the most typically used and representative
cell line model in bladder cancer research, with characteristics of
a short generation time (19 h), high grade, and highly aggressive
and metastatic potential. Therefore, T24 cells were selected as the
representative cell line for subsequent research. Exosomes can
regulate the TME through the transfer of functional small RNAs or
proteins (23). To confirm whether
exosomes derived from T24 cells uptake into macrophages, exosomes
were first collected from conditioned medium of T24 cells. TEM
revealed that the purified exosomes had a cup-shaped morphology
(Fig. 2A). NTA revealed that the
diameters of the particles were within the expected range for
exosomes (40-150 nm) and that the particles expressed the currently
known exosomal markers CD9, CD63 and TSG101, consistent with the
defined features of exosomes (Fig. 2B
and C). To evaluate the biological interactions between
exosomes derived from T24 cells and immune cells, fluorescently
labelled exosomes were cultured with macrophages. After 8 h of
incubation, confocal microscopy revealed the presence of stained
exosomes in the cytoplasm of macrophages (Fig. 2D).
T24 cell-derived exosomes activate a
TAM-like macrophage phenotype that can promote migration and
invasion of T24 cells
To determine whether T24 cell-derived exosomes are
capable of promoting M2 macrophage polarization, the expression of
markers typical of the M2 phenotype was examined. Commonly used
cytokine markers indicating macrophage polarization towards the M2
phenotype include IL-10 and transforming growth factor β (TGF-β)
(24). In addition, a
non-exo-CM-treated group was used to observe the effect of
substances outside exosomes on M2 polarization of macrophages.
RT-qPCR showed that the mRNA levels of IL-10 and TGF-β were
significantly upregulated in exo-treated macrophages compared with
control macrophages, while the levels in non-exo-CM-treated
macrophages did not appear significantly altered (Fig. 3A). M2 macrophages are defined as
having an in vitro phenotype of low IL-12 expression and
high IL-10 expression (25). These
results were further confirmed using ELISA, which revealed the
levels of IL-10 and IL-12 protein secreted into the macrophages
supernatant (Fig. 3B). To further
investigate the effects of exo-treated macrophages on cancer cell
migration and invasion in vitro, macrophages were treated
with exosomes (10 µg/ml) or non-exo-CM for 24 h. Migration
and invasion assays both revealed significant increases in the
numbers of migrated and invasive T24 cells after incubation of the
T24 cells with conditioned medium from exo-treated macrophages
(Fig. 3C). These results suggested
that T24 cell-derived exosomes promoted macrophage polarization
toward the M2 phenotype, thereby enhancing T24 cell migratory and
invasive ability. In addition, the substances outside exosomes do
not participate in M2 differentiation.
 | Figure 3Exosomes derived from T24 cells
induce polarization toward M2 macrophages and can promote bladder
cancer cell migration and invasion. (A) RT-qPCR detection of IL-10
and TGF-β mRNA expression in control, non-exo-CM-treated
macrophages and exo-treated macrophages. (B) IL-10 and IL-12 (p70)
levels measured by ELISA. (C) Migration and invasion assays of T24
cells cultured with supernatants from control, untreated,
non-exo-CM-treated or exo-treated macrophages. Magnification, ×100.
n=3. *P<0.05, **P<0.01,
***P<0.001, ****P<0.01 vs. control
group. RT-qPCR, reverse transcription-quantitative PCR; non-exo-CM,
non-exosome-conditioned medium; exo, exosome. |
miR-21 loaded in T24 cell-derived
exosomes promotes M2 differentiation
Exosomes contain biologically active molecules that
are involved in intercellular communication. Previous studies have
shown that miR-21 is highly expressed in bladder cancer and stromal
cells and is closely related to tumour progression (26). To elucidate whether miR-21 is also
highly expressed in exosomes derived from bladder cancer cells (T24
cells), the expression of miR-21 was examined by RT-qPCR. The
results showed that the expression of miR-21 in T24 cell-derived
exosomes was significantly higher than that in parental cells
(Fig. 4A). It was then examined
whether tumour-derived exosomes could deliver miR-21 to
macrophages. As shown in Fig. 4B,
the level of miR-21 was higher in macrophages treated with T24
cell-derived exosomes compared with in untreated macrophages. To
determine whether T24 cell-derived miR-21 could be directly
transferred from bladder cancer cells to macrophages and affect
macrophage polarization, a co-culture experiment was designed in
which macrophages were co-cultured with T24 cells. RT-qPCR analysis
of miR-21 was performed on the recipient cells. The level of miR-21
was higher in macrophages co-cultured with T24 cells than in
macrophages (Fig. S1A). Compared
with the control, macrophages co-cultured with T24 cells exhibited
significantly higher expression of IL-10 and TGF-β (Fig. S1B). The results suggested that
co-culture with T24 cells promoted macrophage polarization toward
the M2 phenotype. The present study then co-cultured macrophages
with T24 cells that were transiently transfected with a miR-21-5p
inhibitor (anti-miR-21-5p T24 cells). The results showed that
miR-21 expression levels were significantly lower in macrophages
co-cultured with anti-miR-21-5p T24 cells compared with those
co-cultured with T24 cells (Fig.
4C). Furthermore, the mRNA levels of IL-10 and TGF-β were
significantly downregulated in macrophages co-cultured with
anti-miR-21-5p T24 cells compared with those co-cultured with T24
cells (Fig. 4D). The results
showed that co-culture with anti-miR-21-5p T24 cells inhibited M2
phenotypic polarization, indicating that T24 cell-derived miR-21
was a key factor inducing macrophage polarization in thus
co-culture system.
 | Figure 4miR-21 is enriched in T24
cell-derived exosomes and is associated with M2 macrophage
polarization. (A) Relative miR-21 expression in T24 cells and
exosomes. (B) Relative miR-21 expression in macrophages treated
with PBS or exosomes. (C) Relative miR-21 expression in macrophages
co-cultured with T24 cells or anti-miR-21-5p T24 cells. (D) mRNA
levels of IL-10 and TGF-β in macrophages co-cultured with T24 cells
or anti-miR-21-5p T24 cells. (E) miR-21 levels of macrophages
transfected with miR-21-5p mimics or miR-21-5p inhibitor. (F) mRNA
levels of IL-10 and TGF-β in macrophages trans-fected with
miR-21-5p mimics. (G) mRNA levels of IL-10 and TGF-β in macrophages
transfected with miR-21-5p inhibitor. n=3. ▲▲P<0.01
vs. T24 group. *P<0.05, **P<0.01,
***P<0.001 vs. control group. #P<0.05,
##P<0.01, ###P<0.001 vs. NC group.
miR-21, microRNA-21; exo, exosome; Control, macrophages co-cultured
with T24 cells or macrophages; NC, macrophages co-cultured with T24
cells transfected with the appropriate negative control or
macrophages transfected with negative control; anti-miR-21-5p T24,
macrophages co-cultured with T24 cells transfected with a miR-21-5p
inhibitor; Mimics, macrophages transfected with miR-21-5p mimics;
Inhibitor, macrophages transfected with miR-21-5p inhibitor. |
The aforementioned data raise the question of
whether upregulation of endogenous miR-21 can polarize macrophages
toward the M2 phenotype. To answer this question, PMA-treated
macrophages were transiently transfected with miR-21-5p mimics or
miR-21-5p inhibitor, and miR-21 expression levels were evaluated by
RT-qPCR. As presented in Fig. 4E,
miR-21 transcript levels were significantly altered in macrophages
transfected with miR-21-5p mimics or miR-21-5p inhibitor. Compared
with the mimics control, transfection of macrophages with miR-21-5p
mimics significantly upregulated the expression of IL-10 and TGF-β
(Fig. 4F). Transfection of
macrophages with miR-21-5p inhibitor did not significantly decrease
the expression of M2 markers (IL-10 and TGF-β) (Fig. 4G). It was identified that
upregulation of miR-21 in macrophages can polarize macrophages
toward the M2 phenotype, while M2 phenotypic polarization did not
occur in macrophages transfected with miR-21-5p inhibitor.
Therefore, according to the results of the aforementioned
experiments, a conclusion can be made that miR-21 loaded in
T24-derived exosomes can promote M2-differentiation.
miR-21-5p mimics promote T24 cell
migration and invasion by inducing M2 macrophage polarization
Furthermore, the effect of miR-21 expression in
macrophages on T24 cells migration and invasion was investigated.
Transwell and Matrigel assays showed that transfection of
macrophages with miR-21-5p mimics significantly increased the
migration and invasion of T24 cells (Fig. 5A). Compared with control
macrophages and macrophages transfected with a control inhibitor,
miR-21 inhibitor-transfected macrophages showed significant
reductions in the migratory and invasive ability of T24 cells
(Fig. 5B). Therefore, enhancing
miR-21 expression may enable macrophages to promote the migratory
and invasive ability of T24 cells.
PTEN is suppressed by upregulation of
miR-21 in macro- phages
To elucidate the mechanism by which miR-21 mediates
M2 polarization, potential miR-21 targets were searched using the
computational prediction program TargetScan. PTEN deficiency has
been found to facilitate M2 macrophage differentiation in a murine
liver partial warm ischaemia model (27). Therefore, the present study focused
on PTEN, a predicted miR-21 target gene (Fig. 6A). Dual luciferase reporter assays
were performed to examine the direct interaction between miR-21 and
the 3'-untranslated region (3'-UTR) of PTEN. The PTEN 3'-UTR
downstream of a firefly luciferase reporter was cloned and it was
identified that intact miR-21 downregulated the activity of the
resulting reporter construct (Fig.
6B). When a mutant PTEN 3'-UTR with a disrupted miR-21 binding
site was introduced, this downregulation was significantly reversed
(Fig. 6B). The mRNA levels of PTEN
were also determined using RT-qPCR, which revealed that they were
lower in the exo-treated group compared with in the untreated group
(Fig. 6C). RT-qPCR analysis showed
that PTEN gene expression was significantly decreased in
macrophages transfected with miR-21-5p mimics but significantly
increased in macrophages transfected with miR-21-5p inhibitor
(Fig. 6D). Furthermore, the
recently developed PTEN-specific inhibitor SF1670 was used to
observe the role of the PTEN-mediated pathway in regulating
macrophage polarization (28). The
inhibitory effect of SF1670 was tested over 24 h at 500 and 1
µM concentrations by western blotting, which revealed that
SF1670 could inhibit the expression of PTEN in macrophages
transfected with miR-21-5p inhibitor (Fig. 6E). In addition, SF1670 treatment at
the 1 µM concentration significantly increased the
expression of M2 markers in miR-21 inhibitor-transfected
macrophages (Fig. 6F). These
results indicate that PTEN is a direct target gene by which miR-21
induces M2 differentiation.
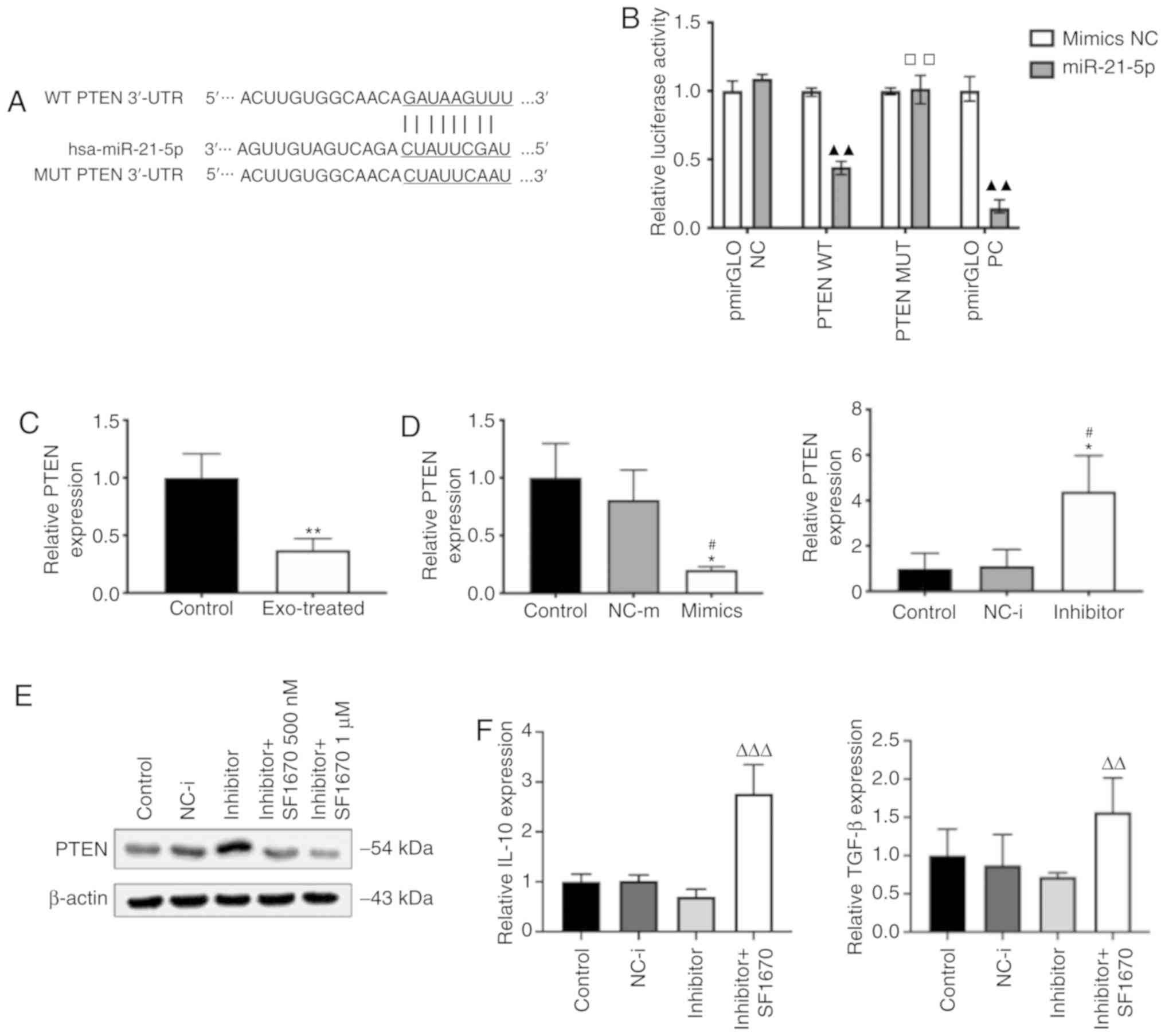 | Figure 6miR-21 downregulates PTEN expression
and activates PI3K/AKT-mediated STAT3 signalling pathways in
macrophages. (A) The binding site of miR-21-5p in the PTEN 3'-UTR
was predicted with TargetScan. (B) M0 macrophages were transfected
with either WT or MUT PTEN coding sequence vectors. Luciferase
activity was measured 24 h after transfection. (C) mRNA levels of
PTEN in macrophages treated with exosomes. (D) PTEN expression of
macrophages transfected with miR-21-5p mimics or miR-21-5p
inhibitor. (E) The PTEN inhibitor SF1670 (1 µM) inhibited
the expression of PTEN in macrophages transfected with miR-21-5p
inhibitor. (F) The levels of IL-10 and TGF-β were increased in
miR-21 inhibitor-transfected cells treated with SF1670 (1
µM). miR-21 downregulates PTEN expression and activates
PI3K/AKT-mediated STAT3 signalling pathways in macrophages. (G) The
expression of PTEN, PI3K, p-AKT and p-STAT3 in exo-treated
macrophages was assessed by western blot analysis. (H) Differences
in the expression of PTEN, PI3K, p-AKT and p-STAT3 between
macrophages co-cultured with T24 cells and those co-cultured with
anti-miR-21-5p T24 cells. (I) Expression of PTEN, PI3K, p-AKT and
p-STAT3 in macrophages transfected with miR-21-5p mimics or
miR-21-5p inhibitor. n=3. ▲▲P<0.01 vs. mimics NC
group. ☐☐P<0.01 vs. PTEN WT miR-21-5p group.
*P<0.05, **P<0.01 vs. control group.
#P<0.05 vs. NC group. ΔΔP<0.01,
ΔΔΔP<0.001 vs. inhibitor group. miR-21, microRNA-21;
pmirGLO NC, pmirGLO negative control; pmirGLO PC, pmirGLO positive
control; NC, negative control; MUT, mutated; PTEN, phosphatase and
tensin homolog; exo, exosome; p-, phosphorylated; 3'-UTR,
3'-untranslated region. |
PI3K/AKT-STAT3 is upregulated following
the suppression of PTEN in macrophages
Notably, STAT3 activation is essential for
differentiation of macrophages into the M2 phenotype (8). A previous report suggested that STAT3
activation is dependent on activation of the PI3K/AKT pathway in
macrophages (29). Therefore,
PI3K/AKT signalling and STAT3 signalling were next in the
exo-treated and untreated groups, by assessing AKT and STAT3
phosphorylation by western blot analysis. The expression of p-AKT
and p-STAT3 was greater in exo-treated macrophages than in
untreated macrophages (Fig. 6G).
Moreover, it was observed that the phosphorylation of AKT and STAT3
in macrophages co-cultured with anti-miR-21-5p T24 cells was lower
than that in macrophages co-cultured with T24 cells (Fig. 6H). It was hypothesized that miR-21
affects macrophage polarization via the STAT3 pathway. To test this
hypothesis, STAT3 phosphorylation status after treatment with
miR-21-5p mimics and miR-21-5p inhibitor was analysed, and STAT3
activation was assessed. In macrophages, the introduction of
miR-21-5p mimics enhanced p-AKT and p-STAT3, while introduction of
miR-21-5p inhibitor attenuated these changes (Fig. 6I). Taken together, these data
suggest that miR-21 directly downregulates PTEN and further
enhances PI3K/AKT-induced STAT3 signalling activity.
Discussion
The present study found that bladder cancer cells
express significantly higher levels of miR-21 compared with M0
macrophages, and that exosomal miR-21 transfer from bladder cancer
cells to M0 macrophages can enable increases in bladder cancer cell
migration and invasion through downregulation of the direct miR-21
target PTEN.
The THP-1 human acute monocytic leukaemia cell line
is the cell line most commonly used to study monocyte/macrophage
differentiation and function (30). However, the cell line THP-1 has
been found to contain a mutation in the N-RAS gene (31). Mutations in the N-RAS gene cause
certain structural changes in RAS proteins, which lead to
inactivation and continue to stimulate cell growth or
differentiation. The present study predominantly investigated the
relationship between T24 cells and macrophages, so macrophages were
the main experimental object rather than THP-1 cells, which were
only used to induce the formation of macrophages in the present
study. To determine whether the N-RAS gene mutations will interfere
with our experiment, we only need to observe whether the THP-1
cells with N-RAS gene mutations can be induced to differentiate
into macrophages. We found that M0 macrophages were successfully
obtained from THP-1 cells after stimulation with PMA for 24 h.
Therefore, it was confirmed that the N-RAS gene mutations in the
cell line THP-1 does not affect PMA-induced THP-1 cell
differentiation into macrophage-like cells.
Accumulating evidence has shown that TAMs are
closely related to tumour progression and metastasis through
intercellular communication with cancer cells (32,33).
A recent study has also highlighted the roles of exosomes in the
regulation of TME (34). Such
regulation may result from exosome-induced effects on macrophages
and phenotypic changes due to alterations in chemokines levels
(35). The present study has shown
that exosomes are capable of polarizing M0 macrophages into the
M2-like phenotype, as demonstrated by the production of specific
cytokines and the phenotypic changes in surface markers. Moreover,
the present data showed that macrophages treated with non-exo CM
could not be polarized into the M2-like phenotype. These results
suggest that cancer cell-derived exosomes, rather than other
substances outside the exosomes, activate the polarization
programme in macrophages. The polarization of macrophages into the
M2-like phenotype can be triggered by multiple signals.
miRNA-mediated TAM regulation is one of the most studied topics in
the field of TAM-related carcinogenesis. Numerous studies have
investigated the effects of exosomal miRNAs on TAM functions,
including macrophage polarization (36-38).
Previous studies have shown that miR-21 is often
over-expressed in different types of human cancer, including
non-small cell lung cancer, colorectal cancer (CRC) and breast
cancer (39-41). Notably, miR-21 expression is not
significantly different between bladder cancer and normal
urothelial tissues, but high expression of miR-21 is associated
with poor overall bladder cancer survival and is a good indicator
of metastasis (42). Moreover,
among numerous miRNAs, miR-21 has been found to be overexpressed in
bladder cancer tissue, urine exosomes and white blood cells
(43). This evidence suggests that
miR-21 can be used as a prognostic marker for bladder tumour
metastasis. A recent study has shown that certain miRNAs are
selectively loaded or retained in exosomes, indicating apparent
exosomal enrichment of certain miRNAs compared with most cellular
miRNAs (44). The present study
found that miR-21 was enriched in T24 cell-derived exosomes
isolated by ultracentrifugation. Furthermore, miR-21 could be
transferred by exosomes from bladder cancer cells to macrophages
and subsequently promote tumour migration and invasion, suggesting
that the increased miR-21 expression in M0 macrophages may confer a
more aggressive phenotype in these cells. miR-21 expression has
been associated with M2 macrophages characterized by elevated IL-10
protein levels (45). Upon
incubating M0 macrophages with bladder cancer cells, the present
study confirmed that the M2 macrophage markers were significantly
upregulated. In addition, the changes in cytokine markers in the
macrophages were blocked by co-culturing miR-21
inhibitor-transfected bladder cancer cells with M0 macrophages
in vitro. This blockade was probably due to the higher
endogenous miR-21 expression in bladder cancer cells than in M0
macrophages, as confirmed by RT-qPCR analyses. Therefore, the
aforementioned evidence may explain why T24 cell-derived exosomes
enriched with miR-21 induce M2 differentiation. Notably, a recent
study reported that M2 macrophage-regulated CRC cell migration and
invasion are dependent on M2 macrophage-derived exosomes, which
display high expression levels of miR-21-5p and miR-155-5p, and
that exosome-mediated CRC cell migration and invasion depended on
these two miRNAs (46). Thus,
exosomal miR-21 seems to be able to participate in the formation of
the TME and play a significant role in subsequent cancer cell
migration and invasion.
The present study also demonstrated, to the best of
our knowledge, for the first time, that downregulation of miR-21 in
macrophages can block polarization of M0 macrophages into M2
macrophages. In the context of cancer cells with genetic defects,
miR-21 can promote the polarization of macrophages into the M1-like
phenotype (47). Thus, miR-21
plays a key role in directing macrophage differentiation.
Additionally, it was revealed that miR-21 overexpression in M0
macrophages can induce STAT3 expression and that inhibition of
miR-21 abolishes this effect. Furthermore, it was found that miR-21
inhibitor-induced STAT3 upregulation could be eliminated by SF1670,
showing the dependence of this STAT3 upregulation on PI3K/AKT
activation. The PI3K/AKT pathway is an upstream signalling pathway
of NF-κB and can activate NF-κB in macrophages (48). A study has shown that enhancing the
NF-κB signalling pathway in macrophages can upregulate IL-6
production and secretion in cells (49). Furthermore, blocking NF-κB
activation inhibits IL-6 expression in macrophages (48). Recently, it has been suggested that
IL-6 and STAT3 play key roles in the progression of ovarian cancer,
possibly by polarizing TAMs (50).
The findings of previous studies regarding the relationships among
the PI3K/AKT/NF-κB/STAT3 signalling pathways can explain the
phenomenon of STAT3 upregulation by exosomal miR-21 in PMA-induced
THP-1 human macrophages. Regarding miR-21-induced signalling, a
previous study has indicated that the STAT3 pathway is the major
upstream pathway of miR-21 in monocytes/macrophages (51). Thus, the exact molecular
interactions by which miR-21 induces STAT3 expression need further
exploration.
Despite its successes, the present study has
limitations. Given the highly invasive nature of T24 cells, the
experiment revealed changes only in this one bladder cancer cell
line. Different levels of invasiveness of bladder cancer cell lines
can reflect differences in the influences of exosomes of different
stages of bladder cancer on the polarization of macrophages. This
phenomenon requires further study.
In conclusion, exosomal miR-21 derived from T24
cells could elevate the expression of M2-type characteristic genes
in macrophages to further promote the invasion and migration of
bladder cancer cells. However, these findings do not exclude the
roles of other exosomal cargo molecules derived from cancer
cells.
Supplementary Data
Funding
This study was supported by a grant from the
Foundation of Chongqing Science and Technology Commission (grant
no. cstc2015shmszx0466).
Availability of data and materials
The datasets generated and/or analysed during the
present study are available from the corresponding author upon
reasonable request.
Authors' contributions
FL, WH and XG designed the experiments. FL and HY
analysed the data. FL, XL and GZ performed the experiments. XG
revised the manuscript. WH prepared the figures. FL wrote the
paper. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Franzen CA, Simms PE, Van Huis AF, Foreman
KE, Kuo PC and Gupta GN: Characterization of uptake and
internalization of exosomes by bladder cancer cells. Biomed Res
Int. 2014:6198292014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ghafouri-Fard S, Nekoohesh L and
Motevaseli E: Bladder cancer biomarkers: Review and update. Asian
Pac J Cancer Prev. 15:2395–2403. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tkach M and Théry C: Communication by
extracellular vesicles: Where we are and where we need to go. Cell.
164:1226–1232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang X, Yuan X, Shi H, Wu L, Qian H and
Xu W: Exosomes in cancer: Small particle, big player. J Hematol
Oncol. 8:832015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kucharzewska P and Belting M: Emerging
roles of extracellular vesicles in the adaptive response of tumour
cells to microenvironmental stress. J Extracell Vesicles. 2:2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
EL Andaloussi S, Mäger I, Breakefield XO
and Wood MJ: Extracellular vesicles: Biology and emerging
therapeutic opportunities. Nat Rev Drug Discov. 12:347–357. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sica A and Bronte V: Altered macrophage
differentiation and immune dysfunction in tumor development. J Clin
Invest. 117:1155–1166. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mantovani A, Marchesi F, Porta C, Sica A
and Allavena P: Inflammation and cancer: Breast cancer as a
prototype. Breast. 16(Suppl 2): S27–S33. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cortés M, Sanchez-Moral L, de Barrios O,
Fernández-Aceñero MJ, Martínez-Campanario MC, Esteve-Codina A,
Darling DS, Győrffy B, Lawrence T, Dean DC and Postigo A:
Tumor-associated macrophages (TAMs) depend on ZEB1 for their
cancer-promoting roles. EMBO J. 36:3336–3355. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cassetta L and Pollard JW: Targeting
macrophages: Therapeutic approaches in cancer. Nat Rev Drug Discov.
17:887–904. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Aldo PB, Craveiro V, Guller S and Mor G:
Effect of culture conditions on the phenotype of THP-1 monocyte
cell line. Am J Reprod Immunol. 70:80–86. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qin Z: The use of THP-1 cells as a model
for mimicking the function and regulation of monocytes and
macrophages in the vasculature. Atherosclerosis. 221:2–11. 2012.
View Article : Google Scholar
|
|
14
|
Baietti MF, Zhang Z, Mortier E, Melchior
A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C,
Vermeiren E, et al: Syndecan-syntenin-ALIX regulates the biogenesis
of exosomes. Nat Cell Biol. 14:677–685. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou L, Shen LH, Hu LH, Ge H, Pu J, Chai
DJ, Shao Q, Wang L, Zeng JZ and He B: Retinoid X receptor agonists
inhibit phorbol-12-myristate-13-acetate (PMA)-induced
differentiation of monocytic THP-1 cells into macrophages. Mol Cell
Biochem. 335:283–289. 2010. View Article : Google Scholar
|
|
16
|
Lobb RJ, Becker M, Wen SW, Wong CSF,
Wiegmans AP, Leimgruber A and Möller A: Optimized exosome isolation
protocol for cell culture supernatant and human plasma. J Extracell
Vesicles. 4:270312015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mehdiani A, Maier A, Pinto A, Barth M,
Akhyari P and Lichtenberg A: An innovative method for exosome
quantification and size measurement. J Vis Exp. 50974:2015.
|
|
18
|
Helwa I, Cai J, Drewry MD, Zimmerman A,
Dinkins MB, Khaled ML, Seremwe M, Dismuke WM, Bieberich E, Stamer
WD, et al: A comparative study of serum exosome isolation using
differential ultracentrifugation and three commercial reagents.
PLoS One. 12:e01706282017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Park EK, Jung HS, Yang HI, Yoo MC, Kim C
and Kim KS: Optimized THP-1 differentiation is required for the
detection of responses to weak stimuli. Inflamm Res. 56:45–50.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fuhrman B, Partoush A, Volkova N and
Aviram M: Ox-LDL induces monocyte-to-macrophage differentiation in
vivo: Possible role for the macrophage colony stimulating factor
receptor (M-CSF-R). Atherosclerosis. 196:598–607. 2008. View Article : Google Scholar
|
|
22
|
Park SY, Lee SW, Kim HY, Lee SY, Lee WS,
Hong KW and Kim CD: SIRT1 inhibits differentiation of monocytes to
macrophages: Amelioration of synovial inflammation in rheumatoid
arthritis. J Mol Med (Berl). 94:921–931. 2016. View Article : Google Scholar
|
|
23
|
Whiteside TL: Exosomes and tumor-mediated
immune suppression. J Clin Invest. 126:1216–1223. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Italiani P and Boraschi D: From monocytes
to M1/M2 macrophages: Phenotypical vs. functional differentiation.
Front Immunol. 5:5142014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Martinez FO and Gordon S: The M1 and M2
paradigm of macrophage activation: Time for reassessment.
F1000Prime Rep. 6:132014. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ohno R, Uozaki H, Kikuchi Y, Kumagai A,
Aso T, Watanabe M, Watabe S, Muto S and Yamaguchi R: Both cancerous
miR-21 and stromal miR-21 in urothelial carcinoma are related to
tumour progression. Histopathology. 69:993–999. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yue S, Rao J, Zhu J, Busuttil RW,
Kupiec-Weglinski JW, Lu L, Wang X and Zhai Y: Myeloid PTEN
deficiency protects livers from ischemia reperfusion injury by
facilitating M2 macrophage differentiation. J Immunol.
192:5343–5353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Y, Prasad A, Jia Y, Roy SG, Loison F,
Mondal S, Kocjan P, Silberstein LE, Ding S and Luo HR: Pretreatment
with phosphatase and tensin homolog deleted on chromosome 10 (PTEN)
inhibitor SF1670 augments the efficacy of granulocyte transfusion
in a clinically relevant mouse model. Blood. 117:6702–6713. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tacke RS, Tosello-Trampont A, Nguyen V,
Mullins DW and Hahn YS: Extracellular hepatitis C virus core
protein activates STAT3 in human monocytes/macrophages/dendritic
cells via an IL-6 autocrine pathway. J Biol Chem. 286:10847–10855.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chanput W, Mes JJ and Wichers HJ: THP-1
cell line: An in vitro cell model for immune modulation approach.
Int Immunopharmacol. 23:37–45. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sheng XM, Kawamura M, Ohnishi H, Ida K,
Hanada R, Kojima S, Kobayashi M, Bessho F, Yanagisawa M and Hayashi
Y: Mutations of the RAS genes in childhood acute myeloid leukemia,
myelo-dysplastic syndrome and juvenile chronic myelocytic leukemia.
Leuk Res. 21:697–701. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Franklin RA, Liao W, Sarkar A, Kim MV,
Bivona MR, Liu K, Pamer EG and Li MO: The cellular and molecular
origin of tumor-associated macrophages. Science. 344:921–925. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu H, Lai W, Zhang Y, Liu L, Luo X, Zeng
Y, Wu H, Lan Q and Chu Z: Tumor-associated macrophage-derived IL-6
and IL-8 enhance invasive activity of LoVo cells induced by PRL-3
in a KCNN4 channel-dependent manner. BMC Cancer. 14:3302014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Berchem G, Noman MZ, Bosseler M, Paggetti
J, Baconnais S, Le Cam E, Nanbakhsh A, Moussay E, Mami-Chouaib F,
Janji B and Chouaib S: Hypoxic tumor-derived microvesicles
negatively regulate NK cell function by a mechanism involving TGF-β
and miR23a transfer. Oncoimmunology. 5:e10629682016. View Article : Google Scholar
|
|
35
|
Jang JY, Lee JK, Jeon YK and Kim CW:
Exosome derived from epigallocatechin gallate treated breast cancer
cells suppresses tumor growth by inhibiting tumor-associated
macrophage infiltration and M2 polarization. BMC Cancer.
13:4212013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen X, Ying X and Wang X, Wu X, Zhu Q and
Wang X: Exosomes derived from hypoxic epithelial ovarian cancer
deliver microRNA-940 to induce macrophage M2 polarization. Oncol
Rep. 38:522–528. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Park JE, Dutta B, Tse SW, Gupta N, Tan CF,
Low JK, Yeoh KW, Kon OL, Tam JP and Sze SK: Hypoxia-induced tumor
exosomes promote M2-like macrophage polarization of infiltrating
myeloid cells and microRNA-mediated metabolic shift. Oncogene.
38:5158–5173. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang X, Luo G, Zhang K, Cao J, Huang C,
Jiang T, Liu B, Su L and Qiu Z: Hypoxic tumor-derived exosomal
miR-301a mediates M2 macrophage polarization via PTEN/PI3Kγ to
promote pancreatic cancer metastasis. Cancer Res. 78:4586–4598.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Markou A, Sourvinou I, Vorkas PA, Yousef
GM and Lianidou E: Clinical evaluation of microRNA expression
profiling in non small cell lung cancer. Lung Cancer. 81:388–396.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen TH, Chang SW, Huang CC, Wang KL, Yeh
KT, Liu CN, Lee H, Lin CC and Cheng YW: The prognostic significance
of APC gene mutation and miR-21 expression in advanced-stage
colorectal cancer. Colorectal Dis. 15:1367–1374. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
miR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar
|
|
42
|
Zaravinos A, Radojicic J, Lambrou GI,
Volanis D, Delakas D, Stathopoulos EN and Spandidos DA: Expression
of miRNAs involved in angiogenesis, tumor cell proliferation, tumor
suppressor inhibition, epithelial-mesenchymal transition and
activation of metastasis in bladder cancer. J Urol. 188:615–623.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Armstrong DA, Green BB, Seigne JD, Schned
AR and Marsit CJ: MicroRNA molecular profiling from matched tumor
and bio-fluids in bladder cancer. Mol Cancer. 14:1942015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mittelbrunn M, Gutiérrez-Vázquez C,
Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ,
Bernad A and Sánchez-Madrid F: Unidirectional transfer of
microRNA-loaded exosomes from T cells to antigen-presenting cells.
Nat Commun. 2:2822011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Caescu CI, Guo X, Tesfa L, Bhagat TD,
Verma A, Zheng D and Stanley ER: Colony stimulating factor-1
receptor signaling networks inhibit mouse macrophage inflammatory
responses by induction of microRNA-21. Blood. 125:e1-e132015.
View Article : Google Scholar :
|
|
46
|
Lan J, Sun L, Xu F, Liu L, Hu F, Song D,
Hou Z, Wu W, Luo X, Wang J, et al: M2 macrophage-derived exosomes
promote cell migration and invasion in colon cancer. Cancer Res.
79:146–158. 2019. View Article : Google Scholar
|
|
47
|
Xi J, Huang Q, Wang L, Ma X, Deng Q, Kumar
M, Zhou Z, Li L, Zeng Z, Young KH, et al: miR-21 depletion in
macrophages promotes tumoricidal polarization and enhances PD-1
immunotherapy. Oncogene. 37:3151–3165. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Guo W, Sun J, Jiang L, Duan L, Huo M, Chen
N, Zhong W, Wassy L, Yang Z and Feng H: Imperatorin attenuates
LPS-induced inflammation by suppressing NF-κB and MAPKs activation
in RAW 264.7 macrophages. Inflammation. 35:1764–1772. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang YC, Hu YW, Sha YH, Gao JJ, Ma X, Li
SF, Zhao JY, Qiu YR, Lu JB, Huang C, et al: Ox-LDL upregulates IL-6
expression by enhancing NF-κB in an IGF2-dependent manner in THP-1
macrophages. Inflammation. 38:2116–2123. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ham S, Lima LG, Chai EPZ, Muller A, Lobb
RJ, Krumeich S, Wen SW, Wiegmans AP and Möller A: Breast
cancer-derived exosomes alter macrophage polarization via
gp130/STAT3 signaling. Front Immunol. 9:8712018. View Article : Google Scholar :
|
|
51
|
Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk
ML and Struhl K: STAT3 activation of miR-21 and miR-181b-1 via PTEN
and CYLD are part of the epigenetic switch linking inflammation to
cancer. Mol Cell. 39:493–506. 2010. View Article : Google Scholar : PubMed/NCBI
|