Introduction
Esophageal cancer is the sixth most common cause of
cancer-associated mortality worldwide (1). Neoadjuvant chemoradiotherapy (CRT)
followed by esophagectomy is the standard treatment for locally
advanced esophageal squamous cell carcinoma (ESCC) in Western
countries, whereas neoadjuvant chemotherapy (CT) followed by
esophagectomy or definitive CRT (CRT alone as a primary therapy)
are the standard treatments in Japan (2). Although neoadjuvant CT and definitive
CRT improve the prognosis of patients with ESCC, the 5-year
survival rate is still 37-55% (2,3).
Local recurrence and metastasis are major causes of poor prognosis.
Nevertheless, the prediction is difficult, creating a need for
predictive factors that select patients who are potentially curable
with definitive CRT.
By comparing microarray profiles among pre- and
post-treatment biopsy specimens of patients with ESCC, our previous
study identified a good responder subtype with cytotoxic
T-lymphocyte signatures that were activated by CRT (4). Clustering analysis of 234 tumor
immunity-related genes in 121 pre-treatment ESCC specimens
distinguished the immune-activated cases, termed I-type, from other
cases. In the I-type, the clinical outcome of cadherin 2
(CDH2)-negative cases was significantly better than that of the
CDH2-positive cases. Notably, CD24, keratin 4 (KRT4)
and SIM bHLH transcription factor 2 (SIM2) were
overexpressed in the CDH2-negative cases (4). The differentiation degree in squamous
cell carcinoma has been reported to influence sensitivity and
prognosis in response to CRT (5,6).
SIM2 is a member of the basic HLH-PER-ARNT-SIM transcription
factors, which is isolated from a Down's syndrome-crucial region
(7-9). Aberrant SIM2 expression has been
reported in several types of cancer (10,11).
Recently, we identified SIM2 as a predictive biomarker for patients
with cervical cancer who were potentially curable with CRT
(12). Furthermore, our previous
study reported that SIM2 in ESCC might be a key transcription
factor involved in tumor differentiation and CRT sensitivity
through downregulation of DNA repair and antioxidant genes.
Therefore, SIM2 may be associated with the response to definitive
CRT (13).
CD24 is a small mucin-like cell surface protein,
which is expressed on lymphocytes and epithelial cells (14), and is also expressed in various
types of cancer, including colorectal, pancreatic, lung, liver,
ovarian and breast cancer (15-18).
These studies also reported that CD24 overexpression is associated
with an aggressive course of the disease. Furthermore, CD24 may
serve a role in the metastasis of breast cancer (19-21),
cervical cancer (18), gastric
cancer (22) and bladder cancer
(23,24). CD24 has also been reported as a
marker for stem cells in pancreatic and ovarian cancer (25,26).
However, the role of CD24 in ESCC remains obscure.
KRT4 encodes a type II cytokeratin,
cytokeratin 4 (CK4), which is specifically found in differentiated
layers of the esophageal epithelia. KRT4 is downregulated in
ESCC and head and neck squamous cell carcinoma compared with in
normal squamous epithelium (27,28).
Its low expression is associated with local recurrence of head and
neck squamous cell carcinoma (29). However, the biological functions
and clinical significance of CK4 and CD24 remain unknown in ESCC.
This study investigated the association between their mRNA and
protein expression levels, and clinicopathological characteristics,
and also investigated the functions of CD24 in SIM2-mediated
tumor differentiation and CRT sensitivity.
Materials and methods
Clinical samples
Patients with ESCC who received definitive CRT or
curative esophagectomy with extended lymph node dissection
(surgery) as an initial treatment at the National Cancer Center
Hospital East (Kashiwa, Japan) between June 2005 and March 2009
were recruited. The eligibility criteria were as follows: i)
Patients pathologically diagnosed, using biopsy specimens, with
squamous cell carcinoma prior to receiving definitive CRT or
surgery; ii) patients with stage II/III ESCC who underwent
definitive CRT or surgery; and iii) patients <75 years old whose
performance status according to the Eastern Cooperative Oncology
Group was 0.1 (30). Clinical
staging before neoadjuvant CT (in the surgery group) or definitive
CRT was determined according to the Union for International Cancer
Control-Tumor-Node-Metastasis classification (6th edition)
(31), based on endoscopic
findings and contrast enhanced computed tomography (CECT). Patients
with prior or concurrent types of cancer were excluded from this
study. In the surgery group, clinical outcomes were determined
following surgery alone or neoadjuvant CT followed by surgery.
However, patients who were not able to receive a scheduled complete
course of definitive CRT were included, because such patients whose
therapeutic responses are unpredictable could not be excluded prior
to treatment.
Cell culture
The ESCC T.Tn cell line was purchased from the
Japanese Collection of Research Bioresources Cell Bank. T.Tn cells
were propagated in DMEM/Ham's F-12 (Wako Pure Chemical Industries,
Ltd.) supplemented with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.) and 1% penicillin-streptomycin (Gibco;
Thermo Fisher Scientific, Inc.), and maintained at 37°C in 95%
humidified air containing 5% CO2. A 35-mm NanoCulture
Plate (SCIVAX Corporation) was used for three-dimensional (3D)
culture (13).
Laser-captured micro-dissection
(LCM)
The human esophagus samples were embedded in
TissueTek O.C.T. Compound (Sakura Finetek Japan) and snap-frozen.
The cryostat sections (8 µm) were dissected using a PixCell
II LCM system (Arcturus Engineering, Inc.). To avoid contamination
with dysplastic or cancerous tissues, normal esophageal mucosa was
obtained from gastric cancer samples with normal esophageal tissue
for semi-quantitative reverse transcription-PCR (RT-PCR) analysis
of the three cell layers (differentiated, parabasal and basal cell
layers).
Microarray analysis
RNA was isolated from the biopsy samples from
patients prior to treatment using ISOGEN lysis buffer (Nippon Gene
Co., Ltd.), and were biotin-labeled followed by hybridization to
microarrays (Human Genome U133 Plus 2.0 Array; Affymetrix, Inc.),
according to manufacturer's protocol. The scanned data of the
arrays were processed by Affymetrix Microarray Suite version 5.0
(Affymetrix, Inc.). All of the microarray data were deposited in a
minimum information about a microarray experiment-compliant
database, Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/); the accession
number is GSE69925 (4).
RT-PCR
Total RNA was isolated from cells using ISOGEN lysis
buffer (Nippon Gene Co., Ltd.) followed by precipitation with
isopropanol. RT was performed using oligo dT primers from the
SuperScript III First-Stand Synthesis system (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. PCR
was carried out using the AccuPrimeTaq DNA Polymerase system
(Thermo Fisher Scientific, Inc.), within the linear range of
amplification, for long isoforms of SIM2 (24 cycles),
CD24 (23 cycles) KRT4 (18 cycles) and β-actin
(ACTB; 22 cycles). The thermocycling conditions were as
follows: Initial denaturation at 95°C for 5 min, followed by the
aforementioned number of cycles at 95°C for 1 min, 56°C for 1 min
and 72°C for 1 min, with a final extension step at 72°C for 10 min.
PCR products were then separated by electrophoresis with 2% agarose
gels and results were visualized using ethidium bromide (Wako Pure
Chemical Industries, Ltd.).
RT-quantitative PCR (RT-qPCR) was carried out for
long isoforms of SIM2, CDH2, vimentin (VIM),
snail family transcriptional repressor 2 (SNAI2), twist
family bHLH transcription factor (TWIST)1, TWIST2, CD24,
KRT4 and ACTB. In accordance with the manufacturer's
protocol, RT was conducted using the SuperScript III First-Stand
Synthesis system (Thermo Fisher Scientific, Rockford, IL) and qPCR
was performed on a Bio-Rad iCycler with iQ SYBR Green Supermix
(Bio-Rad Laboratories, Inc.). The thermocycling conditions were as
follows: Initial denaturation at 95°C for 2 min, followed by 40
cycles at 95°C for 15 sec and 55°C for 30 sec, and a final step at
95°C for 1 min and 55°C for 1 min. Results are presented as
linearized quantification cycle (Cq) values normalized to
ACTB and the indicated reference value (2−ΔΔCq)
(32). Primer sequences are listed
in Table I.
 | Table IPrimer sequences for reverse
transcription-PCR. |
Table I
Primer sequences for reverse
transcription-PCR.
| Gene | Forward primer (5′
to 3′) | Reverse primer (5′
to 3′) |
|---|
| ACTB |
GAAGTCCCTTGCCATCCTAA |
GCACGAAGGCTCATCATTCA |
| CD24 |
GCCTCGACACACATAAACCT |
CTGTTCGATCTGTTTGTTCC |
| SIM2a |
TGCCAACCCTGTGTCACTTA |
ACCCTCGGCTTATTTCCTGT |
| SIM2b |
CTTCCCTCTGGACTCTCACG |
AGGCTGTGCCTAGCAGTGTT |
| KRT4 |
CAGGAGTGTCATCTCCAGAA |
GAAGATTCACCTGCAGATGG |
| SNAI2 |
TAGGAAGAGATCTGCCAGAC |
CCCCAAGGCACATACTGTTA |
| VIM |
GCTTTCAAGTGCCTTTCTGC |
GTTGGTTGGATACTTGCTGG |
| CDH2 |
GGCATAGTCTATGGAGAAGT |
GATTTCACAAGTCTTCACCTG |
| TWIST1 |
GCATTTTACCATGGGTCCTC |
ATACTGGGATCAAACTGGCC |
| TWIST2 |
GAGCCTCTGCATGATTGTTTC |
CACTGCAGTCACTTAGCTTG |
| SOD2 |
ATGATCCCAGCAAGATAATG |
AGGACCTTATAGGGTTTTCAG |
Plasmid transfection
The pCMV6-AC-GFP plasmid containing SIM2 cDNA
was purchased from OriGene Technologies, Inc. T.Tn cells were
plated at 2×106 per 10-cm dish, and transfected with
either pCMV6-AC-GFP-SIM2 or empty pCMV6-neo (OriGene
Technologies, Inc.). Briefly, cells were transfected with 4
µg plasmid DNA in 10 µl Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol, overnight at 37°C. Subsequently, the cells
were plated at 6×105 cells/3.5 cm NanoCulture Plate
(SCIVAX Corporation).
Immunohistochemistry (IHC) and
hematoxylin and eosin (HE) staining
Specimens fixed in 10% formalin at room temperature
for 8-24 h and embedded in paraffin were cut into 4-µm
sections, which were dewaxed and dehydrated for routine HE
staining.
For IHC, the endogenous peroxidase activity of
4-µm sections were cut from paraffin-embedded specimens, and
the endogenous peroxidase activity of the sections was blocked with
3% H2O2 in ethanol for 5 min at room
temperature, followed by additional blocking with 3% BSA-PBS (Roche
Diagnostics GmbH) for 1 h at room temperature. Antigen retrieval
was performed in a microwave oven at 95°C using 10 mM citrate
buffer (pH 6.0) for 20 min (CD24 antigen) or Target Retrieval
Solution (cat. no. S2367; Dako; Agilent Technologies, Inc.; pH 9.0)
for 10 min (CK4 antigen). Anti-CD24 (1:500; cat. no. NB100-64861;
Novus Biologicals, LLC) and anti-CK4 antibodies (1:500; cat. no.
ab9004; Abcam) were diluted at 1:500 and slides were incubated with
them at 4°C overnight. The slides were then incubated with a
horseradish peroxidase (HRP)-labeled secondary antibody (Envision™
Kit/HRP system; cat. No. K4063; Dako; Agilent Technologies, Inc.)
at room temperature for 30 min and visualized by DAB (DAB+ Liquid;
Dako; Agilent Technologies, Inc.). The positive percentage of
cancer cells for each case was determined by a pathologist who was
blinded to the clinical data. IHC and HE staining were detected
under a Nikon ECLIPSE light microscope (Nikon Corporation) and was
analyzed using NIS-Elements BR version 4.10 software (Nikon
Corporation).
Small interfering RNA (siRNA)
transfection
CD24 siRNAs and control siRNA (cat. no.
AM4635) were purchased from Ambion; Thermo Fisher Scientific, Inc.
The sequences were as follows: siRNA s2615, UCA AGU AAC UCC UCC CAG
Att; siRNA s2616, CCA GAG UAC UUC CAA CUC Utt). siRNAs (75 nM) were
introduced into 4×105 T.Tn cells (50% cell confluence)
using DharmaFECT 1 Transfection Reagents (GE Healthcare Dharmacon,
Inc.) and cells were incubated for 3 days at 37°C.
Western blotting
Cells were lysed in Laemmli Sample buffer (Bio-Rad
Laboratories, Inc.) containing DTT and 1% protease inhibitor
cocktail (Sigma-Aldrich; Merck KGaA), and protein concentration was
analyzed using the Protein Quantification Assay (MACHEREY-NAGEL
GMBH & Co. KG). Protein samples (35 µg) were separated
by electrophoresis using a NovexWedge Well 4-20% Tris-Glycine Gel
(Thermo Fisher Scientific, Inc.). Proteins were transferred to
nitrocellulose membranes, which were blocked with 5% Membrane
Blocking Reagent (cat. no. RPN2125; GE Healthcare) for 1 h at room
temperature, and incubated with anti-CD24 (1:200; cat. no.
sc-58999; Santa Cruz Biotechnology, Inc.) at 4°C overnight or with
anti-β-actin (1:3,000; cat. no. 4967; Cell Signaling Technology,
Inc.) at room temperature for 2 h. The membranes were then washed
and incubated with HRP-conjugated anti-mouse immunoglobulin
(1:3,000; cat. no. P0260; Dako; Agilent Technologies, Inc.) or
HRP-conjugated anti-rabbit immunoglobulin (1:3,000; cat. no. P0399;
Dako; Agilent Technologies, Inc.) at room temperature for 2 h.
Bands were visualized with Pierce ECL Plus Western Blotting
Substrate (Thermo Fisher Scientific, Inc.).
H2O2 or cisplatin
(CDDP) treatment
Cells were plated at 1×104 cells/well in
a 96-well NanoCulture Plate (SCIVAX Corporation) after siRNA
transfection. A total of 1 day after plating, cells were treated
with H2O2 (150 µM; Wako Pure Chemical
Industries, Ltd.) or CDDP (5 µM; Sigma-Aldrich; Merck KGaA)
at 37°C for 1 or 3 days, respectively. The number of viable cells
was counted using a CellTiter-Glo Luminescent Cell Viability Assay
(Promega Corporation), according to the manufacturer's
protocol.
TGF-β treatment
T.Tn cells were plated at 8×105
cells/well in a 6-well plate and were incubated at 37°C overnight.
Subsequently, the cells were treated with TGF-β1 (10 ng/ml; R&D
Systems, Inc.) at 37°C for 3 days.
Statistical analysis
RT-qPCR data are expressed as the mean ± SE and were
analyzed using one-way ANOVA followed by Tukey's honestly
significant difference test or Dunnett's multiple comparison test.
Recurrence-free survival (RFS) and overall survival (OS) were
estimated using the Kaplan-Meier method and were compared using the
log-rank test by GraphPad Prism version 7.0a (GraphPad Software,
Inc.). RFS was defined as the period from the date of definitive
CRT or surgery until the date of death or recurrence, which was
clinically confirmed through endoscopy or CECT. OS was defined as
the time from the date of definitive CRT or surgery until the last
confirmed date of survival or death, regardless of the cause of
death. Multivariate analysis with the Cox model was used to
investigate the association between patient background, endoscopic
findings and clinicopathological factors, including death or
recurrence. IBM SPSS statistical software package (version 22.0 for
Mac; IBM Japan Ltd.) and Ekuseru-Toukei 2010 (Social Survey
Research Information Co., Ltd.) were used for statistical analyses.
P<0.05 was considered to indicate a statistically significant
difference.
Results
CD24 and KRT4 are differentiation markers
that are downstream of SIM2
Initially, this study analyzed the semi-quantitative
RT-PCR of CD24, KRT4 and SIM2 in three layers
(differentiated, parabasal and basal cell layers) of normal
esophageal mucosa (23 cycles for CD24, 18 cycles for
KRT4 and 24 cycles for SIM2). CD24 and
KRT4 were highly expressed in differentiated cell layers and
moderately expressed in parabasal cell layers. SIM2 was
highly expressed in parabasal and basal cell layers, and moderately
expressed in differentiated cell layers (Figs. 1A and S1). Subsequently, CD24, CK4 and SIM2
protein expression was detected in normal esophageal mucosa by IHC.
In accordance with the RT-PCR results, CD24 and CK4 were highly
expressed in differentiated and parabasal cell layers, whereas SIM2
was expressed highly in parabasal and basal cell layers (Fig. 1B). These data suggested that CD24
and CK4 are differentiation markers in the stratified squamous
epithelia of the esophagus.
 | Figure 1CD24 and CK2, which is encoded by
KRT4, are differentiation markers regulated by SIM2. (A)
Semi-quantitative RT-PCR of CD24 and KRT4 in three
layers (differentiated, parabasal and basal cell layers) of the
normal esophageal mucosa. (B) Immunohistochemical staining of CD24,
CK4 and SIM2 in the normal esophageal mucosa; representative images
are indicated. (C) RT-PCR of SIM2, CD24 and
KRT4 in 3D-cultured T.Tn cells 5 or 8 days after empty
vector or SIM2 transfection (n=3, mean ± SE).
*P<0.05, **P<0.01 and
***P<0.001. ACTB, β-actin; CK4, cytokeratin 4; HE,
hematoxylin and eosin; KRT4, keratin 4; RT-PCR, reverse
transcription-PCR; SIM2, SIM bHLH transcription factor 2. |
To investigate whether CD24 and KRT4
are downstream genes of the tumor differentiation-inducer
SIM2, a 3D culture system was used, which has been reported
to induce differentiation of ESCC through adhesion restriction
(13). Overexpression of
SIM2 in T.Tn cells followed by 3D culture has been reported
to increase spheroid formation (13); in this study, SIM2
over-expression and 3D culture significantly increased CD24
and KRT4 mRNA expression at day 8 (Fig. 1C). These results of in vitro
3D cell culture suggested that CD24 and KRT4 may be
downstream differentiation markers of SIM2.
Patients with ESCC and high CD24 and KRT4
mRNA expression exhibit a favorable prognosis with definitive
CRT
Clinicopathological characteristics of patients with
ESCC who received definitive CRT (n=81) or surgical resection
(n=63) are shown in Table SI.
Using our previously obtained microarray data (GSE69925) (4), CD24 and KRT4 mRNA
expression was examined in biopsy specimens from 81 patients with
ESCC (clinical stages II and III) prior to definitive CRT. A total
of 15 of the 81 cases (18.5%) were classified into a high
CD24 mRNA expression group, whose CD24 expression was
higher than mean + SD (Fig. 2A).
Similarly, 22 of the 81 cases (27%) were classified into a high
KRT4 mRNA expression group, whose KRT4 expression
signal intensity was >50,000 (Fig.
2A). Kaplan-Meier analysis revealed that RFS and OS of the high
CD24 or KRT4 mRNA expression groups were
significantly longer than those of the low CD24 or
KRT4 mRNA expression groups (CD24, lower than mean-SD
and KRT4, signal intensity was <1,000) (Fig. 2B and C).
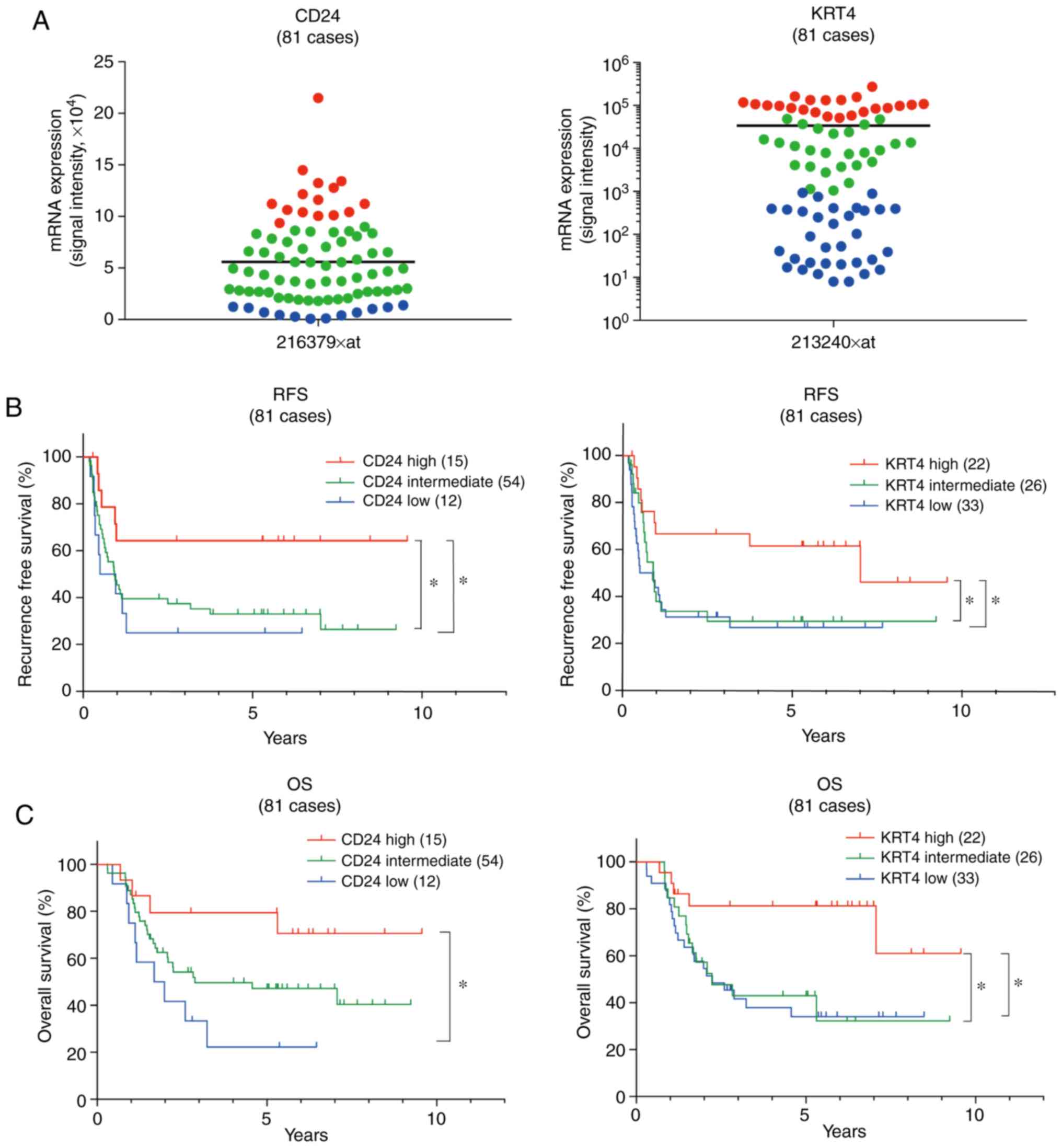 | Figure 2Patients with ESCC and high
CD24 and KRT4 mRNA expression exhibit a favorable
prognosis with definitive CRT. (A) Using our microarray data
(GSE69925), CD24 and KRT4 mRNA expression was
examined in 81 biopsy specimens prior to definitive CRT. A total of
15 of the 81 cases (18.5%) were classified into a high CD24
expression group (red, expression was higher than the mean ± SD).
Similarly, 22 of the 81 cases (27%) were classified into a high
KRT4 expression group (red, expression was >50,000 in
signal intensity). Bar indicates the mean. (B and C) Kaplan-Meier
analysis revealed that RFS and OS of the high CD24 and
KRT4 expression groups were significantly longer than those
of the low CD24 and KRT4 expression groups (blue,
CD24 expression was lower than the mean-SD; KRT4, expression was
<1,000 in signal intensity). *P<0.05. KRT4,
keratin 4; OS, overall survival; RFS, recurrence-free survival. |
Immunohistochemical analyses for
predicting patients with ESCC with a favorable prognosis following
definitive CRT
According to the microarray data, CD24 and
KRT4 mRNA expression may be candidate markers for predicting
patients with ESCC with a favorable prognosis in response to
definitive CRT. The CD24 and KRT4 genes encode CD24
and CK4 proteins, respectively. To verify the results of microarray
analysis, each of these two marker proteins was examined by
immunohistochemical staining in biopsy specimens obtained from 81
patients with ESCC prior to definitive CRT. Representative data are
shown in Fig. 3A. According to the
cut-off values for CD24 and CK4 positivity rates, a sensitivity
test was performed using the hazard ratio (HR) for OS. The minimum
HR was obtained when the cut-off values of 20% CD24-positive and
10% CK4-positive in tumor cells were adopted (CD24: HR, 0.446; 95%
CI, 0.219-0.909; P=0.026 and CK4: HR, 0.176; 95% CI, 0.042-0.728;
P=0.016). High CD24 expression was detected in 26 of the 81
patients (32%), whereas high CK4 expression was detected in 14 of
the 81 patients (17%) (Table II).
As shown in Fig. 3B, RFS and OS of
patients with ESCC and high CD24 or CK4 protein expression were
significantly higher than those of patients with ESCC and low CD24
or CK4 protein expression. Only 10 patients with ESCC exhibited
high expression of both CD24 and CK4, whereas 71 patients with ESCC
exhibited low expression of both CD24 and CK4. Patients with high
CD24 + CK4 expression survived longer than patients with low CD24 +
CK4 expression (Fig. 3B).
 | Table IIMultivariate analysis of RFS and OS
in patients with ESCC undergoing definitive CRT. |
Table II
Multivariate analysis of RFS and OS
in patients with ESCC undergoing definitive CRT.
| Variable | n (%) | RFS
| OS
|
|---|
| Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age | | | | | | | |
| <60 years | 21 (25.9) | Reference | 0.468-1.976 | 0.914 | Reference | 0.776-2.958 | 0.528 |
| ≥60 years | 60 (74.1) | 0.961 | | 0.62 | 0.764 | | |
| Sex | | | | | | | |
| Male | 74 (91.3) | Reference | 0.474-3.470 | 0.624 | Reference | 0.585-5.368 | 0.311 |
| Female | 4 (8.7) | 1.283 | | | 1.772 | | |
| Macroscopic
type | | | | | | | |
| Types 1 and 2 | 50 (61.7) | Reference | 0.572-1.934 | 0.87 | Reference | 0.776-2.958 | 0.224 |
| Type 3 | 31 (38.3) | 1.052 | | | 1.515 | | |
| Tissue type | | | | | | | |
| W/D and M/D | 68 (84.0) | Reference | 0.658-3.518 | 0.327 | Reference | 1.045-7.294 | 0.041a |
| P/D | 13 (16.0) | 1.521 | | | 2.76 | | |
| Location | | | | | | | |
| Ut and Mt | 45 (55.6) | Reference | 0.420-1.441 | 0.425 | Reference | 0.416-1.555 | 0.518 |
| Lt | 36 (44.4) | 0.778 | | | 0.805 | | |
| Circumference | | | | | | | |
| <3/4 | 45 (55.6) | Reference | 0.822-2.761 | 0.185 | Reference | 0.975-3.618 | 0.06 |
| ≥3/4 | 36 (44.4) | 1.507 | | | 1.878 | | |
| c T factor | | | | | | | |
| T2 | 16 (19.8) | Reference | 0.479-2.732 | 0.762 | Reference | 0.544-3.459 | 0.503 |
| T3 | 65 (80.2) | 1.144 | | | 1.372 | | |
| c N factor | | | | | | | |
| Absent | 38 (46.9) | Reference | 0.927-3.608 | 0.082 | Reference | 0.737-3.281 | 0.247 |
| Present | 43 (53.1) | 1.828 | | | 1.555 | | |
| CD24 | | | | | | | |
| Low | 55 (67.9) | Reference | 0.204-0.997 | 0.049a | Reference | 0.108-0.732 | 0.009a |
| High | 26 (32.1) | 0.451 | | | 0.281 | | |
| CK4 | | | | | | | |
| Low | 67 (82.7) | Reference | 0.009-0.960 | 0.043a | Reference | 0.016-0.894 | 0.039a |
| High | 14 (17.3) | 0.289 | | | 0.119 | | |
Multivariate Cox regression analysis in 81 patients
with ESCC revealed that high CD24 or CK4 expression was an
independent favorable prognostic factor in response to definitive
CRT for RFS (CD24: HR, 0.451; 95% CI, 0.204-0.997; P=0.049 and CK4:
HR, 0.289; 95% CI, 0.009-0.960; P=0.043) and OS (CD24: HR, 0.281;
95% CI, 0.108-0.732; P=0.009 and CK4: HR, 0.119; 95% CI,
0.016-0.894; P=0.039) (Table II).
Tumor differentiation type (tissue type) of biopsy specimens was
also revealed to be an independent favorable prognostic factor for
OS, but not for DFS, in response to definitive CRT (Table II). In accordance with CD24 and
CK4 being differentiation markers (Fig. 1), ESCC samples with high CD24 or
CK4 expression, particularly CD24, divided preferentially into well
or moderately differentiated cancer (Table SII).
CD24 and CK4 are predictive biomarkers
for definitive CRT and surgery
Based on the clinicopathological characteristics of
the patients (Table SI), 81
patients with ESCC undergoing CRT were compared with 63 patients
with ESCC undergoing surgery. Kaplan-Meier analyses revealed that
when CD24 was highly expressed, there was no significant difference
in the RFS and OS of 26 patients with ESCC undergoing definitive
CRT compared with the 33 patients with ESCC undergoing surgery.
Conversely, when CD24 was lowly expressed, there was a significant
difference between the RFS and OS of 55 patients with ESCC
undergoing definitive CRT and those of 30 patients with ESCC
undergoing surgery (Fig. 4A).
Although there were more patients with CK4 high expression in the
CRT group, when CK4 was highly expressed, there was no significant
difference in the RFS and OS of patients undergoing definitive CRT
compared with those undergoing surgery (Fig. 4B). Conversely, when CK4 was lowly
expressed, there was a significant difference in the RFS and OS of
patients undergoing CRT compared with those undergoing surgery
(Fig. 4B). As shown in Tables III and IV, multivariate Cox regression analysis
in patients with ESCC and low CD24 or CK4 expression indicated that
there was a significant difference between patients undergoing
definitive CRT and those undergoing surgery in RFS (low CD24 HR,
2.28; 95% CI, 1.182-4.397; P=0.014 and low CK4: HR, 2.142; 95% CI,
1.274-3.599; P=0.004) and OS (low CD24: HR, 3.781; 95% CI,
1.518-9.416; P=0.004 and low CK4: HR, 2.407; 95% CI, 1.317-4.399;
P=0.004). However, in patients with ESCC and high CD24 or CK4,
there was no significant difference between RFS and OS between CRT
and surgery (data not shown). Taken together, in cases with low
CD24 or CK4, surgery was revealed to be a good therapeutic modality
compared with definitive CRT.
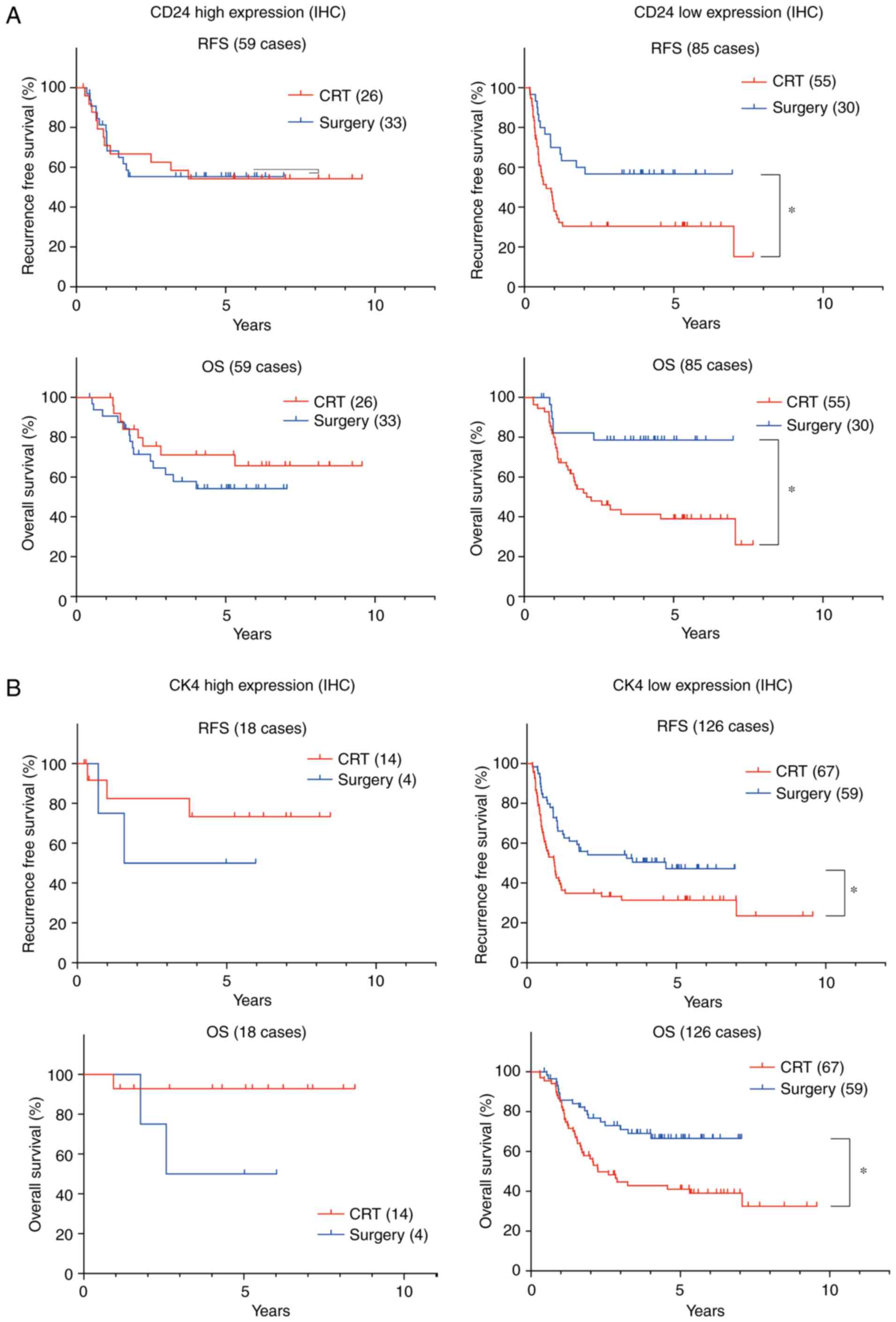 | Figure 4CD24 and CK4 are predictive
biomarkers for definitive CRT and surgery. (A) Based on CD24 and
CK4 protein expression, prognosis was compared between 81 patients
with ESCC undergoing CRT and 63 patients with ESCC undergoing
surgery. In patients with high CD24 expression, there was no
significant difference in RFS and OS between 26 patients undergoing
definitive CRT and 33 patients undergoing surgery, whereas in
patients with low CD24 expression, there was a significant
difference in RFS and OS between 55 patients undergoing definitive
CRT and 30 patients undergoing surgery. (B) Similarly, in patients
with high CK4 expression, there was no significant difference in
RFS and OS between 14 patients undergoing definitive CRT and four
patients undergoing surgery, whereas in patients with low CK4
expression, there was a significant difference in RFS and OS
between 67 patients undergoing definitive CRT and 59 patients
undergoing surgery. *P<0.05. CK4, cytokeratin; CRT,
chemoradiotherapy; IHC, immunohistochemistry; OS, overall survival;
RFS, recurrence-free survival. |
 | Table IIIMultivariate analysis of RFS and OS
in patients with low CD24 expression. |
Table III
Multivariate analysis of RFS and OS
in patients with low CD24 expression.
| Variable | n (%) | RFS
| OS
|
|---|
| Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age | | | | | | | |
| <60 years | 18 (21.2) | Reference | 0.559-2.573 | 0.64 | Reference | 0.453-2.582 | 0.86 |
| ≥60 years | 67 (78.8) | 1.2 | | | 1.082 | | |
| Sex | | | | | | | |
| Male | 75 (88.2) | Reference | 0.421-2.822 | 0.86 | Reference | 0.723-5.161 | 0.189 |
| Female | 10 (11.8) | 1.089 | | | 1.931 | | |
| Macroscopic
types | | | | | | | |
| Types 1 and 2 | 51 (60.0) | Reference | 0.588-1.867 | 0.874 | Reference | 0.930-3.474 | 0.081 |
| Type 3 | 34 (40.0) | 1.048 | | | 1.798 | | |
| Tissue type | | | | | | | |
| W/D and M/D | 75 (88.2) | Reference | 0.488-2.857 | 0.721 | Reference | 0.924-5.869 | 0.073 |
| P/D | 10 (11.8) | 1.181 | | | 2.328 | | |
| Location | | | | | | | |
| Ut and Mt | 78 (91.8) | Reference | 0.707-2.224 | 0.439 | Reference | 0.666-2.493 | 0.452 |
| Lt | 7 (8.2) | 1.254 | | | 1.288 | | |
| Circumference | | | | | | | |
| <3/4 | 49 (57.6) | Reference | 0.858-2.826 | 0.145 | Reference | 0.995-4.040 | 0.052 |
| ≥3/4 | 36 (42.4) | 1.557 | | | 2.005 | | |
| c T factor | | | | | | | |
| T2 | 14 (16.5) | Reference | 0.759-4.734 | 0.171 | Reference | 0.453-3.356 | 0.682 |
| T3 | 71 (83.5) | 1.896 | | | 1.233 | | |
| c N factor | | | | | | | |
| Absent | 41 (48.2) | Reference | 0.601-2.024 | 0.751 | Reference | 0.560-2.274 | 0.736 |
| Present | 44 (51.8) | 1.103 | | | 1.128 | | |
| Treatment | | | | | | | |
| Surgery | 30 (35.3) | Reference | 1.182-4.397 | 0.014a | Reference | 1.518-9.416 | 0.004a |
| CRT | 55 (64.7) | 2.28 | | | 3.781 | | |
 | Table IVMultivariate analysis of RFS and OS
in patients with low CK4 expression. |
Table IV
Multivariate analysis of RFS and OS
in patients with low CK4 expression.
| Variable | n (%) | RFS
| OS
|
|---|
| Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age | | | | | | | |
| <60 years | 30 (23.8) | Reference | 0.689-2.236 | 0.472 | Reference | 0.604-2.218 | 0.659 |
| ≥60 years | 96 (76.2) | 1.241 | | | 1.158 | | |
| Sex | | | | | | | |
| Male | 111 (88.1) | Reference | 0.639-2.698 | 0.459 | Reference | 0.938-4.367 | 0.072 |
| Female | 15 (11.9) | 1.313 | | | 2.023 | | |
| Macroscopic
types | | | | | | | |
| Types 1 and 2 | 78 (61.9) | Reference | 0.626-1.688 | 0.913 | Reference | 0.371-1.100 | 0.106 |
| Type 3 | 48 (38.1) | 1.028 | | | 0.639 | | |
| Tissue type | | | | | | | |
| W/D and M/D | 108 (85.7) | Reference | 0.508-2.086 | 0.935 | Reference | 0.380-1.695 | 0.565 |
| P/D | 18 (14.3) | 1.03 | | | 0.803 | | |
| Location | | | | | | | |
| Ut and Mt | 69 (54.8) | Reference | 0.419-1.126 | 0.137 | Reference | 0.483-1.419 | 0.492 |
| Lt | 57 (45.2) | 0.687 | | | 0.828 | | |
| Circumference | | | | | | | |
| <3/4 | 68 (54.0) | Reference | 0.698-1.793 | 0.64 | Reference | 0.918-2.609 | 0.101 |
| ≥3/4 | 58 (46.0) | 1.119 | | | 1.548 | | |
| c T factor | | | | | | | |
| T2 | 26 (20.6) | Reference | 1.139-4.838 | 0.021a | Reference | 0.711-3.173 | 0.286 |
| T3 | 100 (79.4) | 2.347 | | | 1.502 | | |
| c N factor | | | | | | | |
| Absent | 67 (53.2) | Reference | 0.748-1.967 | 0.434 | Reference | 0.557-1.621 | 0.851 |
| Present | 59 (46.8) | 1.213 | | | 0.95 | | |
| Treatment | | | | | | | |
| Surgery | 59 (46.8) | Reference | 1.274-3.599 | 0.004a | Reference | 1.317-4.399 | 0.004a |
| CRT | 67 (53.2) | 2.142 | | | 2.407 | | |
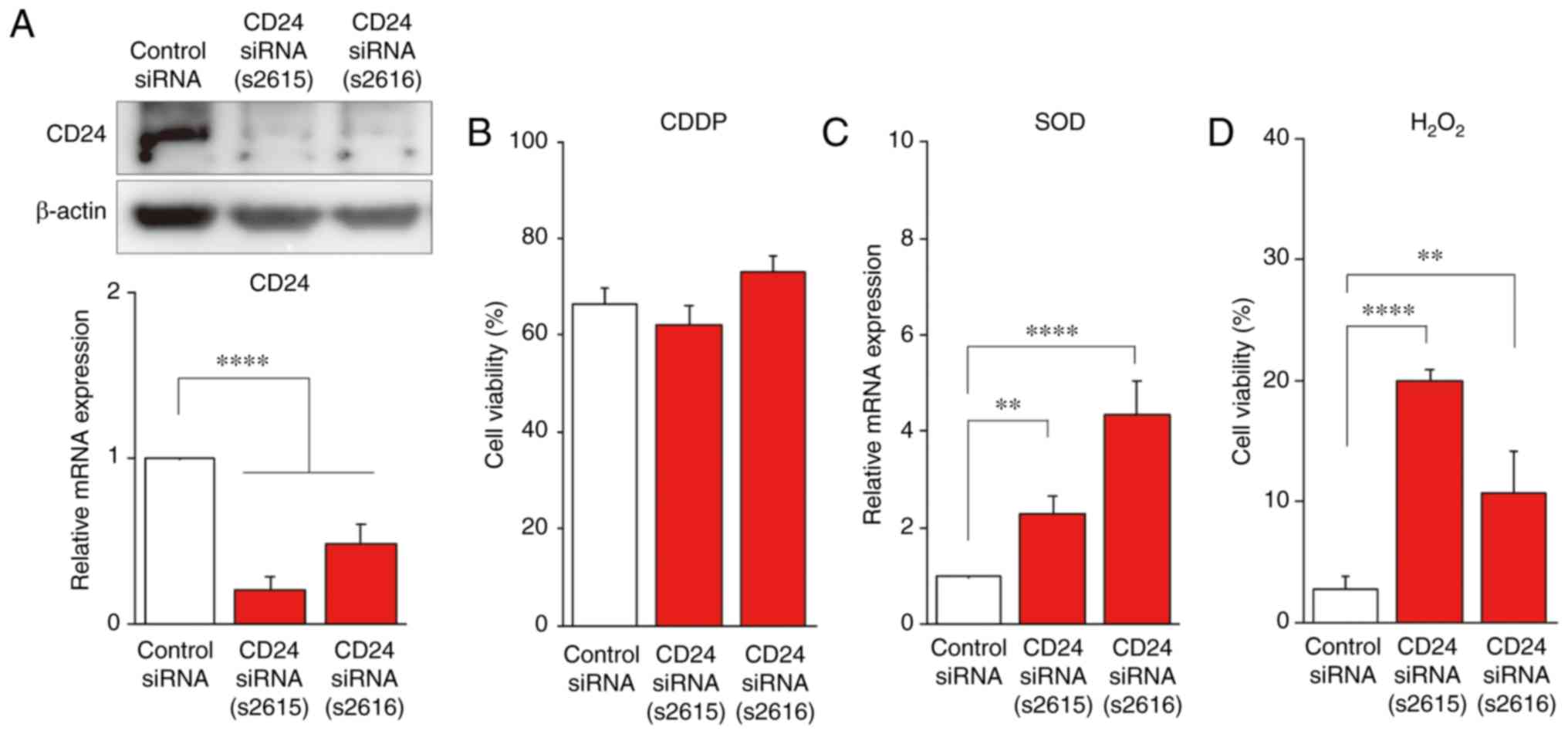
CD24 is associated with radiosensitivity
through superoxide dismutase 2 (SOD2) suppression, but not
chemosensitivity in ESCC cells
In the present study, microarray and IHC analyses of
biopsy specimens from 81 patients with ESCC prior to definitive CRT
revealed that if CD24 mRNA or protein was highly expressed, RFS and
OS were better (Figs. 2 and
3). Furthermore, we recently
reported that SIM2 expression was associated with a favorable
prognosis of patients with ESCC undergoing definitive CRT, and that
SIM2 was involved in chemosensitivity through suppression of
numerous DNA repair genes (X-ray repair cross complementing 5,
BRCA1 DNA repair-associated, FA complementation group D2 and
BRCA1-asssociated RING domain 1) and radiosensitivity through
antioxidant gene (SOD2) suppression (13). These findings indicated that
CD24 may be directly involved in chemosensitivity and/or
radiosensitivity. RT-qPCR was carried out using two CD24
siRNAs (CD24-s2615 and CD24-s2616), and a decrease in CD24
mRNA expression was confirmed (Fig.
5A). Accordingly, CD24 protein expression was also decreased by
CD24 siRNA (Fig. 5A). To
examine the hypothesis that CD24 is involved in CRT sensitivity,
control siRNA-, CD24 siRNA (s2615)- and CD24 siRNA
(s2616)-transfected T.Tn cells were treated with CDDP, which is
used in the standard chemotherapy regimen of ESCC, for 3 days in a
3D culture. The viable ratio of CD24 siRNA (s2615)- or
CD24 siRNA (s2616)-transfected T.Tn cells was not
significantly decreased compared with control siRNA-transfected
T.Tn cells (Fig. 5B), suggesting
that CD24 was not involved in chemosensitivity. However,
CD24 siRNA (s2615)- or CD24 siRNA (s2616)-transfected
T.Tn cells exhibited increased SOD2 mRNA expression compared
with in the control siRNA-transfected T.Tn cells (Fig. 5C). In addition, CD24 siRNAs
were transfected into T.Tn cells and cell viability was
investigated after H2O2 treatment.
CD24 siRNA (s2615)- or CD24 siRNA (s2616)-transfected
T.Tn cells exhibited significantly increased viability following
H2O2 treatment compared with in the control
siRNA-transfected T.Tn cells (Fig.
5D). These findings indicated that CD24 may be involved in
radiosensitivity through SOD2 suppression, but not in
chemosensitivity (Fig. 6).
Discussion
Although definitive CRT improves the prognosis of
patients with ESCC and is an important modality, ~40% of patients
exhibit persistent disease or experience recurrence, resulting in
poor long-term survival (2).
Therefore, predictive biomarkers are needed to select patients who
are potentially curable with definitive CRT. Since preoperative
treatment is increasing for patients with solid tumors, biopsy
specimens of such patients are the only material available that may
be used to predict the effect of neoadjuvant therapy. Great efforts
have been made to identify such predictive biomarkers by numerous
researchers; however, few studies exist that have identifed
biomarkers for definitive CRT using biopsy specimens from patients
with ESCC (4,33). In this study, it was demonstrated
that CD24 and CK4 have great potential to be independent predictive
biomarkers for such patients. Our recent study reported that SIM2
in ESCC was a key transcription factor involved in tumor cell
differentiation and was associated with a good response to CRT
(13). This study revealed that
CD24 and KRT4, which encodes CK4, were
differentiation markers, which were upregulated by SIM2.
Therefore, CD24 and KRT4 may be downstream
differentiation markers of SIM2, and similar to SIM2,
they may serve a role in CRT sensitivity.
Kaplan-Meier analyses revealed that RFS and OS in
the high CD24 and KRT4 mRNA expression groups were
significantly longer than those in the low CD24 and
KRT4 mRNA expression groups. In addition,
immunohistochemical analyses were conducted, and the power of CD24
and CK4 for predicting patients with ESCC and a favorable prognosis
in response to definitive CRT was evaluated. Multivariate Cox
regression analyses revealed that high CD24 or CK4 expression was
an independent favorable prognostic factor in patients undergoing
definitive CRT. Notably, when CD24 or CK4 were highly expressed,
there was no significant difference in RFS and OS between patients
undergoing definitive CRT and those undergoing surgery. However,
when CD24 or CK4 were lowly expressed, there was a significant
difference in RFS and OS between patients undergoing definitive CRT
and those undergoing surgery. Multivariate Cox regression analyses
also indicated a significant difference in RFS and OS between
patients undergoing definitive CRT and those undergoing surgery.
During this study, discrepancies between mRNA and protein levels
were detected in some individual cases. In high or low mRNA
expression groups, these discrepancies are likely decreased if
intermediate cases are removed from these groups, as one microarray
analysis may have variability, particularly in cases with
intermediate mRNA levels; therefore, cases were divided into three
groups with regards to mRNA level (high, intermediate and low). In
summary, for patients with ESCC and low CD24 or CK4 expression, it
may be stated that surgery is preferable to definitive CRT. There
were no significant changes in RFS and OS between patients
undergoing definitive CRT and those undergoing surgery in the high
CD24 or high CK4 groups; however, definitive CRT, which preserves
organs, may be preferable for such patients.
In previous studies, CD24 overexpression has been
reported to be markedly associated with a more aggressive course of
disease (15-18). CD24 may have a role in breast
cancer metastasis (19-21) and has been identified as a
significant poor prognostic factor (34). In ovarian cancer, CD24 is a key
molecule in epithelial-mesenchymal transition (EMT) (35). Furthermore, downregulation of CD24
has been reported to suppress bone metastasis of lung cancer cells
(36). However, the role of CD24
in ESCC remains to be determined.
Our recent studies reported that transfection with
SIM2 reduced the podoplanin (PDPN)-positive basal cell ratio
and improved sensitivity to CDDP (12,13).
Knockdown of PDPN has been reported to reduce resistance to CDDP
(37). In the present study, in
response to CDDP, the number of viable CD24
siRNA-transfected cells was not significantly decreased compared
with the control cells, suggesting that CD24 was not involved in
chemosensitivity. SOD2 is known to efficiently catalyze the
dismutation of reactive oxygen species (38), which are induced by irradiation.
This study demonstrated that CD24 may suppress SOD2
expression and thus reduce resistance to
H2O2. These data indicated that CD24 may be
involved in radiosensitivity through SOD2 suppression, but not in
chemosensitivity (Fig. 6).
Transforming growth factor (TGF)-β is a major
inducer of EMT during embryonic development, as well as the
pathogenesis of fibrotic disorders and cancer progression (39-41).
In ovarian cancer, CD24 and EMT regulators have been reported to be
induced by TGF-β (35). This study
investigated whether TGF-β stimulated the expression of EMT
regulator genes (TWIST1, TWIST2 and SNAI2),
mesenchymal cell marker genes (CDH2 and VIM) and
CD24. As shown in Fig. S2,
TGF-β upregulated CDH2, VIM and SNAI2, but
downregulated CD24, TWIST1 and TWIST2 in T.Tn
cells, suggesting that CD24 was not involved in TGF-β-mediated EMT
in ESCC.
In conclusion, the results of the present study may
foster development of the predictive biomarkers CD24 and CK4 for
selection of the best therapeutic modality, including definitive
CRT, in ESCC. It was hypothesized that IHC of CD24 and CK4 may be
useful for patient stratification; however, biopsy samples are
often too small (2×2 mm) to show a significant difference. For
clinical use, the cut-off values should be determined by future
extensive immunohistochemical analyses using several sections from
multi-institutional cohorts.
Supplementary Data
Funding
This study was supported by the Japan Agency for
Medical Research and Development (Practical Research for Innovative
Cancer Control; grant no. 19ck0106296h0003), Grant-in-Aid for
Scientific Research from the Japan Society for Promotion of Science
(grant nos. 18H03330 and 19K22892), and the National Cancer Center
Research and Development Fund (grant no. 29-A-2).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
KT, HS and TY contributed to the study conception
and design. RK, MK and HS performed the microarray data analyses.
KT, SF, MT and TY performed and evaluated IHC. RK, KT, FC and HS
performed the cell line experiments. KT, TK, HD, KM, MM and TY
analyzed the patient data. KT, SF, RK, FC and HS drafted the
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
participants in this study. All procedures were approved by the
responsible committee on human experimentation at National Cancer
Center East (approval no. 16-97), and were conducted in accordance
with the Helsinki Declaration.
Patient consent for publication
Patients provided informed consent for
publication.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Mr. Richard De Lapp
for editorial comments.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kato K, Muro K, Minashi K, Ohtsu A,
Ishikura S, Boku N, Takiuchi H, Komatsu Y, Miyata Y and Fukuda H;
Gastrointestinal Oncology Study Group of the Japan Clinical
Oncology Group (JCOG): Phase II study of chemoradiotherapy with
5-fluorouracil and cisplatin for stage II-III esophageal squamous
cell carcinoma: JCOG trial (JCOG 9906). Int J Radiat Oncol Biol
Phys. 81:684–690. 2011. View Article : Google Scholar
|
|
3
|
Ando N, Kato H, Igaki H, Shinoda M, Ozawa
S, Shimizu H, Nakamura T, Yabusaki H, Aoyama N, Kurita A, et al: A
randomized trial comparing postoperative adjuvant chemotherapy with
cisplatin and 5-fluorouracil versus preoperative chemotherapy for
localized advanced squamous cell carcinoma of the thoracic
esophagus (JCOG9907). Ann Surg Oncol. 19:68–74. 2012. View Article : Google Scholar
|
|
4
|
Tanaka Y, Aoyagi K, Minashi K, Komatsuzaki
R, Komatsu M, Chiwaki F, Tamaoki M, Nishimura T, Takahashi N, Oda
I, et al: Discovery of a good responder subtype of esophageal
squamous cell carcinoma with cytotoxic T-lymphocyte signatures
activated by chemoradiotherapy. PLoS One. 10:e01438042015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arthur JF and Fenner ML: The influence of
histological grading on prognosis in carcinoma of the tongue (a
computer analysis of 299 cases). Clin Radiol. 17:384–396. 1966.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rowe DE, Carroll RJ and Day CL Jr:
Prognostic factors for local recurrence, metastasis, and survival
rates in squamous cell carcinoma of the skin, ear, and lip.
Implications for treatment modality selection. J Am Acad Dermatol.
26:976–990. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bersten DC, Sullivan AE, Peet DJ and
Whitelaw ML: bHLH-PAS proteins in cancer. Nat Rev Cancer.
13:827–841. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen H, Chrast R, Rossier C, Gos A,
Antonarakis SE, Kudoh J, Yamaki A, Shindoh N, Maeda H, Minoshima S,
et al: Single-minded and Down syndrome? Nat Genet. 10:9–10. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dahmane N, Charron G, Lopes C, Yaspo ML,
Maunoury C, Decorte L, Sinet PM, Bloch B and Delabar JM: Down
syndrome-critical region contains a gene homologous to Drosophila
sim expressed during rat and human central nervous system
development. Proc Natl Acad Sci USA. 92:9191–9195. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Swanson HI, Chan WK and Bradfield CA: DNA
binding specificities and pairing rules of the Ah receptor, ARNT,
and SIM proteins. J Biol Chem. 270:26292–26302. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
DeYoung MP, Tress M and Narayanan R:
Identification of Down's syndrome critical locus gene SIM2-s as a
drug therapy target for solid tumors. Proc Natl Acad Sci USA.
100:4760–4765. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nakamura K, Komatsu M, Chiwaki F, Takeda
T, Kobayashi Y, Banno K, Aoki D, Yoshida T and Sasaki H: SIM2l
attenuates resistance to hypoxia and tumor growth by
transcriptional suppression of HIF1A in uterine cervical squamous
cell carcinoma. Sci Rep. 7:145742017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tamaoki M, Komatsuzaki R, Komatsu M,
Minashi K, Aoyagi K, Nishimura T, Chiwaki F, Hiroki T, Daiko H,
Morishita K, et al: Multiple roles of single-minded 2 in esophageal
squamous cell carcinoma and its clinical implications. Cancer Sci.
109:1121–1134. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kay R, Rosten PM and Humphries RK: CD24, a
signal transducer modulating B cell activation responses, is a very
short peptide with a glycosyl phosphatidylinositol membrane anchor.
J Immunol. 147:1412–1416. 1991.PubMed/NCBI
|
|
15
|
Deng J, Gao G, Wang L, Wang T, Yu J and
Zhao Z: CD24 expression as a marker for predicting clinical outcome
in human gliomas. J Biomed Biotechnol. 2012:5171722012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Su N, Peng L, Xia B, Zhao Y, Xu A, Wang J,
Wang X and Jiang B: Lyn is involved in CD24-induced ERK1/2
activation in colorectal cancer. Mol Cancer. 11:432012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu C, Zheng S, Shen H, Xu K, Chen J, Li
H, Xu Y, Xu A, Chen B, Kaku H, et al: Clinical significance of CD24
as a predictor of bladder cancer recurrence. Oncol Lett. 6:96–100.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tanaka T, Terai Y, Kogata Y, Ashihara K,
Maeda K, Fujiwara S, Yoo S, Tanaka Y, Tsunetoh S, Sasaki H, et al:
CD24 expression as a marker for predicting clinical outcome and
invasive activity in uterine cervical cancer. Oncol Rep.
34:2282–2288. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Baumann P, Cremers N, Kroese F, Orend G,
Chiquet-Ehrismann R, Uede T, Yagita H and Sleeman JP: CD24
expression causes the acquisition of multiple cellular properties
associated with tumor growth and metastasis. Cancer Res.
65:10783–10793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schabath H, Runz S, Joumaa S and Altevogt
P: CD24 affects CXCR4 function in pre-B lymphocytes and breast
carcinoma cells. J Cell Sci. 119:314–325. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vazquez-Martin A, Oliveras-Ferraros C,
Cufí S, Del Barco S, Martin-Castillo B, Lopez-Bonet E and Menendez
JA: The anti-diabetic drug metformin suppresses the
metastasis-associated protein CD24 in MDA-MB-468 triple-negative
breast cancer cells. Oncol Rep. 25:135–140. 2011.
|
|
22
|
Takahashi M, Nakajima M, Ogata H, Domeki
Y, Ohtsuka K, Ihara K, Kurayama E, Yamaguchi S, Sasaki K, Miyachi K
and Kato H: CD24 expression is associated with progression of
gastric cancer. Hepatogastroenterology. 60:653–658. 2013.
|
|
23
|
Overdevest JB, Thomas S, Kristiansen G,
Hansel DE, Smith SC and Theodorescu D: CD24 offers a therapeutic
target for control of bladder cancer metastasis based on a
requirement for lung colonization. Cancer Res. 71:3802–3811. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Thomas S, Harding MA, Smith SC, Overdevest
JB, Nitz MD, Frierson HF, Tomlins SA, Kristiansen G and Theodorescu
D: CD24 is an effector of HIF-1-driven primary tumor growth and
metastasis. Cancer Res. 72:5600–5612. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li C, Heidt DG, Dalerba P, Burant CF,
Zhang L, Adsay V, Wicha M, Clarke MF and Simeone DM: Identification
of pancreatic cancer stem cells. Cancer Res. 67:1030–1037. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gao MQ, Choi YP, Kang S, Youn JH and Cho
NH: CD24+ cells from hierarchically organized ovarian cancer are
enriched in cancer stem cells. Oncogene. 29:2672–2680. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sakamoto K, Aragaki T, Morita K, Kawachi
H, Kayamori K, Nakanishi S, Omura K, Miki Y, Okada N, Katsube K, et
al: Down-regulation of keratin 4 and keratin 13 expression in oral
squamous cell carcinoma and epithelial dysplasia: A clue for
histopathogenesis. Histopathology. 58:531–542. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chung JY, Braunschweig T, Hu N, Roth M,
Traicoff JL, Wang QH, Knezevic V, Taylor PR and Hewitt SM: A
multiplex tissue immunoblotting assay for proteomic profiling: A
pilot study of the normal to tumor transition of esophageal
squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev.
15:1403–1408. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schaaij-Visser TB, Graveland AP, Gauci S,
Braakhuis BJ, Buijze M, Heck AJ, Kuik DJ, Bloemena E, Leemans CR,
Slijper M and Brakenhoff RH: Differential proteomics identifies
protein biomarkers that predict local relapse of head and neck
squamous cell carcinomas. Clin Cancer Res. 15:7666–7675. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Oken MM, Creech RH, Tormey DC, Horton J,
Davis TE, McFadden ET and Carbone PP: Toxicity and response
criteria of the eastern cooperative oncology group. Am J Clin
Oncol. 5:649–655. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sobin LH and Wittekind C: TNM
classification of malignant tumors. 6th edition. Wiley-Liss; New
York, NY: 2002
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
33
|
Ashida A, Boku N, Aoyagi K, Sato H,
Tsubosa Y, Minashi K, Muto M, Ohtsu A, Ochiai A, Yoshida T, et al:
Expression profiling of esophageal squamous cell carcinoma patients
treated with definitive chemoradiotherapy: Clinical implications.
Int J Oncol. 28:1345–1353. 2006.PubMed/NCBI
|
|
34
|
Okabe H, Aoki K, Yogosawa S, Saito M,
Marumo K and Yoshida K: Downregulation of CD24 suppresses bone
metastasis of lung cancer. Cancer Sci. 109:112–120. 2018.
View Article : Google Scholar :
|
|
35
|
Nakamura K, Terai Y, Tanabe A, Ono YJ,
Hayashi M, Maeda K, Fujiwara S, Ashihara K, Nakamura M, Tanaka Y,
et al: CD24 expression is a marker for predicting clinical outcome
and regulates the epithelial-mesenchymal transition in ovarian
cancer via both the Akt and ERK pathways. Oncol Rep. 37:3189–3200.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Moon YW, An HJ, Koo JS, Kim GM, Han H,
Park S, Kim SI, Park HS, Kim S, Kim SK, et al: CD44/CD24 and
aldehyde dehydrogenase 1 in estrogen receptor-positive early breast
cancer treated with tamoxifen: CD24 positivity is a poor
prognosticator. Oncotarget. 9:2622–2630. 2017.
|
|
37
|
Rahadiani N, Ikeda J, Makino T, Tian T,
Qiu Y, Mamat S, Wang Y, Doki Y, Aozasa K and Morii E: Tumorigenic
role of podoplanin in esophageal squamous-cell carcinoma. Ann Surg
Oncol. 17:1311–1323. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zelko IN, Mariani TJ and Folz RJ:
Superoxide dismutase multi-gene family: A comparison of the
CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures,
evolution, and expression. Free Radic Biol Med. 33:337–349. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Taylor MA, Parvani JG and Schiemann WP:
The pathophysiology of epithelial-mesenchymal transition induced by
transforming growth factor-beta in normal and malignant mammary
epithelial cells. J Mammary Gland Biol Neoplasia. 15:169–190. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zavadil J and Böttinger EP: TGF-beta and
epithelial-to-mesenchymal transitions. Oncogene. 24:5764–5774.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee YH, Albig AR, Regner M, Schiemann BJ
and Schiemann WP: Fibulin-5 initiates epithelial-mesenchymal
transition (EMT) and enhances EMT induced by TGF-beta in mammary
epithelial cells via a MMP-dependent mechanism. Carcinogenesis.
29:2243–2251. 2008. View Article : Google Scholar : PubMed/NCBI
|