Introduction
Since the otolaryngologist Danely Slaughter
introduced the concept of field cancerization in 1953 in stratified
squamous epithelia of the oral mucosa of patients suffering from
oral diseases (1), the definition
of this term, also known as the field effect, field defect, or
field carcinogenesis, has changed mainly due to the rapid
development of analytical techniques in molecular biology and
genetics developed over the past decades. First, the original
intent was to describe the occurrence of cancerous cells in
histologically normal tissues at some distances from the primary
lesions, and to explain the multifocality of solid tumors,
particularly in the case of oral cancers, including squamous cell
carcinomas (1,2). These cells are by definition
clinically detectable. By contrast, the current definition of field
cancerization applies to structurally and phenotypically intact
cells residing in histologically normal tissues outside the
confinement of the primary tumor. These cells do not typically
distinguish themselves from their surrounding and thus remain
clinically hidden. The latter points to the second major change of
the term 'field cancerization', i.e., the shift in focus on the
molecular characterization of affected cells and tissues, as
opposed to their phenotypical appearance (3). Consequently, cells that are part of
'field cancerized' tissues are considered to be molecularly altered
in the absence of other visually obvious changes. Furthermore, the
nature of these molecular alterations is typically indicative of a
positively 'primed' or 'committed' status with respect to cell
proliferation, growth, migration and/or survival, essentially
delineating the affected cells as pre-malignant (4-6). The
authors have continuously contributed to the molecular
characterization of field cancerization in prostate tissues by
describing both genetic and biochemical deviations from normalcy
(7-13). This has included the observation of
telomere attrition (9), as well as
the upregulation of protein expression, including the key
transcription factor early growth response 1 (EGR-1) and the
anabolic enzyme fatty acid synthase (FASN) (7,11).
Although the importance of field cancerization in
representing a type of pre-malignancy in tissues that are prone to
tumorigenesis has been recognized and acknowledged, the molecular
and cellular mechanisms underpinning its etiology, while often
discussed, remain largely unknown (4-6).
This is also true for field cancerization in prostatic tissues and
stands in contrast to the growing list of molecular and cellular
markers describing it (13,14).
A recent focus in urological research has been the functional role
of extracellular vesicles released by virtually all types of cells
in the prostate as part of inter-cell and inter-organ compartment
communication (15,16). These extracellular vesicles include
exosomes that have been characterized to be in the range of 30-150
nanometers in diameter. The importance of exosomal function in
normal prostate physiology has been well recognized and is
primarily due to their biologically active 'cargo' that includes
multiple types of RNAs, lipids and proteins (17-20).
Conceptually, the biochemical composition of exosomes reflects the
current physiology of the cell of origin. It is thus not
inconceivable to assume that a specific physiological signature can
be conveyed or transferred to recipient cells. This line of thought
has led to the hypothesis of a potential role of exosome release
and action in the etiology of field cancerization. The authors have
thus begun to test this hypothesis by assessing a potential
correlation in protein expression between the exosomal marker and
tetraspanin, CD9, and the afore-mentioned field cancerization
markers, EGR-1 and FASN, in human prostate tissues. Cultured cell
models of prostate cancer were also used to corroborate these
findings. The results indicate a possible association between
exosomes and the expression of EGR-1 and potentially, that of FASN
in prostate tissues affected by field cancerization. This novel
insight into pathways underlying prostate field effect may lead to
the development of targeted intervention strategies preventing
progression from pre-malignancy to cancer.
Materials and methods
Tissues and cells
Tissue microarrays (TMAs) were purchased from US
Biomax, Inc. (https://www.biomax.us/). No human
tissues from other sources, other than commercially available TMAs,
were used in the present study. The use of any human tissues,
including commercially available TMAs, is covered by the Chapman
University Institutional Review Board (IRB) study #1415H024. For
the present study, the formalin-fixed and paraffin-embedded (FFPE)
TMA BC 19021a was used, featuring 5-µm-thick cancerous,
tumor-adjacent and disease-free (normal) human prostate tissue
cores of 1.5 mm in diameter. Experiments with human tissues was
approved by the Institutional Review Board of Chapman University.
The tissue cohort analyzed in this study consisted of 8
adenocarcinomas, 8 tumor-adjacent tissues, and 6 disease-free
tissues. These were selected for inclusion to represent variation
in age and tumor stage and based on immunofluorescence quality. The
matching status of the tumor-adjacent tissues with the featured
tumors is unknown. Also unknown was the distance from the tumor
margin at which adjacent tissues were resected. However, a common
practice in our own research is resection at a distance of
approximately 1 cm from the visible tumor margin (7-11,21).
The definition of the term 'disease-free' refers to prostate
specimens from autopsy cases from individuals who died due to
conditions unrelated to cancer. The mean age of all cases utilized
was 54.7 years with a range of 21-80 years. The cancer specimens
featured Gleason scores from 4 to 10 and pathological tumor node
metastasis (TNM) stages (according to the American Joint Committee
on Cancer; https://cancerstaging.org/Pages/default.aspx) from
T2aN0M0 to T2N1M1b (Table I).
 | Table IDemographics and clinical parameters
of prostate tissues, and the no. of images analyzed. |
Table I
Demographics and clinical parameters
of prostate tissues, and the no. of images analyzed.
| Prostate
tissues | Age, years | TNMa | Gleason score | No. of images
|
|---|
| CD9 | EGR-1 | FASN |
|---|
| Disease-free | | | | | | |
| 1 | 21 | | | 6 | 6 | 3 |
| 2 | 21 | | | 5 | 5 | 5 |
| 3 | 25 | | | 6 | 10 | 6 |
| | Not applicable | | | | |
| 4 | 25 | | | 6 | 9 | 4 |
| 5 | 27 | | | 6 | 8 | 5 |
| 6 | 27 | | | 6 | 10 | 5 |
| Total | | | | 35 | 48 | 28 |
| Adjacent | | | | | | |
| 1 | 40 | | | 7 | 8 | 5 |
| 2 | 76 | | | 6 | 7 | 3 |
| 3 | 73 | | | 7 | 8 | 7 |
| 4 | 73 | | | 8 | 8 | 7 |
| | Not available | | | | |
| 5 | 62 | | | 6 | 9 | 5 |
| 6 | 62 | | | 6 | 6 | 6 |
| 7 | 72 | | | 7 | 7 | 7 |
| 8 | 80 | | | 6 | 7 | 4 |
| Total | | | | 53 | 60 | 44 |
| Tumor | | | | | | |
| 1 | 76 | T2aN0M0 | 4 (2+2) | 6 | 6 | 5 |
| 2 | 76 | T2aN0M0 | 4 (2+2) | 6 | 6 | 2 |
| 3 | 72 | T2N0M0 | 6 (3+3) | 7 | 8 | 4 |
| 4 | 72 | T2N0M0 | 6 (3+3) | 6 | 7 | 5 |
| 5 | 40 | T2N1M1b | 9 (5+4) | 6 | 6 | 6 |
| 6 | 40 | T2N1M1b | 9 (5+4) | 7 | 5 | 5 |
| 7 | 72 | T2N0M1 | 10 (5+5) | 6 | 6 | 5 |
| 8 | 72 | T2N0M1 | 10 (5+5) | 5 | 7 | 5 |
| Total | | | | 49 | 51 | 37 |
Non-cancerous RWPE-1 and cancerous LNCaP human
prostate epithelial cells were purchased from the American Type
Culture Collection (ATCC) and cultured in serum-free keratinocyte
basal medium containing 4,500 mg/l glucose, 0.05 mg/ml bovine
pituitary extract and 5 ng/ml recombinant epidermal growth factor
(EGF) (for RWPE-1), or in RPMI-1640 medium supplemented with 10%
heat-inactivated (56°C, 1 h) fetal bovine serum (FBS) and 100 U/ml
penicillin/streptomycin (for LNCaP). Cells were maintained at 37°C
in a humidified 5% CO2 atmosphere. Trypsin-EDTA at 0.25%
was used to detach the cells for splitting and re-culturing.
Immunofluorescence
Immunofluorescence was performed as described in
previous studies by the authors on prostate field cancerization
(7,10,11).
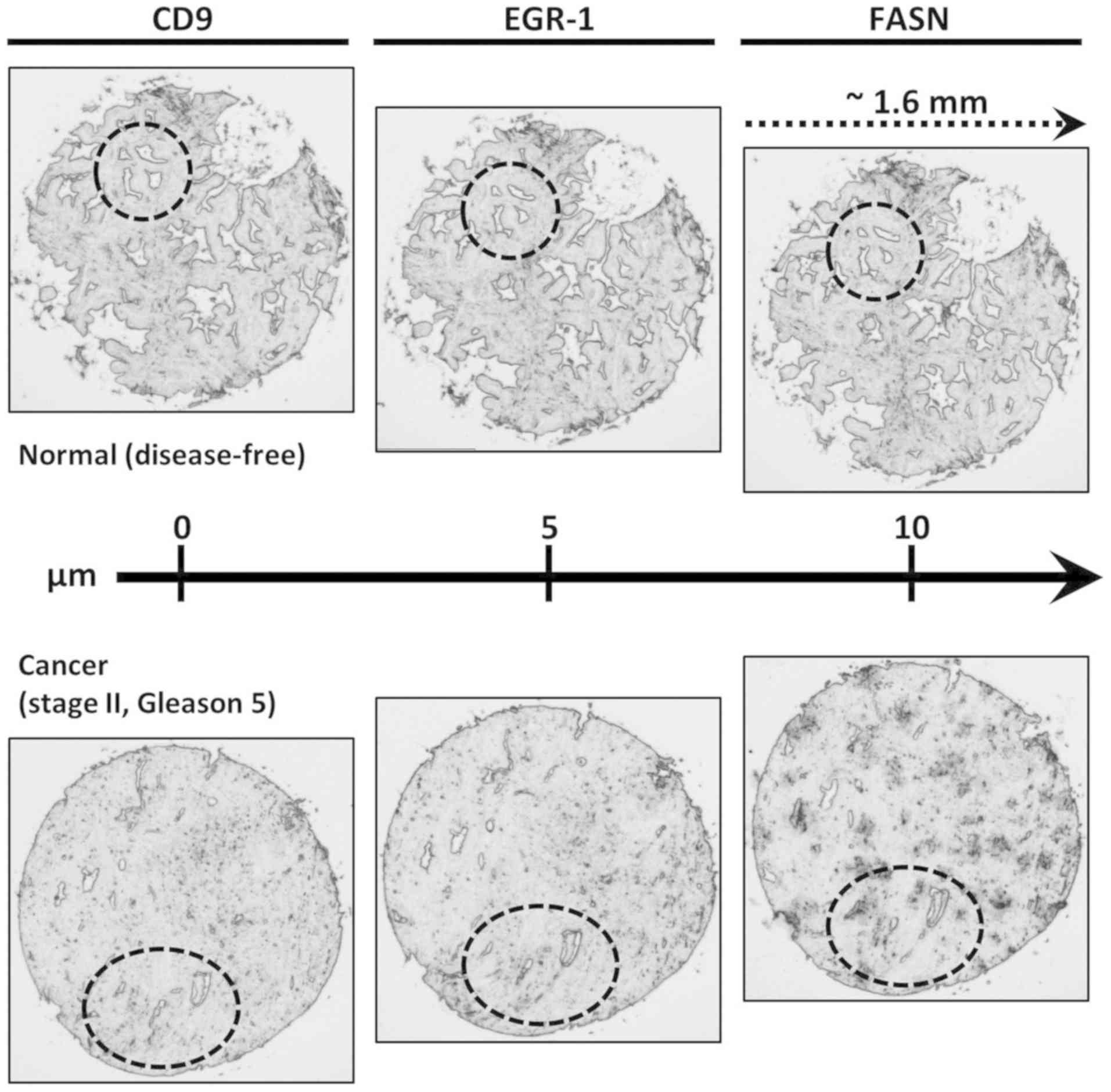
In order to query the same tissue areas for the three protein
markers under investigation (CD9, EGR-1 and FASN), consecutive
sections, each 5 µm apart from each other were used
(Fig. 1). TMAs were subjected to
deparaffinization with xylene and rehydrated with decreasing
concentrations of ethanol. Antigen retrieval was performed in
boiling 10 mM Tris, 1 mM EDTA, 0.05% Tween-20, pH 9.0 (by HCl) for
10 min, washed briefly in tap water, followed by gentle agitation
in Tris buffered saline (TBS; 50 mM Tris, 150 mM NaCl, pH 7.6 by
HCl) containing 0.025% Triton X-100 (TBST). Tissues were blocked in
10% normal goat serum (sc-2040, Santa Cruz Biotechnology) in TBS
containing 1% bovine serum antigen (BSA) for 2 h at room
temperature, then incubated with primary antibodies in TBS
containing 1% BSA at 4°C overnight. These were all mouse monoclonal
antibodies from Abcam used at 3 µg/ml: Anti-CD9 (ab2215),
anti-EGR-1 (ab55160) and anti-FASN (ab218306). The control antibody
to ensure target specificity at the same concentration was normal
mouse IgG (GC270, Millipore). The corresponding secondary antibody,
used at a dilution of 1:750, was Alexa Fluor 594-conjugated goat
anti-mouse IgG (A11005, Life Technologies; Thermo Fisher
Scientific; excitation at 590 nm, emission at 618 nm). Nuclear
counterstaining was performed with diamidino-2-phenylindole (DAPI)
in TBS for 2 min. Fluorescence was preserved using Fluoroshield
solution (Sigma) under coverslips sealed with nail polish. Cells
were cultured and prepared for qualitative immunofluorescence
concomitantly (on the same slide) on Millicell EZ slides
(Millipore) to ensure equal experimental and thus comparative
treatment. The cells were fixed in 4% formaldehyde followed by 3
washes in TBS and stained as for the tissues above.
Fluorescence for both cells and tissues was detected
using an A1R Nikon confocal microscope available at the Chapman
University School of Pharmacy Microscopy Core Facility. For the
tissues, fluorescence was quantified using the NIS-Elements AR
4.30.02/64bit software to analyze acquired digital images.
Consistent with previous studies by the authors (7,10,11),
the fluorescence signal acquisition mode was applied to 2-10 images
per tissue sample. Great care was taken in choosing tissue areas
with as equal as possible numbers of DAPI-stained nuclei from
epithelial compartments to account for equal number of cells
analyzed. In addition, identical areas on the consecutive sections
were imaged for the three specific protein markers (CD9, EGR-1 and
FASN) to allow the reported correlation analyses. For all specific
markers, the acquisition setting was kept identical for all images
taken to ensure the validity of intra- and inter-tissue
comparisons. The number of images amenable to quantitative
fluorescence analysis per individual tissue and protein marker
under investigation is indicated in Table I. In total, 336 images with
associated quantitative immunofluorescence data were available for
the present analysis.
Isolation of exosomes
Exosomes were isolated from cancerous LNCaP and from
non-cancerous RWPE-1 cells according to the Current Protocols in
Cell Biology (John Wiley & Sons, Inc.) using
ultracentrifugation in an Optima XE-90 ultracentrifuge (Beckman
Coulter). Cells grown to 80% confluency in complete growth medium
were washed twice in medium without serum, then incubated at 37°C
for 48 h in medium containing 10% exosome-free FBS. Exosome-free
FBS was prepared by a 2-h long ultracentrifugation of medium/20%
FBS at 100,000 × g at 4°C to pellet exosomes stemming from the FBS.
Upon collection of the medium from the cells, it was subjected to
the following sequential centrifugation procedure (all steps at
4°C): i) 300 × g for 10 min to remove live cells; ii) 2,000 × g for
10 min to remove dead cells; iii) 10,000 × g for 30 min to remove
cell debris; iv) 100,000 × g for 2 h to pellet the exosomes; v) the
resulting pellet was washed in phosphate buffered saline (PBS) to
remove contaminating proteins and recentrifuged at 100,000 × g for
2 h to obtain exosomes of high purity; vi) the final pellet was
resuspended in a small volume of PBS, typically 150 µl for
an original of 40 ml culture supernatant. Exosomes were aliquoted
and stored short-term at −80°C with avoidance of multiple
freeze-thaw cycles. The amount and viability of cells giving rise
to the exosomes was determined by trypan blue exclusion assay and
the cells were frozen at −80°C for western blot analysis (please
see below).
The morphology of the isolated exosomes was further
characterized by atomic force microscopy using an MFP-3D origin
atomic force microscope (AFM; Asylum Research). A 25 µl
aliquot was dropped on amine-functionalized (3-amino-propyl)
trimethoxysilane (APTMS) glass cover slips and dried in air. The
cover slips were washed by sonication with water, acetone, ethanol
and isopropyl alcohol before re-soaked in an ethanol solution of
APTMS for 2 h. Conical-shaped silicon AFM probes with Al reflex
coating (k=42 N/m) were mounted on the cantilever holder and
operated in AC mode. AFM data were processed using MFP3D software
written in an IgorPro environment (Wavemetrics). Exosome dimensions
on the digitized images were quantified using ImageJ software
(https://imagej.nih.gov/ij/).
Treatment of cells with exosomes
RWPE-1 cells were seeded in 6-well plates at a
density of 0.5×106 cells per well and incubated at 37°C
for settlement and growth for 24 h. Either 5 or 50 µg of
LNCaP or 50 µg RWPE-1 exosomes (as a negative control),
corresponded to 1.5×106 and 15.0×106
exosome-producing cells, respectively. The cells were incubated at
37°C for 24 h, washed twice in PBS following the removal of the
supernatant, and collected by either scraping or trypsinization
followed by mild centrifugation at 2,600 × g for 10 min at 4°C.
Cell pellets were snap-frozen in liquid nitrogen to preserve RNA
and protein integrity and were stored short term at −80°C. Scraped
and trypsinized cells were used for analysis by western blot
analysis and reverse transcription-quantitative polymerase chain
reaction (RT-qPCR), respectively (please see below).
Western blot analysis
Western blot analysis was performed on both the
cultured cells and the exosomes derived therefrom. Protein lysates
were generated on ice in lysis buffer: 25 mM Tris, 8 mM
MgCl2, 1 mM DTT, 15% glycerol, 1% Triton X-100 and
protease inhibitor cocktail (Sigma). Insoluble material was removed
by centrifugation of the lysates at 20,000 × g for 10 min at 4°C.
The protein concentration was determined by Bradford assay (Sigma)
against a bovine serum albumin (BSA) standard. In total, 80
µg (for cell lysates) or 20 µg (for exosomal lysates)
total protein were size-separated by sodium dodecyl sulfate
polyacrylamide gel electrophoresis (SDS-PAGE) and
electro-transferred onto polyvinylidene (PVDF) membranes. The
membranes were stained with 0.1% Ponceau S
[3-hydroxy-4-(2-sulfo-4-[4-sulfophenylazo]
phenylazo)-2,7-naphthalenedisulfonic acid; Sigma] in 5% acetic acid
for 5 min at room temperature to visualize the blotted proteins.
Following 2 brief washes in TBS, the membranes were blocked with 5%
milk powder in TBS containing 0.05% Tween-20 (TBST) and probed
overnight at 4°C with the anti-CD9, anti-EGR-1 and anti-FASN
antibodies listed above in the 'Immunofluorescence' paragraph, and
with anti-androgen receptor (AR) antibody (sc-816; AR (N-20), Santa
Cruz Biotechnology), and with anti-β-actin antibody (A1978, Sigma)
(for cell extracts) and anti-actin antibody (A3853, Sigma) (for
exosomes extracts) at typical concentrations of 0.2 µg/ml in
TBST. The detection and chemiluminescent visualization (Clarity ECL
Substrate, Bio-Rad) of target proteins was performed using
secondary horseradish peroxidase-conjugated goat anti-mouse (A0168;
Sigma) and goat anti-rabbit (A0546; Sigma) antibodies used at
1:15,000 dilutions for 1 h at room temperature. Band intensity
(expression level) was quantified by densitometry using ImageJ
software (https://imagej.nih.gov/ij/).
RT-qPCR
RNA was isolated using spin column chromatography
(Qiagen). In total, 1-3 µg of RNA was transcribed into cDNA
using random decamers of the Retroscript RT kit (Life Technologies;
Thermo Fisher Scientific). mRNA expression was quantitated in a CFX
Connect Real Time PCR Detection System from Bio-Rad using the
SYBR-Green PCR Master Mix and SYBR-Green RT-PCR Reagents kit (Life
Technologies; Thermo Fisher Scientific) in 25 µl reactions,
using 100 ng of template cDNA and a final primer concentration of
900 nM. The cycling parameters were 95°C for 5 min followed by 45
cycles of 94°C for 15 sec and 60°C for 1 min. Primers were designed
using Primer Express software (Invitrogen; Thermo Fisher
Scientific) and synthesized by Integrated DNA Technologies. The
following primer sequences (5′ to 3′) were used: EGR-1 forward,
GAGCAGCCCTAC GAGCAC and reverse, AGCGGCCAGTATAGGTGATG; FASN
forward, AGAACTTGCAGGAGTTCTGGGACA and reverse,
TCCGAAGAAGGAGGCATCAAACCT; TATA binding protein (TBP) forward,
CACGAACCACGGCAC TGATT and TBP reverse, TTTTCTTGCTGCCAGTCTGGAC.
RT-qPCR reactions were performed in triplicate. Relative expression
levels were determined by the ΔΔCq method (22) using TBP as the normalization
control after determining that amplification efficiencies were
similar to the ones of the control transcripts.
Statistical analysis
CD9, EGR-1 and FASN expression levels are
represented by signal intensities (sum pixel count per area)
generated by quantitative immunofluorescence analysis (as described
above). Simple, yet straightforward statistical methods were
applied to the datasets using the Microsoft Office Excel software
package. Due to our previously observed and well-known intra- and
inter-specimen heterogeneity in tissue expression studies (7,10,11),
the datasets were inclusive, i.e., all available informative images
were utilized, and no computational calculation was used to
identify potential outliers. The infinite variance due to tissue
heterogeneity is expressed as the coefficient of variation in % in
the text of the 'Results' section. Single factor analysis of
variance (ANOVA) was used to compare multiple datasets with unequal
variances. The post hoc Fishers' least significant difference (LSD)
test was used to determine the significance of the difference
between the means of the datasets. ANOVA followed by LSD was also
used to analyze the results of western blot analysis and RT-qPCR.
Statistical significance in these comparisons of the means was
defined as P≤0.05. The datasets were mined for potential
associations between CD9 and EGR-1 and between CD9 and FASN by
determining the Pearson's correlation coefficient r. The
significance for these observations was determined by first
calculating the t-value of the correlation using the equation
t=r/SQRT(1-r2/n-2), where 'r' is the correlation
coefficient, 'n' is the number of samples, and '2' is the degree of
freedom. The t-value was then used to determine the significance of
'r' by the two-tailed Student t-distribution (TDIST; statistical
significance defined as P≤0.05).
Results
Detection of CD9, EGR-1 and FASN
expression in human prostate cells and tissues
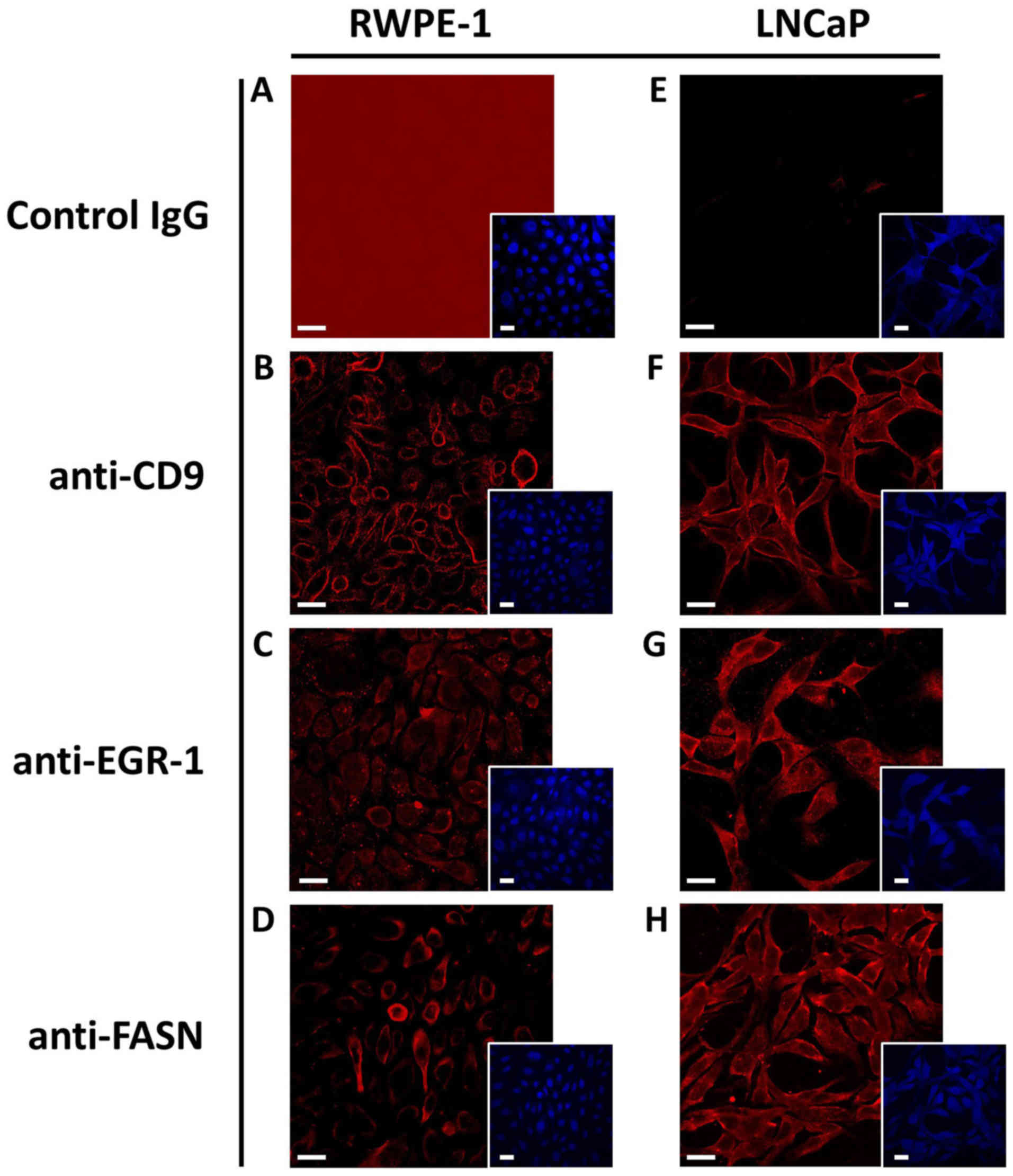
The antibodies used to detect CD9, EGR-1 and FASN by
immunofluorescence in human tissues were first tested in the human
non-cancerous RWPE-1 and in the cancerous LNCaP cell models, which
allowed the illustration of the specificity of the antibodies. As
shown in Fig. 2, CD9 staining was
primarily evident for the cell surface, while staining for FASN was
primarily cytoplasmic for both cell models, as expected. EGR-1
staining seemed to be somewhat more diffuse, in agreement with its
reported possible localization in both the nucleus and the
cytoplasm, depending on cellular type and context (11,23).
In addition, the expression level for all three markers was
slightly higher in the LNCaP than in the RWPE-1 cells. Important
for the use of the antibodies in human tissues, the isotype-matched
unspecific control antibody resulted in minimal, if any, staining
in both cell types.
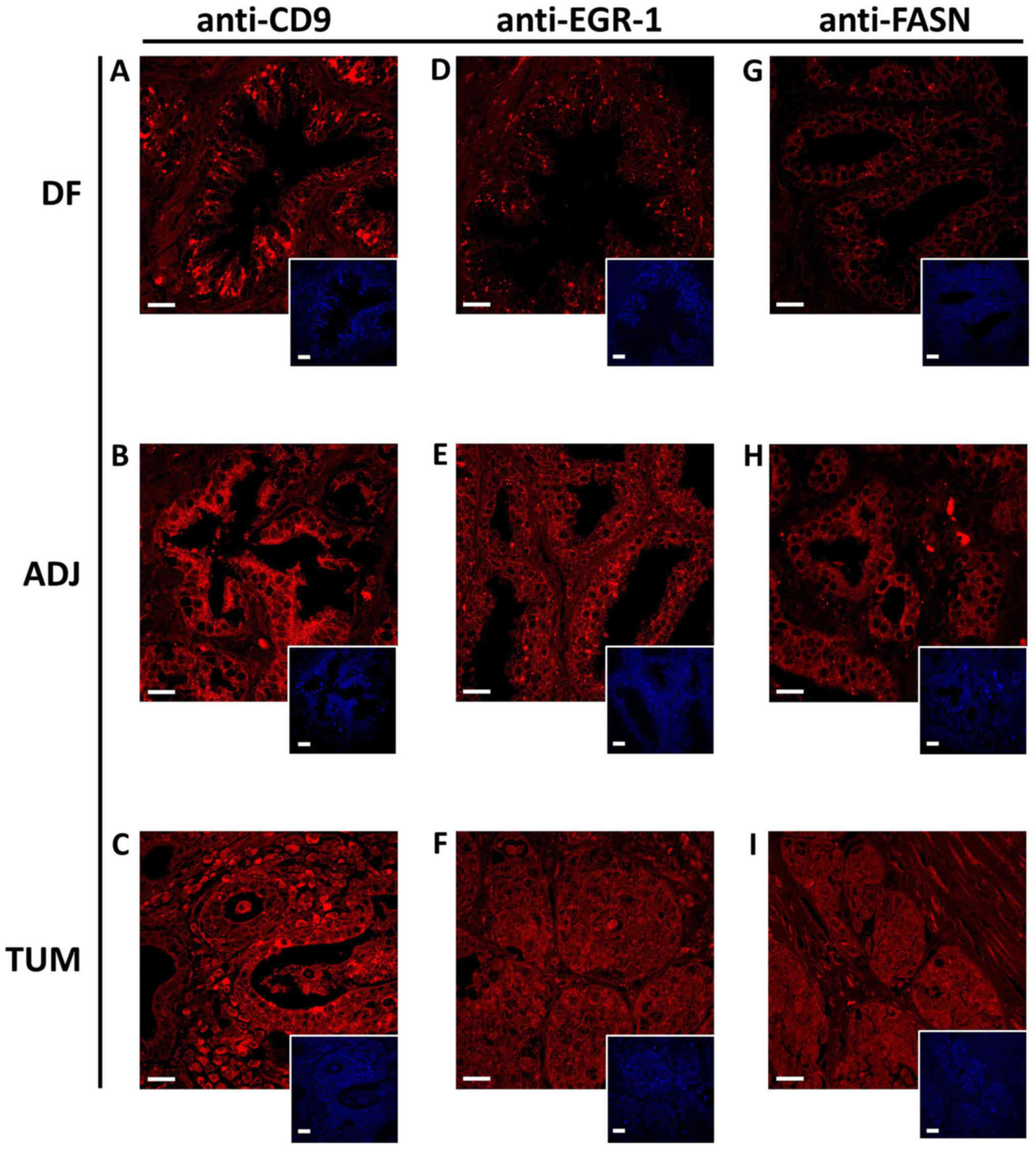
The same antibodies and staining conditions were
used to detect CD9, EGR-1 and FASN in human prostate tissues of the
cancerous type, histologically normal tumor-adjacent, as well as
disease-free specimens, as outlined in Table I. Due to the higher complexity of
human tissues compared to cultured cells, the staining for all
three protein markers was somewhat more diffuse, but nevertheless
typical for the corresponding target, as shown in Fig. 3. A total of 336 digitized confocal
images from 22 individual specimens were used for the
quantification of signal intensity (expression) by computational
detection of sum pixel count per area. The coefficients of
variation ranges for CD9, EGR-1 and FASN expression were 14.5-17.7,
16.1-27.1 and 15.0-21.3%, respectively (data not shown). These are
consistent with previous findings by the authors (7,10,11)
and generally indicate the well-known intra- and inter-tissue
heterogeneity of expression. To acknowledge this heterogeneity, an
inclusive approach was adopted, i.e., any computational
determination of potential outliers was deemed unjustified, and all
available images of sufficient quality were included for the
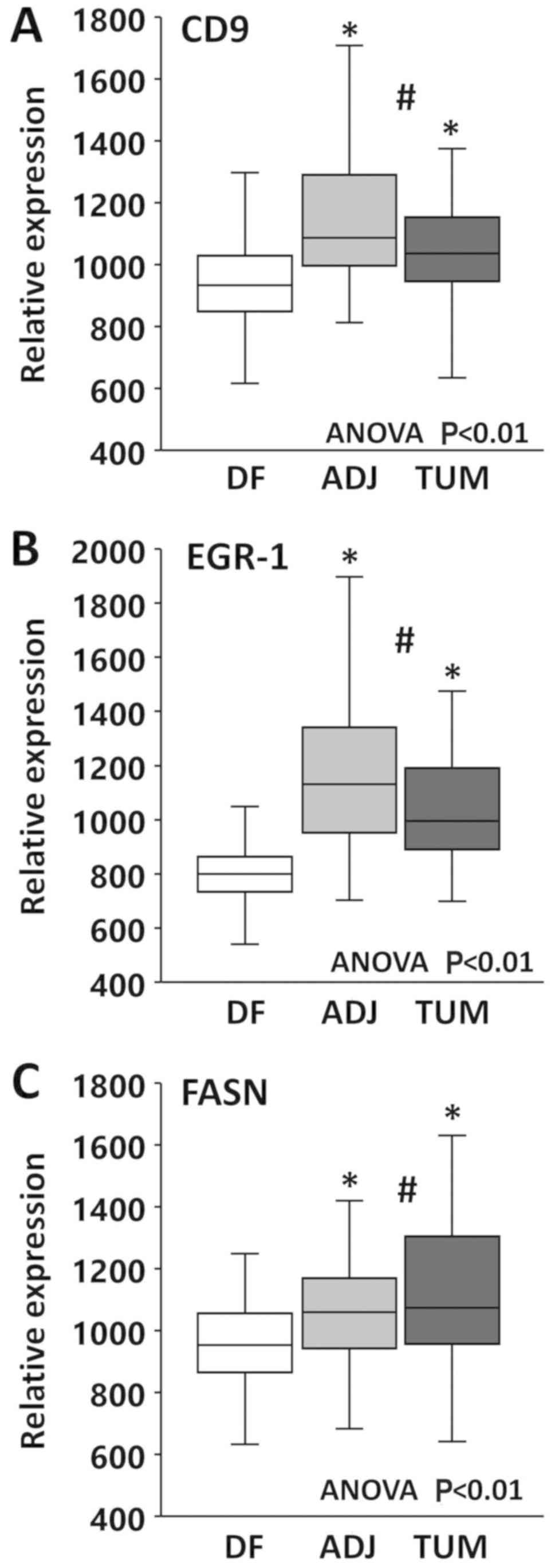
analysis of expression. Group comparisons by ANOVA revealed
significant differences between the types of tissues for all three
markers (P<0.01). As shown in Fig.
4A, CD9 mean expression was slightly, yet significantly higher
in tumor (1.1×) and in tumor-adjacent (1.2×) compared to
disease-free tissues (P<0.01), while it was similar between
tumor and tumor-adjacent tissues (P>0.05). Similarly, as shown
in Fig. 4B, EGR-1 mean expression
in tumor (1.3×) and tumor-adjacent (1.5×) tissues was slightly, yet
significantly elevated compared to disease-free tissues
(P<0.01), while it was similar between tumor and tumor-adjacent
tissues (P>0.05). Finally, the same mean expression pattern was
observed for FASN (Fig. 4C), with
expression levels in tumor (1.2×) and tumor-adjacent (1.1×) being
slightly, yet significantly higher than in disease-free tissues
(P<0.01) and similar between tumor and tumor-adjacent tissues
(P>0.05). Taken together, these results corroborate the field
cancerized nature of tissues adjacent to prostate
adenocarcinomas.
Correlation between CD9, EGR-1 and FASN
expression in human prostate tissues
In a previous study, the authors reported on the
correlation between EGR-1 and other field cancerization markers,
i.e., platelet-derived growth factor A (PDGF-A), macrophage
inhibitory cytokine 1 (MIC-1) and FASN (7), with the question in mind of whether
EGR-1, as a master transcription factor, could be a regulator of
the other three factors. Similarly, one major objective of the
present study was to explore the possibility that exosomes are
effectors of field cancerization in prostate tissues. Thus, this
study aimed to determine a potential association between the
occurrence of CD9 and EGR-1, and between CD9 and FASN within the
individual types of tissues analyzed in this study, and at
determining whether such a correlation changes in these different
types of tissues. This was possible by generating images at the
same position on the TMAs that were consecutive sections
approximately 5 µm apart from each other, as shown in
Fig. 1. Correlations between CD9
and EGR-1, and between CD9 and FASN were determined by Pearson's
correlation analysis, which is by default amenable to both positive
and negative correlations. Of particular interest was to determine
whether possible correlations differ between different types of
tissues, i.e., disease-free, tumor-adjacent and tumor tissues.
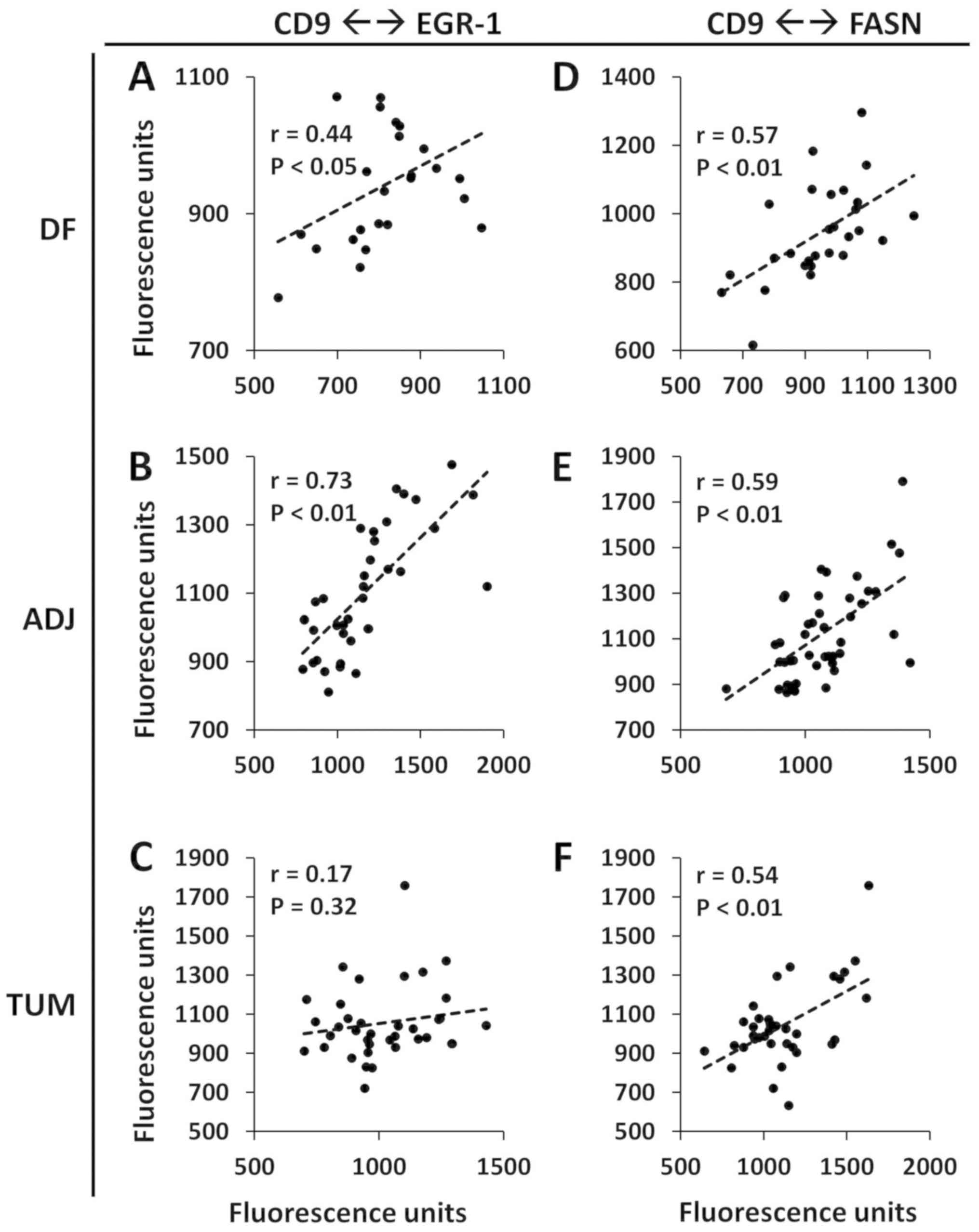
Pearson's correlations between CD9 and EGR-1 were r=0.44
(P<0.05), r=0.73 (P<0.01) and r=0.17 (P=0.32) in
disease-free, tumor-adjacent and tumor tissues, respectively
(Fig. 5A-C). Pearson's
correlations between CD9 and FASN were r=0.57 (P<0.01), r=0.59
(P<0.01) and r=0.54 (P<0.01) in disease-free, tumor-adjacent
and tumor tissues, respectively (Fig.
5D-F). The strongest correlations between CD9 and EGR-1, and
between CD9 and FASN were thus observed in tumor-adjacent tissues.
Given the rather high level of heterogeneity, these values were
deemed to be markedly high. Since CD9 expression represents exosome
formation and excretion (24-26)
and EGR-1 is a marker of field cancerization (7,11),
these results suggest a potential role of exosomes in the formation
of field cancerization.
Effect of exosomes from cancerous cells
on non-cancerous cells
The potential of exosomes derived from cancer cells
to induce the expression of field cancerization markers, such as
EGR-1 and FASN in non-cancerous cells was experimentally examined
using the non-cancerous RWPE-1 and the cancerous LNCaP cell models.
The golden standard method was used, i.e., ultracentrifugation, to
isolate exosomes from LNCaP and RWPE-1 cells and determined their
protein concentration. Consistently, it was calculated that 1
µg exosomal protein was produced by approximately 300,000
cells under standard growth conditions during 24 h of culture.
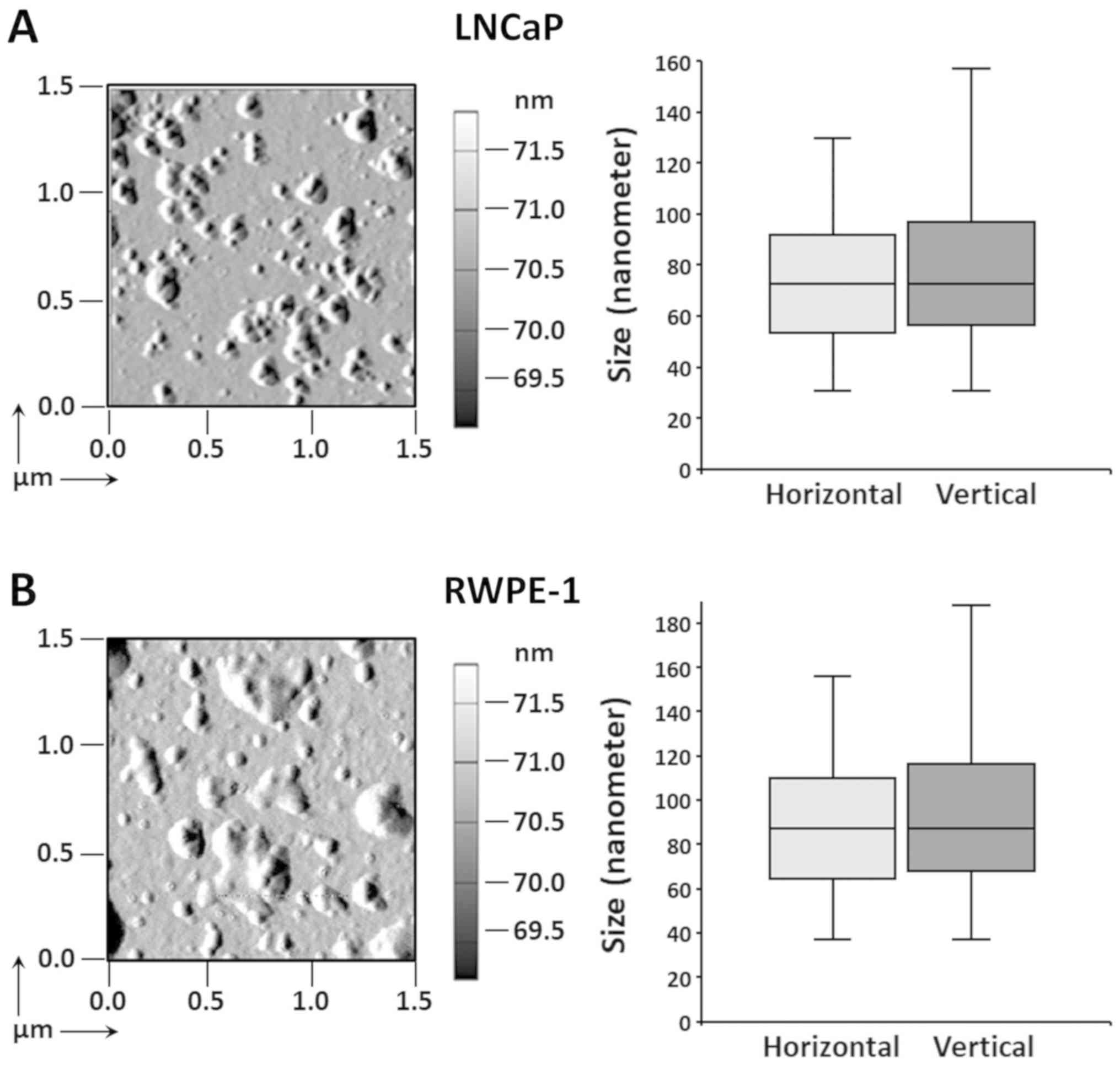
Atomic force microscopic analysis of the LNCaP and RWPE-1 exosomes
(Fig. 6) revealed horizontal and
vertical dimensions of 74.6±26.2 and 78.0±28.4 nm for the LNCaP
cells (Fig. 6A), and 89.6±31.4 and
93.6±34.0 nm for the RWPE-1 cells (Fig. 6B), respectively. This size is in
agreement with that of numerous previous reports on exosomes from
cells of prostatic origin (17-20).
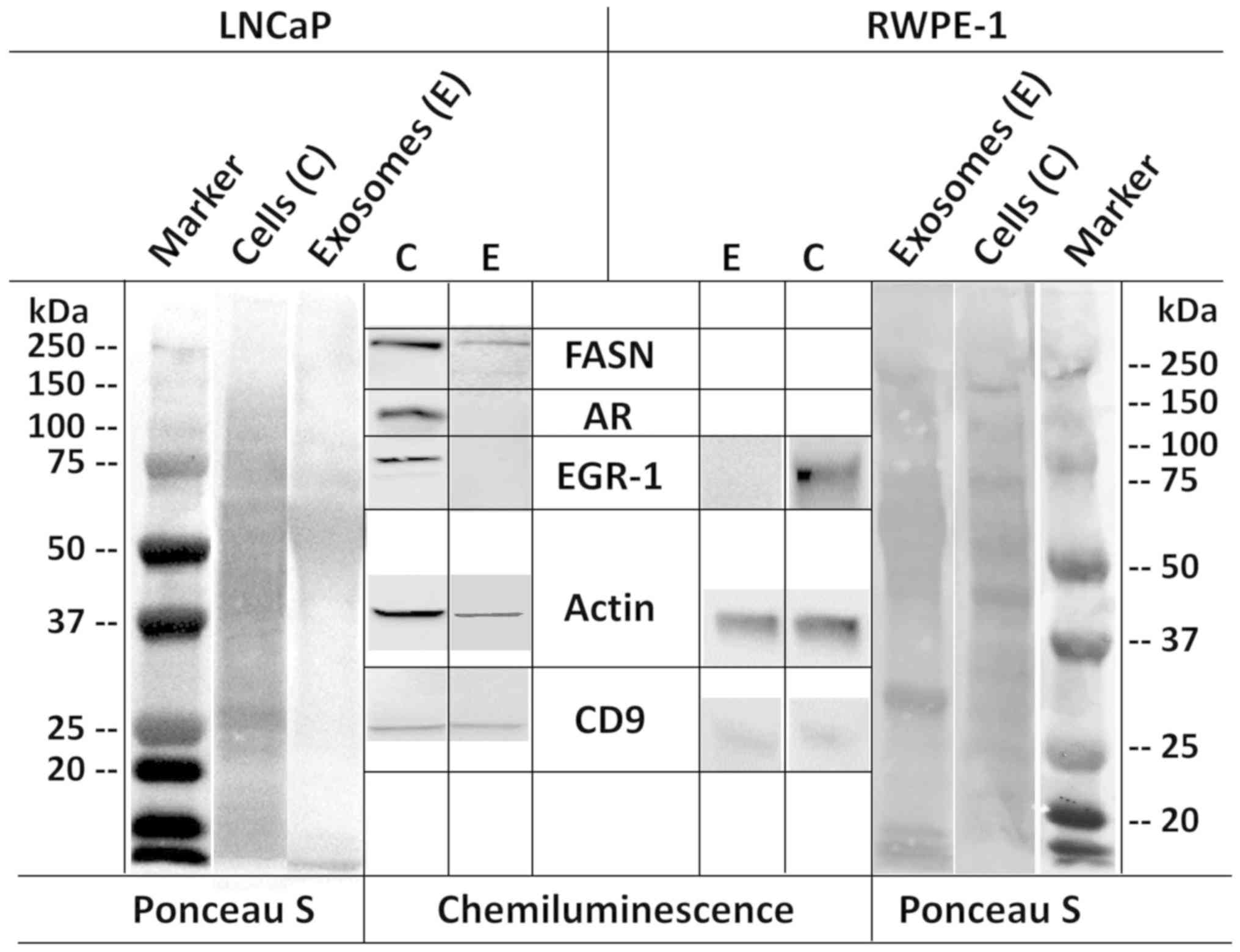
LNCaP and RWPE-1 exosomes were also analyzed biochemically by
western blot analysis, along with the corresponding cells that
secreted them (Fig. 7).
Immunodetection using specific antibodies revealed the presence of
CD9 in both exosomes and cells. In accordance with previously
reported proteomic profiles of exosomes secreted by prostate cells
(24), actin and FASN were also
detected in both LNCaP cells and exosomes, although FASN expression
was under the detection limit for RWPE-1 cells. However, another
study did not report the presence of FASN in LNCaP exosomes, but
instead reported the presence of AR (26), which we did not find in either cell
types. These discrepancies may be due to different culture
conditions, collection times, and other experimental and/or
analytical parameters. EGR-1 was detected in the cellular extracts,
but not in the exosomal lysates, which is congruent with its
absence in the previously reported proteomic profiles (24,26).
Finally, the AR was expressed in LNCaP cells, but was virtually
absent in RWPE-1 cells if not induced by excess androgen as
reported (ATCC) (Fig. 7).
To examine the effect of prostate cancer
cell-derived exosomes on the expression of the two field
cancerization markers EGR-1 and FASN in non-cancerous prostate
cells, the RWPE-1 cells were treated with LNCaP-derived exosomes at
a 3:1 and 30:1 cell-to-cell ratio for 16 h. The protein profiles of
the lysates of the treated RWPE-1 cells did not appear to be
different from the control, suggesting that the exosomes did not
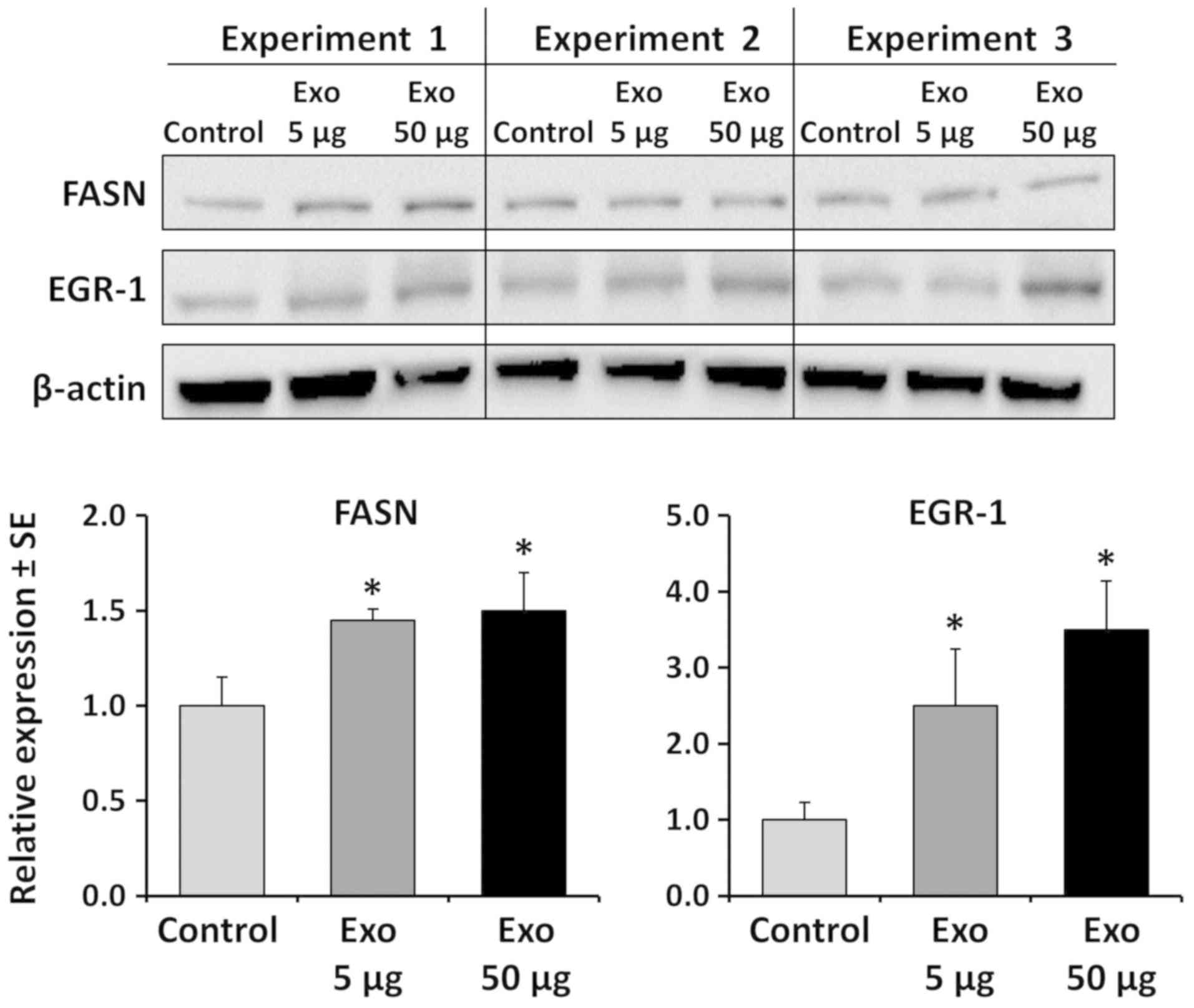
induce major expressional changes (data not shown). Western blot
analysis revealed an inducive effect of up to 3.5-fold, for EGR-1,
but only up to 1.5-fold for FASN (Fig.
8). FASN protein has previously been demonstrated to be part of
the exosomal content released by prostate cells (24). Thus, in order to demonstrate that a
possible inducive expression was due to transcriptional activation,
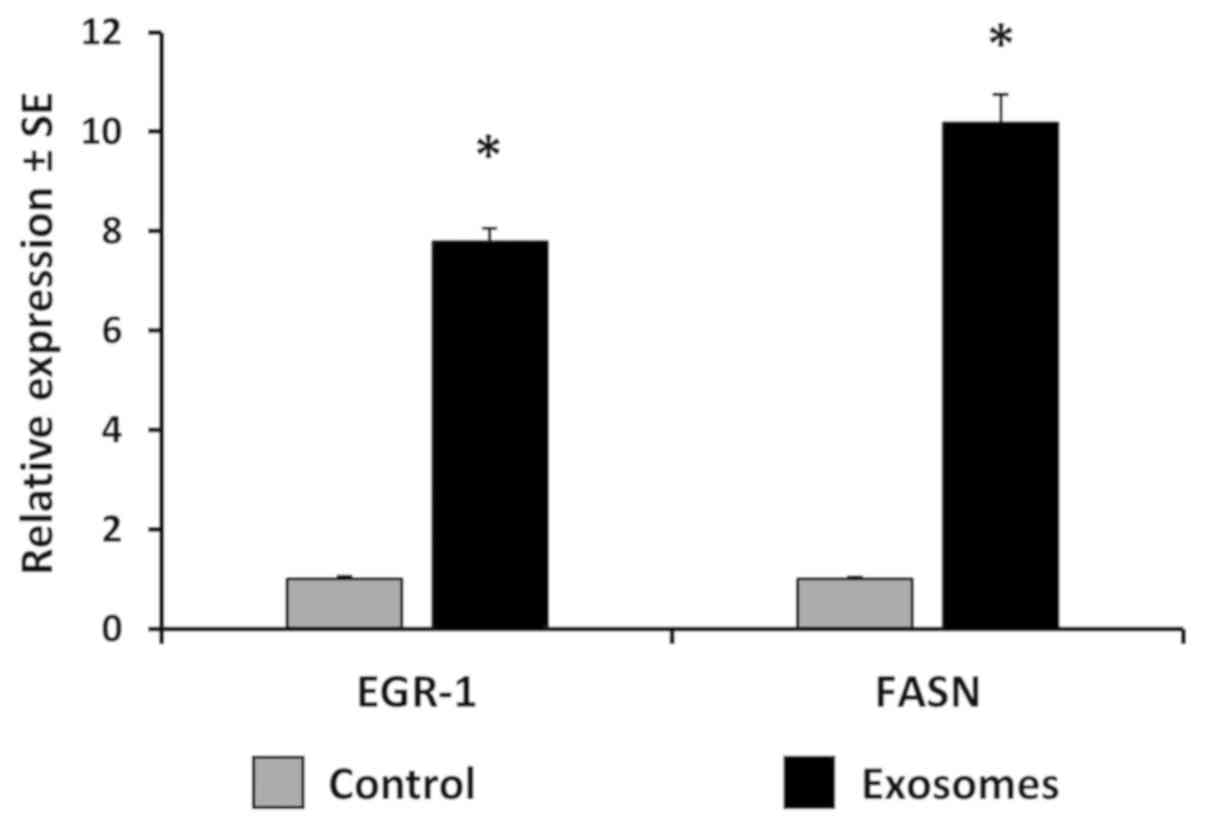
the induction of FASN mRNA was measured by RT-qPCR. At lower
concentrations of exosomes, FASN mRNA transcription was induced
approximately 10-fold. Similarly, EGR-1 mRNA was induced
approximately 8-fold (Fig. 9).
Overall, these results are in good agreement with the observations
made in the tissues and suggest a regulatory association between
exosome release and FASN and EGR-1 expression in human prostate
tissues.
Discussion
Several reviews of molecular pathology known as
field cancerization, or the field effect, have emphasized its
potential to improve the clinical management of solid tumors,
including prostate cancer (6,13,14,27).
In this regard, it is conceivable that the molecular aberrations,
be it of genetic, epigenetic and/or biochemical nature, could act
as biomarkers along the entire development of the disease and/or as
molecular targets for preventative or interventive therapy. The
authors have previously contributed to the identification of
markers of prostate field cancerization and have reported on their
potential clinical uses, thereby validating some of them (7-13).
Accordingly, the authors have previously reported on two recurrent
markers of field cancerization, i.e., the key transcription factor
and master regulator, EGR-1, and the lipogenic enzyme, FASN,
specifically their upregulation at the protein level in
histologically normal tissue adjacent to prostate tumors when
compared to disease-free, truly normal prostatic tissues (7,8,11).
It is not inconceivable that such molecular aberrations could be
used for example, to improve the diagnosis of prostate cancer in
false-negative biopsies (13). The
latter continue to challenge confirmatory diagnoses of prostate
adenocarcinoma following an abnormal prostate-specific antigen
(PSA) test or suspicious digital rectal examination (DRE) (28-31).
In this scenario, tissue affected by field cancerization increases
the clinically informative area under microscopic analysis by the
surgical pathologist when combined with immunological techniques.
This could lead to a reduction of repeat biopsies and thus, a more
effective clinical management. The possibility to predict the
existence of lesions in a tissue without their visual detection has
prompted others to call tissues affected by field cancerization
'TINT', for 'tumor indicating normal tissue' (32). Even in the case of a positive
detection of cancerous tissue in biopsies, markers of field
cancerization could have a meaningful application. The clinical
setting referred to here is active surveillance, which is
increasingly chosen by patients diagnosed with low-risk prostate
cancer, defined as low number of positive biopsy cores, low % of
tissue affected and low Gleason grade (33, 34). Active surveillance programs are
meant to defer more aggressive treatments of curative intent but
with quality of life lowering side effects, including radical
prostatectomy (35,36). These programs include frequent PSA
testing and the histological examination of repeat biopsies to
monitor potential cancer progression. It is thus conceivable that
well-defined areas, markers and parameters of field cancerization
could be monitored during this time. Similarly, the effect of
neoadjuvant therapeutic interventions could be assessed during this
pre-surgical setting (13,37). The extent of field cancerization
could also be indicative of a positive surgical margin, which is an
important clinical parameter in the administration of focal
therapy, a less invasive therapeutic modality on the rise (13,38).
In this scenario, the presence of a field effect at the margin may
be indicative of an elevated risk for progression or of the extent
of tumor multifocality within the prostate (39). Lastly, given the widely accepted
premise that the observed molecular aberrations in histologically
normal tissues constitute a state of pre-malignancy, markers of
field cancerization could represent targets for therapeutic
intervention (13,32).
Regardless of the potential clinical application, it
is widely accepted that the molecular etiology of field
cancerization should be understood in order to fully benefit from
it. It is thus important to identify the distinct cellular and
molecular mechanisms and pathways that result in the molecular
aberrations observed in tissues affected by a field effect. For
example, for the afore-mentioned protein factors EGR-1 and FASN,
the events that lead to their upregulation in histologically normal
tissues adjacent to existing prostate tumors remain unknown. Thus,
it would be of interest to determine the mechanisms through which a
prostate tumor lesion influences its surrounding tissues, thereby
potentially priming it for the induction of multifocal disease. As
is well-known from 2019, the role of exosomes in cell-to-cell and
in tissue-to-tissue communication has been established and proven
to be a major mode of physiological and reciprocal interactions in
multicellular organisms (15,16).
While studies of exosomes of prostatic origin tend to focus on
their application as biomarkers in liquid biopsy schemes and on the
molecular characterization of their content (17-20),
reports on specific exosomal factors on the genotype and phenotype
of recipient cells are now increasing in number. For example,
prostate cancer cell-derived exosomes have been shown to inhibit
and promote osteoclast and osteoblast cell activity, respectively
(40,41), which could promote the overall
osteoblastic phenotype of prostate cancer metastases. Exosomes shed
by prostate cancer cells have been shown to carry integrin αvβ3
which, when deposited in recipient cells, is postulated to increase
their motility (42). Similarly,
the motility of stromal cells has been shown to increase by
prostate cancer exosomes (43).
Hypoxia-induced exosomes lead to increased prostate cell survival
and invasiveness by targeting molecules of the adherens junctions
(44,45), while the exosomal factor and
tetraspanin CD9 promotes prostate cancer cell growth under
androgen-deprived conditions (46). Based on such reports, the existence
of a new link has been hypothesized between cancer cell-derived
exosomes and the induction of field cancerization. Accordingly, the
present study attempted for the first time, at least to the best of
our knowledge, to demonstrate a quantitative association between
the exosomal marker, CD9, and our previously identified markers of
field effect, EGR-1 and FASN. This study on a pilot tissue cohort
indicates a positive correlation between CD9 expression on one side
and EGR-1 and FASN expression on the other, with the link with the
former factor being more significant. The authors have previously
demonstrated that these two factors are upregulated in prostate
tissues resected 1 cm from the visible tumor margin at both the
mRNA and protein level (7,8,11).
The induction of both factors in histologically normal areas of the
prostate by tumor-derived exosomes is congruent with our hypothesis
of a 'priming', potentially tumor-promoting effect. EGR-1 is a
central transcription regulator of a number of molecular pathways
and acts divergently according to the cell context (23,47,48).
However, in prostate tumors it acts as a promoter of cancer
progression (47,49,50).
FASN is equally established in prostate cancer and has been termed
a metabolic oncogene. It promotes tumor cell proliferation through
lipid biosynthesis and the post-translational modification of
proteins and is the focus of ongoing efforts to develop specific
inhibitors of its enzymatic activity (51, 52). Of note, the authors have previously
demonstrated a potential regulatory function of EGR-1 for FASN
expression (7). Overall, the data
presented in this report corroborates in an independent tissue
cohort and data set that EGR-1 and FASN are recurrent markers of
prostate field cancerization. Of note, the magnitude of the
overexpression of EGR-1 and FASN in field cancerized prostate
tissues can vary in independent studies due to tissue heterogeneity
and to the type of antibodies used. Of note however, the present
study independently corroborates the previous findings by the
authors (7,10,11).
In addition, for both factors, future studies are required to
include testing whether their transcriptional and enzymatic
activities, respectively, are also heightened in field cancerized
tissues. As a novel observation, the data of the present study
demonstrate that CD9 expression contributes similarly to this
phenomenon through its change (increase) from disease-free to
tumor-adjacent and to histologically cancerous tissues. The authors
acknowledge the possibility that the observed CD9 staining in the
tissues can be due to its documented expression in prostate gland
epithelial cells (proteinatlas.org). Since CD9 is also a proven exosomal
marker (24), it is not
inconceivable that the observed expression is representative of
exosomal release. However, the findings of this study need to be
corroborated in follow-up studies using additional exosomal
markers, including for example CD63 (53). Importantly, the results of this
study support the notion that exosomes released by cancerous
lesions in the prostate could induce the upregulation of factors
that promote the biochemical transformation of physiologically and
phenotypically normal cells, leading to the formation of
molecularly altered fields. For EGR-1, the data of this study are
congruent with those of other studies in other cell systems, where
exosomes exert their actions through EGR-1 expression and
activation, for example in muscle cells affected by exosomes
released by adipose cells (54).
For the most part, in this study, the in
vitro data using the LNCaP and the RWPE-1 cell models support
the associations made in situ in human tissues, although the link
between CD9 (exosomes) and EGR-1 (field effect) is clearer. With
respect to FASN, this study prompts for caution when comparing
observations made in tissues with those made in cell models. This
is due to the fact that FASN may be part of the 'cargo' of exosomes
released by prostate cancer cells, although conflicting results
exist (24,26). Of note, the results of this study
support the those of the study by Duijvesz et al (24). Thus, an elevated FASN expression in
field cancerization may be, at least in part, directly due to
exosomal FASN delivery. The in vitro experiments of this
study, however, indicate de novo induction of FASN mRNA,
which, for the most part, is accompanied by corresponding protein
levels. The FASN experiments performed herein warrant further
investigations using functional approaches in systems that reflect
the complexity of human tissues. The same may be true for the AR,
the presence of which in prostate exosomes is equally inconclusive
(24,26). It is known that in cultured cells,
the physiological complexity is relatively low and may not reflect
entirely the complex regulatory networks at work in tissues. With
respect to the latter, it can be argued that tissue studies are
static and compromised by sample heterogeneity. However, they are
also physiologically relevant and better reflect the complexity of
cellular and molecular pathways influenced by the environment, for
example in animal models. Importantly, it is demonstrated herein
that when coupled with sophisticated and quantitative data
acquisition methods, they can deliver meaningful indications of
molecular associations in a physiologically relevant in situ
environment, even in the presence of high heterogeneity. As it is
widely accepted that field effect represents a pre-malignant state,
such knowledge may aid in the development of targeted intervention
strategies preventing progression of pre-malignancy to cancer.
Funding
This study was supported by the Department of
Defense Prostate Cancer Research Program grant no. W81XWH-15-1-0056
(to MB), and by grants from the Chapman University Center for
Undergraduate Excellence (to PAP, JPTN and ELC), and a generous
gift from Melinda and Edward Subia of Orange County CA.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
The authors' significant contributions to this study
were the following: FA and PAP were involved in cell and tissue
immunofluorescence and analysis. NSP and JPTN were involved in
exosome isolation and characterization. JPTN and ELC were involved
in western blot analysis and RT-qPCR. ACJ was involved in
immunofluorescence protocol development and study concept design.
MB was involved in the conception and design of the study, the
procurement and management of funding, supervision, data analysis
and in the writing of the manuscript. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
In the present study, tissue microarrays (TMAs)
used. No human tissues from other sources, other than commercially
available TMAs, were used in the present study. The use of any
human tissues, including commercially available TMAs, is covered by
the Chapman University Institutional Review Board (IRB) study
#1415H024. Experiments with human tissues was approved by the
Institutional Review Board of Chapman University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Dr Molla Islam of
the Chemistry and Biochemistry Division of Chapman University
Schmid College of Science and Technology for analyzing the LNCaP
and RWPE-1 exosomes by atomic force microscopy. The departmental
office and staff of the Chapman University Schmid College of
Science and Technology are acknowledged for providing
administrative support.
References
|
1
|
Slaughter DP, Southwick HW and Smejkal W:
Field cancerization in oral stratified squamous epithelium;
clinical implications of multicentric origin. Cancer. 6:963–968.
1953. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Angadi PV, Savitha JK, Rao SS and
Sivaranjini Y: Oral field cancerization: Current evidence and
future perspectives. Oral Maxillofac Surg. 16:171–180. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Braakhuis BJ, Tabor MP, Kummer JA, Leemans
CR and Brakenhoff RH: A genetic explanation of Slaughter's concept
of field cancerization: Evidence and clinical implications. Cancer
Res. 63:1727–1730. 2003.PubMed/NCBI
|
|
4
|
Curtius K, Wright NA and Graham TA: An
evolutionary perspective on field cancerization. Nat Rev Cancer.
18:19–32. 2018. View Article : Google Scholar
|
|
5
|
Takeshima H and Ushijima T: Accumulation
of genetic and epigenetic alterations in normal cells and cancer
risk. NPJ Precis Oncol. 3:72019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lochhead P, Chan AT, Nishihara R, Fuchs
CS, Beck AH, Giovannucci E and Ogino S: Etiologic field effect:
Reappraisal of the field effect concept in cancer predisposition
and progression. Mod Pathol. 28:14–29. 2015. View Article : Google Scholar
|
|
7
|
Gabriel KN, Jones AC, Nguyen JP, Antillon
KS, Janos SN, Overton HN, Jenkins SM, Frisch EH, Trujillo KA and
Bisoffi M: Association and regulation of protein factors of field
effect in prostate tissues. Int J Oncol. 49:1541–1552. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Haaland CM, Heaphy CM, Butler KS, Fischer
EG, Griffith JK and Bisoffi M: Differential gene expression in
tumor adjacent histologically normal prostatic tissue indicates
field cancerization. Int J Oncol. 35:537–546. 2009.PubMed/NCBI
|
|
9
|
Heaphy CM, Fleet TM, Treat EG, Lee SJ,
Smith AY, Davis MS, Griffith JK, Fischer EG and Bisoffi M:
Organ-wide telomeric status in diseased and disease-free prostatic
tissues. Prostate. 70:1471–1479. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jones AC, Antillon KS, Jenkins SM, Janos
SN, Overton HN, Shoshan DS, Fischer EG, Trujillo KA and Bisoffi M:
Prostate field cancerization: Deregulated expression of macrophage
inhibitory cytokine 1 (MIC-1) and platelet derived growth factor A
(PDGF-A) in tumor adjacent tissue. PLoS One. 10:e01193142015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jones AC, Trujillo KA, Phillips GK, Fleet
TM, Murton JK, Severns V, Shah SK, Davis MS, Smith AY, Griffith JK,
et al: Early growth response 1 and fatty acid synthase expression
is altered in tumor adjacent prostate tissue and indicates field
cancerization. Prostate. 72:1159–1170. 2012. View Article : Google Scholar
|
|
12
|
Treat EG, Heaphy CM, Massie LW, Bisoffi M,
Smith AY, Davis MS and Griffith JK: Telomere DNA content in
prostate biopsies predicts early rise in prostate-specific antigen
after radical prostatectomy for prostate cancer. Urology.
75:724–729. 2010. View Article : Google Scholar
|
|
13
|
Trujillo KA, Jones AC, Griffith JK and
Bisoffi M: Markers of field cancerization: Proposed clinical
applications in prostate biopsies. Prostate Cancer.
2012:3028942012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nonn L, Ananthanarayanan V and Gann PH:
Evidence for field cancerization of the prostate. Prostate.
69:1470–1479. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Choi JY, Kim S, Kwak HB, Park DH, Park JH,
Ryu JS, Park CS and Kang JH: Extracellular Vesicles as a Source of
Urological Biomarkers: Lessons Learned From Advances and Challenges
in Clinical Applications to Major Diseases. Int Neurourol J.
21:83–96. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dhondt B, Van Deun J, Vermaerke S, de
Marco A, Lumen N, De Wever O and Hendrix A: Urinary extracellular
vesicle biomarkers in urological cancers: From discovery towards
clinical implementation. Int J Biochem Cell Biol. 99:236–256. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pan J, Ding M, Xu K, Yang C and Mao LJ:
Exosomes in diagnosis and therapy of prostate cancer. Oncotarget.
8:97693–97700. 2017.PubMed/NCBI
|
|
18
|
Vlaeminck-Guillem V: Extracellular
Vesicles in Prostate Cancer Carcinogenesis, Diagnosis, and
Management. Front Oncol. 8:2222018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Panigrahi GK and Deep G: Exosomes-based
biomarker discovery for diagnosis and prognosis of prostate cancer.
Front Biosci. 22:1682–1696. 2017. View
Article : Google Scholar
|
|
20
|
Soekmadji C, Russell PJ and Nelson CC:
Exosomes in prostate cancer: Putting together the pieces of a
puzzle. Cancers (Basel). 5:1522–1544. 2013. View Article : Google Scholar
|
|
21
|
Fordyce CA, Heaphy CM, Joste NE, Smith AY,
Hunt WC and Griffith JK: Association between cancer-free survival
and telomere DNA content in prostate tumors. J Urol. 173:610–614.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Mora GR, Olivier KR, Cheville JC, Mitchell
RF Jr, Lingle WL and Tindall DJ: The cytoskeleton differentially
localizes the early growth response gene-1 protein in cancer and
benign cells of the prostate. Mol Cancer Res. 2:115–128.
2004.PubMed/NCBI
|
|
24
|
Duijvesz D, Burnum-Johnson KE, Gritsenko
MA, Hoogland AM, Vredenbregt-van den Berg MS, Willemsen R, Luider
T, Paša-Tolić L and Jenster G: Proteomic profiling of exosomes
leads to the identification of novel biomarkers for prostate
cancer. PLoS One. 8:e825892013. View Article : Google Scholar
|
|
25
|
Malla RR, Pandrangi S, Kumari S, Gavara MM
and Badana AK: Exosomal tetraspanins as regulators of cancer
progression and metastasis and novel diagnostic markers. Asia Pac J
Clin Oncol. 14:383–391. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mizutani K, Terazawa R, Kameyama K, Kato
T, Horie K, Tsuchiya T, Seike K, Ehara H, Fujita Y, Kawakami K, et
al: Isolation of prostate cancer-related exosomes. Anticancer Res.
34:3419–3423. 2014.PubMed/NCBI
|
|
27
|
Dakubo GD, Jakupciak JP, Birch-Machin MA
and Parr RL: Clinical implications and utility of field
cancerization. Cancer Cell Int. 7:22007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bjurlin MA, Meng X, Le Nobin J, Wysock JS,
Lepor H, Rosenkrantz AB and Taneja SS: Optimization of prostate
biopsy: The role of magnetic resonance imaging targeted biopsy in
detection, localization and risk assessment. J Urol. 192:648–658.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bostanci Y, Kazzazi A and Djavan B:
Optimizing prostate biopsy. Minerva Urol Nefrol. 64:233–243.
2012.
|
|
30
|
Delongchamps NB and Haas GP: Saturation
biopsies for prostate cancer: Current uses and future prospects.
Nat Rev Urol. 6:645–652. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rabbani F, Stroumbakis N, Kava BR, Cookson
MS and Fair WR: Incidence and clinical significance of
false-negative sextant prostate biopsies. J Urol. 159:1247–1250.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Halin S, Hammarsten P, Adamo H, Wikström P
and Bergh A: Tumor indicating normal tissue could be a new source
of diagnostic and prognostic markers for prostate cancer. Expert
Opin Med Diagn. 5:37–47. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mazzucchelli R, Galosi AB, Santoni M,
Lopez-Beltran A, Scarpelli M, Cheng L and Montironi R: Role of the
pathologist in active surveillance for prostate cancer. Anal Quant
Cytopathol Histpathol. 37:65–68. 2015.PubMed/NCBI
|
|
34
|
Pomerantz M: Active Surveillance:
Pathologic and Clinical Variables Associated with Outcome. Surg
Pathol Clin. 8:581–585. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bellardita L, Valdagni R, van den Bergh R,
Randsdorp H, Repetto C, Venderbos LD, Lane JA and Korfage IJ: How
does active surveillance for prostate cancer affect quality of
life? A systematic review. Eur Urol. 67:637–645. 2015. View Article : Google Scholar
|
|
36
|
Kwon O and Hong S: Active surveillance and
surgery in localized prostate cancer. Minerva Urol Nefrol.
66:175–187. 2014.PubMed/NCBI
|
|
37
|
Lou DY and Fong L: Neoadjuvant therapy for
localized prostate cancer: Examining mechanism of action and
efficacy within the tumor. Urol Oncol. 34:182–192. 2016. View Article : Google Scholar
|
|
38
|
Marshall S and Taneja S: Focal therapy for
prostate cancer: The current status. Prostate Int. 3:35–41. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Andreoiu M and Cheng L: Multifocal
prostate cancer: Biologic, prognostic, and therapeutic
implications. Hum Pathol. 41:781–793. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Duan Y, Tan Z, Yang M, Li J, Liu C, Wang
C, Zhang F, Jin Y, Wang Y and Zhu L: PC-3-Derived Exosomes Inhibit
Osteoclast Differentiation by Downregulating miR-214 and Blocking
NF-κB Signaling Pathway. BioMed Res Int. 2019:86508462019.
View Article : Google Scholar
|
|
41
|
Li SL, An N, Liu B, Wang SY, Wang JJ and
Ye Y: Exosomes from LNCaP cells promote osteoblast activity through
miR-375 transfer. Oncol Lett. 17:4463–4473. 2019.PubMed/NCBI
|
|
42
|
Krishn SR, Singh A, Bowler N, Duffy AN,
Friedman A, Fedele C, Kurtoglu S, Tripathi SK, Wang K, Hawkins A,
et al: Prostate cancer sheds the αvβ3 integrin in vivo through
exosomes. Matrix Biol. 77:41–57. 2019. View Article : Google Scholar
|
|
43
|
McAtee CO, Booth C, Elowsky C, Zhao L,
Payne J, Fangman T, Caplan S, Henry MD and Simpson MA: Prostate
tumor cell exosomes containing hyaluronidase Hyal1 stimulate
prostate stromal cell motility by engagement of FAK-mediated
integrin signaling. Matrix Biol. 78-79:165–179. 2019. View Article : Google Scholar
|
|
44
|
Panigrahi GK, Praharaj PP, Peak TC, Long
J, Singh R, Rhim JS, Abd Elmageed ZY and Deep G: Hypoxia-induced
exosome secretion promotes survival of African-American and
Caucasian prostate cancer cells. Sci Rep. 8:38532018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ramteke A, Ting H, Agarwal C, Mateen S,
Somasagara R, Hussain A, Graner M, Frederick B, Agarwal R and Deep
G: Exosomes secreted under hypoxia enhance invasiveness and
stemness of prostate cancer cells by targeting adherens junction
molecules. Mol Carcinog. 54:554–565. 2015. View Article : Google Scholar
|
|
46
|
Soekmadji C, Riches JD, Russell PJ,
Ruelcke JE, McPherson S, Wang C, Hovens CM, Corcoran NM, Hill MM
and Nelson CC; Australian Prostate Cancer Collaboration
BioResource: Modulation of paracrine signaling by CD9 positive
small extracellular vesicles mediates cellular growth of androgen
deprived prostate cancer. Oncotarget. 8:52237–52255. 2016.
|
|
47
|
Gitenay D and Baron VT: Is EGR1 a
potential target for prostate cancer therapy? Future Oncol.
5:993–1003. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pagel JI and Deindl E: Early growth
response 1 - a transcription factor in the crossfire of signal
transduction cascades. Indian J Biochem Biophys. 48:226–235.
2011.PubMed/NCBI
|
|
49
|
Adamson E, de Belle I, Mittal S, Wang Y,
Hayakawa J, Korkmaz K, O'Hagan D, McClelland M and Mercola D: Egr1
signaling in prostate cancer. Cancer Biol Ther. 2:617–622. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Adamson ED and Mercola D: Egr1
transcription factor: Multiple roles in prostate tumor cell growth
and survival. Tumour Biol. 23:93–102. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Baron A, Migita T, Tang D and Loda M:
Fatty acid synthase: A metabolic oncogene in prostate cancer? J
Cell Biochem. 91:47–53. 2004. View Article : Google Scholar
|
|
52
|
Zadra G, Photopoulos C and Loda M: The fat
side of prostate cancer. Biochim Biophys Acta. 1831:1518–1532.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Duijvesz D, Versluis CY, van der Fels CA,
Vredenbregt-van den Berg MS, Leivo J, Peltola MT, Bangma CH,
Pettersson KS and Jenster G: Immuno-based detection of
extracellular vesicles in urine as diagnostic marker for prostate
cancer. Int J Cancer. 137:2869–2878. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Pan J, Alimujiang M, Chen Q, Shi H and Luo
X: Exosomes derived from miR-146a-modified adipose-derived stem
cells attenuate acute myocardial infarction-induced myocardial
damage via downregulation of early growth response factor 1. J Cell
Biochem. 120:4433–4443. 2019. View Article : Google Scholar
|