Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most
common form of pancreatic cancer (1), with an estimated 458,918 diagnoses
and 432,242 deaths from pancreatic cancer globally in 2018
(2), mainly due to its high degree
of invasiveness and metastatic potential (1,3).
Therefore, continued research of the molecular mechanisms
underlying PDAC metastasis are essential for identifying new
targets to prevent tumor progression and recurrence.
Exosome-mediated intercellular communication is an
important factor in regulating malignant cell behavior (4). In particular, exosomal microRNAs
(miRNAs) induce tumor progression by downregulating the expression
of tumor suppressor genes (5,6).
Among the known exosomal miRNAs, miRNA (miR)-21 is a
well-established oncogenic miRNA that serves various
cancer-specific roles, including those associated with metastasis.
For example, exosomes derived from hypoxic oral squamous cell
carcinoma cells deliver miR-21 to normoxic cells to induce a
pro-metastatic phenotype (7); in
addition, exosomal miR-21 has been demonstrated to promote the
migration and invasion of esophageal cancer cells by targeting
programmed cell death-4 (8). The
expression level of miR-21 has been reported to be elevated in
pancreatic stellate cell (PSC)-derived exosomes in patients with
PDAC (9-11). PSCs are a special type of
cancer-associated fibroblasts that are often abnormally activated
in PDAC and are closely associated with PDAC progression; however,
to the best of our knowledge, the specific functions of miR-21 and
its underlying molecular mechanisms in regulating PDAC cell
malignancy have not been elucidated (12-19).
The present study investigated the differences in
miR-21 expression profiles in PDAC and normal tissues based on
public databases, and explored effect and potential mechanism of
PSC-derived exosomal miR-21 on the development of PDAC-related
malignant phenotypes.
Materials and methods
Datasets and analysis
Data on miRNA expression in pancreatic
adenocarcinoma (PAAD) tissue were downloaded from The Cancer Genome
Atlas (TCGA) (http://cancergenome.nih.gov/) (20). Clinical data, survival time and
KRAS mutation status of patients with PAAD were also
downloaded from the cBioPortal database (www.cbioportal.org) (21). Weighted gene correlation network
analysis (WGCNA) was used for miRNA data analysis, and GS was
calculated by R software with the WGCNA package (15). Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway enrichment analysis of miR-21 target genes
was conducted using the Search Tool for the Retrieval of
Interacting Genes database (https://string-db.org) (22). The effects of the KRAS mRNA
expression levels on the survival of patients with PAAD were
further evaluated using the UALCAN database (http://ualcan.path.uab.edu/index.html) (14).
Cell culture and transfection
Human PDAC cell lines PANC-1 and MIAPaCa-2 were
obtained from the American Type Culture Collection and maintained
in DMEM (Corning, Inc.) supplemented with 10% FBS (Corning, Inc.)
and 1% penicillin and streptomycin (100 U/ml; Cell Resource Center
of Peking Union Medical College). Human PSCs were isolated from
three fresh surgically resected pancreatic cancer tissues (three
male patients; mean age, 60.67 years; age range, 56-64 years;
diagnosed as PDAC by pathological examination between January and
August 2017), which were cut into ~1-mm3 sections using sterile
tweezers and scissors, followed by washing twice with PBS (Corning,
Inc.) containing 1% penicillin and streptomycin. The tissue blocks
were seeded in 6-well culture plates in the presence of 1 ml
DMEM/F-12 (1:1; Corning, Inc.) containing 10% FBS and 1% penicillin
and streptomycin (12), and the
culture medium was changed every 24 h. The cells were cultured at
37°C in an atmosphere with 5% CO2.
For small interfering RNA (siRNA) transfection, the
PSCs were transfected with an miR-21 inhibitor or negative-control
(NC) oligonucleotides (Table I;
Shanghai GenePharma Co., Ltd.) using Lipofectamine® 2000
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. The cells were cultured for 24 h at 37°C in an
atmosphere with 5% CO2.
 | Table IPrimers and inhibitors used in the
present study. |
Table I
Primers and inhibitors used in the
present study.
| Name | Type | Sequence
(5′→3′) |
|---|
| miR-21 | Stem-loop |
GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACTCAACA |
| Forward |
GCGGCGTAGCTTATCAGACTGA |
| Reverse |
GTGCAGGGTCCGAGGT |
| U6 | Forward |
CTCGCTTCGGCAGCACA |
| Reverse |
AACGCTTCACGAATTTGCGT |
| NC inhibitor | Sense |
CAGUACUUUUGUGUAGUACAA |
| miR-21
inhibitor | Sense |
UCAACAUCAGUCUGAUAAGCUA |
Isolation and identification of
exosomes
After reaching 90% confluence, PSCs and transfected
PSCs were washed with PBS and incubated in freshly prepared
DMEM/F12 without FBS for 48 h at 37°C. Exosomes were isolated from
the conditioned medium by differential centrifugation (Optima
L-100XP ultracentrifuge; Beckman Coulter, Inc.) as previously
described (13). Following
ultracentrifugation, the isolated exosomes were dropped to the
copper mesh, precipitated for 2 min, and filter paper was used to
absorb the excess sample. After drying the copper mesh on the
filter paper for 5 min, 1% uranyl acetate was added for 2 min and
dried for 40 min. Transmission electron microscopy (TEM; EM-1400
plus; JEOL, Ltd.) was used to assess the morphological
characteristics of exosomes (magnification, ×80,000). The PSCs were
fixed with 2.5% glutaraldehyde at 4°C for 2 h; electron microscopy
was used to observe ultrastructural changes (magnification,
×50,000). The exosome concentration was determined using a
bicinchoninic acid assay (Thermo Fisher Scientific, Inc.) according
to the manufacturer's instructions.
Exosome labeling
Exosomes isolated from PSCs were suspended in 100
µl PBS, followed by the addition of 1 ml Diluent C from the
PKH67 Fluorescent Cell Linker kit (Sigma-Aldrich; Merck KGaA).
PKH67 buffer (2 µl) was then added to the solution, and
following a 3-min incubation at 25°C, 4 ml serum-free DMEM/F12 was
added to terminate the labeling reaction. After an additional 1
min, the medium was added to a sub-confluent layer of PANC-1 or
MIAPaCa-2 cells, which were then incubated at 37°C for 3 h. The
cells were washed twice with PBS and observed under an IX71
fluorescence microscope (Olympus Corporation; magnification,
×400).
Migration assays
For wound-healing assays, PANC-1 or MIAPaCa-2 cells
were seeded into 6-well plates (5×105 cells/well). Upon
reaching ~90% confluence, the monolayer was scratched using a
200-µl pipette tip, followed by culture in 2 ml DMEM without
FBS. The relative scratch width was determined at 0, 24 and 48 h
using an inverted light microscope (Olympus Corporation). ImagePro
Plus software (v.6.0; Media Cybernetics, Inc.) was used to
calculate the average relative scratch width as follows: Relative
scratch width = wound area / wound height.
Transwell assays were performed to validate the
results of the wound-healing assays. PANC-1 and MIAPaCa-2 cells
suspended in 100 µl DMEM containing 0.05% FBS were added to
the upper chambers of Transwell plates (1×105
cells/well; Corning, Inc.), and 600 µl DMEM with 10% FBS was
added to the lower chambers of each well. After incubation at 37°C
for 16 h, the chambers were fixed with 4% PFA for 15 min at 25°C,
stained with 0.5% crystal violet for 10 min at 25°C and washed
twice with PBS. Migrated cells were counted in five randomly
selected fields under an inverted light microscope (Olympus
Corporation).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from PSCs (untreated, and
transfected PSCs), PANC-1 and MIAPaCa-2 cells (treated and
untreated with PSCs) and their exosomes using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. miR-21 was reverse-transcribed to
cDNA using a High-Capacity cDNA Reverse Transcription Kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using a miRNA-specific
stem-loop reverse transcriptase primer (25°C for 1 min, 37°C for
120 min and 85°C for 5 min); the first-strand cDNA was used as a
template for qPCR. PCRs were run using a Maxima SYBR®
Green qPCR master mix (Applied Biosystems, Foster City, CA, USA) on
an ABI 7500 real-time PCR detection system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) using the following thermocycling
conditions: 95°C for 60 sec, followed by 40 cycles of 95°C for 10
sec, 60°C for 30 sec and 72°C for 10 sec. All reactions were run in
triplicate, and the 2−ΔΔCt method (23) was used to determine relative gene
expression levels using U6 as an internal control (Table I).
Western blotting
Different amounts of PSC-derived exosomes (0, 5, 10
or 20 µg) were added to PANC-1 cells. After 48 h, the cells
and exosomes were collected and lysed in radioimmunoprecipitation
assay buffer (Applygen Technologies, Inc.) in the presence of
protease and phosphatase inhibitors (Thermo Fisher Scientific,
Inc.) for 30 min to extract total proteins, which were quantified
by a bicinchoninic acid assay (Thermo Fisher Scientific, Inc.).
Proteins (40 µg/lane) were separated by 10% SDS-PAGE and
transferred to polyvinylidene difluoride membranes (EMD Millipore),
which were blocked with 5% skimmed milk powder (BD Biosciences) for
1 h at 25°C. The membranes were subsequently incubated at 4°C
overnight with the following primary antibodies: CD63 (1:30; cat.
no. D360973), CD9 (1:30; cat. no. D264336; both from Sangon Biotech
Co., Ltd.), ERK1/2 (1:1,000; cat. no. 9102S), phospho-ERK1/2
(Thr202/Tyr204; 1:1,000; cat. no. 9101S), Akt (1:1,000; cat. no.
4691), phospho-Akt (Ser473; 1:1,000; cat. no. 4060P), vimentin
(1:1,000; cat. no. 5741T), N-cadherin (1:1,000; cat. no. 13116),
β-catenin (1:1,000; cat. no. 8480), Snail (1:1,000; cat. no. 3879T)
and E-cadherin (1:1000; cat. no. 3195T; all from Cell Signaling
Technology, Inc.). The membranes were subsequently incubated with
horseradish peroxidase (HRP)-conjugated goat anti-rabbit/mouse
secondary antibodies (cat. nos. ZB-5301 and ZB-5305; 1:10,000;
Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.) at room
temperature for 1 h. Immunocomplexes were detected using Immobilon
western chemiluminescent HRP substrate (EMD Millipore), and images
were captured using the ChemiDoc MP imaging system with Image Lab
Touch software (v.2.0; Bio-Rad Laboratories, Inc.).
Gelatin zymography
PANC-1 cells were seeded in 6-well plates at
5×105 cells/well and incubated in DMEM with 10% FBS at
37°C for 24 h. Subsequently, the medium was replaced with
serum-free DMEM, and the exosomes from untreated PSCs or PSCs
treated with the miR-21 inhibitor were added to the wells. After 24
h, culture supernatants were collected, and 40 µl of each
supernatant per lane was loaded into a 10% SDS-PAGE gel containing
1 mg/ml gelatin (Sigma-Aldrich; Merck KGaA). Following
electrophoresis, the gel was washed twice for 30 min with 2.5%
Triton X-100 at room temperature and incubated in zymography buffer
[50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 10 mM CaCl2 and 1
µM ZnCl2] at 37°C for 24 h. The gel was
subsequently stained for 2 h in 0.25% Coomassie Brilliant Blue
R-250 and destained with destaining buffer (methanol: glacial
acetic acid: distilled water, 3:1:6). A clear white band indicated
the presence of matrix metalloproteinase-2 or -9 (MMP-2/9)
activity. The gels were photographed using the ChemiDoc MP imaging
system (Bio-Rad Laboratories, Inc.).
Statistical analysis
Data were analyzed using SPSS (v.21.0; IBM Corp.), R
software (v.3.4.3; https://www.r-project.org/; 'WGCNA' package) and
GraphPad Prism 5 (GraphPad Software, Inc.). A log-rank test was
used to analyze the Kaplan-Meier curve to evaluate the association
between KRAS expression and the survival rate. The data from
the wound-healing and Transwell assays were analyzed by one-way
analysis of variance followed by Tukey's post hoc test. P<0.05
was considered to indicate a statistically significant
difference.
Results
High miR-21 expression levels are
associated with poor prognosis and Ras pathway activation in
patients with pancreatic cancer
To investigate whether miR-21 serves a key role in
pancreatic cancer pathogenesis and progression, the relationship
between miRNA expression levels in tumor tissues and prognosis in
patients with pancreatic cancer were analyzed using the WGCNA
approach based on TCGA data. In the WGCNA, miR-21 was located in
the turquoise module, which correlated well with overall survival
(OS), disease-free survival (DFS) and tumor grade (P<0.05;
Fig. 1A and B). Specifically, the
gene significance value of miR-21 was positively correlated with
tumor grade and negatively correlated with OS and DFS (P<0.05;
Fig. 1B). KEGG analysis revealed
the primary involvement of miR-21 target genes in the Ras and ERK
pathways (Fig. 1C). In addition,
database analysis revealed that the survival probability of
patients with high KRAS expression was lower compared with
that in patients with low/medium KRAS expression (P=0.023;
Fig. 1D). The frequency of
KRAS mutations was 91% in patients with PAAD (Fig. 1E). These results suggested that
miR-21 may be associated with the activation of the Ras pathway in
pancreatic cancer.
 | Figure 1miR-21 level is a key factor in the
prognosis of patients with PDAC and is associated with the Ras
signaling pathway. (A) miRNA expression in patients with PAAD
according to data from TCGA database (n=131) and WGCNA analysis.
Samples were divided based on the co-expression of the miRNAs into
several subsets by hierarchical clustering, with branches
representing different gene modules (represented by a specific
color) and miRNAs within the modules exhibiting high degrees of
co-expression. (B) The GS of members of the turquoise module was
analyzed by WGCNA, indicating significant correlations with OS, DFS
and tumor grade in patients with PAAD. (C) The primary signaling
pathways involving miR-21 target genes. (D) Effects of KRAS
expression levels on PAAD patient survival time based on data from
the UALCAN and TCGA databases (n=177). (E) KRAS-mutation
status in patients with pancreatic cancer (n=149). miR, miRNA,
microRNA; WGCNA, weighted gene correlation network analysis; PDAC,
pancreatic ductal adenocarcinoma; PAAD, pancreatic adenocarcinoma;
TCGA, The Cancer Genome Atlas; OS, overall survival; DFS,
disease-free survival; GS, gene significance. |
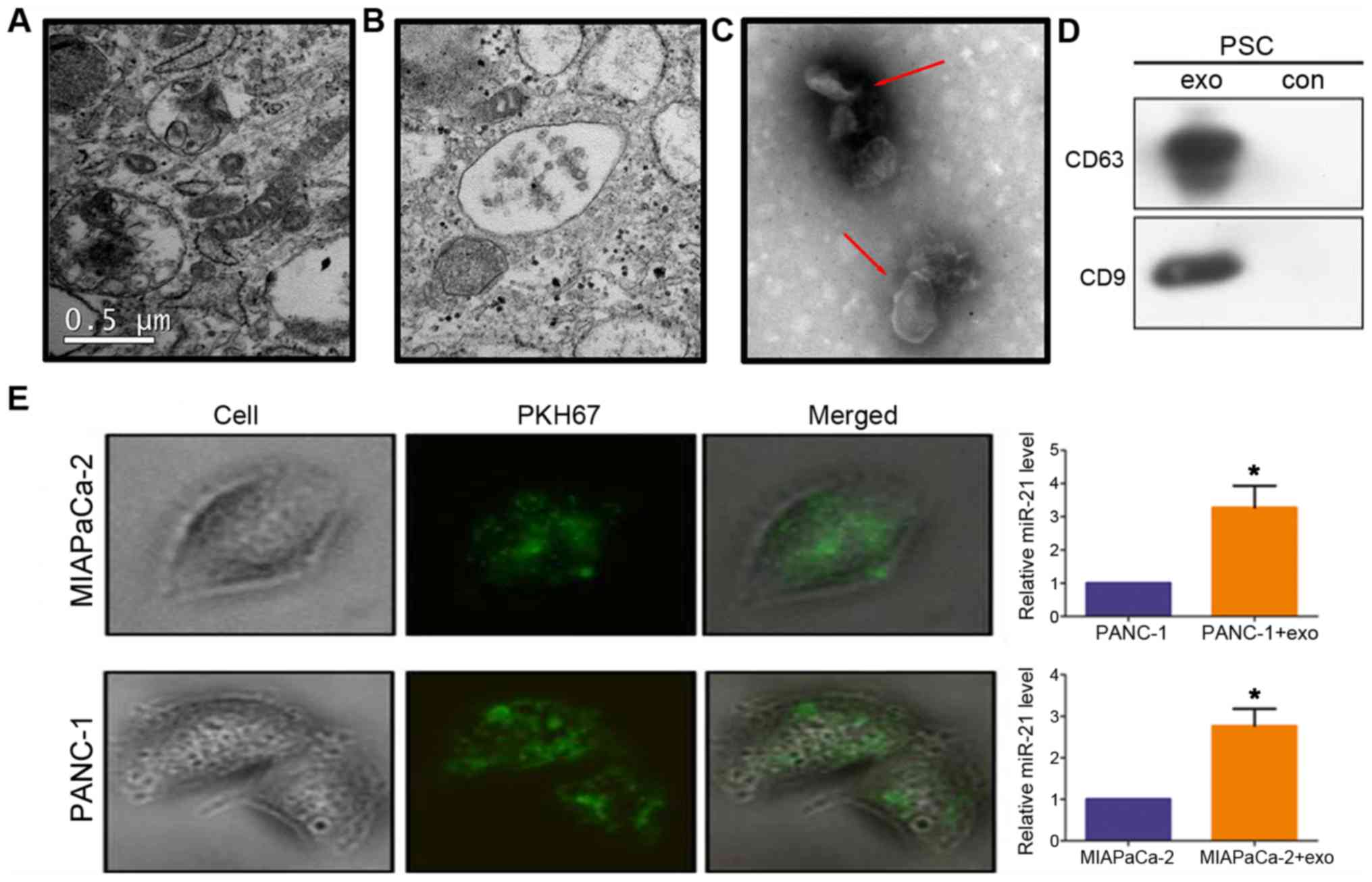
PDAC cells internalize exosomal miR-21
from PSCs
TEM images of the ultrastructure of PSCs revealed
numerous multivesicles in the cytosol containing varying amounts of
vesicles (Fig. 2A and B). Membrane
structures of 40-100 nm in size were observed in the extracellular
vesicles (Fig. 2C, red arrows),
which exhibited strong expression of the exosomal markers CD63 and
CD9 (Fig. 2D). This result
confirmed that the isolated exosomes were obtained from the
conditioned media of PSC cultures. To determine whether PDAC
internalized PSC-derived exosomes, PSC-derived exosomes were
labeled with the dye PKH67 and added to MIAPaCa-2 and PANC-1
cultures. After 3 h, fluorescent exosomes were detected inside
these cells (Fig. 2E), indicating
their effective uptake. In addition, miR-21 levels were increased
in these cells compared with untreated cells (P<0.05; Fig. 2E).
PSC-derived exosomal miR-21 induces PDAC
cell migration
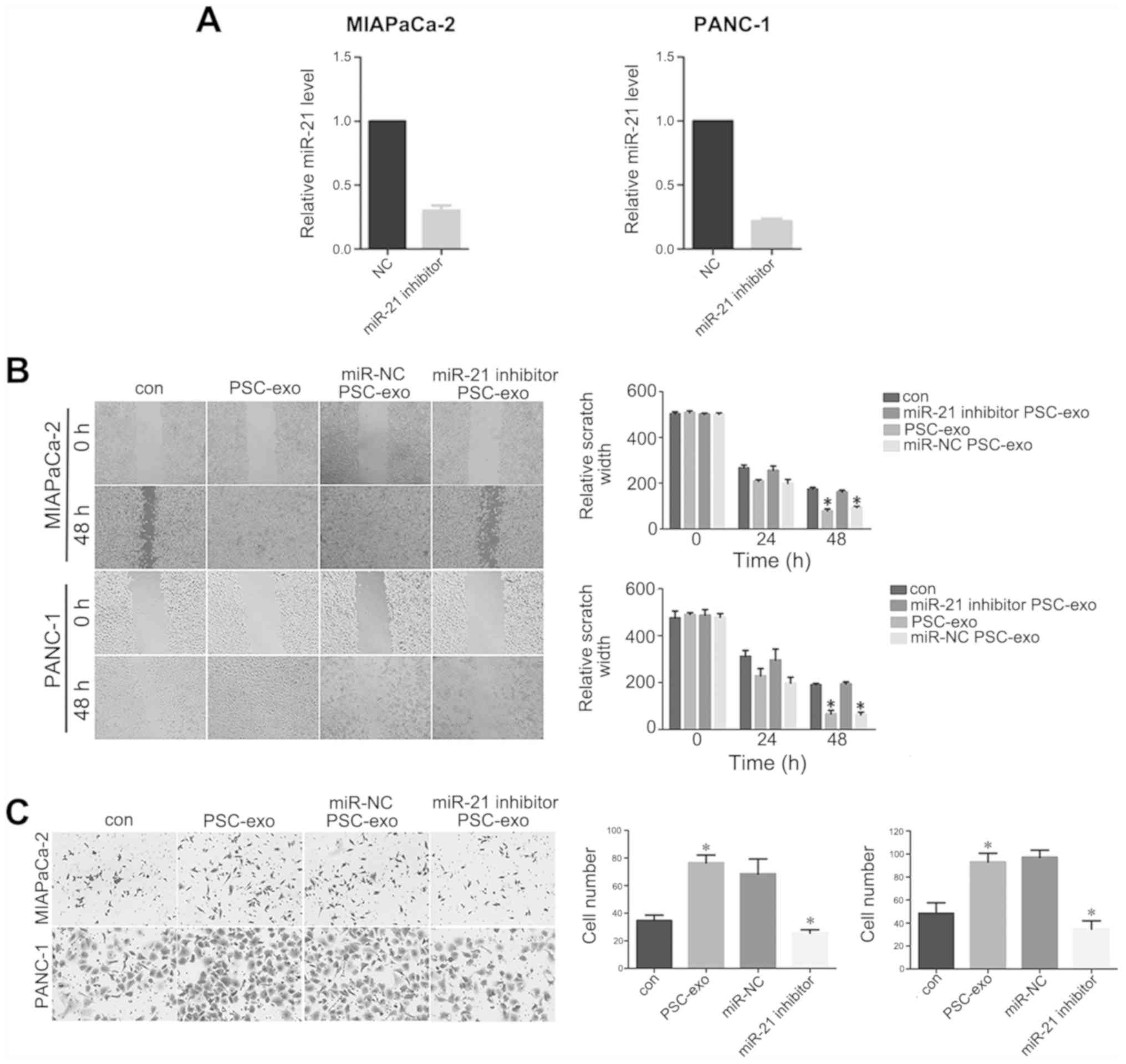
The miR-21 inhibitor was transfected into PANC-1 and
MIAPaCa-2 cells, and relative miR-21 levels were decreased compared
with the negative control group (P<0.05; Fig. 3A), which indicated that miR-21 was
successfully transfected into the two cells lines. To determine
whether the migration-promoting effect of PSCs was mediated by
exosomes, wound-healing and Transwell assays were performed. The
results of the wound-healing assay demonstrated that MIAPaCa-2 and
PANC-1 cells treated with exosomes for 48 h migrated over longer
distances compared with the cells in the respective control groups
(P<0.05; Fig. 3B).
Additionally, Transwell assays demonstrated a higher number of
MIAPaCa-2 and PANC-1 cells migrating across the membrane following
exosome treatment compared with those observed in the control
groups (P<0.05; Fig. 3C). In
addition, the two assays revealed that the migratory ability of
PDAC cells decreased significantly following treatment with
exosomes from PSCs transfected with the miR-21 inhibitor compared
with that observed in cells treated with exosomes derived from
miR-NC-treated PSCs (Fig. 3B and
C).
PSC-derived exosomal miR-21 induces
epithelial-mesenchymal transition (EMT) and promotes MMP-2/9
expression in PDAC cells
The EMT process and MMP-2/9 activity are widely
regarded as vital factors involved in tumor metastasis, and EMT and
pancreatic tumor aggressiveness are reportedly strongly associated
(17,18). Therefore, it was hypothesized that
PSC-derived exosomal miR-21 may induce EMT. To test this
hypothesis, the levels of EMT biomarkers in MIAPaCa-2 and PANC-1
cells were determined following treatment with PSC-derived
exosomes. As demonstrated in Fig. 4A
and B, the levels of N-cadherin, Snail and vimentin were
considerably increased, whereas E-cadherin expression was
downregulated in the cells treated with PSC-derived exosomes. By
contrast, inhibition of exosomal miR-21 decreased N-cadherin, Snail
and vimentin expression levels and increased E-cadherin expression
levels in MIAPaCa-2 and PANC-1 cells (Fig. 4A and B). In addition, MMP-2/9
activity increased in exosome-treated cells compared with untreated
controls and cells treated with the PSC supernatant (NC) (Fig. 4C). Inhibition of PSC-derived
exosomal miR-21 decreased MMP-2/9 activity in PANC-1 cells compared
with that observed in cells treated with exosomes from
miR-NC-treated PSCs (Fig. 4D).
 | Figure 4PSC-derived exosomal miR-21 induces
EMT and increases MMP-2/9 and Ras/ERK/Akt signaling activity. (A
and B) MIAPaCa-2 and PANC-1 cells were treated with exosomes from
untreated PSCs or PSCs treated with the miR-21 inhibitor for 48 h.
Levels of EMT-related proteins were analyzed by western blotting.
(C) MMP-2/9 activity in PANC-1 cells treated with PSC-derived
exosomes. (D) MMP-2/9 activity in MIAPaCa-2 and PANC-1 cells
treated with exosomes from PSCs transfected with an miR-21
inhibitor. (E) The effect of treatment with PSC-derived exosomes
(0, 5, 10 or 20 µg) on phospho-Akt and phospho-ERK levels in
PANC-1 cells. (F and G) Phospho-Akt and phospho-ERK levels in
MIAPaCa-2 and PANC-1 cells following treatment with exosomes
obtained from PSCs treated with the miR-21 inhibitor. (H) Summary
figure of the interaction model and signaling pathways between PSCs
and PDAC. PSC, pancreatic stellate cell; miR, microRNA; EMT,
epithelial-mesenchymal transition; MMP, matrix metalloproteinase;
p-, phospho-; NC, negative control cells treated with the
supernatant from exosome cultures; exo, exosome-enriched
conditioned medium; con, untreated control cells; PDAC, pancreatic
ductal adenocarcinoma. |
Exosomal miR-21 enhances the Ras/ERK and
Ras/Akt signaling pathway activity in PDAC cells
Activation of the K-Ras/Akt and K-Ras/ERK signaling
pathways induced by KRAS alterations has been reported to be
a primary cause of pancreatic cancer progression (24-27).
Since the KEGG pathway analysis suggested that miR-21 was
associated with Ras pathway activation, it was hypothesized that
PSC-derived exosomal miR-21 may be involved in regulating the
activation of the Ras/ERK and Ras/Akt signaling pathways. In
support of this hypothesis, the addition of PSC-derived exosomes to
the PANC-1 cell culture gradually increased the levels of Akt
phosphorylation in proportion with the amount of exosomes added,
with phosphorylation levels peaking at 20 µg (Fig. 4E). The level of ERK phosphorylation
also gradually increased along with increasing exosome number,
although phosphorylation levels peaked at 10 µg rather than
20 µg, which may indicate a dose-dependent effect within a
certain range (Fig. 4E). To
confirm that these signaling pathways were activated by exosomal
miR-21, 10 µg exosomes from PSCs treated or with the miR-21
inhibitor or miR-NC were added to PANC-1 and MIAPaCa-2 cell
cultures and incubated for 48 h. Inhibition of exosomal miR-21
resulted in notable decreases in ERK and Akt phosphorylation levels
in PANC-1 cells compared with those in the NC groups, although this
change was not obvious in MIAPaCa-2 cells (Fig. 4F and G). These results suggested
that exosomal miR-21 may induce the activation of the Ras/ERK and
Ras/Akt signaling pathways.
Discussion
The results of the present study demonstrated that
PSC-derived exosomal miR-21 was internalized by PDAC cells and
subsequently modulated the migratory capacity and motility of PDAC
cells by enhancing the EMT process and increasing MMP-2/9 activity
in addition to enhancing Ras/ERK and Ras/Akt signaling activation
(Fig. 4H). These results suggested
a novel role of exosomal miR-21 in the progression and invasiveness
of pancreatic cancer.
Previous studies have reported that abnormally
activated PSCs contribute to PDAC progression, including the
induction of pancreatic fibrosis (28), collagen production (29) and cell-cell interactions (30). However, the underlying molecular
mechanisms have not been completely elucidated. Recently,
cancer-associated fibroblast-derived exosomes have attracted
attention as regulators of tumor malignant phenotypes (31), which has been validated in ovarian
(13), colorectal (32), breast (33) and other types of cancer. Therefore,
the results of the present study demonstrating the effects of
PSC-derived exosomes in promoting the EMT process and migratory
capacity of PDAC cells are in accordance with previous
observations. Additionally, these results may contribute further
insight into the mechanism of PSC-induced tumor metastasis.
Exosomes mediate intercellular cross-talk under
physiological and pathological conditions through the transfer of
miRNAs (34). To explore the
molecular mechanisms underlying the processes observed in the
present study, exosome-associated miRNAs were analyzed. miR-21 is a
well-known oncogenic miRNA that promotes tumor progression by
inhibiting the expression of tumor-suppressor genes (35,36)
and is associated with high metastatic potential and a poor patient
prognosis (37). miR-21 has been
demonstrated to be significantly upregulated in activated
PSC-derived exosomes (10). In the
present study, WGCNA results revealed that high miR-21 expression
was associated with a poor prognosis in patients with PAAD. KEGG
analysis further showed that miR-21 primarily regulates the Ras and
ERK signaling pathways and activates Ras signaling in patients with
PAAD. Therefore, miR-21 may represent a key factor in pancreatic
cancer onset and development. Accordingly, it was hypothesized in
the present study that PSC-derived exosomal miR-21 may contribute
to the malignant phenotype of pancreatic cancer cells.
In vitro experiments provided partial support
for this hypothesis. Inhibition of miR-21 expression in PSC-derived
exosomes resulted in decreased PDAC cell motility, possibly by
inhibiting EMT and MMP-2/9 activity, demonstrating that exosomal
miR-21 serves an important role in promoting PDAC cell migration.
In line with the bioinformatics analysis, the in vitro
transfer of exosomal miR-21 from PSCs to neighboring cancer cells
led to increased activation of the Ras/ERK and Ras/Akt signaling
pathways and expression of the EMT-associated genes and MMP-2/9. By
contrast, inhibition of exosomal miR-21 decreased the activation of
Ras signaling. These results suggests that PSC-derived exosomes
induced EMT and increased the migratory capacity of PDAC cells via
the activation of Ras/ERK and Ras/Akt signaling. Although previous
studies have reported that miR-21 regulates Ras/ERK and Ras/Akt
signaling by repressing Ras P21 protein activator 1 translation
(38,39), to the best of our knowledge, the
present study is the first to demonstrate that PSC-derived exosomal
miR-21 transfer promotes the activation of these two signaling
pathways in PDAC cells.
This study had some limitations. In addition to
miRNAs, exosomes also contain mRNAs, DNA fragments and various
proteins (5); therefore, the
effects of exosomes on the malignant phenotype observed in PDAC
cells may be more complex and variable than the sole influence of
miR-21. Thus, further studies should assess other potential effects
such as cell proliferation, apoptosis and drug resistance (40). In addition, the roles of other
miRNAs differentially expressed between cancer and healthy tissues
should be explored. Additionally, exosomal miR-21 expression levels
need to be analyzed in a larger panel of PSCs and PDAC cells to
validate the results of the present study. Finally, the effects of
exosomal miR-21 on PDAC metastasis should be further analyzed in
vivo.
In conclusion, the results of the present study
suggested that PSC-derived exosomes were released into the tumor
microenvironment to promote PDAC EMT and migration via
miR-21-induced activation of the Ras/ERK and Ras/Akt signaling
pathways. These results highlighted PSC-derived exosomal miR-21 as
a novel molecular determinant of PSC-PDAC cell communication to
promote EMT induction, PDAC metastasis and progression, suggesting
a potential new target for treatment or prognosis prediction for
patients with pancreatic cancer.
Funding
This work was supported by the National Natural
Science Foundation of China (grant nos. 81502625, 81602162 and
31471366), the National Key Research and Development Program of
China (grant no. 2016YFC0901500) and the Center for Molecular
Pathology, Chinese Academy of Medicine Science and Peking Union
Medical College, Beijing, China (grant no. 2017PT31008).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HWW, ZYL and THL conceived and designed the study.
QM and YX performed the experiments and data analysis. QM and ZYL
wrote, reviewed and revised the manuscript. HWW and YX provided
technical support.
Ethics approval and consent to
participate
This study was approved by the Research Ethics
Boards of Peking Union Medical College Hospital. All patients
provided written informed consent prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Subramani R, Lopez-Valdez R, Arumugam A,
Nandy S, Boopalan T and Lakshmanaswamy R: Targeting insulin-like
growth factor 1 receptor inhibits pancreatic cancer growth and
metastasis. PLoS One. 9:e970162014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McGuigan A, Kelly P, Turkington RC, Jones
C, Coleman HG and McCain RS: Pancreatic cancer: A review of
clinical diagnosis, epidemiology, treatment and outcomes. World J
Gastroenterol. 24:4846–4861. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Razidlo GL, Magnine C, Sletten AC, Hurley
RM, Almada LL, Fernandez-Zapico ME, Ji B and McNiven MA: Targeting
Pancreatic Cancer Metastasis by Inhibition of Vav1, a Driver of
Tumor Cell Invasion. Cancer Res. 75:2907–2915. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang H and Chen L: Tumor microenviroment
and hepatocellular carcinoma metastasis. J Gastroenterol Hepatol.
28(Suppl 1): 43–48. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kahlert C and Kalluri R: Exosomes in tumor
microenvironment influence cancer progression and metastasis. J Mol
Med (Berl). 91:431–437. 2013. View Article : Google Scholar
|
|
6
|
Gajos-Michniewicz A, Duechler M and Czyz
M: MiRNA in melanoma-derived exosomes. Cancer Lett. 347:29–37.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li L, Li C, Wang S, Wang Z, Jiang J, Wang
W, Li X, Chen J, Liu K, Li C, et al: Exosomes Derived from Hypoxic
Oral Squamous Cell Carcinoma Cells Deliver miR-21 to Normoxic Cells
to Elicit a Prometastatic Phenotype. Cancer Res. 76:1770–1780.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liao J, Liu R, Shi YJ, Yin LH and Pu YP:
Exosome-shuttling microRNA-21 promotes cell migration and
invasion-targeting PDCD4 in esophageal cancer. Int J Oncol.
48:2567–2579. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takikawa T, Masamune A, Yoshida N, Hamada
S, Kogure T and Shimosegawa T: Exosomes Derived From Pancreatic
Stellate Cells: MicroRNA Signature and Effects on Pancreatic Cancer
Cells. Pancreas. 46:19–27. 2017. View Article : Google Scholar
|
|
10
|
Ali S, Suresh R, Banerjee S, Bao B, Xu Z,
Wilson J, Philip PA, Apte M and Sarkar FH: Contribution of
microRNAs in understanding the pancreatic tumor microenvironment
involving cancer associated stellate and fibroblast cells. Am J
Cancer Res. 5:1251–1264. 2015.PubMed/NCBI
|
|
11
|
Charrier A, Chen R, Chen L, Kemper S,
Hattori T, Takigawa M and Brigstock DR: Connective tissue growth
factor (CCN2) and microRNA-21 are components of a positive feedback
loop in pancreatic stellate cells (PSC) during chronic pancreatitis
and are exported in PSC-derived exosomes. J Cell Commun Signal.
8:147–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bachem MG, Schneider E, Gross H,
Weidenbach H, Schmid RM, Menke A, Siech M, Beger H, Grünert A and
Adler G: Identification, culture, and characterization of
pancreatic stellate cells in rats and humans. Gastroenterology.
115:421–432. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Au Yeung CL, Co NN, Tsuruga T, Yeung TL,
Kwan SY, Leung CS, Li Y, Lu ES, Kwan K, Wong KK, et al: Exosomal
transfer of stroma-derived miR21 confers paclitaxel resistance in
ovarian cancer cells through targeting APAF1. Nat Commun.
7:111502016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Langfelder P and Horvath S: WGCNA.an R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar
|
|
16
|
Xu Y, Li H, Huang C, Zhao T, Zhang H,
Zheng C, Ren H and Hao J: Wnt2 protein plays a role in the
progression of pancreatic cancer promoted by pancreatic stellate
cells. Med Oncol. 32:972015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wrighton KH: Cell migration: EMT promotes
contact inhibition of locomotion. Nat Rev Mol Cell Biol.
16:5182015. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
El-Ghlban S, Kasai T, Shigehiro T, Yin HX,
Sekhar S, Ida M, Sanchez A, Mizutani A, Kudoh T, Murakami H, et al:
Chlorotoxin-Fc fusion inhibits release of MMP-2 from pancreatic
cancer cells. BioMed Res Int. 2014:1526592014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu LF, Wu ZP, Chen Y, Zhu QS, Hamidi S and
Navab R: MicroRNA-21 (miR-21) regulates cellular proliferation,
invasion, migration, and apoptosis by targeting PTEN, RECK and
Bcl-2 in lung squamous carcinoma, Gejiu City, China. PLoS One.
9:e1036982014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiao Y, Fu Z, Li Y, Meng L and Liu Y: High
EIF2B5 mRNA expression and its prognostic significance in liver
cancer: A study based on the TCGA and GEO database. Cancer Manag
Res. 10:6003–6014. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45(D1): D362–D368. 2017. View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: AAnalysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
24
|
Tiwari P, Sahay S, Pandey M, Qadri SS and
Gupta KP: Preventive effects of butyric acid, nicotinamide, calcium
glucarate alone or in combination during the 7, 12-dimethylbenz (a)
anthracene induced mouse skin tumorigenesis via modulation of
K-Ras-PI3K-AKTpathway and associated micro RNAs. Biochimie.
121:112–122. 2016. View Article : Google Scholar
|
|
25
|
Nussinov R, Muratcioglu S, Tsai CJ, Jang
H, Gursoy A and Keskin O: The Key Role of Calmodulin in KRAS-Driven
Adenocarcinomas. Mol Cancer Res. 13:1265–1273. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vena F, Li Causi E, Rodriguez-Justo M,
Goodstal S, Hagemann T, Hartley JA and Hochhauser D: The MEK1/2
Inhibitor Pimasertib Enhances Gemcitabine Efficacy in Pancreatic
Cancer Models by Altering Ribonucleotide Reductase Subunit-1
(RRM1). Clin Cancer Res. 21:5563–5577. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sahu N, Chan E, Chu F, Pham T, Koeppen H,
Forrest W, Merchant M and Settleman J: Co-targeting of MEK and
PDGFR/STAT3 pathways to treat pancreaticductal adenocar-cinoma. Mol
Cancer Ther. 16:1729–1738. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu Z, Vonlaufen A, Phillips PA, Fiala-Beer
E, Zhang X, Yang L, Biankin AV, Goldstein D, Pirola RC, Wilson JS,
et al: Role of pancreatic stellate cells in pancreatic cancer
metastasis. Am J Pathol. 177:2585–2596. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu Z, Pothula SP, Wilson JS and Apte MV:
Pancreatic cancer and its stroma: A conspiracy theory. World J
Gastroenterol. 20:11216–11229. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Masamune A and Shimosegawa T: Pancreatic
stellate cells: A dynamic player of the intercellular communication
in pancreatic cancer. Clin Res Hepatol Gastroenterol. 39(Suppl 1):
S98–S103. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Prakash J: Cancer-Associated Fibroblasts:
Perspectives in Cancer Therapy. Trends Cancer. 2:277–279. 2016.
View Article : Google Scholar
|
|
32
|
Hu Y, Yan C, Mu L, Huang K, Li X, Tao D,
Wu Y and Qin J: Fibroblast-Derived Exosomes Contribute to
Chemoresistance through Priming Cancer Stem Cells in Colorectal
Cancer. PLoS One. 10:e01256252015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Donnarumma E, Fiore D, Nappa M, Roscigno
G, Adamo A, Iaboni M, Russo V, Affinito A, Puoti I, Quintavalle C,
et al: Cancer-associated fibroblasts release exosomal microRNAs
that dictate an aggressive phenotype in breast cancer. Oncotarget.
8:19592–19608. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Forterre A, Jalabert A, Chikh K, Pesenti
S, Euthine V, Granjon A, Errazuriz E, Lefai E, Vidal H and Rome S:
Myotube-derived exosomal miRNAs downregulate Sirtuin1 in myoblasts
during muscle cell differentiation. Cell Cycle. 13:78–89. 2014.
View Article : Google Scholar :
|
|
35
|
Gao W, Xu J, Liu L, Shen H, Zeng H and Shu
Y: A systematic-analysis of predicted miR-21 targets identifies a
signature for lung cancer. Biomed Pharmacother. 66:21–28. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar :
|
|
37
|
Pfeffer SR, Yang CH and Pfeffer LM: The
Role of miR-21 in Cancer. Drug Dev Res. 76:270–277. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sharma SB, Lin CC, Farrugia MK, McLaughlin
SL, Ellis EJ, Brundage KM, Salkeni MA and Ruppert JM: MicroRNAs 206
and 21 cooperate to promote RAS-extracellular signal-regulated
kinase signaling by suppressing the translation of RASA1 and
SPRED1. Mol Cell Biol. 34:4143–4164. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu H, Huang X, Liu X, Xiao S, Zhang Y,
Xiang T, Shen X, Wang G and Sheng B: miR-21 promotes human nucleus
pulposus cell proliferation through PTEN/AKT signaling. Int J Mol
Sci. 15:4007–4018. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang L, Yao J, Li W and Zhang C:
Micro-RNA-21 Regulates Cancer-Associated Fibroblast-Mediated Drug
Resistance in Pancreatic Cancer. Oncol Res. 26:827–835. 2018.
View Article : Google Scholar
|