Introduction
In recent decades, thyroid cancer has become a very
common type of endocrine malignancy whose incidence has rapidly
increased worldwide (1). As the
most frequent histological type of thyroid cancer, the increased
incidence of papillary thyroid carcinoma (PTC) has been pivotal to
the increased numbers of patients affected by thyroid cancer
(2). More sensitive reliable
methods of diagnostic imaging and surveillance have also
contributed to the higher detection of PTC (3). Despite the low mortality rate due to
PTC, the disease recurrence is high (4). Therefore, exploring the potential
molecular mechanisms responsible for carcinogenesis and the
development of PTC is imperative.
Similar to other malignancies, thyroid cancer is a
complex disease intertwined by multiple genetic and environmental
factors (5). Recently, the reduced
expression of ribosomal S6 kinase 4 (RSK4) has been implicated in
the carcinogenesis of colon, kidney, breast, ovarian and
endometrial cancers, suggesting a role as a putative tumor
suppressor gene (6-10). RSK4 belongs to the 90-kDa S6
ribosomal kinase family, and its coding gene is located on the
X-chromosome (Xq21.1) (9). RSK4
mediates signal transduction downstream of the mitogen-activated
protein kinase (MAPK) cascades, serving as a serine/threonine
kinase (11). Due to this kinase
activity, RSK4 also plays a significant role in the regulation of
cellular survival, division and differentiation (9). It has been reported that RSK4
overexpression is capable of inducing a senescence-like phenotype
in vitro, while its downregulation can induce the escape
from cellular senescence and, consequently, immortalization during
the early stages of tumorigenesis (7). PTC is driven by multiple genetic
elements, in which the BRAF V600E mutation is the most
common oncogenic point mutation, occurring with a prevalence of 45%
in PTC (12). BRAF V600E
mutation aberrantly activates the MAPK pathway, and plays a crucial
role in the carcinogenesis of thyroid cancer (13-15).
Furthermore, RSK4 is likely to act directly on or downstream of
ERK, therefore inhibiting the MAPK signaling cascade (16).
Epigenetics can be described as a heritable
modification of gene function that does not affect the genomic DNA
sequence, although ultimately, it leads to a change in phenotype.
In the mammalian genome, the methylation of gene promoters is one
of the most common epigenetic events (17). As a common feature of
tumorigenesis, hypermethylation can block the activity of certain
genes which function as tumor suppressors, while demethylation can
induce gene reactivation and expression (18). Therefore, the inactivation of gene
transcription by promoter methylation may serve as a potential
platform for the early diagnosis, risk and prognosis assessment,
treatment and management of cancers (19).
The implication of RSK4 methylation in human
PTC has not been previously reported, at least to the best of our
knowledge. Thus, the present study demonstrates that RSK4
hypermethylation partly contributes to the downregulation of RSK4
expression and is related to the tumor size and lymph node
metastasis in PTC. Moreover, the present study investigated the
association between RSK4 and the BRAF V600E mutation
in PTC. The present study indicates that RSK4 potentially
functions as a tumor suppressor and also participates in the
carcinogenesis of PTC.
Materials and methods
Patients and tissue samples
A total of 226 paired papillary thyroid carcinoma
tissues and corresponding paracancerous counterparts were acquired
between January, 2018 and February, 2019, after obtaining informed
consents, from patients at the Affiliated Hospital of Qingdao
University (Qingdao, China). For patients <18 years old, their
written informed consents were provided by their parents. Tissue
samples were collected, immediately frozen in liquid nitrogen and
stored at -80°C until further analysis. All samples were confirmed
by pathological assessment. In this study population, 169 patients
were female and 57 were male. The median age at diagnosis was 45
years (range, 17-76 years). Patients with primary PTC were included
and those who had received chemotherapy or radiotherapy prior to
surgery were excluded. The study design and implementation were
approved by the Ethics Committee of the Affiliated Hospital of
Qingdao University.
Cells and cell culture
Immortalized thyroid cancer cell lines (TPC-1 and
BHT101) and a normal thyroid epithelial cell line (Nthy-ori3-1)
were obtained from ExPASy Bioinformatics Resource Portal and Type
Culture Collection of the Chinese Academy of Sciences,
respectively. TPC-1 and BHT101 cells were cultured in DMEM
supplemented with 10% FBS (both from Gibco; Thermo Fisher
Scientific, Inc.). Nthy-ori3-1 cells were cultured in F12K medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS.
The cells were maintained in incubator at 37°C and 5%
CO2. When required, the TPC-1, BHT101 and Nthy-ori3-1
cells were treated with 10 µM 5-Aza-deoxycytidine (5-Aza;
Sigma-Aldrich; Merck KGaA) for 5 consecutive days.
Reverse transcription-quantitative RCR
(RT-qPCR)
Total RNA was extracted using TRIzol reagent (T9424,
Sigma-Aldrich; Merck KGaA) according to the manufacturer's
protocol. Total RNA (3 µg) was reverse transcribed at 37°C for 15
min and 85°C for 5 sec using the PrimeScript™ RT reagent kit with
gDNA Eraser (Code no. RR047, Takara Bio, Inc.). Subsequently,
single-stranded cDNAs were subjected to qPCR using TB
Green® Premix Ex Taq™ (Code no. RR420, Takara
Bio, Inc.) on a LightCycler® 96 Real-Time PCR System (Cobas z480,
Roche). The amplification conditions were as follows: 95°C for 30
sec, followed by 30 cycles of 95°C for 5 sec and 60°C for 30 sec.
The following gene-specific qPCR primers were used in this study:
RSK4 forward, 5'-GAACAG ACATTTCCAGCTAAAAAGG-3' and reverse,
5'-TGCCTG AAGATCCAGCAACTAA-3'; BRAF forward, 5'-TGGGGA
ACGGAACTGATTTTTC-3' and reverse, 5'-TTTTGTGGT GACTTGGGGTTG-3'.
GAPDH was used as a reference gene (normalizer) and amplified using
the following primers: Forward, 5'-TGCACCACCAACTGCTTAGC-3' and
reverse, 5'- GGC AT GGACT GT GGT CAT GAG-3'. The relative mRNA
expression of RSK4 was analyzed using the 2-AACq
method (20).
Western blot analysis
Cell samples were washed twice with PBS and then
lysed in Laemmli buffer (2% SDS, 10% glycerol, 0.125M Tris-Cl, pH
6.8). The BCA method was used for protein quantification. The
concentrations of separating gel and stacking gel were 10 and 5%.
Equal amounts of total protein (20 µg) were resolved by SDS-PAGE
and transferred onto nitrocellulose membranes. The membranes were
blocked with 5% skim milk and incubated with respective primary
antibodies overnight at 4°C. The membranes were washed 3 times with
TBST buffer (containing 0.1% Tween-20) and then incubated with
corresponding HRP-conjugated secondary antibody for 1 h at room
temperature. The membranes were washed again, 3 times with TBST,
prior to protein detection. Specific protein bands were developed
by chemiluminescence using a commercial ECL kit (Millipore). The
following antibodies were used for western blot analysis: Mouse
anti-RSK4 monoclonal antibody (1:500, 27330, Novus Biologicals);
rabbit anti-GAPDH polyclonal antibody (1:2,000, E-AB-20059,
Elabscience Biotechnology); the secondary antibodies were as
follows: Goat anti-rabbit/goat anti-mouse IgG polyclonal antibody
(1:10,000, 111-035-003/115-005-003, Jackson ImmunoResearch
Laboratories).
DNA extraction and sodium bisulfite
modification
Total genomic DNA was extracted from the tissues and
cells using a Genomic DNA extraction kit (Aidlab). All procedures
followed according to manufacturer's protocol. Genomic DNA samples
(3 µg each) was bisulfite-modified and then purified according to
the manufacturer's protocol. (Qiagen).
Methylation-specific PCR (MSP)
In total, 134 pairs of specimens of PTC patients
were included. Specific primers of RSK4 for the unmethylated
(5'-TTTTTATTGATGTTTGGG TGAT-3' and 5'-AACAACAACCACTCAATAATAAC-3')
and methylated reactions (5'-TTATTGACGTTTGGGTGAC-3' and
5'-GACAACCGCTCGATAATAAC-3') were utilized. The modified DNA was
amplified by PCR using the following conditions: 98°C for 5 min,
followed by 35 cycles of 98°C for 30 sec, 56°C for 30 sec and 72°C
for 30 sec, and a final 5-min incubation at 72°C. PCR products were
isolated by agarose gel electrophoresis.
Bisulfite sequencing (BGS)
In total, 31 pairs of specimens of PTC patients were
included. Modified DNA samples were subjected to PCR amplification
to further analyze the methylation status of the RSK4
promoter. The primers used for bisulfite genomic sequencing were as
follows: Forward, 5'-TTTTAA GTTGGGGAATTTTTATTGA-3' and reverse,
5'-AAAACAACTCTATACTACCTCTCCAAAAAC-3' (21). DNA samples were amplified by PCR
using the following conditions: 98°C for 5min, followed by 35
cycles of 98°C for 30 sec, 56°C for 30 sec and 72°C for 30 sec, and
a final 5-min incubation at 72°C. PCR products were ligated into
the pMD18-T Vector (Takara Bio, Inc.), followed by transformation
into competent the Escherichia coli strain, DH5a (Takara
Bio, Inc.). At least 6 clones from each ligation reaction were
randomly selected. Bacterial solutions were then sequenced by the
Beijing Genomics Institute (China). The percentage of methylated
DNA for each sample was computed according to the number of
methylated CpG dinucleotides/(6×21)×100%.
BRAF V600E mutation analysis
The hot spot region of BRAF V600E mutation
was amplified using Premix Taq™ (Takara, Code No. R004). The
following PCR primers were used: Forward, 5'-GCTTGCTCTGATAGGAAAATG
AG-3' and reverse, 5'-GTAACTCAGCAGCATCTCAGG-3'. Genomic DNA was
amplified by PCR using following conditions: 95°C for 3 min,
followed by 38 cycles of 95°C for 30 sec, 57°C for 30 sec and 72°C
for 45 sec, and a final 10-min incubation at 72°C. Respective PCR
products were sequenced by the Beijing Genomics Institute to
confirm the BRAF V600E mutation.
Cell transfection
siRNAs targeting BRAF (5'-GCAUCAAUG GAUACCGUUA-3')
and control siRNAs (5'-UUCUCCGAA CGUGUCACGU-3') were synthesized by
GenPharma. To knockdown BRAF V600E, siRNAs were transfected
into the BHT101 cells using HiPerFect Transfection Reagent
(Qiagen), according to the manufacturer's protocol. Until
transfection, the cells were incubated at 37°C and 5%
CO2, and the transfection complexes which had been
incubated at room temperature for 10 min were then added to the
cells. After 24 h, the cells were harvested and total RNA was
extracted.
Cell proliferation assay
The proliferation of the TPC-1 and BHT101 cells was
measured using the cell counting kit-8 (MedChemExpress). For this,
cells were seeded into 96-well culture plates (2×103
cells/well) and cultured accordingly for 24 h. CCK-8 reagent was
added (10 µl/well) at the 0, 24, 48, 72, 96 h followed by
incubation for an additional 2 h at 37°C. The absorbance was
measured at 450 nm using an enzyme immunoassay analyzer (Gen5
software, BioTek Eliasa).
Statistical analysis
Statistical analysis was performed in silico using
SPSS (version 23.0. IBM) and Prism 7 (GraphPad Software Inc.). Data
with normal distribution are presented as the means ± SEM.
Otherwise, data are expressed as the median with the first and
third interquartile range (Q1, Q3). Paired samples from the same
patients were compared using the paired Student's t-test. An
unpaired t-test or two-tailed Mann Whitney U test was used to
analyze the difference between 2 groups. Multiple groups (>3)
were compared using one-way analysis of variance ANOVA and the
Bonferroni multiple comparisons test. Spearman's correlation
analysis was used to analyze the correlation between RSK4
mRNA expression and the methylation rates in PTC tissues. The
difference of rate was analyzed using the Chi-square test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Reduced RSK4 expression and
hypermethylation of its promoter region in PTC
To verify whether the inactivation of RSK4 may
contribute to thyroid carcinogenesis, RSK4 expression in PTC
tissues and paracancerous counterparts was analyzed by RT-qPCR. The
RSK4 mRNA levels were notably reduced in the PTC tissues
when compared with their counterparts (n=226, P<0.0001)
(Fig. 1A).
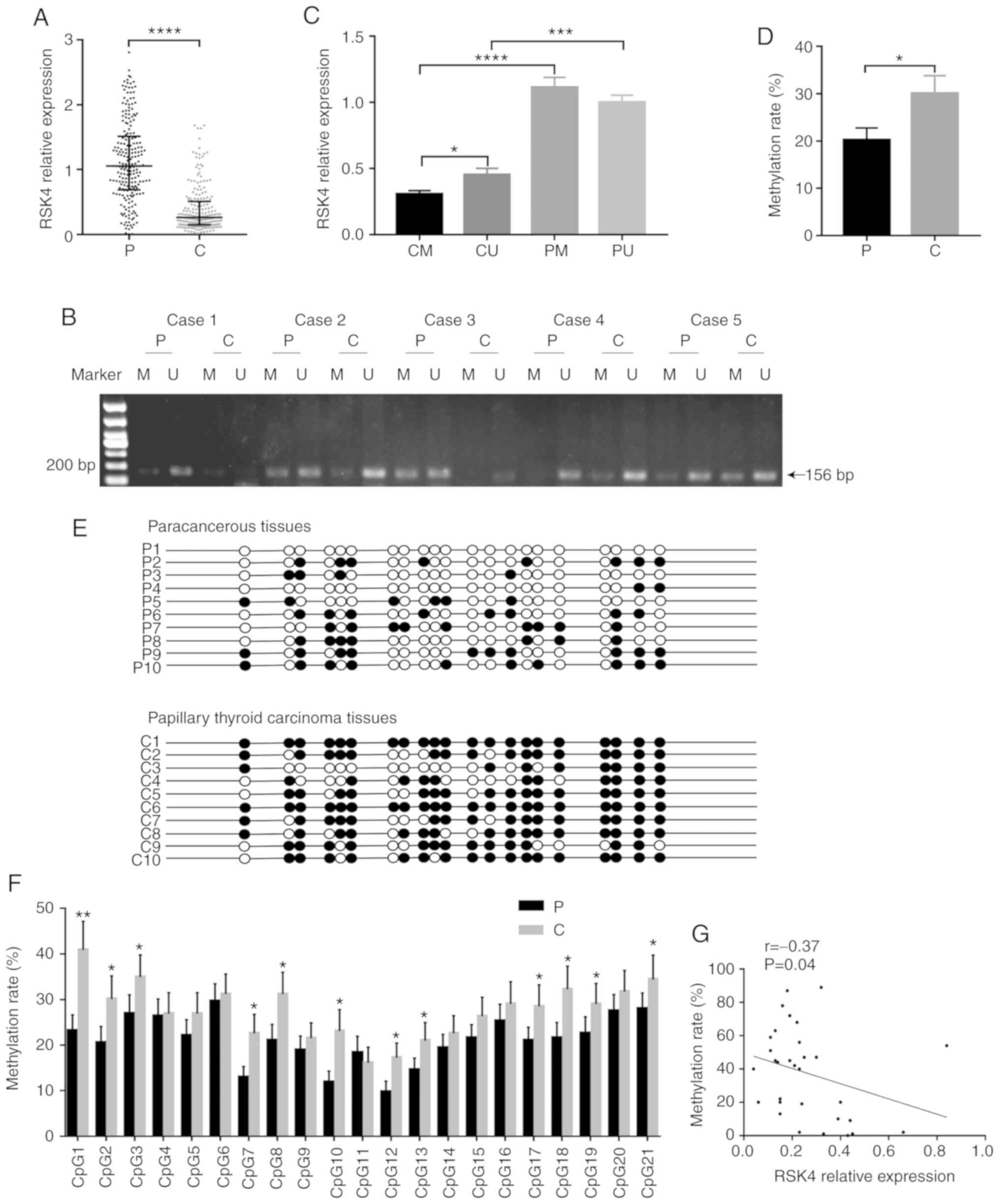 | Figure 1.RSK4 is downregulated and its
promoter region is hypermethylated in PTC tissues. (A) RSK4
expression in PTC tissues and paired paracancerous tissues,
analyzed by RT-qPCR (n=226). (B) Representative agarose gel
electrophoretogram showing different types of MSP products (P,
paracancerous tissues; C, PTC tissues; U, unmethylated; M,
methylated). (C) RSK4 expression in different groups, analyzed by
RT-qPCR (CU, unmethylated PTC tissues; CM, methylated PTC tissues;
PU, unmethylated paracancerous tissues; PM, methylated
paracancerous tissues). (D) Methylation rate in PTC tissues and
paired paracancerous tissues, analyzed by BGS. (E) BGS analysis of
methylation in RSK4 promoter. A total of 21CpG sites of 10
patients were randomly selected from 31 patients analyzed by BGS.
The black and white circles indicate the methylated and
unmethylated CpG sites, respectively. (F) Methylation ratio of each
CpG sites, identified in the sequenced RSK4 promoter region
from PTC and paired paracancerous tissues. (G) Analysis of
correlation between RSK4 mRNA expression and the methylation
rates in PTC tissues. *P<0.05; **P<0.01; ***P<0.001;
****P<0.0001, compared to paracancerous tissues or as indicated.
RSK4, ribosomal S6 kinase 4; PTC, papillary thyroid carcinoma; MSP
methylation-specific PCR; BGS, bisulfite genomic sequencing. |
In order to assess the possible mechanism of the
down- regulation of RSK4 levels in PTC, the methylation
status of its gene was examined. A 240-bp region, including 21 CpG
sites located in the RSK4 promoter, was analyzed. MSP and
BGS analyses were performed to investigate whether a
promoter-specific CpG methylation was associated with the decreased
RSK4 expression. MSP analysis revealed that the methylation rates
of RSK4 in the PTC tissues (93/134, 69.40%) were higher than
those in the paracancerous tissues (75/134, 55.97%, P<0.05).
RSK4 expression in methylated PTC tissues (n=93) was reduced
compared with that in unmethylated PTC tissues (n=41, P<0.05)
(Fig. 1C), with no significant
difference in expression in the paracancerous tissues (P>0.05).
RSK4 expression in methylated PTC tissues was lower than that in
methylated paracancerous tissues (n=74, P<0.0001). Moreover,
RSK4 expression in unmethylated PTC tissues was also lower than
that in unmethylated paracancerous tissues (n=60, P<0.001)
(Fig. 1C).
BGS analysis also provided evidence that the
methylation rates were higher in PTC tissues (n=31, P<0.05)
(Fig. 1D and E). In the majority
of CpG sites, the methylation rates of the RSK4 promoter
were higher in PTC than in paracancerous tissues (Fig. 1F). In addition, it was found that
RSK4 mRNA expression negatively correlated with the
methylation rates in PTC tissues (r=-0.37, P<0.05) (Fig. 1G). Taken together, these results
indicate that the methylation rates of the RSK4 promoter are
increased in PTC, leading to the downregulation of RSK4
expression.
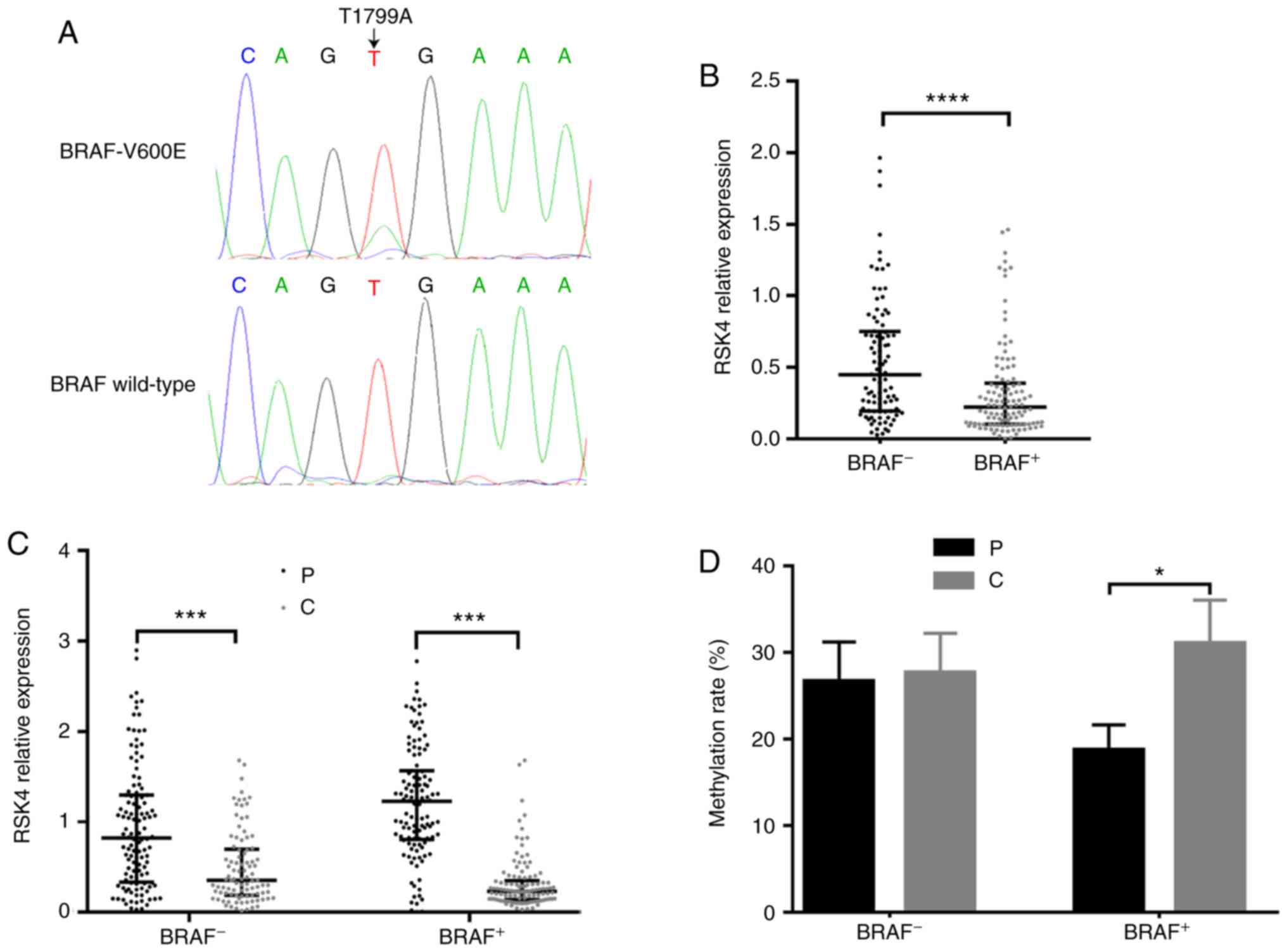
Association between BRAF V600E mutation,
and the expression and methylation of RSK4 in PTC
Previous studies have revealed that RSK4 is involved
in MAPK signaling transduction and, therefore, it can regulate ERK
activation. Thus, the present study investigated whether there was
any association between the BRAF V600E mutation with RSK4
expression and methylation in PTC. The chromatograms of DNA
sequencing are presented in Fig.
2A, denoting BRAF T1799A and wild-type BRAF. It
was found that the RSK4 mRNA levels (PTC relative to the
paracancerous tissues) in PTC patients with BRAF V600E
mutation (n=124) were significantly lower than those with wild-type
BRAF (n=102, P<0.0001, Fig.
2B). Independently of the BRAF mutation status, the
RSK4 mRNA levels in PTC tissues were lower than those in
paracancerous tissues (Fig. 2C).
However, only in patients with BRAF V600E mutation, the
methylated rates in the PTC tissues were higher than those in
paracancerous tissues, as detected by MSP (73.23 vs. 50.7%,
P<0.001) and BGS (P<0.05) (Fig.
2D). Taken together, these findings indicate that BRAF
V600E mutation may be one of reasons for the downregulation of
RSK4 mRNA levels due to the increasing methylation rates in
PTC.
Association between RSK4 methylation
levels and the clinicopathological characteristics of patients with
PTC
To further explore the role of RSK4
hypermethylation in PTC, 134 (MSP) and 31 (BGS) PTC patients with
available information were analyzed. BGS analysis revealed that the
methylation rates were higher in tumors with lymph node metastasis
(Table I). MSP analysis revealed
that the methylation rates in PTC tumors >1.0 cm in diameter
(34/41, 82.93%) were higher than those in PTC tumors <1.0 cm in
diameter (18/30, 60%) in patients with BRAF V600E mutation
(Table II). In addition, the
methylation rates were higher in tumors with lymph node metastasis
(36/44, 81.82%) compared with those without metastasis (16/27,
59.26%) in patients with BRAF V600E mutation (Table II). PTC patients with Hashimoto
thyroiditis were also included to examine whether there was an
association between the methylation status of RSK4 and
Hashimoto thyroiditis in PTC; it was found that the methylation
rate of RSK4 was not related to Hashimoto thyroiditis
(Tables I and II). These findings are in accordance
with a tumor suppressor role for RSK4 in inhibiting tumor
growth and metastasis in PTC.
 | Table I.Association between the RSK4
methylation rate and clinicopathological features of patients with
PTC. |
Table I.
Association between the RSK4
methylation rate and clinicopathological features of patients with
PTC.
| Variable | No. of
patients | Methylation rate in
PTC tissues | P-value |
|---|
| Sex |
| Male | 16 | 29.86±4.784 | 0.1324 |
| Female | 15 | 18.47±5.503 | |
| Age (years) |
| <55 | 25 | 28.63±4.134 | 0.7537 |
| ≥55 | 6 | 31.75±10.490 | |
| Tumor size
(cm) |
| ≤1 | 13 | 26.68±5.405 | 0.5785 |
| >1 | 18 | 31.08±5.392 | |
| Lymphatic
metastasis |
| Present | 17 | 37.93±6.038 | 0.0369 |
| Absent | 14 | 22.08±4.304 | |
| Invasion |
| Present | 8 | 24.01±8.946 | 0.4293 |
| Absent | 23 | 31.06±4.176 | |
| TNM stage |
| I | 20 | 30.42±3.795 | 0.4296 |
| II | 11 | 21.23±16.31 | |
| Hashimoto
thyroiditis |
| Present | 9 | 31.48±8.938 | 0.8512 |
| Absent | 22 | 29.00±4.163 | |
 | Table II.Association between the RSK4
methylation status and clinicopathological features of patients
with PTC with or without BRAF V600E mutation. |
Table II.
Association between the RSK4
methylation status and clinicopathological features of patients
with PTC with or without BRAF V600E mutation.
| Variable | BRAF+
| BRAF-
|
|---|
| No. of
patients | Methylated | Unmethylated | P-value | No. of
patients | Methylated | Unmethylated | P-value |
|---|
| Sex | | | | | | | | |
| Male | 23 | 15 | 8 | 0.2906 | 12 | 5 | 7 | 0.0586 |
| Female | 48 | 37 | 11 | | 51 | 36 | 15 | |
| Age (years) | | | | | | | | |
| <55 | 60 | 44 | 16 | 0.9667 | 55 | 36 | 19 | 0.8699 |
| ≥55 | 11 | 8 | 3 | | 8 | 5 | 3 | |
| Tumor size
(cm) | | | | | | | | |
| ≤1 | 30 | 18 | 12 | 0.0311 | 36 | 22 | 14 | 0.4455 |
| >1 | 41 | 34 | 7 | | 27 | 19 | 8 | |
| Lymphatic
metastasis | | | | | | | | |
| Present | 44 | 36 | 8 | 0.0371 | 31 | 22 | 9 | 0.3346 |
| Absent | 27 | 16 | 11 | | 32 | 19 | 13 | |
| Invasion | | | | | | | | |
| Present | 16 | 10 | 6 | 0.2703 | 9 | 4 | 5 | 0.1607 |
| Absent | 55 | 42 | 13 | | 54 | 37 | 17 | |
| TNM stage | | | | | | | | |
| I | 58 | 42 | 16 | 0.7399 | 55 | 36 | 19 | 0.8699 |
| II | 13 | 10 | 3 | | 8 | 5 | 3 | |
| Hashimoto
thyroiditis | | | | | | | | |
| Present | 10 | 6 | 4 | 0.3076 | 19 | 11 | 8 | 0.4318 |
| Absent | 61 | 46 | 15 | | 44 | 30 | 14 | |
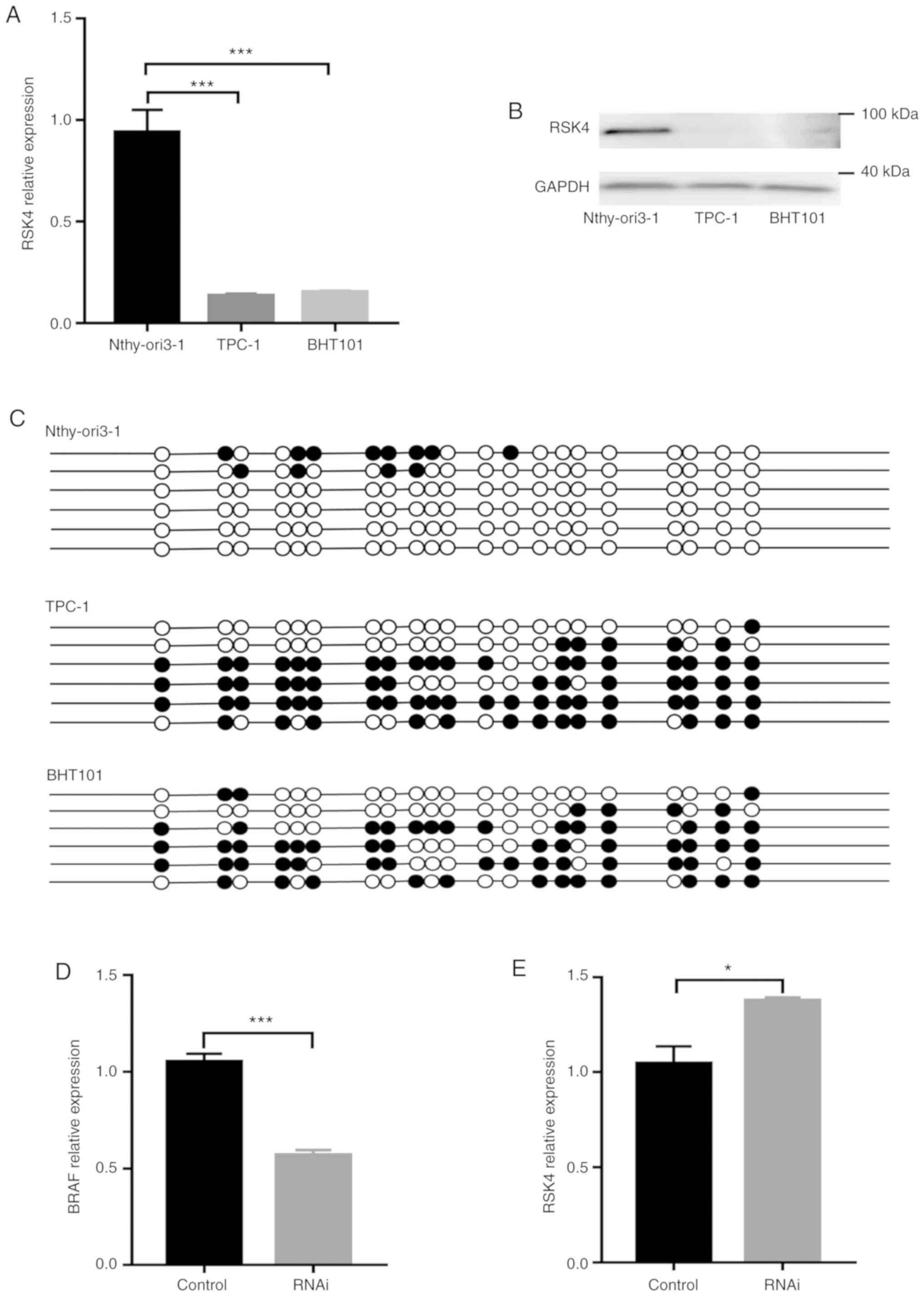
RSK4 expression associated with BRAF
V600E is down- regulated and hypermethylated in thyroid cancer
cells
RSK4 levels and the methylation status of its
promoter were examined in vitro, using a normal
(Nthy-ori3-1) and thyroid cancer cell lines (TPC-1 and BHT101). The
results revealed higher methylation rates of the RSK4
promoter in TPC-1 and BHT101 when compared with Nthy-ori3-1 cells
(Fig. 3C). By contrast, both the
RSK4 mRNA and protein levels were lower in the TPC-1 and
BHT101 cells when compared with the Nthy-ori3-1 cells (Fig. 3A and B). As shown in Fig. 3D and E, BRAF expression was
significantly downregulated and RSK4 expression was significantly
upregulated following the knockdown of BRAF V600E in the
BHT101 cells. These findings suggest that RSK4 expression is
negatively associated with the methylation status of its promoter
and is also influenced by BRAF V600E in thyroid cancer
cells. These in vitro data are consistent with the results
obtained with the human tissue specimens mentioned above.
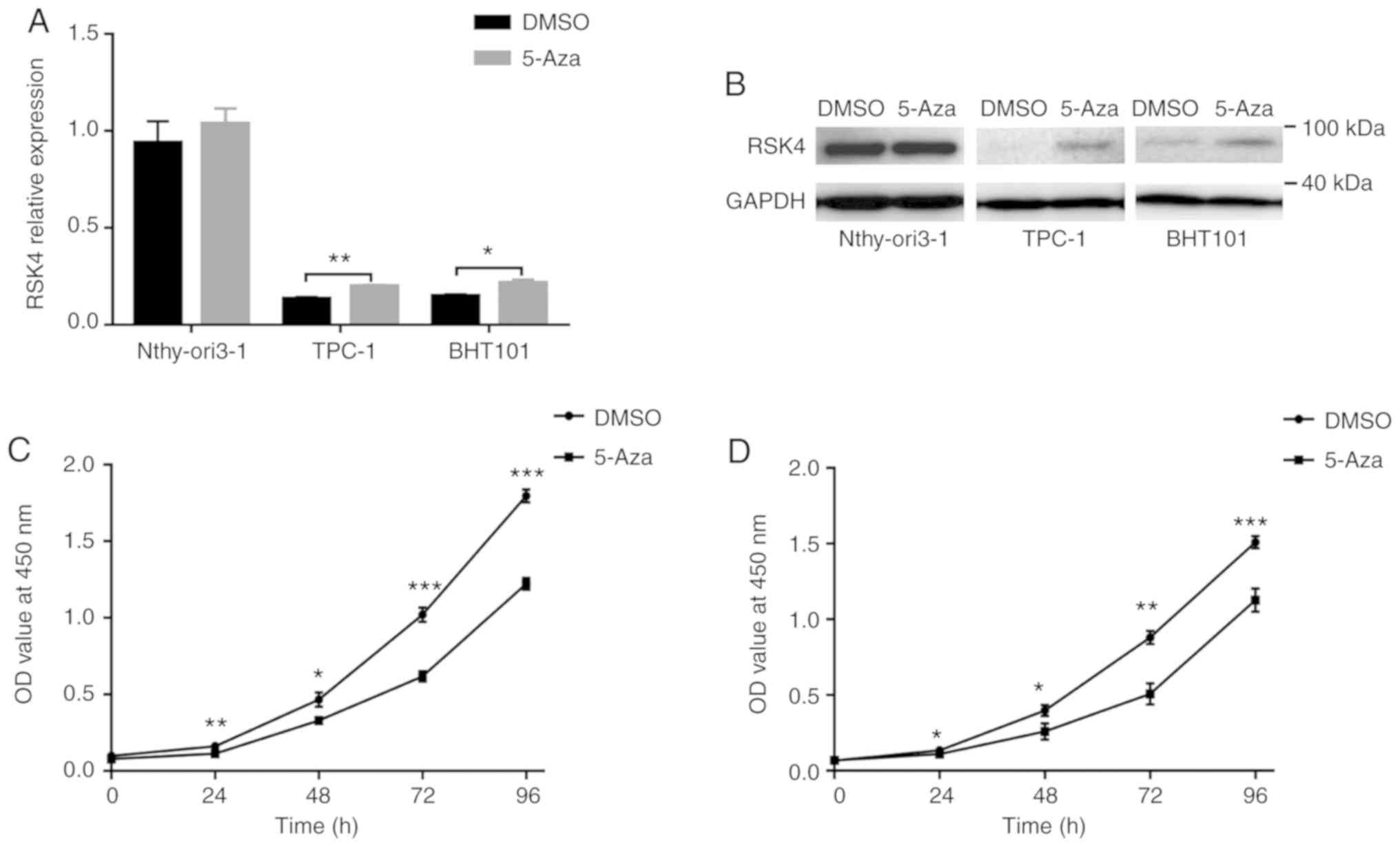
RSK4 expression is enhanced by 5-Aza and DNA
demethylation inhibits the proliferation of thyroid cancer cells.
To further investigate the role of promoter hypermethylation
in the regulation of RSK4 expression, the TPC-1, BHT101 and
Nthy-ori3-1 cells were treated with the demethylating agent, 5-Aza.
The results revealed that the RSK4 mRNA and protein levels
were restored in the TPC-1 and BHT101 cells, but not in the
Nthy-ori3-1 cells (Fig. 4A and B).
Furthermore, the proliferative ability of the TPC-1 and BHT101
cells was significantly suppressed by treatment with 10 µM 5-Aza
(Fig. 4C and D). These data
suggest that the hypermethylation of the RSK4 promoter can
reduce gene expression and, importantly, DNA demethylation can lead
to the repression of thyroid cancerous cell proliferation.
Discussion
Papillary thyroid cancer (PTC) accounts for 80-90%
of all thyroid cancers and is the most common endocrine malignancy,
with a rapidly increasing incidence worldwide (22). Although thyroid cancer has been
frequently associated with a relatively indolent progress and a
good prognosis, some patients become incurable with notable
morbidity and mortality (3,23).
In spite of the overall 10-year survival rate (approximately 90%),
approximately 10-20% of patients with PTC with stage I or II
disease suffer from disease recurrence (24). The molecular pathogenesis of
thyroid cancer involves genetic alterations of multiple oncogenes,
such as BRAF, TERT and RAS, and tumor suppressor
genes, such as PTEN and RASAL1. Their potential
association with clinicopathologic aggressiveness and the poor
prognosis of thyroid cancer has been partially demonstrated
(12,14,25).
Advances in the molecular pathogenesis of thyroid cancer have
exhibited great prospect toward more effective diagnosis and
treatment strategies for this disease (12). Moreover, the understanding of these
genetic alterations may further lead to a reduction in unnecessary
treatments for indolent PTC, and may also improve the prognosis of
patients affected by clinically aggressive cancers (3).
RSK4 is a functional ribosome protein kinase that
belongs to the p90 RSK family. Recent studies have demonstrated its
apparent significance in tumorigenesis, since a low or no RSK4
expression has been associated with malignant tumors of the colon,
kidney, ovary, pancreas, hematological system and endometrium
(8,9,26-28).
Specifically, it has been demonstrated that RSK4 may drive cellular
senescence by modulating Rb and p21, since the upregulation of RSK4
expression can increase p21 levels (6,7,29).
These studies have indicated that the downregulation of RSK4 may be
an important element in promoting cell transformation and,
therefore, may support the notion of the role of RSK4 in regulating
senescence as tumor suppressor protein (6-8,16).
Similarly, the present study found that RSK4 expression was
significantly decreased in the majority of PTC tissues and in a
PTC-derived cell line.
Promoter hypermethylation has been recognized as
standard mechanism that leads to the transcriptional inactivation
of tumor suppressor genes (TSGS) in a variety of cancer types
(30). Some studies have indicated
that the hypermethyl- ation of the RSK4 promoter can lead to
a decrease in its own expression in several malignancies, including
breast, endometrial, oral and ovarian cancers. Accordingly, the
decreased RSK4 expression has been restored by DNA
methyltransferase inhibitors in several cancer cell lines (21,27,30,31).
However, no studies to date have reported the methylation status of
RSK4 in PTC and paracancerous tissues, at least to the best
of our knowledge. Hence, the authors conducted MSP and BGS to
finally examine the methylation status of RSK4. Both
approaches were able to demonstrate that the methylation rates of
RSK4 promoter were consistently higher in PTC than in
paracancerous tissues. This increased methylation status negatively
correlated with RSK4 expression in PTC tissues. Upon methylation,
the RSK4 mRNA levels in PTC were lower than those in
paracancerous tissues. Under non-methylating conditions, the
RSK4 mRNA levels were less prominently decreased in PTC when
compared with those in paracancerous tissues. As stated, it appears
that RSK4 methylation may coexist in both PTC and
paracancerous tissues, where promoter methylation may be one of the
various mechanisms that can lead to a reduction in RSK4 expression.
According to the clinicopathological data, this study also
demonstrated the association between RSK4 promoter methylation with
lymph node metastasis and tumor size, providing evidence that
RSK4 methylation may play a role in the development and
prognosis of PTC. Indeed, previous studies have indicated that
RSK4 can suppress tumor growth, invasiveness and metastatic
ability of other cancer cell lines (7,8).
Strikingly, the aberrant RSK4 expression in PTC
cells could be reversed by broad-spectrum demethylated drugs, and
DNA demethylation inhibited the proliferation of thyroid cancer
cells. In other words, the tumor suppressor effect of RSK4,
to a certain extent, could be restored by demethylation, suggesting
that targeted demethylation drugs may be an attractive option for
tumor therapy in the near future.
An extensive number of genetic alterations have been
involved in the tumorigenesis of various thyroid cancers. The
T1799A transverse point mutation of BRAF is a prototypical
example, which leads to the expression of a BRAF V600E
mutant protein and contributes to constitutive activation of this
serine/threonine kinase. BRAF V600E mutation can activate
the MAPK signaling pathway, which is a key feature of PTC (12,32).
A previous comprehensive and multicenter study demonstrated a
strong association between BRAF V600E with the poor
clinicopathological outcomes of PTC, including aggressive
pathological characteristics, disease recurrence, loss of
radioiodine avidity and unfavorable prognosis (23,33-35).
Previous studies have demonstrated that RSK4 can negatively
modulate the MAPK pathway at the ERK or the downstream levels,
depending on its kinase activity (10,26).
The present study analyzed the status of BRAF mutations in
226 PTC tissues and, as a result, it was found that BRAF
V600E mutation rates were considerably high. Furthermore, this
study indicates that BRAF V600E mutation may influence RSK4
expression, leading to a more significant difference on mRNA levels
between PTC tissues and paracancerous tissues. Moreover, it was
found that this mutation can influence the methylation status of
RSK4, thereby influencing the clinicopathological outcomes
of patients with PTC, which reiterates the putative role of
RSK4 as a cancer suppressor gene in thyroid tumors.
To conclude, the present study demonstrates that the
hypermethylation of CpG islands is one prominent mechanism that can
lead the downregulation of RSK4 expression. The hypermethylation of
the RSK4 promoter is associated with tumor proliferation and
lymph node metastasis. BRAF V600E mutation also has an
effect on RSK4 mRNA levels and gene methylation status,
supporting the distinctive role of RSK4 in the development,
progression and prognosis of PTC.
Funding
The present study was supported by the Key research
and Development program of Shandong Province of China
(2018GSF118051); the China Postdoctoral Science Foundation Grant
(2019M662303); the Natural Science Foundation of Shandong Province
of China (ZR2016HM29); and the Funded program of Science and
Technology Development of Shinan District, Qingdao, Shandong
Province of China (2018-4-013-YY).
Availability of data and materials
The data that support the findings of this study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YY, YW, JC and WS designed the study and wrote the
manuscript. YY, JC, KC and WS performed the majority of the
experiments. YW, JH, HH, AD, JW, QZ, JY, SZ, YZ, PW and FW
participated in the experimentation. WS provided the patients and
participated in the discussions. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of Affiliated Hospital of Qingdao University (QDFY WZ
2018-1-14-02) and was conducted in accordance with the Helsinki
Declaration of 1964 and its later amendments or comparable ethical
standards. All patients provided written informed consent. For
patients <18 years old, their written informed consents were
provided by their parents.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank all of the
investigators for their involvement in this study.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Haugen BR, Alexander EK, Bible KC, Doherty
GM, Mandel SJ, Nikiforov YE, Pacini F, Randolph GW, Sawka AM,
Schlumberger M, et al: 2015 American thyroid association management
guidelines for adult patients with thyroid nodules and
differentiated thyroid cancer: The American thyroid association
guidelines task force on thyroid nodules and differentiated thyroid
cancer. Thyroid. 26:1–133. 2016. View Article : Google Scholar :
|
|
3
|
Cabanillas ME, McFadden DG and Durante C:
Thyroid cancer. Lancet. 388:2783–2795. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tuttle RM, Ball DW, Byrd D, Dilawari RA,
Doherty GM, Duh QY, Ehya H, Farrar WB, Haddad RI, Kandeel F, et al:
Thyroid carcinoma. J Natl Compr Canc Netw. 8:1228–1274. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Riesco-Eizaguirre G and Santisteban P:
ENDOCRINE TUMOURS: Advances in the molecular pathogenesis of
thyroid cancer: Lessons from the cancer genome. Eur J Endocrinol.
175:R203–R217. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
López-Vicente L, Pons B, Coch L, Teixidó
C, Hernández-Losa J, Armengol G, Ramon Y and Cajal S: RSK4
inhibition results in bypass of stress-induced and oncogene-induced
senescence. Carcinogenesis. 32:470–476. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lopez-Vicente L, Armengol G, Pons B, Coch
L, Argelaguet E, Lleonart M, Hernandez-Losa J, de Torres I and
Ramon y Cajal S: Regulation of replicative and stress-induced
senescence by RSK4, which is down-regulated in human tumors. Clin
Cancer Res. 15:4546–4553. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Arechavaleta-Velasco F, Zeferino-Toquero
M, Estrada-Moscoso I, Imani-Razavi FS, Olivares A, Perez-Juarez CE
and Diaz-Cueto L: Ribosomal S6 kinase 4 (RSK4) expression in
ovarian tumors and its regulation by antineoplastic drugs in
ovarian cancer cell lines. Med Oncol. 33:112016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dewdney SB, Rimel BJ, Thaker PH, Thompson
DM Jr, Schmidt A, Huettner P, Mutch DG, Gao F and Goodfellow PJ:
Aberrant methylation of the X-linked ribosomal S6 kinase RPS6KA6
(RSK4) in endometrial cancers. Clin Cancer Res. 17:2120–2129. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang Y, Ye X, Ji Y, Zhou X, Yang H, Wei W
and Li Q: Aberrant expression of RSK4 in breast cancer and its role
in the regulation of tumorigenicity. Int J Mol Med. 40:883–890.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Anjum R and Blenis J: The RSK family of
kinases: Emerging roles in cellular signalling. Nat Rev Mol Cell
Biol. 9:747–758. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xing M: Molecular pathogenesis and
mechanisms of thyroid cancer. Nat Rev Cancer. 13:184–199. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin JD, Fu SS, Chen JY, Lee CH, Chau WK,
Cheng CW, Wang YH, Lin YF, Fang WF and Tang KT: Clinical
manifestations and gene expression in patients with conventional
papillary thyroid carcinoma carrying the BRAF(V600E) mutation and
BRAF pseudogene. Thyroid. 26:691–704. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xing M: Genetic-guided risk assessment and
management of thyroid cancer. Endocrinol Metab Clin North Am.
48:109–124. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang T, Chen C, Pan NF, Sun LY, Jiang XL,
Li JN, Tang Y and Jiang Y: BRAF V600E mutation and TERT promoter
mutation in papillary thyroid carcinomas and their association with
clinicopathological characteristics. Sichuan Da Xue Xue Bao Yi Xue
Ban. 50:919–924. 2019.In Chinese. PubMed/NCBI
|
|
16
|
Myers AP, Corson LB, Rossant J and Baker
JC: Characterization of mouse Rsk4 as an inhibitor of fibroblast
growth factor-RAS-extracellular signal-regulated kinase signaling.
Mol Cell Biol. 24:4255–4266. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peixoto P, Grandvallet C, Feugeas JP,
Guittaut M and Hervouet E: Epigenetic control of autophagy in
cancer cells: A key process for cancer-related phenotypes. Cells.
8:E16562019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Skvortsova K, Stirzaker C and Taberlay P:
The DNA methylation landscape in cancer. Essays Biochem.
63:797–811. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Curtis CD and Goggins M: DNA methylation
analysis in human cancer. Methods Mol Med. 103:123–136. 2005.
|
|
20
|
Kubista M, Andrade JM, Bengtsson M,
Forootan A, Jonak J, Lind K, Sindelka R, Sjöback R, Sjögreen B,
Strömbom L, et al: The real-time polymerase chain reaction. Mol
Aspects Med. 27:95–125. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Niskakoski A, Kaur S, Staff S,
Renkonen-Sinisalo L, Lassus H, Järvinen HJ, Mecklin JP, Bützow R
and Peltomäki P: Epigenetic analysis of sporadic and
Lynch-associated ovarian cancers reveals histology-specific
patterns of DNA methylation. Epigenetics. 9:1577–1587. 2014.
View Article : Google Scholar
|
|
22
|
Mao Y and Xing M: Recent incidences and
differential trends of thyroid cancer in the USA. Endocr Relat
Cancer. 23:313–322. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu R, Bishop J, Zhu G, Zhang T, Ladenson
PW and Xing M: Mortality risk stratification by combining BRAF
V600E and TERT promoter mutations in papillary thyroid cancer:
Genetic duet of BRAF and TERT promoter mutations in thyroid cancer
mortality. JAMA Oncol. 3:202–208. 2017. View Article : Google Scholar
|
|
24
|
Tsumagari K, Abd Elmageed ZY, Sholl AB,
Friedlander P, Abdraboh M, Xing M, Boulares AH and Kandil E:
Simultaneous suppression of the MAP kinase and NF-kB pathways
provides a robust therapeutic potential for thyroid cancer. Cancer
Lett. 368:46–53. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu D, Yang C, Bojdani E, Murugan AK and
Xing M: Identification of RASALl as a major tumor suppressor gene
in thyroid cancer. J Natl Cancer Inst. 105:1617–1627. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Khalaileh A, Dreazen A, Khatib A, Apel R,
Swisa A, Kidess-Bassir N, Maitra A, Meyuhas O, Dor Y and Zamir G:
Phosphorylation of ribosomal protein S6 attenuates DNA damage and
tumor suppression during development of pancreatic cancer. Cancer
Res. 73:1811–1820. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Q, Jiang Y, Wei W, Ji Y, Gao H and Liu
J: Frequent epigenetic inactivation of RSK4 by promoter methylation
in cancerous and non-cancerous tissues of breast cancer. Med Oncol.
31:7932014. View Article : Google Scholar
|
|
28
|
Rafiee M, Keramati MR, Ayatollahi H,
Sadeghian MH, Barzegar M, Asgharzadeh A and Alinejad M:
Down-regulation of ribosomal S6 kinase RPS6KA6 in acute myeloid
leukemia patients. Cell J. 18:159–164. 2016.PubMed/NCBI
|
|
29
|
LLeonart ME, Vidal F, Gallardo D,
Diaz-Fuertes M, Rojo F, Cuatrecasas M, López-Vicente L, Kondoh H,
Blanco C, Carnero A, et al: New p53 related genes in human tumors:
Significant downregulation in colon and lung carcinomas. Oncol Rep.
16:603–608. 2006.PubMed/NCBI
|
|
30
|
Xiao K, Yu Z, Shi DT, Lei Z, Chen H, Cao
J, Tian W, Chen W and Zhang HT: Inactivation of BLU is associated
with methylation of Sp1-binding site of BLU promoter in gastric
cancer. Int J Oncol. 47:621–631. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Foy JP, Pickering CR, Papadimitrakopoulou
VA, Jelinek J, Lin SH, William WN Jr, Frederick MJ, Wang J, Lang W,
Feng L, et al: New DNA methylation markers and global DNA
hypomethylation are associated with oral cancer development. Cancer
Prev Res (Phila). 8. pp. 1027–1035. 2015, View Article : Google Scholar
|
|
32
|
Xing M, Alzahrani AS, Carson KA, Shong YK,
Kim TY, Viola D, Elisei R, Bendlova B, Yip L, Mian C, et al:
Association between BRAF V600E mutation and recurrence of papillary
thyroid cancer. J Clin Oncol. 33:42–50. 2015. View Article : Google Scholar :
|
|
33
|
Xing M, Westra WH, Tufano RP, Cohen Y,
Rosenbaum E, Rhoden KJ, Carson KA, Vasko V, Larin A, Tallini G, et
al: BRAF mutation predicts a poorer clinical prognosis for
papillary thyroid cancer. J Clin Endocrinol Metab. 90:6373–6379.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang F, Zhao S, Shen X, Zhu G, Liu R,
Viola D, Elisei R, Puxeddu E, Fugazzola L, Colombo C, et al: braf
V600E confers male sex disease-specific mortality risk in patients
with papillary thyroid cancer. J Clin Oncol. 36:2787–2795. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kimura ET, Nikiforova MN, Zhu Z, Knauf JA,
Nikiforov YE and Fagin JA: High prevalence of BRAF mutations in
thyroid cancer: Genetic evidence for constitutive activation of the
RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma.
Cancer Res. 63:1454–1457. 2003.PubMed/NCBI
|